Abstract
As a core component of the adherens junction, α-catenin stabilizes the cadherin/catenin complexes to the actin cytoskeleton for the mechanical coupling of cell-cell adhesion. α-catenin also modulates actin dynamics, cell polarity, and cell-migration functions that are independent of the adherens junction. We have determined the solution structures of the α-catenin monomer and dimer using in-line size-exclusion chromatography small-angle X-ray scattering, as well as the structure of α-catenin dimer in complex to F-actin filament using selective deuteration and contrast-matching small angle neutron scattering. We further present the first observation, to our knowledge, of the nanoscale dynamics of α-catenin by neutron spin-echo spectroscopy, which explicitly reveals the mobile regions of α-catenin that are crucial for binding to F-actin. In solution, the α-catenin monomer is more expanded than either protomer shown in the crystal structure dimer, with the vinculin-binding M fragment and the actin-binding domain being able to adopt different configurations. The α-catenin dimer in solution is also significantly more expanded than the dimer crystal structure, with fewer interdomain and intersubunit contacts than the crystal structure. When in complex to F-actin, the α-catenin dimer has an even more open and extended conformation than in solution, with the actin-binding domain further separated from the main body of the dimer. The α-catenin-assembled F-actin bundle develops into an ordered filament packing arrangement at increasing α-catenin/F-actin molar ratios. Together, the structural and dynamic studies reveal that α-catenin possesses dynamic molecular conformations that prime this protein to function as a mechanosensor protein.
Introduction
α-Catenin is a necessary component of the cadherin-catenin complexes that stabilize the cell-cell adherens junctions (AJ) (1, 2, 3). The AJs are specialized eukaryotic cell-cell anchoring junctions found in epithelial, endothelial, and neuronal tissues of higher organisms and are essential for embryonic morphogenesis, tissue integrity and remodeling, and wound healing. The AJs are composed of transmembrane cadherins that bind extracellularly in a homotypic and calcium-dependent manner with the cadherins on adjacent cells, and cadherins interact intracellularly with the actin microfilaments (4). Linkage of cadherins to microfilaments is accomplished by the β-catenin/α-catenin complex (5, 6, 7), with α-catenin interacting directly with the actin filaments in a cell-cell contact and mechanical-stress-dependent fashion (8, 9). The cadherin/β-catenin/α-catenin complex can also interact with the actin cytoskeleton indirectly by binding to other actin-binding proteins, including α-actinin, vinculin, and eplin (10, 11, 12), which form an alternative linkage of the cadherin complex to the actin cytoskeleton. α-Catenin is thought to undergo a significant conformational change in response to mechanical stress to associate with the microfilaments and with other actin-binding proteins; recent biophysical studies have identified the changes that occur to α-catenin and vinculin under pN force levels (13). Because of the role that α-catenin plays in facilitating the physical-stress-dependent interactions of the cadherin-catenin complex, α-catenin is considered a mechanosensor (9, 14, 15, 16, 17).
Additionally, α-catenin is localized outside the adherens junction complex and regulates actin dynamics that is independent of the AJs (18, 19, 20). The homodimers of α-catenin bind to F-actin and bundle the actin filaments (5, 21), which inhibit Arp2/3-complex-mediated actin polymerization, and alters lamellipodia architecture and cell migration (18, 22). Recent studies show that α-catenin homodimerization is sufficient to control its cortical localization and that the α-catenin homodimers are recruited to the phosphoinositide-activated filopodia membrane protrusion to promote cell-cell adhesion (19).
α-Catenin is a 906-residue protein that has six functional domains (see Fig. 1 A). The N-terminal homodimerization N1 and N2 domains overlap with β-catenin binding sites (7, 23, 24). The middle M-fragment composed of three four-bundle helical domains M1, M2, and M3. M1 binds vinculin in a mechanical-stress-dependent manner (25, 26, 27), and the C-terminal actin-binding domain (ABD) is composed of a five-helix bundle plus a disordered C-terminal region that also binds to the tight-junction protein ZO-1 (28). Crystallographic studies have determined the structures of several constructs of α-catenin, including the homodimerization domain that overlaps with the heterodimerization with β-catenin (29), the M-fragment (30), a larger N domain plus the M domains as well as a portion of the actin-binding domain (27), and the heterodimer structure of a chimera of the N1 domain with a β-catenin-binding peptide (23). The nearly full-length α-catenin homodimer provides a complete view of an asymmetric α-catenin dimer at atomic resolution (31). However, the crystal structures are significantly compact, and it is difficult to explain some of the functional properties of α-catenin such as the reversible dynamic conformational changes under force (32, 33). The molecular dynamics allowing α-catenin to function as a mechanosensor is thus largely unknown.
Figure 1.
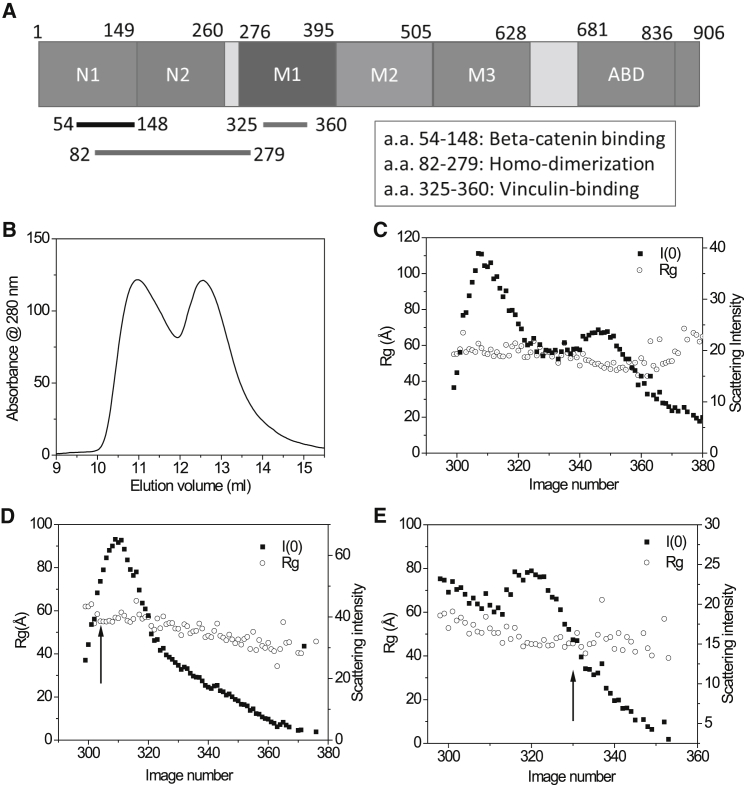
α-Catenin and SEC-SAXS analysis. (A) The primary structure and domains in α-catenin are shown. (B) SEC-ultraviolet of α-catenin is shown. The fractions at elution volume 10.9 and 12.5 mL correspond to dimeric and monomeric α-catenin, respectively. (C) SEC-SAXS of the dimer and monomer peaks is shown. (D) SEC-SAXS of the dimer peak is shown. (E) SEC-SAXS of the monomer peak is shown. Note that because (E) used a different SEC column from (C) and (D), the peak positions are shifted. The arrows in (D) and (E) mark the fractions of SAXS data selected for further analysis as shown in Fig. 2.
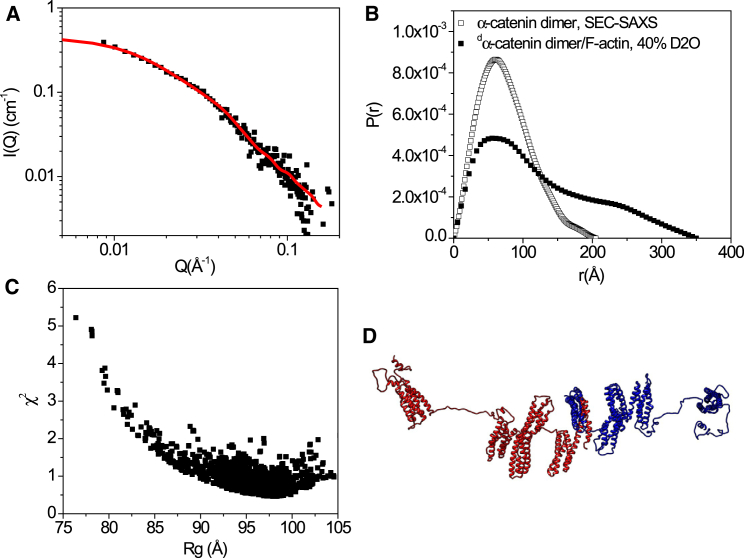
Using in-line size-exclusion chromatography small-angle x-ray scattering (SEC-SAXS), we are able to separate the α-catenin monomer and dimer fractions and perform SAXS to determine the solution structures of the full-length α-catenin monomer and dimer, respectively. The SEC-SAXS experiments reveal flexible structures of the α-catenin monomer in solution, in particular with flexible linkers between the N2 and M1 domains and between the M-fragment and the ABD. The solution structure of the homodimer is more expanded than the crystal structure, with ABDs being well separated from the main fragment of each protomer. We have also performed contrast-matching small angle neutron scattering (SANS) on deuterium-labeled α-catenin in complex with F-actin and showed that α-catenin adopts an even more extended conformation than in solution. At increasing α-catenin/actin molar ratios, α-catenin starts to assemble F-actin filaments into bundles, with ordered filament arrangement and an interfilament spacing of αβωνθ 180 Å.
Because our solution structural analyses suggest flexible conformations of α-catenin, we posit that the dynamic conformations of α-catenin play roles in binding to F-actin and for α-catenin to function as a mechanosensor. Here, we report the first analysis, to our knowledge, of the nanoscale dynamics of the α-catenin dimer using neutron spin-echo spectroscopy (NSE) to identify the mobile regions in the α-catenin dimer on submicrosecond time scales. The NSE data and our theoretical analyses suggest that the dynamic regions are the disordered C-terminal tails of ABDs in each protomer, which have previously been implicated as essential in stabilizing cell-cell adhesion and in normal brain tissue development (34, 35).
Materials and Methods
Protein expression and purification
The human cDNA encoding the full-length human αE-catenin was subcloned into the pET32a vector (EMD Biosciences, San Diego, CA), with a 6× His plus thioredoxin fusion tag followed by a tobacco etch virus protease (TEV) cleavage sequence at the amino-terminal end of the expressed proteins. The expression plasmid was transformed into the Rosetta 2 (DE3) competent cells (EMD Biosciences). The bacterial cells were grown in Luria-Bertani medium in the presence of 0.1 mg/mL ampicillin and 0.034 mg/mL chloramphenicol. At the cell optical density of ∼0.7 at 600 nm, the cells were induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside, and the cells were grown at 20°C overnight. The cells were harvested, pelleted, and stored at −80°C.
For protein purification, the cell pellet was suspended in 50–80 mL lysis/binding buffer containing 50 mM sodium phosphate (pH 7.5), 300 mM NaCl, 20 mM imidazole, 0.25 mM tris (2-carboxyethyl) phosphine hydrochloride, and 0.1 mM phenylmethylsulfonyl fluoride. The cell suspension was subjected to sonication cycles on ice, and the lysed cell was centrifuged at 9000 rotations per minute for 20 min at 4°C. The supernatant was purified by a HiTrap Chelating Sepharose HP (GE Healthcare Life Sciences, Wauwatosa, WI) precharged with Ni2+ and pre-equilibrated with the lysis/binding buffer. The protein was then further purified and analyzed by SEC using an ÄKTA fast-protein liquid chromatograph (GE Healthcare Life Sciences) with a Superdex 200 Increase 10/30 column (GE Healthcare Life Sciences). The SEC running buffer is 25 mM Tris-HCl (pH 8.0), 300 mM NaCl, 3 mM ethylenediaminetetra acetic acid, and 0.25 mM tris (2-carboxyethyl) phosphine hydrochloride. The fusion tag was incubated with 20 μM TEV protease on ice for 24 hr. After TEV cleavage, only a glycine residue remains at the N-terminus of α-catenin. A second round of SEC was used to separate α-catenin from the fusion tag. The protein concentrations were determined by ultraviolet absorbance at 280 nm using the extinction coefficient calculated using the ProtParam program (36) on the ExPASy Proteomics Server (https://us.expasy.org/).
For producing deuterated proteins for SANS studies, the α-catenin expression plasmid was transformed in Rosetta 2(DE3) cells, and the cells were grown in M9 medium containing 85% D2O using our published protocols (37, 38, 39, 40, 41). Purification of the deuterated protein was the same as that of the undeuterated proteins.
Actin polymerization and reconstitution of α-catenin/F-actin complex
To polymerize actin, 10 mg of lyophilized nonmuscle G-actin (from Cytoskeleton, Denver, CO) was resuspended by adding 900 μL of G-actin buffer of 20 mM Tris-HCl (pH 7.5), 0.2 mM CaCl2, 0.2 mM ATP, and 0.2 mM 1,4-dithiothreitol. A 100 μL 10× F-actin polymerization buffer of 500 mM KCl, 20 mM MgCl2, and 10 mM ATP was added to achieve a final concentration of 1× F-actin buffer to initiate actin polymerization. Actin was allowed to polymerize at room temperature for 1 hr. Before SANS experiments, the deuterated α-catenin (dα-catenin) was mixed with a stock solution of F-actin of 10 mg/mL (239 μM) at an actin/α-catenin molar ratio of 15.1, 7.6, and 3.8, respectively, while the actin concentration was kept at 81.5 mg/mL. The dα-catenin/F-actin complex was then dialyzed against 40 and 100% D2O buffer of 20 mM dTris-DCl (Cambridge Isotope Lab, Tewksbury, MA) (pD 7.5), 100 mM NaCl, 50 mM KCl, and 0.2 mM ATP exhaustively at 8°C before SANS experiments. F-actin was dialyzed against 100% D2O buffer exhaustively at 8°C before SANS experiments.
In-line SEC-SAXS experiments
The SEC-SAXS experiments were performed at the Stanford Synchrotron Radiation Light Source (SSRL) Bio-SAXS beamline 4-2 in a similar manner as recently reported (42, 43, 44). The experimental setup and structural parameters are summarized in Table S1. Briefly, two same SEC columns (Superdex 200 Increase 3.2/300 column; GE Healthcare Life Sciences) were employed for high-throughput tandem SEC-SAXS data collection. The columns were equilibrated with the SEC running buffer (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 3 mM ethylenediaminetetra acetic acid, 1mM 1,4-dithiothreitol) before use. A 10 μL of 9.5 mg/mL α-catenin was applied to the column. In total, 500 images were recorded with 1-s exposure every 5 s at 0.05 mL/min flow rate.
The program SasTool (http://ssrl.slac.stanford.edu/∼saxs/analysis/sastool.htm) was employed for data reduction, including scaling, azimuthal integration, averaging, and background subtraction. The first 100 images at the early part of the void volume were averaged and used as a buffer-scattering profile for the background subtraction. After taking the first 100 images or so, the x-ray shutter was closed until the main elution peak so as to keep the sample cell clean from the radiation-damaged sample. The data were then presented as I(Q) vs. Q, where Q = 4πsin(θ)/λ, 2θ is the scattering angle, and λ is the wavelength of the x ray. The script hplcplots, available at SSRL beamline 4-2, was used for consecutive Guinier analysis, for implementing the program AUTORG [3], and for assessing data quality (e.g., radiation damage and cleanliness of the sample cell), by providing the radius of gyration (Rg), I(0), and an experimental intensity at a low Q value. Because marginal interparticle interactions (concentration dependence) were observed over the peak, the average profiles, image number 300th–304th for the α-catenin dimer and 330th–334th for the α-catenin monomer, were generated, scaled, and merged for further analyses.
SANS experiments
SANS data were collected using the EQ-SANS instrument (45), which is a time-of-flight SANS instrument, located at the Spallation Neutron Source of Oak Ridge National Laboratory. A single instrument configuration with a 4-m sample-to-detector distance was employed. The beam was defined with a 25-mm-diameter source aperture and a 10-mm-diameter sample aperture. Two instrument configurations were used for measuring the complex. One configuration was that the instrument choppers ran at 30 Hz with frame skipping and a minimal wavelength of 2 Å. This configuration provided a Q-range of 0.004–0.40 Å−1. The other configuration was that the instrument choppers’ ran at 60 Hz, which provided a minimal wavelength of 4 Å. The configuration spans a Q-range from 0.007–0.27 Å−1. SANS experiments were performed at 10.0 ± 0.1°C.
The program Mantid (46) was used to reduce the data from the samples and from the backgrounds using standard procedures that correct for incident flux spectrum, sample transmission, and detector sensitivity, as well as the detector dark current, which represents electronic noise and natural sources of radiation. Then, the data were azimuthally averaged to provide I(Q) vs. Q. Absolute intensity scaling for both configurations was done with a calibrated standard (47). The sample scattering was then corrected for the solvent scattering by subtracting the one-dimensional profiles to produce the final, reduced data.
SAXS and SANS data analysis and computational modeling
SAXS and SANS data analyses to extract protein conformational changes is described in previous studies (38, 48, 49, 50). The length distribution function P(r) was generated using the program GNOM (51) to obtain the radius of gyration Rg and the maximal dimension Dmax. The Monte Carlo simulation module in the program suite SASSIE (52) was used to generate the all-atomic models of the α-catenin monomer and dimer. Monte Carlo simulations were performed at 300 K. The maximal angle that each torsion in each of the flexible regions can sample is 30°. About 10,000 trial attempts were performed for each round of simulations. The SASCAL module in SASSIE or the program CRYSOL or CRYSON (53) was used to compute the SAXS or SANS curves, and the ANALYZE module in SASSIE was used to compare and fit the SAXS or the SANS data. Structural models with the minimal χ2 values were selected, and a representative model is shown in the figures.
Before using SASSIE for Monte Carlo simulations, the disordered regions missing in the crystal structure of the nearly full-length α-catenin (31) (Protein Data Bank (PDB): 4IGG) were reconstructed using the homology modeling program SAXSTER (54). These include residues (amino acids (a.a.) 18–81) that were reconstructed against the crystal structure of mouse α-catenin N-terminal domain (PDB: 4P9T) (55), the linker regions a.a. 636–665 between the M3 domain and the ABD domain, and a.a. 862–907 at the C-terminal end. The full-length α-catenin structure dimer model was generated by match making to the crystal structure of α-catenin dimer, PDB: 4IGG or 4K1N (27), using UCSF Chimera (56). These full-length protein monomer and dimer models were used to compute the SAXS curves of the monomer and dimer using the program CRYSOL (53) and compared with the actual SEC-SAXS results of monomer and dimer fractions, respectively.
NSE spectroscopy experiments and data analysis
Before the NSE experiments, α-catenin was exchanged in 1× phosphate buffered saline (pH 7.4) (PBS) D2O buffer. Before making the 1× PBS D2O buffer, the PBS powder was dissolved in D2O and dried in a heated vacuum oven for at least three cycles to exchange H into D. α-Catenin at 11.0 mg/mL concentration was used for NSE experiments.
The NSE experiments were performed with the upgraded high-resolution IN15 instrument at the Institut Laue Langevin (ILL) (B.F., P. Falus, I. Hoffmann, M. Gradzielski, F. Thomas, unpublished data). We used three different wavelengths: 6, 10, and 14 Å covering 0.5–42, 0.2–194, and 0.6–532-ns time intervals. The covered Fourier time scales with the third power of the wavelength, but the incoming flux also drops roughly with the fourth power. The choice of the wavelength was made by optimizing the compromise between the resolution need and the incoming neutron flux. The beam monochromatization in each case was 15% full width at half-maximum as given by the neutron velocity selector. The samples were filled in quartz cells with 2-mm sample thickness, and the temperature was controlled at 10.0 ± 0.1°C. Instrumental resolution was measured from the standard Grafoil (GrafTech, Lakewood, OH), which gives a strong, elastic, coherent small-angle scattering. The background was measured on the PBS D2O buffer, and the sample spectra were corrected using the relative transmissions following the standard procedures.
The effective diffusion constant Deff(Q) as a function of Q, which is obtained from the normalized intermediate scattering function I(Q,t)/I(Q,0), is as follows:
To determine the center-of-mass diffusion constant Do of α-catenin, dynamic light scattering (DLS) experiments were performed using an ALV CGS3 instrument (ALV GmbH, Langen, Germany) with a fixed 90° scattering angle configuration and a 632.8 nm laser. DLS experiments were performed on the same sample used for NSE experiments and with a series of dilutions in PBS D2O buffer (see Fig. S2). DLS experiments were performed at 10.0 ± 0.1°C. Details of theoretical analysis of NSE are provided in the Supporting Materials and Methods and in (39, 40, 57, 58, 59)
Negative staining and electron microscopy
The stock solution of F-actin filament or α-catenin/F-actin was ∼1 mg/mL in F-actin buffer. Samples at various dilutions were applied to glow-discharged, carbon-coated copper grids, blotted, negatively stained with 1% uranyl acetate, and air dried. The prepared grids were visualized at the City College Electron Microscopy Facility on a JEOL 2100 transmission electron microscope (JEOL USA, Peabody, MA).
Results
Expanded solution structure of α-catenin monomer
Previous studies show that α-catenin exists as a mixture of monomer and dimer in solution (5, 6, 7, 23, 26). We have analyzed the conformation of α-catenin dimer and monomer using SEC-SAXS. SEC-SAXS shows two peaks similar to those results reported previously (see Fig.1, B and C). The first peak corresponds to a dimeric α-catenin, and the second peak is the monomeric α-catenin. The independence of Rg versus elution volume within the dimer fraction peak or the monomer peak suggests that the species detected within each fraction is homogeneous and that the intermolecular interaction effects are negligible (Fig. 1, D and E). SEC can thus separate the dimer and monomer fractions well, and the SAXS data of each peak is from the dimer and monomer fractions of α-catenin, respectively (see Fig. 2, A and B).
Figure 2.
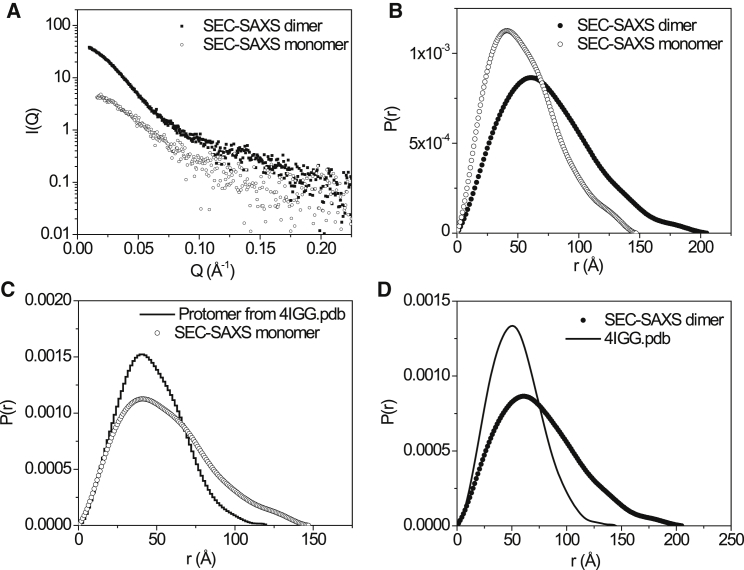
(A) SEC-SAXS of the dimer (filled black dots) and monomer fractions (open black circles). (B) The P(r) functions of dimer (filled black dots) and monomer (open black circles) calculated from SAXS data are shown. (C) A comparison of the P(r) of the SEC-SAXS monomer fraction (open black circles) with that calculated from crystal structure (one of the PDB: 4IGG protomers) after the disordered regions were reconstructed (black line). (D) A comparison of the P(r) of the SEC-SAXS dimer fraction (filled black dots) with that calculated from crystal structure (PDB: 4IGG) after the disordered regions were reconstructed in the PDB file (black line).
Table 1 summarizes the Rg and Dmax of the monomer and dimer α-catenin obtained from the SEC-SAXS experiments, respectively, as well as those parameters computed from the crystal structures (PDB: 4IGG) (31) after the missing disordered regions are reconstructed by homology modeling (see Materials and Methods). In solution, the length distribution function P(r) of the α-catenin monomer is significantly more expanded than that computed from either protomer of the α-catenin dimer crystal structure, with Rg of 43.7 ± 1.1 Å and Dmax = 147 Å (see Fig. 2 C). The Rg and Dmax of the α-catenin monomer are similar to that of the Danio rerio αE-catenin and that of Caenorhabditis elegans α-catenin homolog HMP-1 that exist as monomers in solution (60, 61) but smaller than the αN-catenin reported earlier (27).
Table 1.
Summary of Rg and Dmax of α-Catenin in Solution and in Complex to F-actin
| Rg (Å) | Dmax (Å) | |
|---|---|---|
| SEC-SAXS monomer | 43.7 ± 1.1 | 147 |
| SEC-SAXS dimer fraction | 58.8 ± 1.6 | 205 |
| In complex with F-action from contrast-matching SANS in 40% D2O | 99.7 ± 2.1 | 350 |
| Monomer crystal structure | 35.6a | 120 |
| Dimer crystal structure | 42.3a | 150 |
Calculated from crystal structure coordinates (PDB: 4IGG) after the missing disordered regions were reconstructed by homolog modeling; see Materials and Methods.
The SAXS data of dimer fraction indicates the α-catenin dimer has Rg of 58.8 ± 1.6 Å and Dmax = 205 Å, which are notably larger than those computed from the crystal structure (see Fig. 2 D and Table 1). P(r) of the dimer fraction is also more expanded than that computed from the crystal structure of the α-catenin dimer (Fig. 2 D). The comparison suggests that both the α-catenin monomer and dimer adopt conformations in solution that are quite different from the crystal structure.
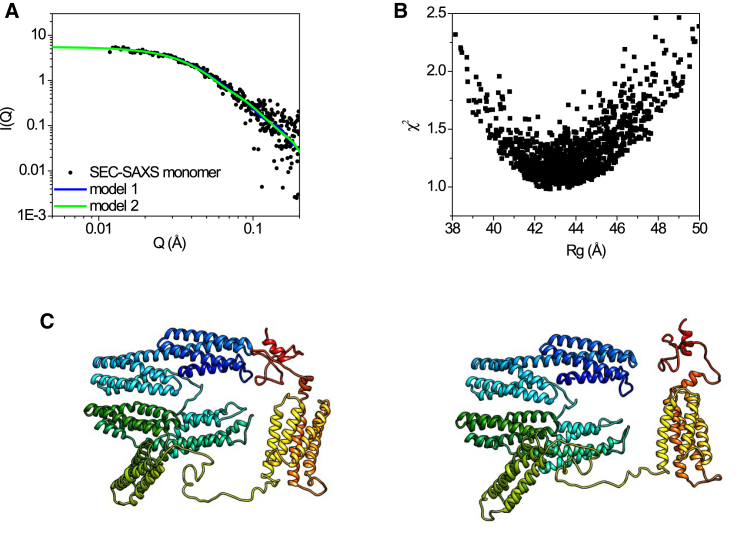
Using the SEC-SAXS data of the monomer and dimer fractions, respectively, as constraints, we have performed structural modeling of the α-catenin monomer using the program suite SASSIE (62). With SASSIE, a large number of all atomic structural models are generated by Monte Carlo simulations, and the generated models are fitted against experimental SAXS or SANS data. For the α-catenin monomer, we have used one of the protomers of the crystal structure as the reference or starting structure for Monte Carlo simulation after reconstructing the disordered regions that are missing in the crystal structure using homology modeling (see Materials and Methods). Those Monte Carlo-generated models that best fit the experimental SAXS data of the α-catenin monomer fraction are selected. Because the crystal structure of α-catenin is tightly packed, at least two rounds of simulation are needed to generate structures whose calculated small-angle scattering profiles fit the experimental SAXS data. For the first-round Monte Carlo simulation, the N1-M fragment (a.a. 1–629) and the ABD (a.a. 680–906) were treated as rigid independent domains, and the linker (a.a. 630–675) connecting M3-ABD was treated as a flexible linkage that can adopt multiple conformations. The first-round simulation generated structures that fit the experimental SAXS data of α-catenin with χ2 > 10. One of the structural models with the lowest χ2 was selected as the reference structure for the second-round simulation, which assumes flexible linkage between the N2 and M1 domains (a.a. 259–275), linkage between M3 and ABD (a.a. 630–675), and a flexible C-terminal tail (a.a. 836–906). Fig. 3 shows two representative structures generated by the simulations that best fit the monomer SEC-SAXS data with χ2 < 0.98.
Figure 3.
Solution structures of α-catenin monomer. (A) A SAXS curve computed from structural models generated from SASSIE Monte Carlo simulation (green and blue lines) with best fit to the monomer fraction of experimental SEC-SAXS data (black square) is shown, χ2 < 0.98. (B) A χ2 vs. Rg plot of the fits of SASSIE generated models to the monomer SEC-SAXS data is shown. (C) Two representative structural models of open and closed α-catenin monomer with minimized χ2 fit to the experimental SAXS data are shown.
Overall, the monomer structures that best fit the SAXS data (Fig. 3 C) resemble those of a flat and round disc or a clamp-like structure. The solution structures are more expanded than either protomer in the crystal structure of an α-catenin dimer (31). In contrast to the crystal structure, the helical bundle of N2 is not in close contact with the helical bundle of M1, and the linker connecting N2-M1 can undergo restricted motions. The M fragment remains compact and unchanged, like that shown in the crystal structures (27, 31).
Unlike the crystal structure, in which the ABD is collapsed on the M fragment, the solution structural models of a monomer show that the ABD is not in contact with the M fragment. Instead, the ABD is in proximity to the N1 domain because of the clamp-like structure of the full-length α-catenin monomer (see Fig. 3 C). In the clamp-like monomer structure, the disordered C-terminal tail of the ABD (a.a. 836–906) can adopt a closed conformation by folding back to be close to the N1 helix bundle (Fig. 3 C, left panel); this disordered C-terminal tail can also move away from the N1 helix bundle to adopt a more open conformation (Fig. 3 C, right panel). This observation suggests that the α-catenin monomer may possess a dynamic head-to-tail intramolecular autoregulatory activity. The closed form of an α-catenin monomer thus resembles the autoinhibited structure of vinculin (63, 64, 65, 66). The dynamic autoregulation in the α-catenin monomer may explain previous observations that the α-catenin monomer does not bind F-actin as well as the α-catenin dimer (5). Previous biochemical studies have shown that the disordered tail (a.a. 836–906) is critical for α-catenin binding to F-actin (34).
Expanded asymmetric solution structure of α-catenin dimer
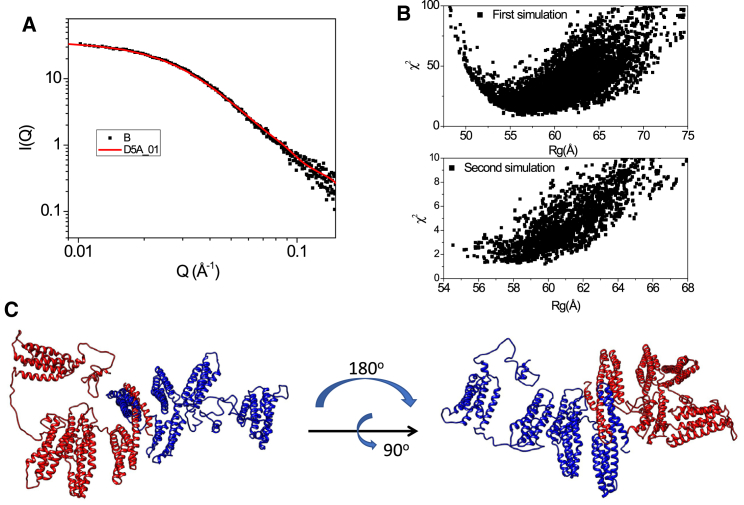
Using the structural model of α-catenin monomer shown in Fig. 3 C (left panel), we have first constructed the full-length dimer model of α-catenin by superposition of the dimerization domain (a.a. 82–171) of each monomer to the crystal structure of α-catenin dimer (PDB: 4IGG) using the MatchMaking tool in UCSF Chimera (56). This reference dimer structural model is subjected to Monte Carlo simulations in SASSIE. During simulations, the linkers connecting N2-M1, M3-ABD, and the C-terminal tail of a.a. 836–906 are assumed to be flexible in each protomer. The simulations generated structures that fit the dimer fraction of SEC-SAXS data with a χ2 < 1.21 (see Fig. 4, A and B). A representative dimer structure that best fits the SEC-SAXS data of α-catenin dimer is shown in Fig. 4 C.
Figure 4.
Solution structure of α-catenin dimer. (A) A SAXS curve of an α-catenin dimer model generated from SASSIE Monte Carlo simulation (red line) with best fit to the dimer fraction of SEC-SAXS data (black square) is shown, χ2 < 1.21. (B) χ2 vs. Rg plots of the fits of SASSIE generated models to the dimer SEC-SAXS data are shown. Two rounds of simulations are performed to minimize the χ2. (C) A representative α-catenin dimer structural model with minimized χ2 is shown.
In solution, the SAXS data of dimeric α-catenin is consistent with structural models that are flat, resembling a rectangular disc with N and M fragments from each protomer forming the core of the disc, and the ABD is well separated from the main body of the disc (Fig. 4 C). The flat disc shape of the dimer is different from the crystal structure, mainly due to the shift of the linker region connecting N2 and the M fragments. The significant difference between the solution structure and the crystal structure is in the locations of ABD in the dimer. In the crystal structure, the ABD is located within the dimer and in tight contact with the M fragment. In the solution structure, the ABD of each protomer is located outside the core disc and is ready to bind to actin filaments. As a result, the overall solution dimer structure is more expanded than the crystal dimer. The separation of both ABDs from the main body of the disc makes the ABDs available for binding, thus explaining why the α-catenin dimer can bind to F-actin robustly and bundle actin filaments (5, 21).
Notably, the solution dimer structure of α-catenin is asymmetric. The P(r) of each protomer in the solution dimer is different. The arrangement of M domains in each subunit is different, and the ABDs adopt different orientations in each protomer. The asymmetry in solution agrees with the asymmetric dimer structures observed previously by biophysical experiments and crystallography (7, 27, 31). However, each protomer in the solution dimer structure is more expanded than the crystal structure (see Fig. S1). We suggest that the asymmetric dimer solution structure is due to independent domain motions in each protomer that are not correlated in the entire dimeric structure.
Structure of α-catenin dimer in complex to F-actin
When combined with selective deuteration and contrast-variation, SANS can elucidate the conformation of each component in a macromolecular complex (37, 48, 49, 50, 67, 68, 69). We have determined the conformation of dα-catenin in complex to F-actin, and the spatial arrangement of actin filaments in the α-catenin assembled bundle. In 40% D2O, the scattering length density of the buffer matches that of hydrogenated F-actin (so F-actin is “invisible”) (38), and the molecular conformation of dα-catenin in the complex can thus be revealed (see Fig. 5). In the complex, the conformation of dα-catenin becomes more extended than the dimer form in solution, with Rg = 99.7 ± 2.1 Å and Dmax =350 Å (see Fig. 5 B and Table 1). Using the full-length α-catenin dimer structure in solution shown in Fig. 4 C as the starting structure, SASSIE Monte Carlo simulation requires the assumption of flexible linkers connecting the N1-M1 and M3-ABD domains and the C-terminal tail a.a. 836–906 in ABD to fit the 40% D2O SANS data with 1.0 (Fig. 5 C). The models that best fit the SANS data are elongated homodimers with the ABD further separated from the M fragment and the M fragment further separated from the N2 domain than that of the dimer in the solution state (Fig. 5 D). Within such an extended dimer structure, the M fragment is well exposed to solvent as compared to the monomer or dimer in solution states, even though the M fragment remains a compact structure. Previous studies have shown that mechanical force is required for the hidden vinculin-binding sites to be exposed to bind to vinculin (13), thus requiring dynamics in an essential way.
Figure 5.
Structure of dα-catenin dimer in complex to F-actin in 40% D2O at the contrast matching point of F-actin. (A) SANS on the dα-catenin/F-actin complex in 40% D2O buffer at the matching point of F-actin (black square) is shown. The red line is the fit of the structural model shown in (D). The concentration of dα-catenin is 1.74 mg/mL. The concentration of actin is ∼5 mg/mL. The actin/dα-catenin molar ratio is 6.8:1. (B) The P(r) of dα-catenin/F-actin complex in 40% D2O that corresponds to the conformational of dα-catenin (filled black square) is shown, as compared to that of α-catenin dimer in solution from SEC-SAXS (open black square). (C) A χ2 vs. Rg plot of the fit of SASSIE Monte Carlo generated structures to the SANS data in 40% D2O buffer is shown. (D) A representative structural model of dα-catenin with a best fit to the experimental SANS data is shown, χ2 < 0.9.
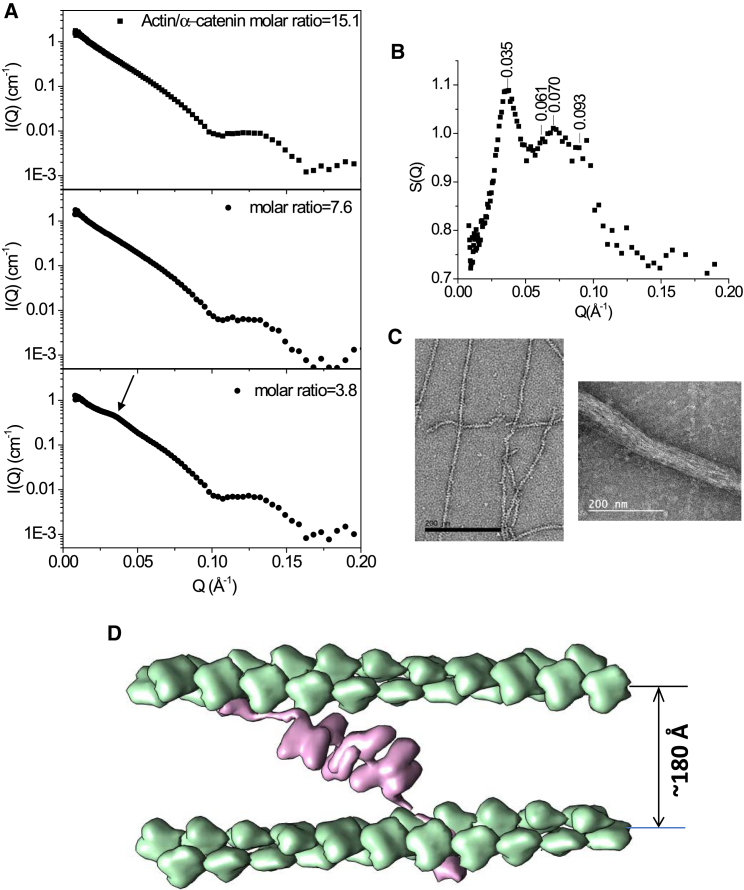
In 100% D2O at which the scattering length density of the buffer matches that of the dα-catenin, dα-catenin is invisible, and only the scattering of hydrogenated F-actin in the complex is detected. Fig. 6 A shows the scattering of dα-catenin in complex to F-actin in 100% D2O buffer, with the actin concentration kept at 81.5 μΜ while the molar ratio of actin monomer/dα-catenin is changed (see Fig. 6 A). When the molar ratio of actin/dα-catenin is lowered to 3.8:1, a correlation peak at Q = 0.035 Å−1 appears (Fig.6 A bottom panel), suggesting the assembly of ordered F-actin filaments by the dα-catenin dimer into bundles with defined interfilament spacing. Such a correlation peak was not observed at other contrasts, such as in 0% D2O buffer at which both F-actin and dα-catenin contribute to the scattering. The molar ratio of actin/dα-catenin = 3.8:1 corresponds to one dα-catenin dimer per 7.6 actin monomers.
Figure 6.
(A) Contrast-matched SANS on the dα-catenin/F-actin complex in 100% D2O buffer at the matching point of dα-catenin. The three panels are the I(Q) of the complex at different actin/dα-catenin molar ratios, with a distinct correlation peak appearing at molar ratio = 3.8 (see arrow). The actin concentration is kept at 81.5 μM in all three panels. (B) The structure factor S(Q) of the F-actin bundle, generated by normalizing the I(Q) at molar ratio = 3.8 (bottom panel) by the I(Q) at molar ratio = 15.1 (top panel), is shown. (C) The left panel shows a negative-staining electron microscopy image of F-actin filament mixed with ezrin, a protein that does not bundle F-actin. The right panel shows a negative-staining electron microscopy image of an α-catenin-assembled F-actin bundle. (D) A structural model of the dα-catenin dimer between two actin filaments is shown.
Normalizing the scattering I(Q) shown in the bottom panel of Fig. 6 A by the I(Q) of the top panel of Fig. 6 A gives the structure factor S(Q) (70), which represents only the spatial arrangement of the actin filaments (see Fig. 6 B). S(Q) shows several peaks with the fundamental peak at Q = 0.035 Å−1, corresponding to a center-of-axial distance d = 179.5 Å between two F-actin filaments. An instrument resolution convoluted Gaussian fit to the fundamental peak, using the Gaussian peak fit program in SASview (http://www.sasview.org/), gives the full width at half maximum as 0.0120 Å−1, indicating that the cross-sectional diameter of bundle is ∼520 Å. A secondary peak at Q = 0.070 Å−1 is apparent in Fig. 6 B with position of 1:2 relative to the fundamental peak. The appearance of these two S(Q) peaks at high dα-catenin/actin molar ratios suggests a development into F-actin bundles with ordered filament alignment. Fig. 6 C shows the negative-stain electron microscope images of F-actin filaments alone (left panel) and the α-catenin assembled F-actin filament bundles (right panel). A measure of the cross sections of the α-catenin F-actin bundle in the EM image, using the program ImageJ (71), gives an average diameter of ∼560 Å.
The other peaks in Fig. 6 B, with peak positions relative to the fundamental peak, may suggest the development of the F-actin filaments in the dimeric dα-catenin assembled bundles into a hexagonal phase (72). However, these peaks are less distinguishable as compared to the 1:2 peak, which prevents us from drawing a definitive conclusion of a hexagonal packing of the filaments. The assembly of ordered hexagonal F-actin bundles has been observed in the complex of other F-actin cross-linker proteins such as α-actinin and epsin (73, 74).
Using the F-actin structure determined from electron cryomicroscopy (75) and the dα-catenin dimer structure in complex to F-actin in 40% D2O shown in Fig. 5 D, we constructed a model of the complex of dα-catenin with F-actin (see Fig. 6 D). In the α-catenin-assembled F-actin bundle, the maximal dimension of dα-catenin is 350 Å, as shown in Fig. 5 B, but the interfilament spacing between two filaments is 179.5 Å. We thus posit that α-catenin is not bound orthogonally to the actin filament but forms a diagonal angle of ∼30° with the filament to fit between two filaments. Forming a diagonal angle with the actin filament may make the α-catenin dimer more effective in transducing both the longitudinal (from myosin pulling of the filament) and transverse (filament bending fluctuations) forces from the filament, as the α-catenin connector has vector components in both the longitudinal and transverse directions.
Nanoscale dynamics of the C-terminal tail of α-catenin by NSE
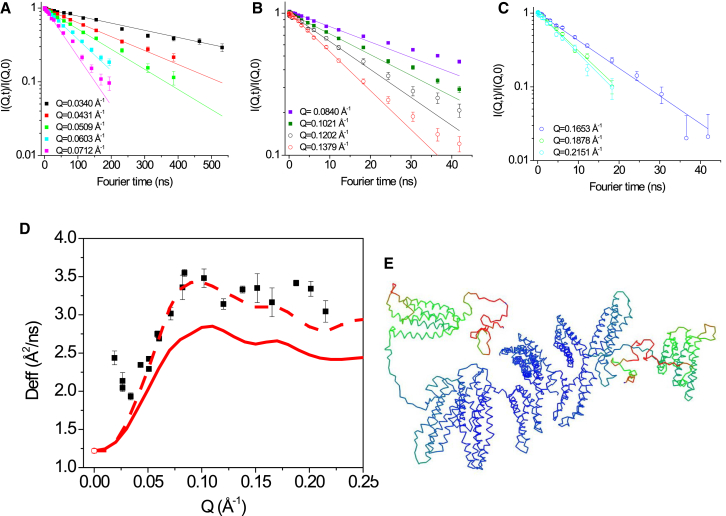
Using NSE, we have determined the nanoscale dynamics of the α-catenin dimer. This specific NSE experiment measures nanoscale dynamics on timescales of 1–550 ns, and on length scales from 0.018 to 0.21 Å−1 that are comparable to the length scales probed by a small-angle scattering experiment. NSE experiments were performed at 11.0 mg/mL α-catenin concentration after α-catenin was exchanged into D2O PBS buffer. SANS data of the same sample used for the NSE experiment shows that under this experimental condition, α-catenin is a dimer (see Fig. S4). The intermediate scattering functions, I(Q,t)/I(Q,0), are shown in Fig. 7, A–C. Notably, in the Q range of 0.0712–0.1379 Å−1, I(Q,t)/I(Q,0) deviates from a single exponential behavior (see Fig. 7, A and B). We thus have used the initial slope fit of ln[I(Q,t)/I(Q,0)] of <10 ns in this Q range to obtain the decay rate and the effective diffusion constant Deff(Q) (see Fig. 7 D). Previously, we have developed a theoretical framework and have shown that comparing NSE experiments with theoretical calculations can pinpoint which part of the full-length protein is moving (39, 59, 76). Here, once again, we apply our analysis to NSE data to locate the nanoscale moving parts of a protein.
Figure 7.
NSE spectroscopy of α-catenin dimer. (A), (B) and (C) show intermediate correlation function I(Q,t)/I(Q,0) as a function of Fourier time at different Q values. The lines are single exponential fits to the initial slope of I(Q,t)/I(Q,0). Note that at Q between 0.0509–0.1379 Å−1I(Q,t)/I(Q,0), shows non-single exponential behavior. (D) The effective diffusion constants Deff(Q) (black squares) obtained from the initial slopes of the spectra in (A), (B) and (C) are shown. The solid red line is the theoretical Deff(Q) using the coordinates of α-catenin dimer shown in Fig. 4C, assuming the α -catenin dimer is a rigid body. The dashed red line is the theoretical Deff(Q), assuming amino acid residues 836–906 in each protomer of the dimer are moving. (E) The dynamics of α-Catenin dimer colored by normal mode analysis, with blue representing low mobility and red high mobility, is shown.
Using the structural coordinates of α-catenin dimer shown in Fig. 4 C, we are able to calculate and compare the theoretical effective diffusion constant Deff(Q) with the experimental data for various models of the mobility tensor of the protein. When assuming a rigid-body mobility tensor for the α-catenin dimer, the theoretical Deff(Q) is noticeably lower than the experimental value (Fig. 7 D). However, when we use a mobility tensor for a model in which residues 836–906 of each constituent protomer in the dimer form separate mobile domains, substantial improvement with the NSE data occurs. This improved result when internal motion is included implies that the segment of a.a. 836–906 that is the disordered C-terminal tail in the ABD moves separately from the rest of the structure on the nanoscales measured by NSE. As a second control, we calculate the Deff(Q) expected when the entire ABD domain (a.a. 680–906) of each protomer forms a mobile subunit. As shown in Fig. S3, the calculated Deff(Q) is significantly higher than the experimental value for this larger subunit. This comparison of mobility tensor models with data illustrates how NSE reveals the nanoscale motion of the disordered C-terminal tail. In particular, NSE delimits the boundary of the moving segment, with the disordered C-terminus residues 836–906 forming the mobile subunit of each protomer and not the total ABD domain residues 680–906. Normal mode analysis using IMOD (77) also predicts that this disordered C-tail is more mobile than the rest of the dimer (see Fig. 7 E). Comparison of the values of Deff(Q) evaluated as a function of Fourier time suggests that the internal mode has a decay rate of order 10 ns.
The nanoscale dynamics of the C-terminal tail of α-catenin is likely to be functionally important. Our previous study shows that the nanoscale dynamics of disordered proteins modulates the recognition kinetics of target binding partners (76). The nanoscale motion of the disordered C-terminal tail could be necessary for the α-catenin dimer to engage and to dock with F-actin before the five-helix bundle binds to F-actin and thus should influence the kinetics of α-catenin binding to F-actin. In support of this hypothesis, previous studies show that direct interaction of residues 866–906 with F-actin is necessary for stabilizing cell-cell adhesion (34) and that mice with a truncation mutation lacking this C-terminal tail of αN-catenin have abnormal cerebellar development (35).
Discussion
As an essential component of the adherens junction, α-catenin modulates the interaction of cadherin with the actin cytoskeleton in a mechanical-force-dependent manner and thereby plays important roles in maintaining cell-cell adhesion and tissue remodeling. This study presents a revised view of the α-catenin monomer and dimer structures in that this multidomain protein is flexible in solution. The flexible structure may be important for the multifaceted functions of α-catenin and for this protein to function as a mechanically sensitive adaptor.
The solution structures show that both the monomer and the dimer are more expanded in solution than in the crystal structures, with the ABD adopting substantially different configurations. Additional movements of the M fragment relative to N domains are required in both the monomer and the dimer structures to satisfy the constraint of the SAXS data. The solution structure from SAXS or SANS is an average of an ensemble of structures that a protein can adopt. The more expanded solution structure indicates the multidomain α-catenin can sample a variety of conformations dynamically from extended to compact states. Binding to the cadherin/catenin complex may shift α-catenin to a state that reduces its binding to the actin filament, whereas mechanical stress changes α-catenin to a more open state to make the vinculin and actin binding sties available. Indeed, even when in complex to cadherin/β-catenin, α-catenin is a dynamic structure that retains some F-actin and vinculin binding activity (78, 79).
The monomeric α-catenin forms a flat clamp-like structure, with the ABD being somewhat autoregulated by the homodimerization domain through dynamic intramolecular interactions. However, in the solution structure of dimeric α-catenin, the ABD is well separated from the main body of the dimer, making the ABD available for F-actin binding. These structural features explain previous biochemical results that the α-catenin monomer does not bind F-actin as easily as the dimeric protein and that α-catenin dimers can bundle F-actin filaments (5, 21).
In both the solution structures of the monomer and the dimer, the M fragment remains a compact core without changes in the arrangement of the M1, M2, and M3 domains. Previous studies show that vinculin-binding sites are masked by the inter-helix-bundle interactions in the M fragment (9, 13, 25, 26, 27, 80). Thus, the population of α-catenin monomer or dimer conformations with open vinculin-binding sites is low, and α-catenin is not expected to bind to vinculin spontaneously. When in complex with the F-actin filaments, the α-catenin dimer becomes more extended, and the M fragment may start to open at this point. However, the flexural rigidity of an actin filament is less than 1 pN × nm2 (81). About 5 pN force is required to open the binding site in α-catenin for vinculin-binding. Thus, the bending fluctuations of the actin filament alone may only be on the margin for opening the M fragment and exposing the vinculin binding sites. The pulling force of myosin on the filament may be required to open the vinculin-binding sites in α-catenin for α-catenin to bind vinculin spontaneously.
NSE is a novel technique that can determine protein dynamics on nanoscale-length scales that are difficult to access by other methods (57, 82, 83, 84, 85, 86, 87). We report the first study of the nanoscale dynamics in an α-catenin dimer by NSE to our knowledge. Our analysis of NSE data reveals that the nanoscale motion of α-catenin is specifically located at the disordered C-terminal tail of α-catenin. Previous studies have shown that this C-terminal tail of the ABD is critical for stabilization of cell-cell adhesion and in nervous tissue and brain development (34, 35). Our previous study has shown that the nanoscale dynamics of a disordered protein affects its binding kinetics to a target protein (76). Future studies will test the hypothesis that the disordered tail of the ABD influences the kinetics of engagement of α-catenin with the F-actin filament.
We further present the first view of the nanostructure of α-catenin dimer in complex with F-actin filaments to our knowledge. The development from randomly oriented F-actin filament into an ordered hexagonal bundle at relatively high α-catenin to actin molar ratio is likely to be important for the morphogenesis of filipodium protrusion. The cellular pool that α-catenin modulates is an important component of the polarized architecture of lamellipodia and is essential for filopodia protrusions in migrating cells and to promote the initial engagement of cell-cell contacts (1, 19, 20, 88). In concert with other actin-bundling proteins such as formins and Ena/VASP (74), α-catenin may contribute to the transition of branched and cross-linked actin networks in lamellipodia to the parallel actin bundles in filopodia.
Author Contributions
I.D.N., D.J.E.C., and Z.B. designed and performed the experiments, analyzed the data, and wrote the manuscript. T.M., T.M.W., C.B.S., W.T.H., A.M., and B.F. contributed the resources, performed the experiments, and contributed to data analysis.
Acknowledgments
The authors thank Jorge Morales for helping with the transmission electron microscopy image collection. The authors thank Carrie Gao for technical support during the SANS experiments. The authors thank Ralf Schwens at the ILL for access and helping to use the dynamical light scattering setup (ALV CGS3 instrument) at the Partnership for Soft Condensed Matter labs at the ILL.
This research was funded by National Science Foundation grant MCB-1817684 to Z.B. and 2G12 RR003060 from the National Center for Research Resources to City College of New York. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. A portion of the research conducted at Oak Ridge National Laboratory’s Spallation Neutron Source was sponsored by the Scientific User Facilities Division, Office of Basic Energy Sciences, U.S. Department of Energy. I.D.N. acknowledges the support of the ILL for costs for travel and subsistence.
Editor: Enrique De La Cruz.
Footnotes
Supporting Materials and Methods, four figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30769-0.
Contributor Information
David J.E. Callaway, Email: dcallaway@ccny.cuny.edu.
Zimei Bu, Email: zbu@ccny.cuny.edu.
Supporting Material
References
- 1.Kobielak A., Fuchs E. Alpha-catenin: at the junction of intercellular adhesion and actin dynamics. Nat. Rev. Mol. Cell Biol. 2004;5:614–625. doi: 10.1038/nrm1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gumbiner B.M. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 3.Takeichi M. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 2014;15:397–410. doi: 10.1038/nrm3802. [DOI] [PubMed] [Google Scholar]
- 4.Brasch J., Harrison O.J., Shapiro L. Thinking outside the cell: how cadherins drive adhesion. Trends Cell Biol. 2012;22:299–310. doi: 10.1016/j.tcb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drees F., Pokutta S., Weis W.I. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai R., Sarpal R., Tepass U. Monomeric α-catenin links cadherin to the actin cytoskeleton. Nat. Cell Biol. 2013;15:261–273. doi: 10.1038/ncb2685. [DOI] [PubMed] [Google Scholar]
- 7.Koslov E.R., Maupin P., Rimm D.L. Alpha-catenin can form asymmetric homodimeric complexes and/or heterodimeric complexes with beta-catenin. J. Biol. Chem. 1997;272:27301–27306. doi: 10.1074/jbc.272.43.27301. [DOI] [PubMed] [Google Scholar]
- 8.Buckley C.D., Tan J., Dunn A.R. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 2014;346:1254211. doi: 10.1126/science.1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yonemura S., Wada Y., Shibata M. alpha-catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- 10.Weiss E.E., Kroemker M., Rüdiger M. Vinculin is part of the cadherin-catenin junctional complex: complex formation between α-catenin and vinculin. J. Cell Biol. 1998;141:755–764. doi: 10.1083/jcb.141.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abe K., Takeichi M. EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc. Natl. Acad. Sci. USA. 2008;105:13–19. doi: 10.1073/pnas.0710504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knudsen K.A., Soler A.P., Wheelock M.J. Interaction of alpha-actinin with the cadherin/catenin cell-cell adhesion complex via alpha-catenin. J. Cell Biol. 1995;130:67–77. doi: 10.1083/jcb.130.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao M., Qiu W., Yan J. Force-dependent conformational switch of α-catenin controls vinculin binding. Nat. Commun. 2014;5:4525. doi: 10.1038/ncomms5525. [DOI] [PubMed] [Google Scholar]
- 14.Han M.K., de Rooij J. Converging and unique mechanisms of mechanotransduction at adhesion sites. Trends Cell Biol. 2016;26:612–623. doi: 10.1016/j.tcb.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Huveneers S., de Rooij J. Mechanosensitive systems at the cadherin-F-actin interface. J. Cell Sci. 2013;126:403–413. doi: 10.1242/jcs.109447. [DOI] [PubMed] [Google Scholar]
- 16.Barry A.K., Tabdili H., Leckband D.E. α-catenin cytomechanics--role in cadherin-dependent adhesion and mechanotransduction. J. Cell Sci. 2014;127:1779–1791. doi: 10.1242/jcs.139014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leckband D.E., de Rooij J. Cadherin adhesion and mechanotransduction. Annu. Rev. Cell Dev. Biol. 2014;30:291–315. doi: 10.1146/annurev-cellbio-100913-013212. [DOI] [PubMed] [Google Scholar]
- 18.Benjamin J.M., Kwiatkowski A.V., Nelson W.J. AlphaE-catenin regulates actin dynamics independently of cadherin-mediated cell-cell adhesion. J. Cell Biol. 2010;189:339–352. doi: 10.1083/jcb.200910041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood M.N., Ishiyama N., Gottardi C.J. α-Catenin homodimers are recruited to phosphoinositide-activated membranes to promote adhesion. J. Cell Biol. 2017;216:3767–3783. doi: 10.1083/jcb.201612006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vassilev V., Platek A., Takeichi M. Catenins steer cell migration via stabilization of front-rear polarity. Dev. Cell. 2017;43:463–479.e5. doi: 10.1016/j.devcel.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 21.Rimm D.L., Koslov E.R., Morrow J.S. Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl. Acad. Sci. USA. 1995;92:8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen S.D., Kwiatkowski A.V., Nelson W.J. αE-catenin actin-binding domain alters actin filament conformation and regulates binding of nucleation and disassembly factors. Mol. Biol. Cell. 2013;24:3710–3720. doi: 10.1091/mbc.E13-07-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pokutta S., Weis W.I. Structure of the dimerization and beta-catenin-binding region of alpha-catenin. Mol. Cell. 2000;5:533–543. doi: 10.1016/s1097-2765(00)80447-5. [DOI] [PubMed] [Google Scholar]
- 24.Obama H., Ozawa M. Identification of the domain of alpha-catenin involved in its association with beta-catenin and plakoglobin (gamma-catenin) J. Biol. Chem. 1997;272:11017–11020. doi: 10.1074/jbc.272.17.11017. [DOI] [PubMed] [Google Scholar]
- 25.Choi H.J., Pokutta S., Weis W.I. αE-catenin is an autoinhibited molecule that coactivates vinculin. Proc. Natl. Acad. Sci. USA. 2012;109:8576–8581. doi: 10.1073/pnas.1203906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rangarajan E.S., Izard T. The cytoskeletal protein α-catenin unfurls upon binding to vinculin. J. Biol. Chem. 2012;287:18492–18499. doi: 10.1074/jbc.M112.351023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishiyama N., Tanaka N., Ikura M. An autoinhibited structure of α-catenin and its implications for vinculin recruitment to adherens junctions. J. Biol. Chem. 2013;288:15913–15925. doi: 10.1074/jbc.M113.453928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itoh M., Nagafuchi A., Tsukita S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to α catenin and actin filaments. J. Cell Biol. 1997;138:181–192. doi: 10.1083/jcb.138.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pokutta S., Weis W.I. The cytoplasmic face of cell contact sites. Curr. Opin. Struct. Biol. 2002;12:255–262. doi: 10.1016/s0959-440x(02)00318-4. [DOI] [PubMed] [Google Scholar]
- 30.Yang J., Dokurno P., Barford D. Crystal structure of the M-fragment of alpha-catenin: implications for modulation of cell adhesion. EMBO J. 2001;20:3645–3656. doi: 10.1093/emboj/20.14.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rangarajan E.S., Izard T. Dimer asymmetry defines α-catenin interactions. Nat. Struct. Mol. Biol. 2013;20:188–193. doi: 10.1038/nsmb.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim T.J., Zheng S., Wang Y. Dynamic visualization of α-catenin reveals rapid, reversible conformation switching between tension states. Curr. Biol. 2015;25:218–224. doi: 10.1016/j.cub.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrick S., Li J., Leckband D. Salt bridges gate α-catenin activation at intercellular junctions. Mol. Biol. Cell. 2018;29:111–122. doi: 10.1091/mbc.E17-03-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pappas D.J., Rimm D.L. Direct interaction of the C-terminal domain of α-catenin and F-actin is necessary for stabilized cell-cell adhesion. Cell Commun. Adhes. 2006;13:151–170. doi: 10.1080/15419060600726142. [DOI] [PubMed] [Google Scholar]
- 35.Park C., Falls W., Ackerman S.L. Deletion in Catna2, encoding alpha N-catenin, causes cerebellar and hippocampal lamination defects and impaired startle modulation. Nat. Genet. 2002;31:279–284. doi: 10.1038/ng908. [DOI] [PubMed] [Google Scholar]
- 36.Gasteiger E.H.C., Gattiker A., Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker J.M., editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 37.Li J., Callaway D.J., Bu Z. Ezrin induces long-range interdomain allostery in the scaffolding protein NHERF1. J. Mol. Biol. 2009;392:166–180. doi: 10.1016/j.jmb.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jayasundar J.J., Ju J.H., Bu Z. Open conformation of ezrin bound to phosphatidylinositol 4,5-bisphosphate and to F-actin revealed by neutron scattering. J. Biol. Chem. 2012;287:37119–37133. doi: 10.1074/jbc.M112.380972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farago B., Li J., Bu Z. Activation of nanoscale allosteric protein domain motion revealed by neutron spin echo spectroscopy. Biophys. J. 2010;99:3473–3482. doi: 10.1016/j.bpj.2010.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Callaway D.J., Bu Z. Essential strategies for revealing nanoscale protein dynamics by neutron spin echo spectroscopy. Methods Enzymol. 2016;566:253–270. doi: 10.1016/bs.mie.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 41.Bhattacharya S., Dai Z., Bu Z. A conformational switch in the scaffolding protein NHERF1 controls autoinhibition and complex formation. J. Biol. Chem. 2010;285:9981–9994. doi: 10.1074/jbc.M109.074005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsui T., Gu S., Jin R. Structural basis of the pH-dependent assembly of a botulinum neurotoxin complex. J. Mol. Biol. 2014;426:3773–3782. doi: 10.1016/j.jmb.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edwards A.L., Matsui T., Khosla C. Architectures of whole-module and bimodular proteins from the 6-deoxyerythronolide B synthase. J. Mol. Biol. 2014;426:2229–2245. doi: 10.1016/j.jmb.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martel A., Liu P., Tsuruta H. An integrated high-throughput data acquisition system for biological solution X-ray scattering studies. J. Synchrotron Radiat. 2012;19:431–434. doi: 10.1107/S0909049512008072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao J.K., Gao C.Y., Liu D. The extended Q-range small-angle neutron scattering diffractometer at the SNS. J. Appl. Cryst. 2010;43:1068–1077. [Google Scholar]
- 46.Arnold O., Bilheux J.-C., Gigg M. Mantid—data analysis and visualization package for neutron scattering and μ SR experiments. Nucl. Instrum. Meth. Phys. Res. A. 2014;764:156–166. [Google Scholar]
- 47.Wignall G.D., Bates F.S. Absolute calibration of small-angle neutron-scattering data. J. Appl. Cryst. 1987;20:28–40. [Google Scholar]
- 48.Ali Khajeh J., Ju J.H., Bu Z. Molecular conformation of the full-length tumor suppressor NF2/Merlin--a small-angle neutron scattering study. J. Mol. Biol. 2014;426:2755–2768. doi: 10.1016/j.jmb.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X., Khajeh J.A., Bu Z. Phosphatidylinositol 4,5-bisphosphate clusters the cell adhesion molecule CD44 and assembles a specific CD44-Ezrin heterocomplex, as revealed by small angle neutron scattering. J. Biol. Chem. 2015;290:6639–6652. doi: 10.1074/jbc.M114.589523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho D.L., Byrnes W.M., Bu Z. Structure-specific DNA-induced conformational changes in Taq polymerase revealed by small angle neutron scattering. J. Biol. Chem. 2004;279:39146–39154. doi: 10.1074/jbc.M404565200. [DOI] [PubMed] [Google Scholar]
- 51.Semenyuk A.V., Svergun D.I. GNOM - A program package for small-angle scattering data-processing. J. Appl. Cryst. 1991;24:537–540. [Google Scholar]
- 52.Perkins S.J., Wright D.W., Curtis J.E. Atomistic modelling of scattering data in the collaborative computational project for small angle scattering (CCP-SAS) J. Appl. Cryst. 2016;49:1861–1875. doi: 10.1107/S160057671601517X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Svergun D., Barberato C., Koch M.H. CRYSOL–a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 54.dos Reis M.A., Aparicio R., Zhang Y. Improving protein template recognition by using small-angle x-ray scattering profiles. Biophys. J. 2011;101:2770–2781. doi: 10.1016/j.bpj.2011.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shibahara T., Hirano Y., Hakoshima T. Structure of the free form of the N-terminal VH1 domain of monomeric α-catenin. FEBS Lett. 2015;589:1754–1760. doi: 10.1016/j.febslet.2015.05.053. [DOI] [PubMed] [Google Scholar]
- 56.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 57.Callaway D.J., Bu Z. Visualizing the nanoscale: protein internal dynamics and neutron spin echo spectroscopy. Curr. Opin. Struct. Biol. 2017;42:1–5. doi: 10.1016/j.sbi.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Callaway D.J., Bu Z. Nanoscale protein domain motion and long-range allostery in signaling proteins- a view from neutron spin echo sprectroscopy. Biophys. Rev. 2015;7:165–174. doi: 10.1007/s12551-015-0162-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bu Z., Biehl R., Callaway D.J. Coupled protein domain motion in Taq polymerase revealed by neutron spin-echo spectroscopy. Proc. Natl. Acad. Sci. USA. 2005;102:17646–17651. doi: 10.1073/pnas.0503388102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller P.W., Pokutta S., Kwiatkowski A.V. Danio rerio αE-catenin is a monomeric F-actin binding protein with distinct properties from Mus musculus αE-catenin. J. Biol. Chem. 2013;288:22324–22332. doi: 10.1074/jbc.M113.458406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang H., Bang I., Choi H.J. Structural and functional characterization of Caenorhabditis elegans α-catenin reveals constitutive binding to β-catenin and F-actin. J. Biol. Chem. 2017;292:7077–7086. doi: 10.1074/jbc.M116.769778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Curtis J.E., Raghunandan S., Krueger S. SASSIE: a program to study intrinsically disordered biological molecules and macromolecular ensembles using experimental scattering restraints. Comput. Phys. Commun. 2012;183:382–389. [Google Scholar]
- 63.Demali K.A. Vinculin--a dynamic regulator of cell adhesion. Trends Biochem. Sci. 2004;29:565–567. doi: 10.1016/j.tibs.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 64.Bakolitsa C., Cohen D.M., Liddington R.C. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–586. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- 65.Ziegler W.H., Liddington R.C., Critchley D.R. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 66.Izard T., Brown D.T. Mechanisms and functions of vinculin interactions with phospholipids at cell adhesion sites. J. Biol. Chem. 2016;291:2548–2555. doi: 10.1074/jbc.R115.686493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trewhella J. Small-angle scattering and 3D structure interpretation. Curr. Opin. Struct. Biol. 2016;40:1–7. doi: 10.1016/j.sbi.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 68.Krueger S. Designing and performing biological solution small-angle neutron scattering contrast variation experiments on multi-component assemblies. Adv. Exp. Med. Biol. 2017;1009:65–85. doi: 10.1007/978-981-10-6038-0_5. [DOI] [PubMed] [Google Scholar]
- 69.Heller W.T. Small-angle neutron scattering and contrast variation: a powerful combination for studying biological structures. Acta Crystallogr. D Biol. Crystallogr. 2010;66:1213–1217. doi: 10.1107/S0907444910017658. [DOI] [PubMed] [Google Scholar]
- 70.Higgins J.S., Benoit H.C. Clarendon Press; Oxford, UK: 1994. Polymers and Neutron Scattering. [Google Scholar]
- 71.Schindelin J., Arganda-Carreras I., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Jeu W.H. Oxford University Press; Oxford, UK: 2016. Basic X-ray Scattering for Soft Matter. [Google Scholar]
- 73.Claessens M.M., Semmrich C., Bausch A.R. Helical twist controls the thickness of F-actin bundles. Proc. Natl. Acad. Sci. USA. 2008;105:8819–8822. doi: 10.1073/pnas.0711149105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Purdy K.R., Bartles J.R., Wong G.C. Structural polymorphism of the actin-espin system: a prototypical system of filaments and linkers in stereocilia. Phys. Rev. Lett. 2007;98:058105. doi: 10.1103/PhysRevLett.98.058105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Avery A.W., Fealey M.E., Egelman E.H. Structural basis for high-affinity actin binding revealed by a β-III-spectrin SCA5 missense mutation. Nat. Commun. 2017;8:1350. doi: 10.1038/s41467-017-01367-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Callaway D.J.E., Matsui T., Bu Z. Controllable activation of nanoscale dynamics in a disordered protein alters binding kinetics. J. Mol. Biol. 2017;429:987–998. doi: 10.1016/j.jmb.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lopéz-Blanco J.R., Garzón J.I., Chacón P. iMod: multipurpose normal mode analysis in internal coordinates. Bioinformatics. 2011;27:2843–2850. doi: 10.1093/bioinformatics/btr497. [DOI] [PubMed] [Google Scholar]
- 78.Biswas K.H., Hartman K.L., Groves J.T. Sustained α-catenin activation at E-cadherin junctions in the absence of mechanical force. Biophys. J. 2016;111:1044–1052. doi: 10.1016/j.bpj.2016.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wickline E.D., Dale I.W., Kwiatkowski A.V. αT-catenin is a constitutive actin-binding α-catenin that directly couples the cadherin·catenin complex to actin filaments. J. Biol. Chem. 2016;291:15687–15699. doi: 10.1074/jbc.M116.735423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas W.A., Boscher C., Dufour S. α-Catenin and vinculin cooperate to promote high E-cadherin-based adhesion strength. J. Biol. Chem. 2013;288:4957–4969. doi: 10.1074/jbc.M112.403774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gittes F., Mickey B., Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Callaway D.J., Farago B., Bu Z. Nanoscale protein dynamics: A new frontier for neutron spin echo spectroscopy. Eur. Phys. J. E Soft Matter. 2013;36:76. doi: 10.1140/epje/i2013-13076-1. [DOI] [PubMed] [Google Scholar]
- 83.Lal J., Maccarini M., Makowski L. Modulation of hemoglobin dynamics by an allosteric effector. Protein Sci. 2017;26:505–514. doi: 10.1002/pro.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lal J., Fouquet P., Makowski L. Neutron spin-echo studies of hemoglobin and myoglobin: multiscale internal dynamics. J. Mol. Biol. 2010;397:423–435. doi: 10.1016/j.jmb.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Katava M., Maccarini M., Sterpone F. Thermal activation of ‘allosteric-like’ large-scale motions in a eukaryotic Lactate Dehydrogenase. Sci. Rep. 2017;7:41092. doi: 10.1038/srep41092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu Y. Intermediate scattering function for macromolecules in solutions probed by neutron spin echo. Phys. Rev. E. 2017;95:020501. doi: 10.1103/PhysRevE.95.020501. [DOI] [PubMed] [Google Scholar]
- 87.Bu Z., Callaway D.J. Proteins move! Protein dynamics and long-range allostery in cell signaling. Adv. Protein Chem. Struct. Biol. 2011;83:163–221. doi: 10.1016/B978-0-12-381262-9.00005-7. [DOI] [PubMed] [Google Scholar]
- 88.Vasioukhin V., Bauer C., Fuchs E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell. 2001;104:605–617. doi: 10.1016/s0092-8674(01)00246-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.