Abstract
The metastasis in which the cancer cells degrade the extracellular matrix (ECM) and invade to the surrounding and far tissues of the body is the leading cause of mortality in cancer patients. With a lot of advancement in the field, yet the biological cause of metastasis are poorly understood. The microfluidic system provides advanced technology to reconstruct a variety of in vivo-like environment for studying the interactions between tumor cells (TCs) and endothelial cells (ECs). This review gives a brief account of both two-dimensional models and three-dimensional microfluidic systems for the analysis of TCs-ECs co-culture as well as their applications to anti-cancer drug screening. Furthermore, the advanced methods for analyzing cell-to-cell interactions at single-cell level were also discussed.
Keywords: Microfluidic, Cell analysis, Cell co-culture, Cell interaction, Review
1. Introduction
Metastasis causes about 90% of the cancer-associated mortality. The cancer cells with the attempt to metastasize undergo an invasion-metastasis cascade (Fig. 1) which is a multistep process consisting of two major phases, the physical translocation and colonization [1], [2], [3], [4]. During this process the cancers cells detach from the primary tumor mass and enter the blood or lymph circulation system (intravasation). Approximately 1 × 10-7% of all tumor cells enter the bloodstream [5]. The circulatory tumor cells (CTCs) arising from a solid tumor are exposed to a novel micro-environment of the circulatory system. In circulatory system depending on the size of the blood vessel, the blood flow velocity can reach 0.03–40 cm/s [6], with arterial hemodynamic shear-force of 4.0–30.0 dyn/cm2 and venous shear-force of 0.5–4.0 dyn/cm2. Therefore, these cells must bear hemodynamic forces and overcome the effects of fluid shear [7], [8], [9]. In addition, CTCs in the bloodstream also collide with red blood cells or adhere to leukocytes, platelets, and microphages [10]. The CTCs that survived in the blood vessel then enter into the microvessels of distant sites through the bloodstream. One CTC floating with the blood flow needs to adhere to the endothelium near the endothelial wall. It passes through the transitions from rolling to crawling migration before anchoring to the endothelium, and then transmigrates the endothelial wall using one of the perivascular migration, transcellular migration or a mosaic process mechanism [11]. The CTCs then arrest and extravasate through vascular walls into the surrounding microenvironment (extravasation). The migration to surrounding tissues occurs actively or passively in the result of a complicated crosstalk with the surrounding components. The collision between a CTC and a vessel wall may lead to transient or persistent adhesion as a result of ligand–receptor interactions [9]. The arrest of CTCs on a specific site of endothelial cells (ECs) and transport cells through vascular system is a critical step in metastatic cancer [1], [12], [13], [14], [15]. The CTCs finally organize in the new tissue and form a micro-metastatic colony in the distant parenchyma and may proliferate to form microscopic colonies. After colonization, the CTCs usually remain dormant, while in some cases the dormancy is broken and leads to a lethal macrometastasis [16], [17]. Such specific interactions between CTCs and ECs are proposed to control patterns of metastasis in lung, breast, and other common solid cancers [18]. Many distant metastases are considered to be established by hematogenous spread of these CTCs, but every CTC is not capable of a potential future metastasis [19]. Each step in the metastatic cascade is closely related to the interaction between tumor cells (TCs) and the elements of microenvironment [20], [21], [22]. These interactions occur either directly or indirectly through stable cell-cell junctions or secreting signal molecules. Folkman et al. [23] revealed that the interaction between TCs and ECs could influence the growth and progression of tumors through paracrine or juxtacrine. This interaction also determines the critical process of angiogenesis, which is considered to be a hallmark of tumorigenesis [23]. Moreover, the complex interconnections between TCs and ECs contribute to the modifications in the gene expression profile of ECs [24] and their activation causes angiogenesis and promotes drug resistance [25]. Similarly, the crosstalk between TCs and ECs could induce drug resistance during the cancer-therapy [26], [27], [28], [29].
Fig. 1.
The metastatic cascade can be envisioned as a process that occurs in two major phases: physical translocation of cancer cells from the primary tumor to a distant organ and colonization of the translocated cells within that organ. (A) To begin the metastatic cascade, cancer cells within the primary tumor acquire an invasive phenotype. (B) Cancer cells can then invade into the surrounding matrix and toward blood vessels, where they intravasate to enter the circulation, which serves as their primary means of passage to distant organs. (C) Cancer cells traveling through the circulation are CTCs. They display properties of anchorage-independent survival. (D) At the distant organ, CTCs exit the circulation and invade into the microenvironment of the foreign tissue. (E) At that foreign site, cancer cells must be able to evade the innate immune response and also survive as a single cell (or as a small cluster of cells). (F) To develop into an active macrometastatic deposit, the cancer cell must be able to adapt to the microenvironment and initiate proliferation. Figure was adapted from Ref. [1].
The majority of patients with advanced metastatic disease have rare exception to be cured by the current therapeutic regimens. A large number of findings have revealed detailed pathogenesis leading to primary tumor formation, yet the biological underpinnings of metastasis remain poorly understood. To establish assays for studying TCs-to-ECs interaction, co-culture model of two or more cell types to simulate the tumor microenvironment niche was conducted, in which the exchange of diffusible factors can be realized. In vitro models of TCs and ECs are typically carried out under static biochemically homogeneous conditions. These static models are not accurate enough to simulate the real situation in blood vessels, because blood flow changes the cell characteristics, such as gene expression [30], mechanical properties [31] and the adhesion capability [32], [33]. In addition, cells cultured in a static well are exposed to a homogeneous biochemical environment, but the concentration gradients of signal molecules at different human anatomical locations are varied [34]. Although, animal models reproduce the main physiological characteristics of human metastatic tumors, but still it is difficult to selectively operate and control the experimental conditions in this model. The common shortcomings of conventional animal models are also time-consuming, labor-intensive and expensive.
For over two decades, the microfluidic-based technologies have being emerged in biomedical research for advanced in vitro cell analysis [35]. Cancer biology research comprises a wide range of research fields, including the common goal to establish tumor tissue with greater in vivo relevance for drug development, drug metabolites, monitoring effect and optimization studies. It is particularly important that microfluidic tumor research pays close attention to the interaction between tumor cells and various types of target cells under physiologically relevant conditions, including TCs-ECs, TCs-stromal cells, TCs-immune cells and others [36], [37], [38], [39]. Microfluidic system provides a simulation scenario with definition and reproducibility, so that the cell behavior in an environment to mimic mechanical forces within living tissues can be reliably studied. In this review, we present the progress, limitations, and future directions in microfluidic based exploration of TCs-to-ECs interactions, categorized as two-dimensional (2D) and three-dimensional (3D) devices for cultivation of tumor cells, anticancer drug screening, and tumor analysis at single-cell level. We first introduce 2D models [40], which are used to investigate the development and progression of cancer, and then shift to a more complex 3D models for studying cancer biology and drug testing. In the last section, we focus on the application of these models in drug screening aimed at improving the effectiveness of cancer treatment.
2. Two-dimensional (2D) models
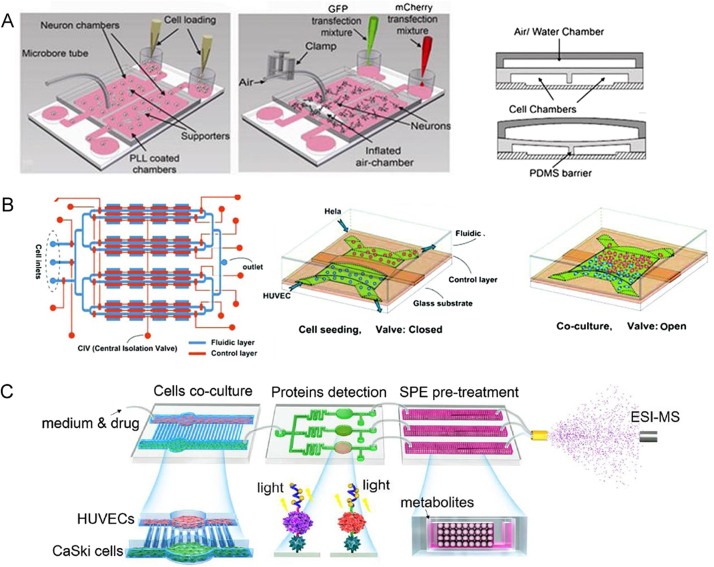
The 2D microfluidic chip has been proven to be a powerful tool for studying the tumor metastasis and cancer progression. In these devices, the co-culture microfluidic chambers were separated and the complex interaction between TCs and ECs was mediated by a number of cytokines, such as vascular endothelial growth factor (VEGF) and transforming growth factor β (TGF- β), which can enhance the drug resistance and induce angiogenesis [40], [41]. Our group reported an integrated microfluidic platform for CaSki cells and human umbilical vein endothelial cells (HUVECs) co-culture with various amount of oxygen (Fig. 2A) [42]. The secreted protein VEGF165 was on-line qualitatively and semi-quantitatively analyzed by functional nucleic acid. Mao et al. [43] proposed a 2D cell co-culture model to simulate the epinephrine communication between 293 and L-02 cells through connecting two channels for cell culture (Fig. 2B). The stimulated 293 cells on the upstream produced epinephrine, and affected L-02 cells on the downstream chamber, and thus enhanced its glucose secretion. The final signaling molecule and metabolites were successfully detected by electrospray ionization-quadrupole time-of-flight mass spectrometer (ESI-Q-TOF-MS) after on-chip solid-phase extraction. This work provided a powerful microfluidic platform for studying the effects of intercellular communication on metabolic pathways. To create a hemodynamic background and investigate synergistic influences of mechanical and biochemical stimulation, Huang et al. [44] designed a microfluidic tumor extravasation model (Fig. 2C). The blood mixed with HeLa cells was introduced to the microchannels consisting of adhered endothelial cells. Meanwhile, flow shear stress (FSS) and cyclic stretch (CS) were induced in this device to simulate the real hemodynamic conditions. The FSS and CS, similar to the hemodynamic conditions of human capillaries, were beneficial to the adhesion of HeLa cells to endothelial cells. In addition, the medium with TNF-α destroyed the endothelial cells monolayer, promoting the adhesion and infiltration of tumor cells. This model also monitored the drug effect of reducing ROS (reactive oxygen species) by platinum nanoparticles (Pt-NPs). They verified that the integrity of the endothelial cells monolayer was important to prevent tumor cells invading.
Fig. 2.
2D microfluidic cell co-culture models for cell-to-cell (TCs-ECs) interaction (A) Cell co-culture microfluidic system under different oxygen gradients, (i) Oxygen induced cell-cell communication; (ii) Illustration of microchip structure; (iii) Microvalve constituted by micropillars; (iv) Two-layer cell co-culture device under low oxygen condition; (v) Schematics of cell secretion detection. Reprinted with permission from Ref. [42]. (B) Integrated microfluidic device coupled with mass spectrometry for detecting signaling to study cell-to-cell communication. Reprinted with permission from Ref. [43]. (C) Integrated FSS and CS within the microfluidic chip mimicing vascular microenvironment to investigate the interaction of TCs-ECs. Reprinted with permission from Ref. [44].
The 2D microfluidic cell migration and interaction systems integrated with microvalves have been proven suitable for improving throughput and facilitating the automation. Gao et al. [45] introduced a microfluidic platform for cell co-culture in vitro, on which a pneumatic microvalve system was integrated to separate different types of cells in different microfluidic channels (Fig. 3A). In this micro-device, the TCs-ECs communication directly occurred when the microvalve was opened. Live-cell imaging was used to observe the cross-migration of TCs (murine 4T1 mammary tumor cells) and ECs (human dermal microvascular endothelial cells, HDVECs). The 4T1s and HDVECs migrated towards each other under normoxic conditions, whereas HDVECs migrated to 4T1s only with hypoxic treatment induced by cobalt chloride (CoCl2). Similarly, Zheng et al. [46] cultured HeLa and HUVEC cells in two adjacent chambers separated by a distance of 300 µm and a pneumatic microvalve (Fig. 3B). The microvalve helped to record the starting position and time of cells migration. Once the valve was open, the interaction between the TCs and ECs was studied. They found that the TCs co-cultured with ECs were more migratory than those cultured separately. They also observed the withdrawal of HUVECs in the presence of HeLa cells, which indicated that soluble factors secreted from HeLa repelled endothelial cells. To understand TCs-ECs interaction, it is necessary to develop an integrated and versatile platform. Lin et al. [47] developed an integrated microfluidic system with three individual components, the cell co-culture component, the protein detection component, and the pre-treatment component (Fig. 3C). This device has the ability to observe cells phenotype, and realize real-time detection of signal molecules and drug metabolites.
Fig. 3.
Complex 2D microfluidic cell migration and interaction systems. (A) The cell-to-cell contact between tumor cells and endothelial cells was controlled with integrated pneumatic valves. Reprinted with permission from Ref. [45]. (B) Cell-migration was monitored in 4 × 4 interconnected micro-chambers simultaneously by pneumatic controlled valves. Reprinted with permission from Ref. [46]. (C) Integrated microfluidic platform with multiple functions to probe the tumor-endothelial cell interaction. Reprinted with permission from Ref. [47].
The extensive application of the 2D co-culture model benefits from its low cost, reproducibility, accessibility and easy operation. However, 2D models are insufficient to recapitulate physiological systems [48], for example spatial cell-cell interaction and extra cellular matrix (ECM) [49], changes in the complex tumor microenvironment and the common effects of various components [50], dynamic metabolic needs and hypoxia induced by tissue growth [51]. These parameters are responsible for the variety in protein expression [52], [53], cell susceptibility to death signals [54], [55], cell proliferation [56], differentiation [57], metabolism [58], shape and fate [59], and signal transduction and responsiveness to external stimulation [60]. In addition, 2D models with low level of physiological correlation may cause misunderstandings of the cytotoxicity, leading to poor prediction of in vivo behavior. These limitations may account for commonplace discrepancies between drug screening in monolayer models and clinical trial efficacy of new molecules [61], [62].
3. Three-dimensional (3D) models
The 3D microfluidic platform simulates cell function and the geometry of tissues and organs, which expresses the complex interactions between tumor and its micro-environment. A growing number of microfluidic devices featuring 3D co-culture of cancer spheroids mimic cancer environment for a variety of TCs-ECs interactions [63]. Song et al. [64] reported a new microfluidic vasculature system to imitate interactions between circulating breast TCs with microvascular ECs at potential sites of metastasis. On this platform, a site-specific stimulation of ECs with a chemokine (CXCL12) was devised through a sandwich device (Fig. 4A). On the top breast TCs layer, a funnel-shaped inlet was intersected with perpendicularly-oriented channels in the bottom PDMS layer filled with CXCL12. In the middle, a thin and porous polyester membrane was sandwiched between top and bottom channels, which could permit both diffusion of biomolecule and adhesion of ECs. This model could mimic serial interactions of CTCs with ECs and enable real-time imaging to investigate the process of cell adhesion and metastases. Utilizing this device, it was confirmed that: i) prolonged stimulation of CXCL12 could work as a high level and directional chemokine in target organs for metastatic cancers. ii) region-specific endothelium stimulation with chemokines would affect CTCs adhesion to ECs. To simulate the morphological features of the blood vessels more truly (round channel morphology like a cellular lumen), Liu et al. [65] established a novel 3D tumor-microvascular structure for in vitro study of antioxidants effects on malignant glioma cells. In this model, a 3D hydrogel containing lumen was constructed to co-culture TCs and ECs to mimic microvascular environment. The U87 cells were first infused with TG-gelatin hydrogel followed by polymerization, and then HUVEC suspensions were injected and adhered on the inner surface of the lumen generated by pulling out the PU fiber (Fig. 4B). In the following experiment, three typical antioxidants (α-lipoic acid, catechins and ascorbic acid) were selected to investigate the antioxidant effects of glioma cells in the simulative tumor microenvironment. From this culture platform, it was obvious that the HUVEC cells exhibited excellent talent of transportation and penetrable functions. In addition, α-lipoic acid, a strong antioxidant, showed higher selectivity to U87 cells than HUVEC cells. Moreover, in order to indicate the important role of immune cells and soluble factors in the progression of cancer via intravasation, Zervantonakis et al. [66] constructed a 3D microfluidic model based on triple co-culture channels for ECs, macrophages and cancer cells (Fig. 4C). The study provided evidence that biochemical factors are key parameters at TCs-ECs interface. The author found that tumor necrosis factor-alpha (TNF-α) secreted by macrophages was the main promoting force that damaged the tightness of the endothelial barrier and permitted TCs to invade into the vascular channel. Endothelial barrier could also be impaired with the increasing number and dynamics of TC-EC interactions.
Fig. 4.
On-chip cell migration and cancer in three-dimensional models. (A) Microfluidic vasculature enables region-specific activation of endothelium under physiological flow conditions. (i) Schematic of the microfluidic device demonstrating multi-layer fabrication with a thin, porous polyester membrane sandwiched by the top and bottom PDMS layers;(ii) Chemokine in the bottom channel activates the endothelium from the basal face while cancer cells flow in the top channel above the endothelium. Reprinted with permission from Ref. [64]. (B) 3D tumor-microvascular structure simulated on a microfluidic chip for study of antioxidants effects on malignant glioma cells in vitro. Reprinted with permission from Ref. [65]. (C) Microfluidic tumor-vascular interface model. (i) Endothelial channel (green), tumor channel (red), and 3D ECM (dark gray) between the two channels; (ii) Phase contrast image showing the fibrosarcoma cells (HT1080, red) invading through the ECM (gray) toward the endothelium (MVEC, green). A single 3D ECM hydrogel matrix region is outlined with the white dashed square. Reprinted with permission from Ref. [66].
Particularly, the movement of tumor cells across vascular barrier has attracted wide attention for the study of metastasis. Although various hydrogels and matrices have successfully created 3D vessels, yet the huge challenge for analytical applications on vascular bio-chip is to form microvasculature allowing continue perfusion. However, with the current level of technological development, endothelialization PDMS channels are easier to modify than complex 3D bioengineered microvasculatures. The greater challenge is to establish standardized and commercialized 3D models, so that hospitals can provide personalized anti-cancer drug screening platform for cancer patients.
4. Anti-cancer drugs screening
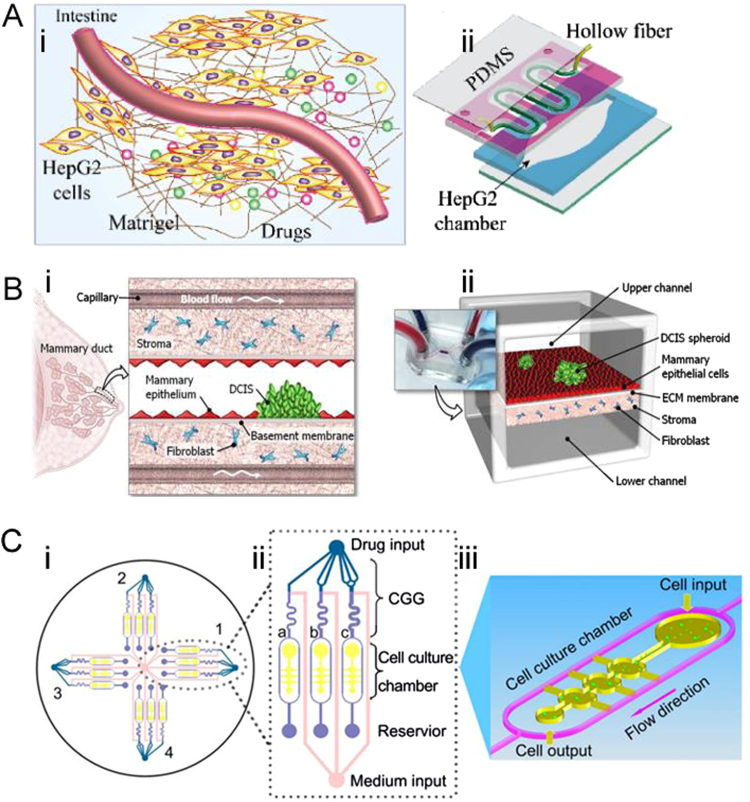
Microfluidic based co-cultured model is a growing platform for drug screening to get better therapeutic output. Jie et al. [67] developed a double-layer microfluidic chip integrated with a hollow fiber (HF) to reconstruct intestine-liver behavior for investigating the absorption and metabolism of combined drugs (Fig. 5A). The HF with Caco-2 cells adhered to its cavity was incorporated into the bottom elliptical chamber cultured with HepG2 cells. Caco-2 cells were cultured in a curved HF cavity to simulate intestinal absorption drugs. The HepG2 cells were incubated in the bottom chamber to simulate the metabolic process of the liver. The HF was coated by homogeneous matrix gel around its outer wall. The drug flowed through the HF and penetrated into the bottom chamber through the micropores on the fiber wall. The HepG2 cells in the bottom chamber absorbed the substances from the upper layer. Genistein and dacarbazine were selected as model drugs. Combined treatment of drugs affects cell survival, liver toxicity, and cell cycle. The results indicated that the viability of HepG2 cells was not obviously suppressed when the combined concentration was below − 100 μg/mL, and HepG2 cells maintained the ability of drug metabolism. High drug concentration (above 250 μg/mL) induced HepG2 cells apoptosis. Mass spectrometry (ESI-IT-TOF-MS and ESI-Q-TOF-MS) was used to elucidate the metabolic pathway and the metabolites. This co-culture chip successfully presented a platform for long-term observation of drug absorption, transport and metabolism, and could be a useful in vitro simulation model for further drug screening. Similarly, Choi et al. [68] proposed a compartmentalized 3D microfluidic device (Fig. 5B) for drug screening with co-cultured breast tumor spheroids and human mammary duct epithelial cells as well as mammary fibroblasts. They studied the impact of the clinical anticancer drug paclitaxel related to the size of ductal carcinoma in situ (DCIS). Results have shown that paclitaxel arrested tumor cells proliferation and inhibited the growth of DCIS in the microenvironment. This micro-engineered disease model represented the complexity of breast cancer pathophysiology and could be used as a favorable tool to systematically study the invasive of DCIS in the breast cancer microenvironment. To create a higher throughput system, Xu et al. [69] established a platform comprising 4 × 3 hydrogel culture chambers for anticancer drug sensitivity test (Fig. 5C). The human non-small cell lung cancer cell line (SPCA-1), and human lung fibroblast cell line (HFL-1) cells from fresh tumor tissues of lung cancer were fed into a 3D chip under the condition of continuous flow medium to simulate the in vivo tumor microenvironment. Those cells were treated with anticancer drugs in three different concentrations produced automatically by concentration gradient generators (CGG) to screen the appropriate chemotherapy schemes. They successfully measured the sensitivity of different anticancer drugs in parallel and recommended appropriate doses and single/combination chemotherapy schemes for eight patients. With the advantages of simplicity, reliability and high throughput, this microfluidic platform was claimed to be a promising tool for screening individualized treatment.
Fig. 5.
Microfluidic cell co-culture systems applied to anti-cancer drug screening. (A) Schematic illustration of the microfluidic intestine-liver model. (i) The overall schematic representation of the microfluidic intestine-liver chip; (ii) Schematic illustration of the double-layer microchip. Reprinted with permission from Ref. [67]. (B) 3D co-culture model for drug screening.(i) DCIS is embedded in a mammary duct consisting of the mammary epithelium and a basement membrane surrounded by stromal tissue that contains fibroblasts; (ii)The microarchitecture of DCIS and the surrounding tissue layers is reproduced in the breast cancer-on-a-chip micro-device. Reprinted with permission from Ref. [68]. (C) A microfluidic3D co-culture system for drug sensitivity testing of a lung cancer model. Reprinted with permission from Ref. [69].
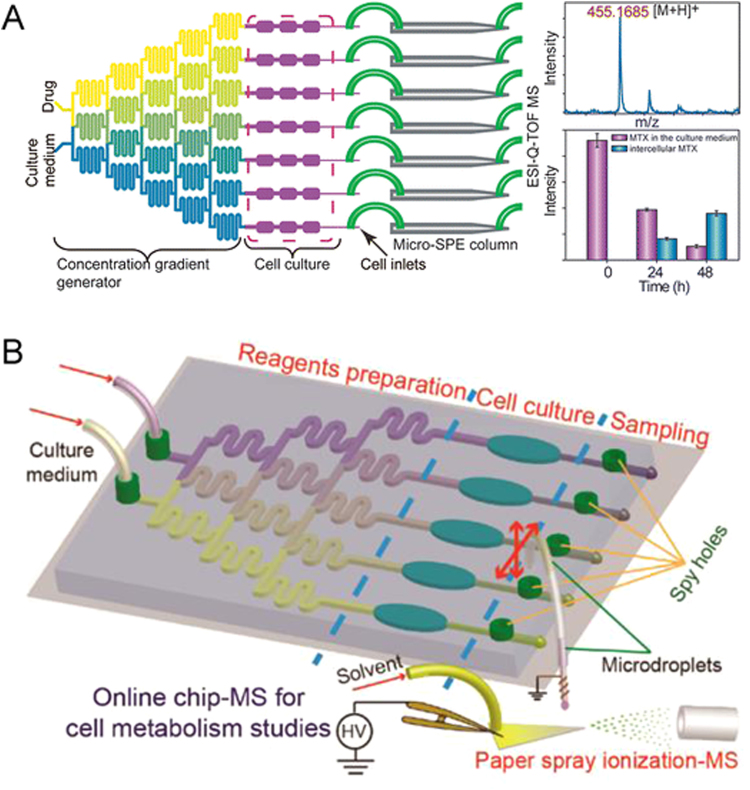
It is necessary to detect the drug metabolism with microfluidic chip, which is based on cell-to-cell interaction to screen drugs with high throughput. The detectors commonly used in the microfluidic analysis platform include electrochemistry [70], fluorescence microscopy [71], and mass spectrometry [72], [73]. Preferred characteristics for a detector are good specificity, high sensitivity, low cost and time saving, especially for microanalysis in complex systems. For a more comprehensive discussion about detectors suitable for biospecies detection, readers can refer to a previous review [74]. In general, mass spectrometry (MS) is the most powerful tool in metabolic spectrum analysis, because it provides abundant structural information and simultaneous detection of multiple analytes by different mass to charge (m/z) ratio. The MS can detect most biomolecules ranging from small molecules to peptides, proteins, and nucleic acids. Recently, the rapid development of high resolution MS has reduced the limit of detection (LOD) to fmol [75]. Our laboratory has set up a lot of work on “Chip-MS” based on co-culture microfluidic device for cell or drug metabolism detection. As shown in Fig. 6A, an integrated co-culture microfluidic platform was designed for high-throughput drug screening with online ESI-Q-TOF MS [76]. This device consisted of two different functional components that could be connected by polyethylene tubes. One component was drug gradient generators followed by cell co-culture chambers, in which a multi process event could be achieved: liquid diffusion and mixing, cell co-culture, drug stimulation, drug absorption and drug induced cytotoxicity test; another component was with on-chip solid-phase extraction (SPE) columns for performing sample cleanup and concentration prior to MS analysis. Thus, this combination system could simultaneously realize characterization of drug absorption and evaluation of cytotoxicity. With the increase of drug concentration, the proportion of apoptotic cells (HepG2 and Caco-2) was dose-dependent. That is to say, higher drug concentration induced increased cell cytotoxicity, which was also proved by the results of ESI-Q-TOF MS analysis. The “Chip-MS” model provided a simple method for drug screening, which had the characteristics of high detecting speed, low sample consumption, high throughput and good sensitivity. Liu et al. [77] reported a “Chip-MS” microfluidic device (Fig. 6B) with paper spray ionization MS, which did not require SPE columns to clean up and concentrate the sample. With this online multichannel “Chip-MS” microfluidic platform, lactate efflux from normal cells and cancer cells was tested under hypoxia condition. Besides, differential inhibitory effects and dose−response information about α-cyano-4-hydroxycinnamate on different cancer cells were also monitored. Although the system provided a rapid evaluation of cellular metabolism and drug screening but lacked the investigation of the cells co-culture.
Fig. 6.
“Chip-MS” based on microfluidic device for high-throughput drug screening with online (A) ESI-Q-TOF MS. Reprinted with permission from Ref. [76], and (B) paper spray ionization MS. Reprinted with permission from Ref. [77].
Scientists have developed a number of microchip models to explore tumor and drug screening. Most of these works have tried to achieve high throughput and sensitivity and it is believed that these could provide a useful platform for effective drug screening. However, still microfluidic chips for drug screening cope with low throughput, low sensitivity, and complex design. Moreover, microfluidic systems face huge challenges covering rebuilding complex 3D models to mimic in vivo-like microenvironments surrounding a variety of different types of cancer and exploring high-content and high-throughput analysis for testing new anti-cancer drugs. Vascular bio-chip model serves as a basic research in angiogenesis. Similarly, screening biological factors promoting angiogenesis and regeneration are of great significance for the developmental biology and the screening of inhibitory drugs could provide a valuable research for cancer biology.
5. Cell-to-cell heterogeneity
The biochemical samples prepared by traditional methods are usually obtained from a large amount of cells. However, the seemingly identical cells in a cellular environment exhibit genetic and phenotypic heterogeneity [78]. Single-cell technology provides an important strategy to understand cell behavior, cell metabolism, and also identifies the cellular heterogeneity within a single cancer type. Cell adhesion is one of the most vital features related to cell spreading area, cell size, cytoactive, and intercellular metabolites [79]. Especially, it is also very important to find out the process of CTCs adhesion to ECs. However, traditional methods for cell adhesion test are commonly characterized by counting cells number, which produce statistical results failing to represent the cell adherence ability of individual cells [80], [81]. Recently, our group reported a microfluidic chip-based live single-cell extractor (LSCE), which could extract an adhered single-cell without deterioration of cellular components [82]. This design contained two parallel micro-channels, which realized the control of micro-region fluid in open environment (Fig. 7). Trypsin in the micro-region acted locally on a single-cell below the chip, achieving the extraction of single cell by disrupting the connecting matrix between the substrate and the cell. This LSCE system successfully revealed the cell heterogeneity between cell adhesion and cell morphology, and provided a region-selective, non-contact, and scatheless single-cell isolation method. The technique has an excellent performance in the study of specific cell groups or specific single cells in a large number of cells. It could extract any adhered single cell in situ, and keep the cell activity at the same time. We believe that this technique could provide a reliable method to study the interaction of tumor cells adhered to endothelial cells at single cell levels.
Fig. 7.
Microfluidic chip-based live single-cell extractor (LSCE). (A) Illustration of LSCE and single-cell extraction system; (B) Illustration of microjet applied on alive single-cell; (C) Live single-cell detachment process. Reprinted with permission from Ref. [82].
6. Conclusions and future directions
In this review, we described the advances in microfluidic co-culture systems for studying the TCs-ECs interaction. Microfluidic models are emerging as powerful tools for the investigation of cell-to-cell interactions. The 2D microfluidic devices have the advantages such as low cost, reproducibility, accessibility, and easy operation, but fail to reiterate physiological systems. The 3D microfluidic platform enabled the simulation of natural cell environment but faced challenges of low throughput, integration of several components, and complex operation for analytical applications. Moreover, most of these efforts are focused on the theoretical proof rather than the development of a universal scheme that can replace the existing models. To adopt microfluidic models for clinical application, it is necessary to combine the primary in vitro cell models with the standardized micro-devices. In the future, more efforts should be made in selection and combination of standardized components and their applications within microfluidic chips to generate more human-like biological microenvironment. Commercialized microfluidic chips may be standardized to produce a wide application prospect, which could be readily available for research purposes and but also can be applied to clinical treatment and screening of individualized treatment prescription for cancer patients.
Acknowledgments
Conflicts of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
We acknowledge the financial support from National Natural Science Foundation of China (Nos. 214350002, 21727814 and 21621003).
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
References
- 1.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Fidler I.J. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat. Rev. Cancer. 2013;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 3.Gupta G.P., Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Talmadge J.E., Fidler I.J. AACR Centennial Series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70:5649–5669. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer A.H. Circulating tumor cells: seeing is believing. Arch. Pathol. Lab. Med. 2009;133:1367–1369. doi: 10.5858/133.9.1367. [DOI] [PubMed] [Google Scholar]
- 6.McCarty O.J.T., Ku D., Sugimoto M. Dimensional analysis and scaling relevant to flow models of thrombus formation: communication from the SSC of the ISTH. J. Thromb. Haemost. 2016;14:619–622. doi: 10.1111/jth.13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell M.J., King M.R. Computational and experimental models of cancer cell response to fluid shear stress. Front. Oncol. 2013;3:44. doi: 10.3389/fonc.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phillips K.G., Kuhn P., McCarty O.J. Physical biology in cancer. 2. The physical biology of circulating tumor cells. Am. J. Physiol. Cell Physiol. 2014;306:C80–C88. doi: 10.1152/ajpcell.00294.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wirtz D., Konstantopoulos K., Searson P.C. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat. Rev. Cancer. 2011;11:512–522. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitrugno A., Tormoen G.W., Kuhn P. The prothrombotic activity of cancer cells in the circulation. Blood Rev. 2016;30:11–19. doi: 10.1016/j.blre.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rejniak K.A. Circulating tumor cells: when a solid tumor meets a fluid microenvironment. Adv. Exp. Med. Biol. 2016;936:93–106. doi: 10.1007/978-3-319-42023-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ali S., Lazennec G. Chemokines: novel targets for breast cancer Metastasis. Cancer Metastasis. Rev. 2007;26:401–420. doi: 10.1007/s10555-007-9073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert A.W., Pattabiraman D.R., Weinberg R.A. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greene J.M., Levy D., Herrada S.P. Mathematical modeling reveals that changes to local cell density dynamically modulate baseline variations in cell growth and drug response. Cancer Res. 2016;76:2882–2890. doi: 10.1158/0008-5472.CAN-15-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izumchenko E., Chang X., Michailidi C. The TGFβ-miR200-Mig6 pathway orchestrates the EMT-associated kinase switch that induces resistance to EGFR inhibitors. Cancer Res. 2014;74:3995–4005. doi: 10.1158/0008-5472.CAN-14-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massague J., Obenauf A.C. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sosa M.S., Bragado P., Aguirre-Ghiso J.A. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat. Rev. Cancer. 2014;14:611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomax-Browne H., Brooks S. Interactions between cancer cell glycans and endothelial cells during adhesion and transmigration events in metastasis. Comp. Biochem. Physiol. Part A. 2008;150:S191–S192. [Google Scholar]
- 19.Thiele J.A., Bethel K., Kralickova M. Circulating tumor cells: fluid surrogates of solid tumors. Annu. Rev. Pathol. 2017;12:419–447. doi: 10.1146/annurev-pathol-052016-100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joyce J.A., Pollard J.W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeung K.T., Yang J. Epithelial–mesenchymal transition in tumor metastasis. Mol. Oncol. 2017;11:28–39. doi: 10.1002/1878-0261.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folkman J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat. Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 24.Khodarev N.N., Yu J., Labay E. Tumour-endothelium interactions in co-culture: coordinated changes of gene expression profiles and phenotypic properties of endothelial cells. J. Cell. Sci. 2003;116:1013–1022. doi: 10.1242/jcs.00281. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann O.I., Ilmberger C., Magosch S. Impact of the spheroid model complexity on drug response. J. Biotechnol. 2015;205:14–23. doi: 10.1016/j.jbiotec.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 26.Lu J., Ye X., Fan F. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of jagged-1. Cancer Cell. 2013;23:171–185. doi: 10.1016/j.ccr.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo F., Li P., French J.B. Controlling cell–cell interactions using surface acoustic waves. Proc. Natl. Acad. Sci. 2015;112:43–48. doi: 10.1073/pnas.1422068112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye L.-Y., Chen W., Bai X.-L. Hypoxia-induced epithelial-to-mesenchymal transition in hepatocellular carcinoma induces an immunosuppressive tumor microenvironment to promote metastasis. Cancer Res. 2016;76:818–830. doi: 10.1158/0008-5472.CAN-15-0977. [DOI] [PubMed] [Google Scholar]
- 29.Ham B., Wang N., D'Costa Z. TNF receptor-2 facilitates an immunosuppressive microenvironment in the liver to promote the colonization and growth of hepatic metastases. Cancer Res. 2015;75:5235–5247. doi: 10.1158/0008-5472.CAN-14-3173. [DOI] [PubMed] [Google Scholar]
- 30.Chen B.P., Li Y.S., Zhao Y. DNA microarray analysis of gene expression in endothelial cells in response to 24-h shear stress. Physiol. Genom. 2001;7:55–63. doi: 10.1152/physiolgenomics.2001.7.1.55. [DOI] [PubMed] [Google Scholar]
- 31.Weinbaum S., Tarbell J.M., Damiano E.R. The structure and function of the endothelial glycocalyx layer. Annu. Rev. Biomed. Eng. 2007;9:121–167. doi: 10.1146/annurev.bioeng.9.060906.151959. [DOI] [PubMed] [Google Scholar]
- 32.Dong C., Slattery M., Liang S. Micromechanics of tumor cell adhesion and migration under dynamic flow conditions. Front. Biosci. 2005;10:379–384. doi: 10.2741/1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomes N., Berard M., Vassy J. Shear stress modulates tumour cell adhesion to the endothelium. Biorheology. 2003;40:41–45. [PubMed] [Google Scholar]
- 34.Wang J., Loberg R., Taichman R.S. The pivotal role of CXCL12 (SDF-1)/CXCR4 axis in bone metastasis. Cancer Metastasis. Rev. 2006;25:573–587. doi: 10.1007/s10555-006-9019-x. [DOI] [PubMed] [Google Scholar]
- 35.Rothbauer M., Zirath H., Ertl P. Recent advances in microfluidic technologies for cell-to-cell interaction studies. Lab Chip. 2018;18:249–270. doi: 10.1039/c7lc00815e. [DOI] [PubMed] [Google Scholar]
- 36.Menon N.V., Chuah Y.J., Cao B. A microfluidic co-culture system to monitor tumor-stromal interactions on a chip. Biomicrofluidics. 2014;8:70–78. doi: 10.1063/1.4903762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charwat V., Rothbauer M., Tedde S.F. Monitoring dynamic interactions of tumor cells with tissue and immune cells in a lab-on-a-chip. Anal. Chem. 2013;85:11471–11478. doi: 10.1021/ac4033406. [DOI] [PubMed] [Google Scholar]
- 38.Rothbauer M., Ertl P., Theiler B.A. Biointerfaces: anisotropic crystalline protein nanolayers as multi-functional biointerface for patterned co-cultures of adherent and non-adherent cells in microfluidic devices. Adv. Mater. Interfaces. 2015;2:4557–4558. [Google Scholar]
- 39.Konstantopoulos K., Thomas S.N. Cancer cells in transit: the vascular interactions of tumor cells. Annu. Rev. Biomed. Eng. 2009;11:177–202. doi: 10.1146/annurev-bioeng-061008-124949. [DOI] [PubMed] [Google Scholar]
- 40.Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- 41.Whiteside T.L. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27:5904–5912. doi: 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin X., Chen Q., Liu W. Oxygen-induced cell migration and on-line monitoring biomarkers modulation of cervical cancers on a microfluidic system. Sci. Rep. 2015;5:9643. doi: 10.1038/srep09643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao S., Zhang J., Li H. Strategy for signaling molecule detection by using an integrated microfluidic device coupled with mass spectrometry to study cell-to-cell communication. Anal. Chem. 2013;85:868–876. doi: 10.1021/ac303164b. [DOI] [PubMed] [Google Scholar]
- 44.Huang R., Zheng W., Liu W. Investigation of tumor cell behaviors on a vascular microenvironment-mimicking microfluidic chip. Sci. Rep. 2015;5:17768. doi: 10.1038/srep17768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao Y., Majumdar D., Jovanovic B. A versatile valve-enabled microfluidic cell co-culture platform and demonstration of its applications to neurobiology and cancer biology. Biomed. Microdevices. 2011;13:539–548. doi: 10.1007/s10544-011-9523-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng C., Zhao L., Chen Ge. Quantitative study of the dynamic tumor–endothelial cell interactions through an integrated microfluidic coculture system. Anal. Chem. 2012;84:2088–2093. doi: 10.1021/ac2032029. [DOI] [PubMed] [Google Scholar]
- 47.Lin L., Lin X., Feng Q. Integrated microfluidic platform with multiple functions to probe tumor-endothelial cell interaction. Anal. Chem. 2017;89:10037–10044. doi: 10.1021/acs.analchem.7b02593. [DOI] [PubMed] [Google Scholar]
- 48.Xu X., Farach-Carson M.C., Jia X. Three-dimensional in vitro tumor models for cancer research and drug evaluation. Biotechnol. Adv. 2014;32:1256–1268. doi: 10.1016/j.biotechadv.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamichhane S.P., Arya N., Kohler E. Recapitulating epithelial tumor microenvironment in vitro using three dimensional tri-culture of human epithelial, endothelial, and mesenchymal cells. BMC Cancer. 2016;16::581. doi: 10.1186/s12885-016-2634-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cavo M., Fato M., Peñuela L. Microenvironment complexity and matrix stiffness regulate breast cancer cell activity in a 3D in vitro model. Sci. Rep. 2016;6::35367. doi: 10.1038/srep35367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klimkiewicz K., Weglarczyk K., Collet G. A 3D model of tumour angiogenic microenvironment to monitor hypoxia effects on cell interactions and cancer stem cell selection. Cancer Lett. 2017;396:10–20. doi: 10.1016/j.canlet.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 52.Fischbach C., Chen R., Matsumoto T. Engineering tumors with 3D scaffolds. Nat. Methods. 2007;4:855–860. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 53.Kenny P.A., Lee G.Y., Myers C.A. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu T., Lin B., Qin J. Carcinoma-associated fibroblasts promoted tumor spheroid invasion on a microfluidic 3D co-culture device. Lab Chip. 2010;10:1671–1677. doi: 10.1039/c000022a. [DOI] [PubMed] [Google Scholar]
- 55.Hsiao A.Y., Torisawa Y.S., Tung Y.C. Microfluidic system for formation of PC-3 prostate cancer co-culture spheroids. Biomaterials. 2009;30:3020–3027. doi: 10.1016/j.biomaterials.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang F., Weaver V.M., Petersen O.W. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc. Natl. Acad. Sci. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hosseinkhani H., Hosseinkhani M., Tian F. Osteogenic differentiation of mesenchymal stem cells in self-assembled peptide-amphiphile nanofibers. Biomaterials. 2006;27:4079–4086. doi: 10.1016/j.biomaterials.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 58.Rhodes N.P., Srivastava J.K., Smith R.F. Metabolic and histological analysis of mesenchymal stem cells grown in 3-D hyaluronan-based scaffolds. J. Mater. Sci. Mater. Med. 2004;15:391–395. doi: 10.1023/b:jmsm.0000021108.74004.7e. [DOI] [PubMed] [Google Scholar]
- 59.Asghar W., El Assal R., Shafiee H. Engineering cancer microenvironments for in vitro 3-D tumor models. Mater. Today. 2015;18:539–553. doi: 10.1016/j.mattod.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zschenker O., Streichert T., Hehlgans S. Genome-wide gene expression analysis in cancer cells reveals 3D growth to affect ECM and processes associated with cell adhesion but not DNA repair. Plos One. 2012;7:11. doi: 10.1371/journal.pone.0034279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jain R.K., Duda D.G., Clark J.W. Lessons from phase III clinical trials on anti-VEGF therapy for cancer. Nat. Clin. Pract. Oncol. 2006;3:24–40. doi: 10.1038/ncponc0403. [DOI] [PubMed] [Google Scholar]
- 62.Pampaloni F., Reynaud E.G., Stelzer E.H. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 63.Zhang W., He Z., Yi L. A dual-functional microfluidic chip for on-line detection of interleukin-8 based on rolling circle amplification. Biosens. Bioelectron. 2018;102:652–660. doi: 10.1016/j.bios.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 64.Song J.W., Cavnar S.P., Walker A.C. Microfluidic endothelium for studying the intravascular adhesion of metastatic breast cancer cells. Plos One. 2009;4:e5756. doi: 10.1371/journal.pone.0005756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu H., Jie M., He Z. Study of antioxidant effects on malignant glioma cells by constructing a tumor-microvascular structure on microchip. Anal. Chim. Acta. 2017;978:1–9. doi: 10.1016/j.aca.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 66.Zervantonakis I.K., Hughes-Alford S.K., Charest J.L. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl. Acad. Sci. 2012;109:13515–13520. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jie M., Lin H., He Z. An on-chip intestine-liver model for multiple drugs absorption and metabolism behavior simulation. Sci. China Chem. 2018;61:236–242. [Google Scholar]
- 68.Choi Y., Hyun E., Seo J. A microengineered pathophysiological model of early-stage breast cancer. Lab Chip. 2015;15:3350–3357. doi: 10.1039/c5lc00514k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu Z., Gao Y., Hao Y. Application of a microfluidic chip-based 3D co-culture to test drug sensitivity for individualized treatment of lung cancer. Biomaterials. 2013;34:4109–4117. doi: 10.1016/j.biomaterials.2013.02.045. [DOI] [PubMed] [Google Scholar]
- 70.Jie X., Yang H., Wang M. A peroxisome-inspired chemiluminescent silica nanodevice for the intracellular detection of biomarkers and its application to insulin-sensitizer screening. Angew. Chem. Int. Ed. 2017;129:14788–14793. doi: 10.1002/anie.201708958. [DOI] [PubMed] [Google Scholar]
- 71.Tan M., Yamaguchi S., Yamahira S. Quantitative image cytometry for analyzing intracellular trafficking of G protein-coupled receptors on a chemical-trapping single cell array. Lab Chip. 2017;17:1933–1938. doi: 10.1039/c7lc00198c. [DOI] [PubMed] [Google Scholar]
- 72.Lee S.J., Yoon H.S., Xuan X. A patch type non-enzymatic biosensor based on 3D SUS micro-needle electrode array for minimally invasive continuous glucose monitoring. Sens. Actuators B. 2016;222:1144–1151. [Google Scholar]
- 73.Windmiller J.R., Zhou N., Chuang M.C. Microneedle array-based carbon paste amperometric sensors and biosensors. Analyst. 2011;136:1846–1851. doi: 10.1039/c1an00012h. [DOI] [PubMed] [Google Scholar]
- 74.Yaling L., Shunqiang W., Yu S. Biospecies capture and detection at low concentration. Micro Nanosyst. 2012;4:254–272. [Google Scholar]
- 75.Lin L., Lin J.-M. Development of cell metabolite analysis on microfluidic platform. J. Pharm. Anal. 2015;5:337–347. doi: 10.1016/j.jpha.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao D., Li H., Wang N. Evaluation of the absorption of methotrexate on cells and its cytotoxicity assay by using an integrated microfluidic device coupled to a mass spectrometer. Anal. Chem. 2012;84:9230–9237. doi: 10.1021/ac301966c. [DOI] [PubMed] [Google Scholar]
- 77.Liu W., Lin J.-M. Online Monitoring of lactate efflux by multi-channel microfluidic chip-mass spectrometry for rapid drug evaluation. ACS Sens. 2016;1:344–347. [Google Scholar]
- 78.Jiang Y., Zhao H., Lin Y. Colorimetric detection of glucose in rat brain using gold nanoparticles. Angew. Chem. Int. Ed. 2010;49:4800–4804. doi: 10.1002/anie.201001057. [DOI] [PubMed] [Google Scholar]
- 79.Yu H., Huang S., Chokhawala H. Highly efficient chemoenzymatic synthesis of naturally occurring and non-natural α-2,6-linked sialosides: A.P. Damsela α-2,6-sialyltransferase with extremely flexible donor–substrate specificity. Angew. Chem. Int. Ed. 2006;118:4042–4048. doi: 10.1002/anie.200600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nordström T., Knekt M., Nordström E. A microplate-based fluorometric assay for monitoring human cancer cell attachment to cortical bone. Anal. Biochem. 1999;267:37–45. doi: 10.1006/abio.1998.2971. [DOI] [PubMed] [Google Scholar]
- 81.Pagel M., Hassert R., John T. Multifunktionale Beschichtung verbessert Zelladhäsion auf titan durch kooperativ wirkende peptide. Angew. Chem. Int. Ed. 2016;128:4907–4911. [Google Scholar]
- 82.Mao S., Zhang W., Huang Q. In Situ Scatheless cell detachment reveals correlation between adhesion strength and viability at single-cell resolution. Angew. Chem. Int. Ed. 2018;57:236–240. doi: 10.1002/anie.201710273. [DOI] [PubMed] [Google Scholar]