Abstract
Background and purpose
With extensive research efforts in place to address the clinical relevance of cerebral microbleeds (CMBs), there remains a need for fast and accurate methods to detect and quantify CMB burden. Although some computer-aided detection algorithms have been proposed in the literature with high sensitivity, their specificity remains consistently poor. More sophisticated machine learning methods appear to be promising in their ability to minimize false positives (FP) through high-level feature extraction and the discrimination of hard-mimics. To achieve superior performance, these methods require sizable amounts of precisely labelled training data. Here we present a user-guided tool for semi-automated CMB detection and volume segmentation, offering high specificity for routine use and FP labelling capabilities to ease and expedite the process of generating labelled training data.
Materials and methods
Existing computer-aided detection methods reported by our group were extended to include fully-automated segmentation and user-guided CMB classification with FP labelling. The algorithm's performance was evaluated on a test set of ten patients exhibiting radiotherapy-induced CMBs on MR images.
Results
The initial algorithm's base sensitivity was maintained at 86.7%. FP's were reduced to inter-rater variations and segmentation results were in 98% agreement with ground truth labelling. There was an approximate 5-fold reduction in the time users spent evaluating CMB burden with the algorithm versus without computer aid. The Intra-class Correlation Coefficient for inter-rater agreement was 0.97 CI[0.92,0.99].
Conclusions
This development serves as a valuable tool for routine evaluation of CMB burden and data labelling to improve CMB classification with machine learning. The algorithm is available to the public on GitHub (https://github.com/LupoLab-UCSF/CMB_labeler).
Keywords: Cerebral microbleeds, Lesion, Vascular injury, Magnetic resonance imaging, Susceptibility weighted imaging, Brain tumor, Radiation therapy, Machine learning, Algorithm, Automated
Highlights
-
•
We modified our existing semi-automated microbleed detection method
-
•
Using our new method, specificity is increased and detection time is decreased
-
•
The inter-rater variability for detecting microbleeds is reduced with our method
-
•
Automatically labelled microbleed data can be used in machine learning methods
-
•
Our method has successfully detected microbleeds in multiple clinical populations
1. Introduction
Cerebral microbleeds (CMBs) are size-varying collections of parenchymal hemosiderin induced by microhemorrhage in the brain (Martinez-Ramirez et al., 2014). The CMB foci appear round and hypointense on magnetic resonance (MR) T2⁎-weighted gradient echo (GRE) magnitude images, as a result of susceptibility-related signal loss. Heightened CMB contrast and subsequent improved CMB detection can be achieved with susceptibility-weighted imaging (SWI) and at increasing field strengths where the susceptibility effect is greater (Bian et al., 2014). These imaging developments have driven the exploration of the clinical and prognostic relevance of CMBs in diseases such as cerebral amyloid angiopathy (Haley et al., 2014), stroke (Tang et al., 2011; Gregoire et al., 2012), vascular dementia (Van der Flier and Cordonnier, 2012), traumatic brain injury (TBI) (Lawrence et al., 2017), and radiation therapy (RT) induced injury in brain tumor patients (Lupo et al., 2012). Recent literature has shown increasing evidence that CMBs are linked to cerebrovascular damage caused by microvascular disease (Martinez-Ramirez et al., 2014; Haley et al., 2014), TBI (Lawrence et al., 2017), and RT. (Lupo et al., 2012) Studies in stroke and vascular dementia patients have further demonstrated associations between CMBs, frontal-lobe executive impairments, and increased risk for intracerebral hemorrhage (Tang et al., 2011; Gregoire et al., 2012; Van der Flier and Cordonnier, 2012). Nonetheless, the pathophysiology of CMBs and their role in neurocognitive function and risk of future cerebrovascular events are still under evaluation along with implications for clinically managing patients.
In addition to exploring the clinical relevance of CMBs, simultaneous research efforts have been dedicated to the development of fast and accurate methods for CMB detection and quantification. Visual inspection of CMBs on MR images is a tedious, time-consuming, and often an impractical method for routine examination or exploratory research. In the case of RT-induced CMBs, radii are often <2 mm and can accumulate to over 300 CMBs distributed widely throughout the brain, thus making manual lesion counting nearly impossible. Detection is further complicated by the presence of normal anatomical structures (e.g. intracranial veins) that mimic the shape and contrast of CMBs, introducing substantial intra-rater and inter-rater variability. To improve detection accuracy and minimize the burden of manual lesion counting, several computer-aided CMB detection methods have been proposed in the literature to classify candidate CMBs based on hand-crafted, intensity-based, and geometric-based features (Bian et al., 2013; Seghier et al., 2011; Kuijf et al., 2012; Barnes et al., 2012; van den Heuvel et al., 2016; Fazlollahi et al., 2014). This includes the semi-automated CMB detection algorithm by Bian et al. (Bian et al., 2013) that utilizes a 2D Fast Radial Symmetry Transform (FRST) to initially detect putative CMBs before performing 3D region growing and 2D geometric feature examination to eliminate falsely identified CMBs (Bian et al., 2013). The algorithm was optimized and evaluated in a cohort of 15 glioma patients presenting with RT-induced CMBs on minimum-intensity projected (mIP) SWI images acquired at 3 Tesla (3 T). Compared to prior developments, our algorithm achieved superior performance with a sensitivity of 86.5% and computation time of under 1 min. Despite the improved sensitivity of our method, nearly 45 false positives (FP) were reported on average per patient regardless of the total number of true CMBs identified. Other computer-based methods similarly reported sizable amounts of falsely identified CMBs, eluding to the idea that more sophisticated, data driven approaches are required to accurately distinguish CMBs from their hard mimics.
Chen et al. and Dou et al. were the first to incorporate 2D and 3D deep convolution neural networks (CNNs) to improve CMB detection and classification (Chen et al., 2015; Dou et al., 2016). CNNs have shown a long range of success in their ability to automatically learn hierarchical feature representations from natural images, demonstrating impressive performance in array of recognition tasks including object detection, segmentation, and classification (Sa et al., 2016; Kheradpisheh et al., 2016; Wan et al., 2014; Gupta et al., 2014). In Dou et al.’s proposed method (Dou et al., 2016), the use of a 3D versus 2D convolution kernel provided more contextual information for identifying candidates and discriminating true CMBs from hard mimics. Although their 12-layer cascade framework outperformed prior methods achieving a high sensitivity of 93.16% (for a receptive field of 20x20x16), with reasonable computation time of roughly 1 min per subject, their precision was only 42.69%. This is likely due to either the limited size of their training database which included 320 manually annotated SWI volumes from stroke and normal aging patients to generate approximately 3910 samples or lack of labelled mimics. The limited availability of patient data and manually labelled datasets to establish the ground truth is a common challenge for recognition tasks in the medical image domain that often results in overfitting and poor generalization. While some CNN-related frameworks have been proposed in the literature to handle small-data (Pasini, 2015; Shaikhina and Khovanova, 2017) using strategies such as data augmentation (Huang et al., 2017) and transfer learning (Shin et al., 2016), there still remains a need to evaluate CNNs on larger amounts of real rather than synthetic data before CNNs can be considered reliable clinical decision support tools.
The goal of this study was to develop a research tool that can: 1) expedite the process of generating labelled datasets, 2) obtain a higher specificity than prior approaches and 3) accurately segment detected CMBs for volume quantification. This efficient tool for routine evaluation of CMB burden is a natural extension of our existing computer-aided CMB detection algorithm, providing user-guidance for accurate semi-automated CMB detection with high specificity, volume segmentation, and FP labelling to generate labels of CMB mimics for CNN training.
2. Materials and methods
2.1. Algorithm design
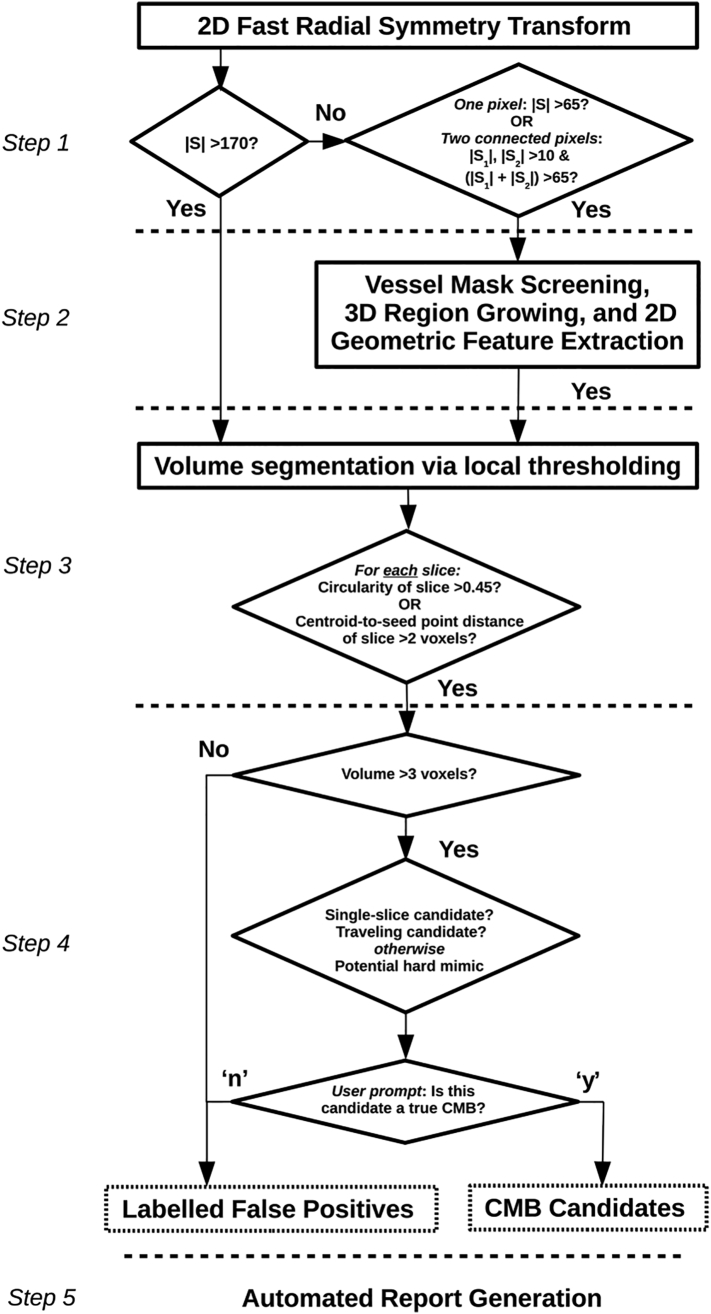
The schematic flowchart in Fig. 1 illustrates the overall design of our algorithm. It initiates with the prior CMB detection algorithm and follows with more recent updates permitting fully automated volume segmentation, user-guided CMB classification, and simultaneous FP labelling. The algorithm is implemented in MATLAB (Natick, MA, USA) and accepts a single, non-projected volumetric T2*-weighted or SWI dataset as the input. Although we demonstrate its performance using non-projected SWI datasets as the input, it is flexible to work on other images of similar contrast.
Fig. 1.
Schematic of the algorithm's architecture. Steps 1–2 are part of the existing algorithm (Bian et al., 2013), while steps 3–5 correspond to recent additions.
2.1.1. Initial Automated CMB detection
As described in Bian et al. and illustrated in Fig. 1, the first step of CMB detection employs a 2D fast radial symmetry transform (FRST) initially proposed by Loy et al. (Loy and Zelinsky, 2003) to identify all possible points of interest (namely, putative CMBs) based on local radial symmetry, |S|. Candidate CMBs are thresholded according to |S|, then undergo vessel mask screen followed by 3D region growing and 2D geometric feature extraction to remove some FPs. At this stage the original detection algorithm would normally terminate, retaining a considerable number of FPs. As a natural extension from this termination point, Section 2.1.2 to Section 2.1.4 describe the new Steps 3 through 5 of the algorithm that are central to the proposed research tool.
2.1.2. Automated CMB volume segmentation
From each seed point identified n Step 2, a cube containing the candidate CMB is extracted from SWI images. The cube boundaries are defined by a voxel search number, computed as some maximum radius value (mm) divided by the voxel size (mm). We found that for a voxel size of 0.5 × 0.5 × 1.0 mm, a maximum radius of 4 mm was sufficient for capturing candidates with relatively large radii.
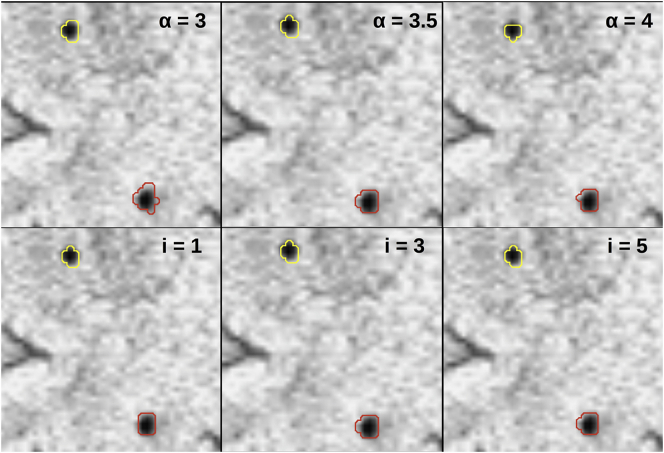
Within the image cube, a local threshold is applied based on an iterative threshold value calculated as, mean_intensitylocalvoxels – α*(stdev_intensitylocalvoxels), where α is the threshold degree. The number of iterations and α were empirically chosen to be 3 and 3.5, respectively. Fig. 2 illustrates the effects of both parameters on segmentation results. Fewer iterations resulted in worse segmentation performance, while an increased number of iterations increased computation time and had little effect on results. A 0.5 increase or decrease in α also resulted in poor segmentation performance.
Fig. 2.
Effect of alpha on segmentation results (top row: i = 3; bottom row: alpha = 3.5).
Because the voxel search number is fixed, image cubes containing candidate CMBs with small radii may be contaminated by the presence of falsely thresholded objects such as other candidate CMBs or elongated vessels. To solve this problem, the segmented region on each slice of an image cube undergoes 2D geometric feature examination to eliminate potential mimics. In the uncommon event that multiple regions are segmented on a single slice of an image cube, the region closest to the seed point is retained. Otherwise, for each segmented region, k, circularity is computed by, 4π*Area(k)/(Perimeter(k) + π) (Bian et al., 2014), along with the centroid-to-seed point distance. If the circularity is <0.45 or the centroid-to-seed point distance is >2 voxels, the region is removed. These cutoffs were chosen empirically, with the intention of using conservative values to achieve good segmentation results and maintain sensitivity. The final output of this third step is a binary mask containing segmented volumes of all candidate CMBs.
2.1.3. User-guided CMB classification and FP labelling
Step 4 involves an interactive component where the user must classify candidate CMBs either as a true positive or FP. A “de-noising” filter is first applied, to eliminate a large portion of FPs with unusually small radii, thereby also minimizing the number of candidates requiring revision. Candidates with a volume < 3 voxels are removed, and subsequently labelled on a “false positive” mask, each with a pixel value of 1. The optimal cutoff value of 3 was solely based on visual inspection.
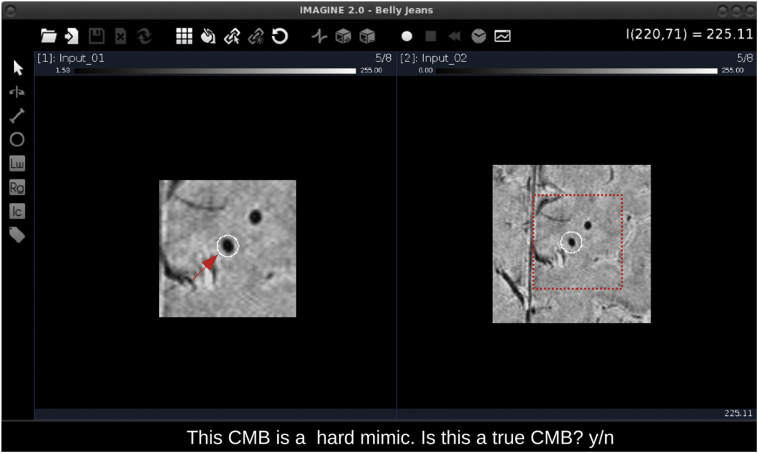
Following de-noising, the remaining candidates are presented to the user one at a time, in consecutive pop-up windows. We opted to use a MATLAB-based 3D image viewer (IMAGINE; Christian Wuerslin, Stanford University) to display candidate CMBs. The interface offers many attractive features that enhance the user's ability to evaluate each candidate CMB, including simultaneous viewing of multiple image volumes, easy navigation between slices, zooming functionality, and real-time resampling of image data to change the orientation.
A screenshot of the complete user interface is provided in Fig. 3. The pop-up window consists of two side-by-side panels each containing an image stack centered on the candidate of interest which is highlighted by a white circle (Fig. 3). The image stacks are conveniently presented at different magnifications for quick and easy evaluation of the candidate CMB, however, the zoom function can be used to further adjust the magnification if desired. In addition to the pop-up window, the user is presented with one of three sentences that classifies the candidate CMB as either a candidate occupying a single slice, as a “travelling” candidate (based on a centroid shift >1 voxel), or as a potential hard mimic. The sentence description provides the user with some context about the candidate to aid their assessment. The user is then asked to return the letter ‘y’ if the candidate appears to be a true CMB, otherwise, the user should respond with the letter ‘n’ if the candidate appears to be a FP. Each FP is labelled on the existing “false positive” mask with a pixel value of either 2, 3, or 4, according to their initial classification as either a single-slice candidate, a travelling candidate, or as a potential hard mimic, respectively. The final FP mask thus contains pixel values ranging from 0 to 4, with 0 representing a null value, and can collectively be interpreted as the total number of FPs removed after Step 2. Additionally, a mask of final CMB candidates is output based on the total number of candidates the user classified as true CMBs.
Fig. 3.
Screen capture of the user interface.
2.1.4. Automated report generation
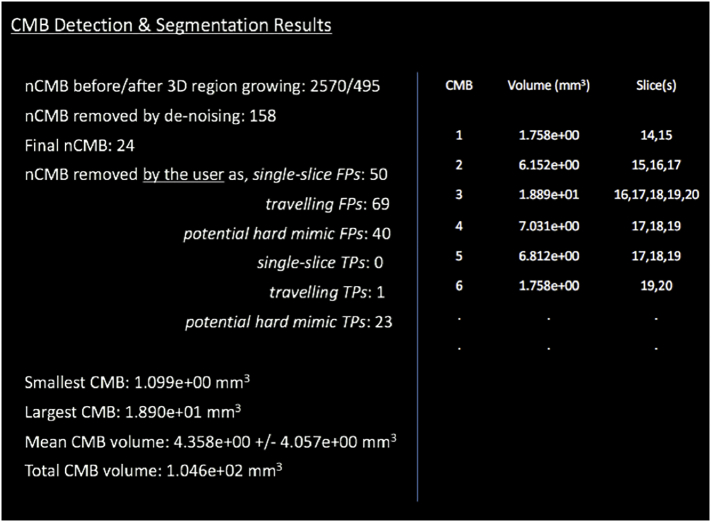
Upon the completion of Steps 1 through 4, volumes are calculated for each true CMB in the final candidate mask. Volume is calculated here as the total number of 3D locally connected voxels (26-connectivity) in a region containing all detected voxels of a candidate, multiplied by the known volume of a single voxel in millimeters.
A text file of the results is automatically generated at this stage and includes the volume and slice location for each true CMB along with the range of volumes, average volume, and total CMB volume (Fig. 4). This information is preceded by the total number of candidates identified before and after 3D regions growing, the final number of true CMBs and FPs, as well as a summary of the user's responses.
Fig. 4.
Example of the report automatically generated in the final step of the algorithm.
2.2. Performance evaluation
2.2.1. Patient population
Parameter optimization and performance evaluation were completed using the same patient dataset as described in Bian et al. (Bian et al., 2013). This included a retrospective cohort of fifteen patients with radiographic evidence of a glioma, who provided informed consent to undergo T2*-weighted MR imaging on our research-dedicated 3 T scanner. To be deemed eligible for participation in the study, each patient had to have had received fractionated external beam radiation therapy at least 2 years prior to MR imaging, leading to vascular injury in the form of CMBs and have evidence at least 10 potential CMBs on initial visual inspection. Of the 15 patients recruited, 5 were randomly assigned to the training set for parameter optimization. The remaining 10 patients comprised the test set on which the performance of the algorithm was evaluated.
2.2.2. Image acquisition and generating SWI images
Patients were imaged using a 3 T MRI system (GE Healthcare, Waukesha, WI) equipped with an 8-channel phased array receiver coil (Nova Medical, Wilmington, MA). A 3D flow-compensated spoiled GRE sequence was used to perform high resolution T2*-weighted axial imaging (repetition time (TR)/echo time (TE)/flip angle (Ɵ) = 56 ms/28 ms/20°; FOV = 24x24cm; 40 slices; slice thickness = 2 mm, in-plane resolution = 0.5 × 0.5 mm). A GRAPPA-based parallel imaging acquisition (acceleration factor (R) = 2) was used to achieve a total acquisition time of under 7 min for supratentorial brain coverage.
SWI series were created according to the method described in Lupo et al. (Lupo et al., 2009) FSL's brain extraction tool (Smith, 2002) was used to create a brain mask from the combined magnitude image, which was subsequently applied to the SWI images in order to remove the skull and background. Intensity normalization was then performed by forcing the range of pixel intensity values to be between 0 and 255, whereby minimum and maximum intensity values were taken as the 0 and 98th percentile intensity of the original image. Although the original algorithm included an additional step to generate continuous mIP SWI images for CMB identification, we have since found that (although more visually appealing) the projection processing can accidently project CMBs in or close to dark structures that do not surround them in actual anatomy, leading to decreased FRST response or leakage in region growing, and ultimately an increased number of FNs. Our added 3D volume segmentation also needed to be performed in the original space.
2.2.3. Visual assessment of CMB burden
Previously labelled datasets by one subspecialty-certified neuroradiologist (CPH) and one experienced reader (JML) were used to establish the ground truth of CMB burden. The raters identified true CMBs on minimum intensity projected SWI images from the test set through both visual inspection and automated detection where extra true CMBs may be revealed. Areas such as the ventricles and tumor cavity were excluded from the assessment given the unlikelihood that a true CMB would occur in either location. ‘Definite’ versus ‘possible’ CMBs were also distinguished through differences in shape, contrast, and location, using a scoring system similar to what has previously been reported (Conijn et al., 2011; de Bresser et al., 2013; Gregoire et al., 2009)
For the proposed algorithm, we opted to consider definite and possible CMBs as a whole, rather than differentiating between the two groups. This decision is supported by the fact that our prior work has already shown superior performance for the detection of definite versus possible CMBs, and that raters tend to agree very well on those CMBs classified as definite, with poorer agreement on CMBs classified as possible. Taken together, when assessed as a whole, one can expect much of the variation in performance and agreement metrics will reflect those CMBs characterized as ‘possible’ rather than ‘definite’ CMBs.
Because CMBs were initially labelled on mIP SWI images, and the proposed algorithm outputs a mask of segmented CMBs candidates in the original space, labels were projected back into the original space and revised by one experienced reader (MAM) resulting in a total of 248 CMBs from 304. These ground truth data were used to evaluate the algorithm's sensitivity limit.
2.2.4. User-guided assessment of CMB burden
A total of three raters including one junior neuroradiology staff (SP) and two experienced readers (MAM, YC), used the proposed algorithm to assess total CMB burden within the test set. Table 1 reports the total number of candidates at various steps of the algorithm (Fig. 1) for the three raters. The intra-rater agreement in CMB detection was evaluated using a measure of the true positive rate for one rater (MAM) who assessed CMB burden across the test set both manually (i.e. ground truth labelling) and with the computer aid. The inter-rater agreement was measured using the Intra-class Correlation Coefficient (ICC) adapted for a fixed set of raters (Shrout and Fleiss, 1979).
Table 1.
Total number of candidates at various steps in the algorithm.
| Rater |
Step 1 |
Step 2 |
Step 4 |
FPs removed |
|||
|---|---|---|---|---|---|---|---|
| >3 voxels | Travelling | Occupying single slice | Potential hard mimic | ||||
| 1 | 9920 | 1228 | 266 | 279 | 147 | 190 | 97 |
| 2 | 9920 | 1228 | 187 | 279 | 175 | 170 | 135 |
| 3 | 9920 | 1228 | 193 | 279 | 171 | 159 | 125 |
2.2.5. Validation of automated volume segmentation
A fourth rater (MS) validated the volume segmentation results in the test set using in-house visualization tools that allow for the modification of individual CMB regions of interest (ROIs) slice-by-slice, as needed (Fig. 5). The degree of overlap between the revised datasets and original segmentation results was measured using a Jaccard Index (Real and Vargas, 1996) computed as the area of the intersection divided by the area of the union of the two masks.
Fig. 5.
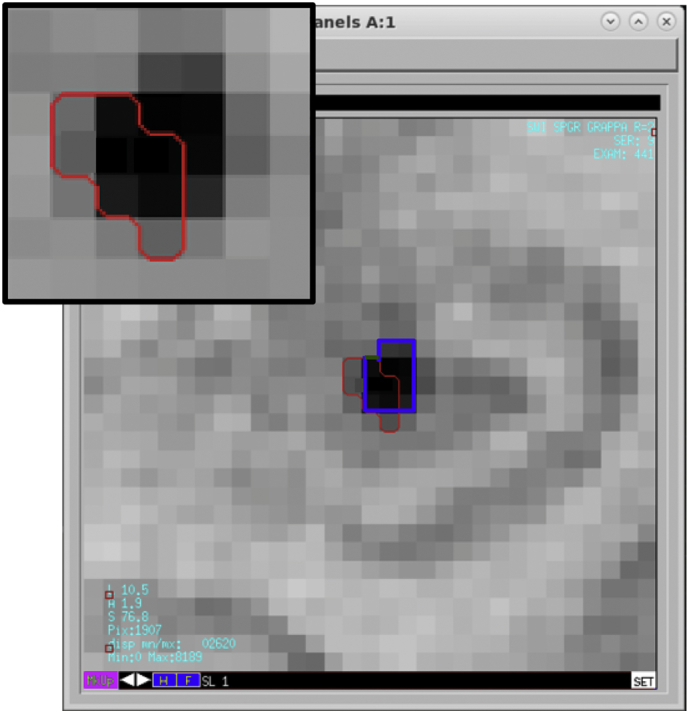
Revision of a CMB ROI on one magnified image slice. The red outline corresponds to the original segmentation result. The blue outline illustrates the revised result which involves an expansion of the ROI to include adjacent pixels of similar intensity value.
3. Results
3.1. Sensitivity
The sensitivity of the original algorithm was maintained at 86.7%. Of the 248 labelled CMBs, 215 were detected by the algorithm, while the remaining 33 CMBs were FNs. On average, this equates to approximately 3.3 CMBs missed per patient.
3.2. Intra-rater agreement
A high intra-rater agreement was found between CMBs labelled by one rater (M.A.M) via 1) manual labelling based on visual inspection and 2) user-guided classification using the proposed algorithm. The spatial agreement between the two datasets was 90.7%, with an average of 7 new candidates identified as true CMBs per subject, when using the proposed algorithm for visual guidance.
3.3. Inter-rater variability and specificity
Amongst the three different raters, an average of 215±44 candidates were classified as true CMBs using the proposed algorithm. It took between 9 and 22 min to evaluate each patient in the test set; this time varied with total CMB burden and with the rater's experience using the algorithm. On average, it took 14.5 min to evaluate approximately 70 candidates, including the time for pre-processing, detection, and segmentation. Compared with manual labelling which can take up to 2 h per patient without including volume segmentation, this result demonstrates a substantial improvement in rating time.
The ICC for between-rater CMB classification was 0.97 with a 95% confidence interval of 0.92 to 0.99, indicating strong agreement between raters and low inter-rater variability. The raters eliminated a total of 464±22 candidates as FPs in step 4 of the algorithm (Fig. 1), where as in the original algorithm a total of 449 FPs were included in the final candidate mask. It is worth noting that a greater number of candidates (including FPs) are typically detected on non-projected versus mIP SWI images, explaining the difference in these outcomes. Nonetheless, the current result demonstrates a substantial reduction in FPs, down to the level of variation amongst raters. For any one rater, the perceived number of FPs is zero. Table 2 compares differences in efficiency and capability amongst our proposed and original algorithms.
Table 2.
Comparison of original and proposed CMB detection algorithm.
| Algorithm |
True CMBs |
Sensitivity |
False positives |
Computation |
Image Type |
Features |
||
|---|---|---|---|---|---|---|---|---|
| Total | Mean | Total | Per patient | |||||
| Bian et al. | 304 | 30.4 | 86.5% | 449 | 44.9 | 1 min | mIP SWI | – |
| Morrison et al. | 248 | 24.8 | 86.7% | 0a | 0a | 9–22 min avg. 14.5 min |
SWI | segmentation FP labelling |
zero as perceived by any one rater, however small variations exist relative to another rater.
3.4. Validation of automated volume segmentation
As summarized in Table 3, the fully automated segmentation results were found to be highly accurate. The mean Jaccard Index was 0.98±0.01 indicating high similarity between the original and revised CMB ROIs.
Table 3.
Summary of results.
| Measure | Metric | Result |
|---|---|---|
| Intra-rater agreement in detectiona | True positive rate | 0.91 |
| Inter-rater variability in detection | ICC | 0.97 CI[0.92,0.99] |
| Volume segmentation accuracy | Jaccard Index | 0.98±0.01 |
Between manual and computer-guided labelling of CMBs for one rater.
4. Discussion
Towards improving our understanding of CMBs and their clinical relevance, there remains a need for fast and accurate methods to detect, segment, and quantify CMBs. While several computer-based methods have been proposed (Bian et al., 2013; Seghier et al., 2011; Kuijf et al., 2012; Barnes et al., 2012; van den Heuvel et al., 2016; Fazlollahi et al., 2014), many of these prior developments have demonstrated poor specificity, thereby limiting their use. The present work describes our modifications to a previously developed CMB detection algorithm, enabling the generation of highly specific and quantitative CMB-labelled datasets, with substantially reduced computation time compared to manual labelling. The FRST-based approach automatically detects CMBs with high sensitivity; and.
human-level classification is achieved through the incorporation of a real-time interface that guides the user to candidate CMBs. Both the intra-rater and inter-rater variability were low, and segmentation results were highly accurate. Collectively, these results demonstrate the efficiency of our tool for evaluating CMB burden.
In designing the algorithm, we opted to include FP labelling capabilities to obtain a mask of CMB mimics that can be used in CNN-based detection and classification methods currently being developed by our group. While such methods appear promising in their ability to accurately differentiate true CMBs from their hard mimics and accelerate the CMB labelling process, they are often viewed as black-box type-problems that lack transparency. In this regard, our tool is advantageous because it offers transparency and user control, and could therefore serve as a valuable adjunct to future CNN-based methods, enabling a more thorough evaluation of CMB burden such as in cases where data quality is poor or when additional intracranial pathologies are present in the brain.
Although the present work illustrates the tool's capabilities and performance specifically for RT-induced CMBs in patients with brain tumors, this development can certainly be applied to other clinical populations affected by CMBs such as patients with cerebral amyloid angiopathy, cerebral vascular malformations, and TBI. Improvements in the algorithm's performance are predicted in these populations because their CMBs tend to appear larger and more pronounced than RT-induced CMBs. The algorithm can also be applied to different iron-sensitive imaging data acquired at various MRI field strengths. In our experience, we have found that CMBs are best detected from SWI images acquired at 7 T because of the increased spatial resolution and enhanced iron sensitivity (Bian et al., 2014).
To our knowledge only one other group has reported on a user-guided interface for CMB detection in traumatic brain injury patients (van den Heuvel et al., 2016). Their computer-aided system initiates with automated CMB detection via a supervised machine learning approach, and follows with the user-guided interface to review and revise classification results. Similar to our experience, the majority of FPs generated by their automated detection system were eliminated after applying the user-guided interface, and there was an observed increase in sensitivity with computer-aid compared to manual labelling solely based on visual inspection. In comparison to prior developments however, their system requires more computation time. They reported 17 min required for pre-processing (e.g. brain extraction, normalization), feature extraction, classification, and CMB segmentation, with an additional 13 min, on average, to review approximately 80 candidates using the guided interface. This is nearly double the computation time required for our algorithm to achieve the same result with comparable sensitivity. In our case, a total of 14.5 min, on average, was needed for pre-processing, detection, segmentation, and user-guided classification of approximately 70 candidates. We achieved a high sensitivity of 86.7% using a simple FRST-based approach with hand-crafted features, whereas their algorithm based on a much more sophisticated machine learning approach, only slightly increased sensitivity to 89%, and still generated enough FPs to warrant the use of a user-guided interface. While this model was trained and classified on one type of data acquired on the same MRI scanner and with consistent acquisition parameters, our parameters were trained to accommodate multiple types of data including less sensitive GRE images as well as over 130 datasets of single- and multi-echo SWI images acquired at 3 T and 7 T (Bian et al., 2014), corresponding to patients between ages 10 and 71 who were treated with RT for an adult or pediatric brain tumor at least 8 months prior to scan. Based on our initial experience, the algorithm appears to be robust, performing well across our heterogeneous database. As a result, we have made our algorithm available to the public in a GitHub repository (https://github.com/LupoLab-UCSF/CMB_labeler). A test set comprised of SWI images acquired on our 3 T and 7 T scanners is also available in the repository.
The main limitation of our algorithm is the lack of ability to accurately track serial changes in CMB development within individual patients. Serial tracking of CMBs is of great interest because it provides insight into the timeline of CMB development relative to treatment and other changes in the brain, as well as their spatial growth pattern, and rate of change of volume. Differences in data quality, brain coverage, and slice positioning may influence how the algorithm and user characterize and classify the same CMB on two datasets acquired at different time points. Newly developing CMBs that often appear as tiny, low-contrast foci, may be rejected by the algorithm or overlooked by the user. Intra-rater variability, though small, also makes it difficult to accurately label the same set of CMBs over multiple serial scans, even with the aid of a user-guided interface. Future studies will incorporate a serial tracking feature into our algorithm, that will first align the datasets and then perform automated CMB detection on the most recent serial scan where we expect to see the highest CMB burden, as well as utilize the labelled FP mimics in a CNN to eliminate the need for user-guided false positive removal and create a fully automated tool. Similar to the current functionality of our guided interface, a montage of interactive windows, localized to the ROIs, will be presented to the user for quick evaluation and between-scan comparison. We predict that these features will substantially reduce the variability in CMB detection across serial data, and minimize human intervention.
5. Conclusions
An efficient tool has been developed for the routine assessment of CMB burden. The algorithm utilizes a 2D fast radial symmetry transform and handcrafted features to automatically detect and segment CMBs with high sensitivity and incorporates a user-guided interface to achieve human-level classification and maximum specificity in much less time than manual labelling. The algorithm also measures local CMB volumes and records FP candidates to facilitate the process of generating labelled training data for more sophisticated machine learning approaches. Performance was evaluated on a test set of 10 patients treated with RT for a brain tumor. Both inter-rater and intra-rater variability in CMB classification were low, and volume segmentation results were highly accurate. The tool has already been successfully applied to over 130 datasets acquired with various imaging parameters at different MR field strengths, demonstrating the algorithms flexibility and robustness to heterogeneous imaging data.
Acknowledgments
Acknowledgements
The authors would like to acknowledge the assistance of the research staff, MRI technicians, and nurses. A special thanks to Angela Jakary, Kim Semien, Kimberly Yamamoto, and Mary Mcpolin, for consenting and scanning the subjects reported in this work.
Grant support
This work was support by NIH grants R01HD079568 and U54NS065705 and GE Healthcare.
References
- Barnes S.R.S., Haacke M.E., Ayaz M. Vol. 29. 2012. Semi-Automated Detection of Cerebral Microbleeds in Magnetic Resonance Images; pp. 844–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian W., Hess C.P., Chang S.M. Computer-aided detection of radiation-induced cerebral microbleeds on susceptibility-weighted MR images. NeuroImage Clin. 2013;2:282–290. doi: 10.1016/j.nicl.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian W., Hess C.P., Chang S.M. Susceptibility-weighted MR imaging of radiation therapy- induced cerebral microbleeds in patients with glioma: a comparison between 3T and 7T. Neuroradiology. 2014;56:91–96. doi: 10.1007/s00234-013-1297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Yu L., Dou Q. Automatic detection of cerebral microbleeds via deep learning based 3D feature representation. Proc. IEEE-ISBI Conf. 2015:764–767. [Google Scholar]
- Conijn M.M., Geerlings M.I., Biessels G.J. Cerebral microbleeds on MR imaging: comparison between 1.5 and 7 T. AJNR. 2011;32:1043–1049. doi: 10.3174/ajnr.A2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bresser J., Brundel M., Conijn M.M. Visual cerebral microbleed detection on 7T MR imaging: reliability and effects of image processing. AJNR. 2013;34:61–64. doi: 10.3174/ajnr.A2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Q., Chen H., Yu L. Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans. Med. Imaging. 2016;35:1182–1195. doi: 10.1109/TMI.2016.2528129. [DOI] [PubMed] [Google Scholar]
- Fazlollahi A., Meriaudeau F., Villemagne V.L. Efficient machine learning framework for computer-aided detection of cerebral microbleeds using the Radon transform. Proc. IEEE-ISBI Conf. 2014:113–116. [Google Scholar]
- Gregoire S.M., Chaudhary U.J., Brown M.M. The microbleed anatomical rating scale (MARS): reliability of a tool to map brain microbleeds. Neurology. 2009;73:1759–1766. doi: 10.1212/WNL.0b013e3181c34a7d. [DOI] [PubMed] [Google Scholar]
- Gregoire S.M., Smith K., Jäger H.R. Cerebral microbleeds and long-term cognitive outcome: longitudinal cohort study of stroke clinic patients. Cerebrovasc. Dis. 2012;33:430–435. doi: 10.1159/000336237. [DOI] [PubMed] [Google Scholar]
- Gupta S., Girshick R., Arbelaez P. European Conf. on Comput. Vis. Springer; 2014. Learning rich features from rgb-d images for object detection and segmentation; pp. 345–360. [Google Scholar]
- Haley K.E., Greenberg S.M., Gurol M.E. Vol. 15. 2014. Cerebral Microbleeds and Macrobleeds: Should They Influence Our Recommendations for Antithrombotic Therapies? pp. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Shan J., Vaidya V. Lung nodule detection in CT using 3D convolutional neural networks. In Proc. IEEE-ISBI Conf. 2017:379–383. [Google Scholar]
- Kheradpisheh S.R., Ghodrati M., Ganjtabesh M. Humans and deep networks largely agree on which kinds of variation make object recognition harder. Front. Comput. Neurosci. 2016;10:92. doi: 10.3389/fncom.2016.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijf H.J., de Bresser J., Geerlings M.I. Efficient detection of cerebral microbleeds on 7.0T MR images using the radial symmetry transform. NeuroImage. 2012;59:2266–2273. doi: 10.1016/j.neuroimage.2011.09.061. [DOI] [PubMed] [Google Scholar]
- Lawrence T.P., Pretorius P.M., Ezra M. Early detection of cerebral microbleeds following traumatic brain injury using MRI in the hyper-acute phase. Neurosci. Lett. 2017;655:143–150. doi: 10.1016/j.neulet.2017.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy G., Zelinsky A. Fast radial symmetry for detecting points of interest. IEEE-TPAMI. 2003;25:959–973. [Google Scholar]
- Lupo J.M., Banerjee S., Hammond K.E. GRAPPA-based susceptibility-weighted imaging of normal volunteers and patients with brain tumor at 7T. Magn. Reson. Med. 2009;27:480–488. doi: 10.1016/j.mri.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo J.M., Chuang C., Chang S.M. 7 tesla susceptibility-weighted imaging to assess the effects of radiation therapy on normal appearing brain in patients with glioma. Int. J. Radiat. Oncol. Biol. Phys. 2012;82:493–500. doi: 10.1016/j.ijrobp.2011.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Ramirez S., Greenberg S.M., Viswanathan A. Cerebral microbleeds: overview and implications in cognitive impairment. Alzheimers Res. Ther. 2014;6:33. doi: 10.1186/alzrt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini A. Artificial neural networks for small dataset analysis. J Thorac Dis. 2015;7:953–960. doi: 10.3978/j.issn.2072-1439.2015.04.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real R., Vargas J.M. The probabilistic basis of Jaccard's index of similarity. Syst. Biol. 1996;45:380–385. [Google Scholar]
- Sa I., Ge Z., Dayoub F. DeepFruits: A Fruit Detection System Using Deep Neural Networks. Sensors (Basel) 2016;16:1222. doi: 10.3390/s16081222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seghier M.L., Kolanko M.A., Leff A.P. Microbleed detection using automated segmentation (MIDAS): a new method applicable to standard clinical MR images. PLoS One. 2011;6:1–9. doi: 10.1371/journal.pone.0017547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikhina T., Khovanova N. Handling limited datasets with neural networks in medical applications: a small-data approach. Artif. Intell. Med. 2017;75:51–63. doi: 10.1016/j.artmed.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Shin H., Roth H.R., Gao M. Deep convolution neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging. 2016;35:1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout P.E., Fleiss J.L. Intraclass correlations: uses in assessing rater reliability. Psychol. Bull. 1979;86:420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- Smith S.M. Fast robust automated brain extraction. Hum. Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.K., Chen Y.K., Lu J. Cerebral microbleeds and quality of life in acute ischemic stroke. Neurol. Sci. 2011;32:449–454. doi: 10.1007/s10072-011-0571-y. [DOI] [PubMed] [Google Scholar]
- van den Heuvel T.L.A., van der Eerden A.W., Manniesing R. Automated detection of cerebral microbleeds in patients with Traumatic Brain Injury. NeuroImage Clin. 2016;12:241–251. doi: 10.1016/j.nicl.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Flier W.M., Cordonnier C. Microbleeds in vascular dementia: clinical aspects. Exp. Gerontol. 2012;47:853–857. doi: 10.1016/j.exger.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Wan J., Wang D., Hoi S.C.H. Deep learning for content-based image retrieval: A comprehensive study. Proc. 22nd ACM Conf. 2014:157–166. [Google Scholar]