Abstract
The phytohormone auxin is the information carrier in a plethora of developmental and physiological processes in plants1. It has been firmly established that canonical, nuclear auxin signaling acts through regulation of gene transcription2. Here we combined microfluidics, live imaging, genetic engineering and computational modeling to reanalyze the classical case of root growth inhibition3 by auxin. We show that Arabidopsis roots react to addition and removal of auxin by extremely rapid adaptation of growth rate. This process requires intracellular auxin perception but not transcriptional reprogramming. The formation of the canonical TIR1/AFB-Aux/IAA co-receptor complex is required for the growth regulation hinting to a novel, non-transcriptional branch of this signaling pathway. Our results challenge the current understanding of root growth regulation by auxin and suggest another, presumably non-transcriptional signaling output of the canonical auxin pathway.
Plant roots are positively gravitropic4. This is essential for plants to colonize land and for water and nutrient uptake. The columella cells of the root tip perceive gravity5,6, and following gravistimulation redirect the flux of the phytohormone auxin to the lower side of the root7. In response to auxin, epidermal cells inhibit their elongation3,8. The differential auxin distribution and cell elongation thus leads to bending of the root8 along the gravity vector (Fig.1A). Root growth inhibition in response to auxin therefore forms the basis for the gravitropic bending enabling land plants to anchor in soil and utilize its resources. The effect of auxin on root growth has been instrumental to identify auxin signaling mechanism. Canonical auxin signaling starts with auxin binding to the TIR1/AFB-Aux/IAA co-receptor complex9 leading to ubiquitination and degradation of Aux/IAA proteins2 that act as transcriptional repressors; the TIR1/AFB-Aux/IAA pathway thus acts via modulation of gene transcription. In the shoot, this nuclear signaling cascade seems to be sufficient to regulate auxin-mediated growth promotion and upward growth towards the light10,11. Components of the TIR1/AFB-Aux/IAA pathway are also crucial for the root growth inhibition by auxin12–14, with roots of corresponding mutants being largely resistant to auxin. A decades-long controversy has argued that auxin’s effect on root growth is too rapid15,16 to be explained by the TIR1-mediated transcriptional regulation. A current model attempting to reconcile this contradiction is that auxin is perceived by an unknown cell surface receptor that mediates the rapid, non-transcriptional growth inhibition, which is later reinforced by the TIR1-mediated transcriptional regulation1,16. Thus, the mechanism of auxin action on root growth remains unknown. Here we established an extensive experimental toolkit including a vertical microfluidic confocal microscopy setup to re-examine the auxin-induced growth inhibition with high time resolution, minimal interference and superior control of experimental conditions.
Fig. 1.
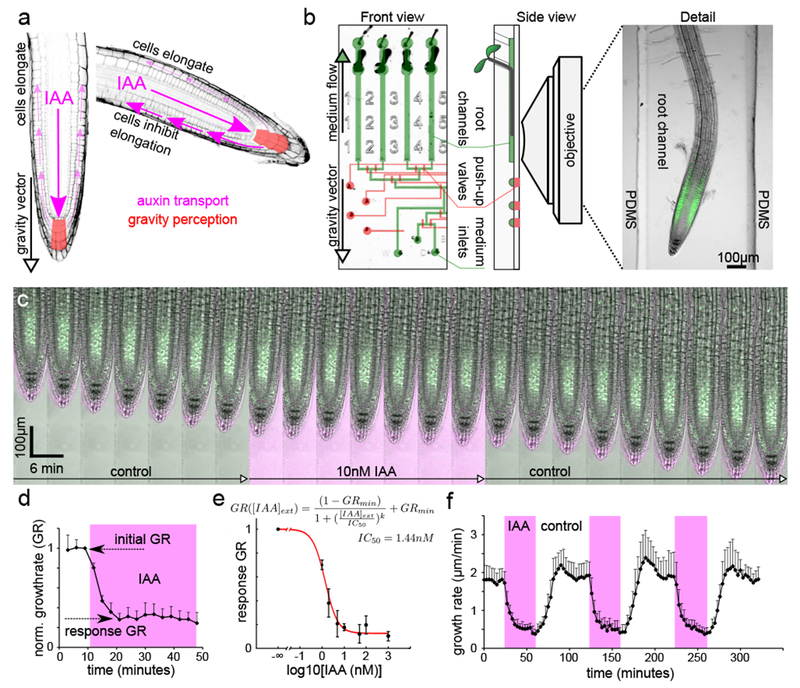
Nanomolar concentrations of auxin reversibly inhibit root growth. (A). Schematics of auxin fluxes in the root tip during gravitropism. (B). The vRootChip device with flow channels and pressure valves are colored in green and red, respectively (left). The side view of the device positioned in the vertical microscope (middle) and a root growing in the channel (right). (C). A timelapse of a DII-Venus root treated with 10nM IAA; IAA medium contains a fluorescent tracer shown in magenta. (D). Quantification of root growth rate (GR) during addition of 10nM IAA, shown in magenta. Normalized to GR precedent to IAA addition, mean of 4 roots, +SD. (E). The dependence of the response growth rate on [IAA]ext. See D for “response GR”. Data points are means of N =3,8,9,3,16,7,16 roots for [IAA]ext =1,2,5,10,50,100,1000nM ±SD. Best fit (red): IC50=1.44nM, GRmin=0.13 (F). Quantification of a repetitive treatment of roots with 10nM IAA (magenta). Mean of 7 roots +SD.
To enable quantitative analysis of root growth response to auxin, roots need to grow along the gravity vector, and any treatment must be extremely gentle to avoid stress that would perturb root growth. We have developed a vertical vRootChip (Fig.1B; Fig.S1A), a microfluidic device with pressure-actuated microvalves derived from the RootChip device17, and performed the experiments using a vertical-stage confocal laser scanning microscope setup18 (Fig.S1B). Roots of Arabidopsis thaliana grew vigorously in the microfluidic channels, and in response to the natural auxin indole-3-acetic acid (IAA) inhibited growth seemingly very rapidly (Fig.1C,D; Movie S1). Growth inhibition was triggered by nanomolar concentrations of IAA (Fig.1E), and the rapid response showed a dose response similar to that of roots grown in presence of IAA over longer periods of time (Fig.S3A), suggesting that both might involve one and the same mechanism. Timing of the growth response was comparable to what was reported for calcium and surface pH response15. These low concentrations well match the physiological levels of IAA that act during gravitropic bending19. The DII-Venus auxin reporter20 reacted to addition of auxin with a very similar dynamics as the growth response (Fig.1C, Fig.S1C), however, the epidermal cells of the elongation zone that are key in auxin reaction did not accumulate this marker in sufficient intensities, and so the DII-Venus dynamics couldn’t be easily compared to the growth response.
Gravitropism is a dynamic process requiring a transient response, and so the ability to recover from the growth inhibition is crucial. Therefore, we analyzed the root’s reaction to withdrawal of auxin from the medium. Surprisingly, following IAA removal from the medium, the roots started to resume the growth rate without any detectable lag phase (Fig.1C,F; Movie S1). The roots reacted to repetitive addition and removal of IAA by repetitive growth inhibition and resumption, respectively, with very reproducible dynamics (Fig.1F). In summary, the growth rate of roots is an extremely sensitive and rapid readout of IAA presence in the medium.
Further, we analyzed the response to 10nM IAA addition and removal in a high temporal resolution. For this, we added a fluorescent tracer to determine exactly the moment when IAA reaches the root surface. This approach unequivocally determined that roots initiated the growth inhibition in less than 30 seconds after auxin reached the root surface (Fig.2A). Following auxin removal, roots started to resume their growth rate within two minutes after the medium was replaced by the auxin-free one (Fig.2B). Even though the resuming response was more variable then the reaction to IAA application, it was very rapid and faster than previously assumed3. We extended the treatment up to 80 minutes of constant auxin exposure (Fig.2C,D), and still roots reacted by rapid (less than 3 minutes) growth resumption when auxin was removed from the medium. These observations do not support any long-term reprogramming of root growth by auxin but suggests a rapidly reversible mechanism of growth regulation.
Fig. 2.
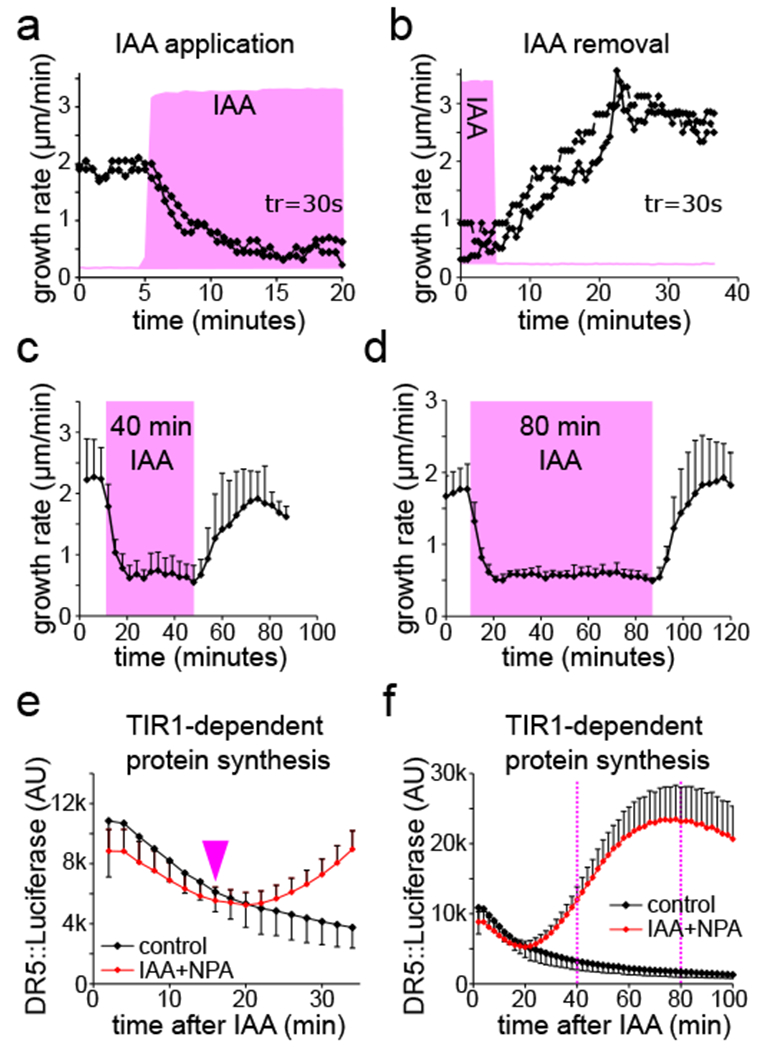
Roots rapidly adapt growth rate to auxin application and removal. (A,B).The reaction of two individual roots to the application (A) and removal (B) of 10nM IAA. The presence of IAA shown as the intensity of the fluorescent tracer (magenta), temporal resolution is 30s. (C,D). Roots rapidly resume the growth rate after 40 minutes (C) and 80 minutes (D) of 10nM IAA treatment. Means of 4 and 6 roots for (C,D), respectively, +SD. (E,F). TIR1-dependent protein synthesis in the root tips was determined by the luminescence intensity of DR5::luciferase. The first detectable increase in the signal is marked by arrowhead (E). At 40 and 80 minutes (dotted lines) the transcriptional response is fully activated (F). 100nM IAA with 1µM NPA was added at timepoint 0. (E) is a zoomed region from (F); means of 5 root tips, ±SD.
The speed of the reaction effectively rules out the possibility that transcription and translation of auxin-induced genes participate in the response, as RNA polymerization, mRNA processing, transport and translation require minutes in eukaryotes21 and, accordingly, the earliest auxin-induced mRNA could be detected approximately 5 minutes after auxin addition22. To directly compare the dynamics of TIR1 transcriptional response with the growth reaction, we harnessed the synthetic auxin-responsive promoter DR5 driving the firefly luciferase enzyme23. The earliest TIR1-dependent gene expression response could be detected approximately 15 minutes after addition of auxin to the roots (Fig.2E) with a full activation only after 40 to 80 minutes of the treatment (Fig.2F). Thus, the dynamics of the auxin-mediated regulation of root growth did not at all temporally correlate with transcriptional activation, therefore arguing against involvement of the transcriptional reprogramming in the growth inhibition response. This contrasts to the situation in the hypocotyl, where the DR5::LUC response matches perfectly with the growth response10.
To further test the requirement of new protein synthesis for the growth inhibition response, we inhibited protein synthesis by the drug cycloheximide. This experiment needed optimization, because cycloheximide on its own influenced root growth (Fig.S2A,B), which can be attributed to its reported rapid effect on ion uptake and respiration in plant tissues24 and to the importance of short-lived proteins in sustaining root growth. Nonetheless, in the presence of lower concentrations of cycloheximide, which were still sufficient to almost completely inhibit the DR5::luciferase response, auxin was fully capable of rapidly inhibiting root growth (Fig.S2B-E). These results show that the auxin-mediated growth inhibition does not involve transcriptome reprogramming not only during the initiation of the response, but also during its duration, which enables a very rapid reversal of the response when auxin concentration decreases. This means that the rapid growth inhibition response is conceptually different from what we thought: it has no distinct “fast” and “slow” component; instead, auxin acts as a constantly fast, non-transcriptional growth brake, on the timescale of tens of seconds to hours. It is important to note that protein synthesis is absolutely required for root growth as such (Fig.S2A,B), and it is probable that the components of the rapid growth response are under control of the transcriptional auxin pathway25 that will lead to alteration of the response sensitivity on the longer time scales.
Next, we addressed where in the cell is the auxin that mediates root growth inhibition perceived. IAA is transported into cells by influx carriers and partly by diffusion; export out of the cell requires efflux carriers7 (Fig.3A). AUX1 influx carrier is expressed in the cells that execute the gravitropic bending (Fig.3B)26, and roots that lack AUX1 are agravitropic and resistant to auxin-mediated growth inhibition in the long term27. In our setup, the aux1 was almost completely insensitive to low IAA concentrations, while it showed a control-like reaction to higher concentrations (Fig.3C; Movie S2). Thus the aux1 mutant’s dose response was shifted to higher IAA concentrations (Fig.3D), similarly to the classical experiment on agar surface (Fig.S3A). As expected, the aux1 mutant showed a normal reaction to the membrane-permeable auxin analog NAA28 (Fig.3E, Fig.S3B), confirming that the mutant is capable of auxin response, but defective in IAA import. These results show that auxin must pass the plasma membrane to trigger the growth inhibition response. To test whether it’s the auxin flux across the membrane itself or the intracellular accumulation of auxin that triggers the response, we pharmacologically inhibited auxin efflux7 (Fig.3A). This resulted in a rapid growth inhibition, similar to IAA application but delayed ca. by 3 minutes (Fig.3F). It was recently claimed that H+ - IAA symport through AUX1 carriers contributes to membrane depolarization or signaling in the root hair cells29. We cannot exclude that the very initial phase of the growth inhibition might be connected directly with flux of the auxin molecules across the plasma membrane, however, altogether the results with the aux1 mutant, membrane permeable NAA and auxin efflux inhibitor point to intracellular auxin concentration ([IAA]cell) as the basis for growth inhibition.
Fig.3.
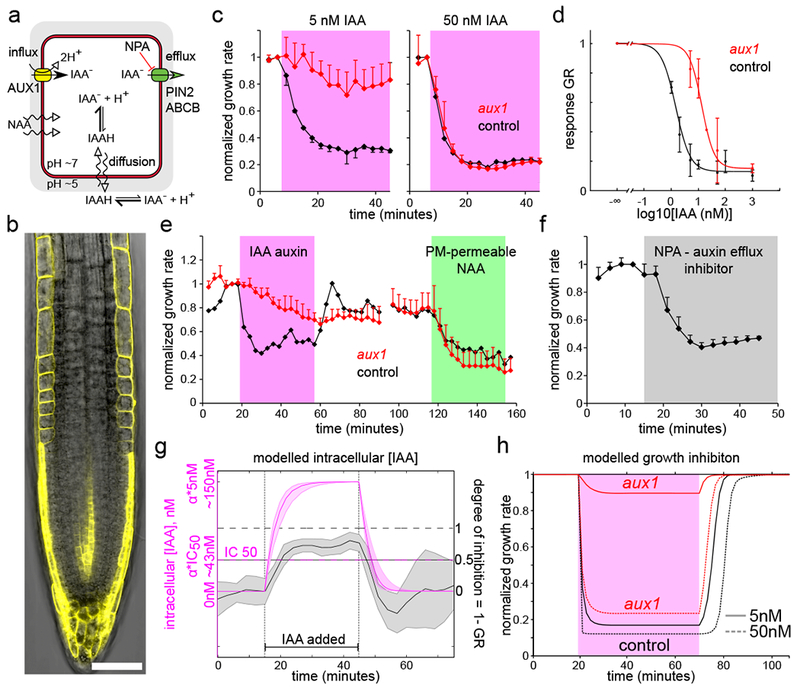
The rapid root growth inhibition depends on auxin levels inside the cell. (A). A schematics of auxin fluxes in a root epidermal cell. (B). Auxin influx carrier AUX1-YFP is expressed in the root epidermal cells. Scale bar = 50µm. (C). The aux1 mutant is resistant to 5nM IAA but reacts to 50nM IAA. Mean normalized values of 3 (5nM), 2 (50nM control) and 5 (50nM aux1) roots, ±SD. (D). The dose response of aux1 is shifted towards higher IAA concentrations by ~10 fold (control is the same as in Fig.1E). Fit: IC50aux1=17.2nM = 11.8*IC50col . Data points are means of N = 10,4,2,9,7 roots for [IAA]ext =5,10,20,50,1000nM respectively +-SD; the 1000nM was done on agar plate. (E). The aux1 mutant reacts little to 5nM IAA (magenta) but its reaction to membrane permeable NAA (100nM, green) is comparable to control roots. Mean normalized values of 5 (aux1) and 2 (control) roots, ±SD. (F). The auxin efflux inhibitor NPA (5µM, gray) triggers root growth inhibition. Mean normalized values of 5 roots, +SD. (G). Auxin accumulation (magenta, simulated) is faster than growth inhibition during application of 5nM IAA (gray, mean experimental values of 1-GR). Note time delay between [IAA]cell = αcol · IC50 and (1 – GR) = 0.5. Accumulation ratio αcol ≅ 30 (Supp.text). Shaded regions correspond to 1 SD (gray, n=6 roots) or variation due to SD of parameter values (magenta, simulation results). (H). The modelled theoretical inhibition for aux1 and control roots.
To understand the dynamics of IAA accumulation, we constructed a computational model of auxin transport in the epidermal cells (Fig.S4 A-C; Sup. text), which incorporated known auxin transport properties of the plasma membrane and capacities of auxin carriers. After IAA addition to the exterior of the modelled wild-type root, the [IAA]cell rapidly reached a steady state concentration approximately 30× higher than the external auxin concentration ([IAA]ext), and removal of auxin from the medium resulted in a rapid decrease of [IAA]cell (Fig.3G; Fig.S4D-F). The timing of IAA accumulation was faster than the growth response, making [IAA]cell a plausible input for the response (Fig.3G). Further, we removed the AUX1 influx carriers to simulate the aux1 mutant. The model predicted steady state [IAA]cell in aux1 being only ~ 2× of [IAA]ext (Fig.S4G,H; Movie S3) which implies that aux1 growth inhibition should be equal to control if [IAA]ext is 30:2 times higher. This prediction matches well the dose response shift of aux1 (11.8± 3.8) which we determined experimentally (Fig.3D). This explains why at low [IAA]ext, aux1’ growth is largely insensitive while at [IAA]ext ≥~50nM aux1 accumulates enough IAA to react similarly to control roots (Sup. Text): the response is already close to saturation at these concentrations. This also clarifies the discrepancy with previous reports that stated that aux1 reacted to IAA almost normally15,16.
Finally, we used the model and fitted dose response curves to test if [IAA]cell exactly maps into the growth rate at any time point (Fig.3H; Sup. text). However, theoretical slopes of growth inhibition and resumption were always steeper than experimental curves, both for aux1 and control, indicating a short delay between signaling and the full execution of the response. Collectively, these results demonstrate that the rapid auxin-mediated root growth inhibition depends on the intracellular IAA accumulation. The auxin receptor that triggers the growth inhibition must therefore be located inside the cell.
Our observations show that the auxin-triggered growth inhibition is a process that starts with intracellular auxin perception but does not involve transcriptome reprogramming, typically associated with the nuclear, TIR1-mediated pathway. However, previous work clearly shows that the TIR1/AFB-Aux/IAA signaling module is required for the auxin-mediated root growth inhibition; the key components of this pathway were even identified based on the auxin-insensitivity of the root growth in the corresponding mutants12–14. How do these facts fit together? We analyzed the auxin response of tir1–1/afb2–1/afb3–1 triple mutant (tir triple)30 that lacks 3 of the 6 auxin co-receptors, but its roots still grow rather well. In the vRootChip, the tir triple mutant had a less steep immediate response to IAA (Fig.4A) and it showed a dose response shift towards higher IAA concentrations (Fig.4B, Fig.S3A), hinting to a direct involvement of the TIR1/AFB in the rapid response to auxin. This was not due to the lack of auxin import, because the tir triple mutant was resistant to the membrane-permeable auxin analogue NAA as well (Fig.S3B). We further took the advantage of the auxin antagonist PEO-IAA31 that binds TIR1 and blocks the formation of the TIR1-auxin-Aux/IAA complex. Addition of PEO-IAA to growing roots resulted to an increase in growth rate (Fig.S5A). However, the most striking effect was observed in roots where growth was partially inhibited by low concentration of IAA: addition of PEO-IAA caused rapid increase in growth rate (Fig.4C). Parallel monitoring of the dynamics of the DII-Venus marker and root growth revealed a direct correlation between the formation of the TIR1/AFB-auxin-Aux/IAA complex and the root growth rate (Fig 1C, Fig.4C). These observations hint at an unexpected involvement of the TIR1-based auxin signaling in the non-transcriptional regulation of root growth.
Fig.4.
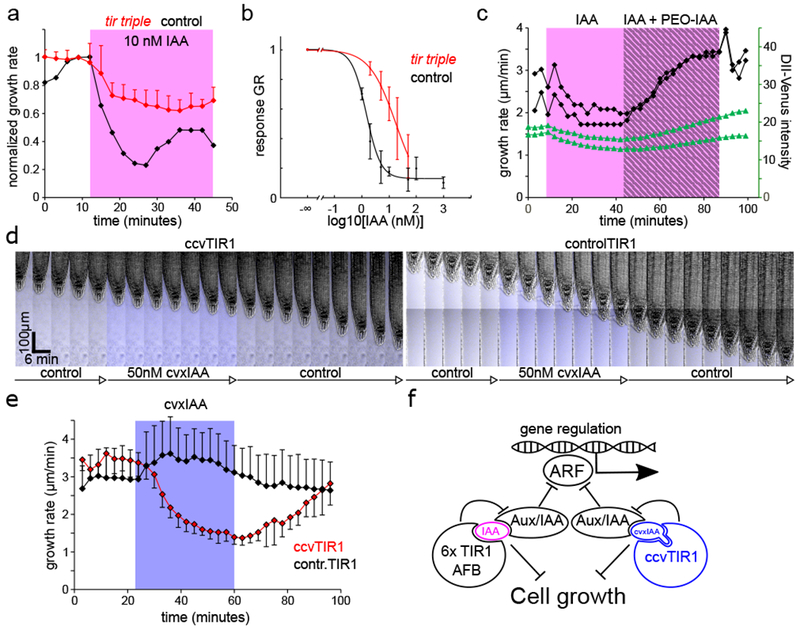
The root growth inhibition requires the TIR1/AFB-Aux/IAA auxin co-receptor. (A). tir triple mutant shows a defect in the response to 10nM IAA; compare to Fig.1 D, F. Mean normalized growth rate, 5 and 2 roots for tir triple and control, +SD. (B). The dose response of the tir triple mutant is shifted towards higher IAA concentrations: IC50=17.8nM (control is the same as in Fig.1E). Data points are means of N= 4,5,4,8,9 roots for [IAA]ext =2,5,10,20,50nM respectively ±SD. (C). Addition of PEO-IAA results in a rapid reversion of the growth inhibition and an increase in the DII-Venus marker intensity. IAA 1nM is shaded in magenta, addition of PEO-IAA (10µM) is marked by gray stripes. DII-Venus intensity (green) was measured in the lateral root cap cells. Growth rates and DII-Venus intensities of two independent roots are shown. (D). cvxIAA causes root growth inhibition only in the synthetic ccvTIR1 line. Addition of cvxIAA (50nM) is marked by the tracer fluorescence in blue. (E). Quantification of the ccvTIR1 (red) and controlTIR1 (black) roots to 50nM cvxIAA (blue). Means of 5 roots for each genotype, ±SD. (F). The formation of the TIR1/AFB-auxin-Aux/IAA complex negatively regulates root growth, likely by a signaling branch that is distinct form the transcriptional regulation function. The synthetic ccvTIR1 receptor and the cvxIAA ligand are shown in blue.
To definitively test the involvement of the TIR1 receptor mechanism in the growth inhibition response directly, we used a synthetic biology approach in which an artificial ligand, convex-IAA (cvxIAA) binds to an engineered concaveTIR1 (ccvTIR1)11. The cvxIAA has no other binding site in the cell, and the ccvTIR1 receptor is unable to bind the natural auxin IAA. Indeed, application of cvxIAA to control roots caused no growth inhibition but cvxIAA application to the ccvTIR1 roots resulted in growth inhibition very similar to the native IAA – control situation (Fig.4D, Movie S4). The cvxIAA was effective at higher concentration (Fig.S5B) and both the inhibition and resuming response were somewhat delayed (~3 minutes) and more gradual (Fig.4E) presumably due to the expected lower affinity of the cvxIAA towards the auxin transporters as compared to native IAA. Regardless, this shows that the growth inhibition could be triggered directly from the synthetic cvxIAA-ccvTIR1 pair demonstrating that the specific activation of the TIR1/AFB-Aux/IAA co-receptor is sufficient to mediate the rapid root growth inhibition.
This study reconciles a long-standing controversy about the rapid versus slow, transcriptional versus non-transcriptional auxin signaling mechanism regulating growth. Our vRootChip-based analysis revealed that the root growth response to auxin is a constant, reversible and rapid non-transcriptional brake proportional to the intracellular auxin levels. This growth break is mediated by the formation of the nuclear TIR1/AFB-auxin-Aux/IAA co-receptor complex, which has been so far associated with auxin-mediated transcriptional regulation1. It implies that this canonical signaling pathway regulates root growth via an unknown, non-transcriptional signaling branch (Fig.4F). Recently, rapid, auxin-dependent depolarization of plasma membrane in root hair cells has been shown also to require AUX1 auxin import, functional TIR1/AFB-Aux/IAA pathway, and the CNGC14 channel29. It was shown previously that rapid auxin response in the root that triggers pH changes and calcium signaling also requires the CNGC14 channel16, but apparently not the TIR1/AFB receptor15. Therefore, it remains to be confirmed whether these distinct rapid auxin effects are mediated by the same non-transcriptional signaling branch triggered from the TIR1/AFB-Aux/IAA receptor.
With the exception of the extremely low concentrations of auxin that were shown to promote growth3, root growth rate is negatively correlated with the formation of the TIR1/AFB-auxin-Aux/IAA complex, and therefore we speculate that free Aux/IAA proteins promote root growth, ubiquitinated Aux/IAA inhibit it, or there is an unknown interactor or substrate of the TIR1/AFB receptor that might regulate growth in either direction. There has been an ongoing debate over ‘fast and slow’ TIR1-dependent and independent auxin responses1. Our observations reveal that the classical auxin-mediated process, the root growth inhibition involves fast and TIR1-dependent mechanism at the same time and force us to broaden our understanding of (nuclear) auxin signaling.
Methods
Plant materials and growth conditions
Arabidopsis thaliana Col-0 was used as control background; in the vRootChip experiments, DII-Venus were often used as control. The following lines originate from these publications: DII-Venus20, DR5::luciferase23, AUX1::AUX1-YFP32, tir triple 30, ccvTIR1 and controlTIR111. aux1 is aux1–100 T-DNA insertion line that was genotyped using these primers: AGCTGCGCATCTAACCAAGT, GTTTCACACCTTCCGCCTAA and the SALK LBb1.3 primer.
For the vRootChip experiments, seeds were surface-sterilized with chlorine gas, sown into cut pipette tips (5mm long) filled with 1/2MS medium with 1% sucrose and 0.8% plant agar that were placed in a round petri dish, according to33. Sown seeds were stratified for 2 days in 4°C, and then placed for 4 days into growth room with 21°C, long day. After that, those seedlings that were just reaching the opening of the pipette tip were placed into the prepared vRootChip (see below). In the chip, 1/4MS, 1/2MS or full MS medium without sucrose (to avoid contamination) was used overnight to 24h, during this time, the roots grew into the microfluidic channels. 1–2hours prior to the experiment, the medium was supplemented with 0.1% sucrose to increase the growth of the roots. During the experiments, medium was flown against the roots with a pressure of 0.2 – 0.4 bar and the seedlings were illuminated with blue and red LED light source as described18.
For other experiments, surface-sterilized seeds were sown on the surface of 1/2MS agar with 1% sucrose and 0.8 % agar in squared petri dishes, stratified for 2 days in 4°C, and then placed vertically for 4 days into growth room with 21°C, long day. For growth measurements over the course of 6 h, seedlings were transferred to 1/2MS with 1% sucrose and 1.5% phytagel and the appropriate treatment and imaged using a flatbed scanner placed vertically, as described34.
Treatments
Treatments in the microfluidic device were achieved by alternating the control medium with a medium with the desired drug or hormone. During the experiments, roots were continuously flushed with medium, to ensure constant composition of the solution and concentration of the treatment molecule (otherwise the roots quickly change the solution and use up the treatment molecule due to the small volume of the channel). The treatment solution was supplemented with a cell-impermeable dextranated fluorescent dye, mostly Tetramethylrhodamine isothiocyanate–Dextran, average mol. weight 20000 (Sigma). The 10mg/ml water stock of the dye was diluted 500–1000× so that it was weakly visible with low 561nm excitation laser intensities. IAA and NAA were from Sigma and were used as 10mM EtOH stocks, PEO-IAA (Olchemim) used as 10mM EtOH or DMSO stock. NPA (Sigma) cvxIAA11, and cycloheximide (Sigma)were used as 10, 10 and 50mM DMSO stocks, respectively.
Imaging and image analysis
The vRootChip was imaged on a vertical-stage laser scanning confocal Zeiss 700, described here:18. Most images were taken with a 20×/0.8 Plan-Apochromat M27 objective, the dextranated reference dye was excited by the 561nM, Venus and YFP with the 488nm laser line.
Root growth measurements were performed using a vertically-positioned flatbed scanner EPSON perfection v370, as described here34. Luciferase bioluminescence was analyzed as described in detail here34.
Root growth rate was measured automatically by stabilizing the drift of the root tip and calculating the growth from the drift vector using MATLAB and Fiji programs. Growth rate in the timepoint n is thus the distance that the root tip travelled between timepoints n-1 and n. DII-Venus intensity was measured in a region of interest drawn around the lateral root cap and stele cells in image series stabilized for growth drift using the StackReg Fiji plugin in the ‘translation’ or ‘rigid body’ option35. All images and movies were viewed, processed and analyzed using Fiji36,37. Data was analyzed in Microsoft Excel, and Figures were assembled in Inkscape, boxplots were made by BoxPlotR (http://shiny.chemgrid.org/boxplotr/ )
vRootChip fabrication protocol
Photomasks:
Photomask designs are exported to DXF from CorelDraw X8 and converted to Gerber using Linkcad 9. We use high-resolution transparency photomasks ~10 microns (JD photo, UK). Valve crossovers have a size of 20 microns. We use a custom-made mask aligner with a 365nm UV LED (Thor Labs) and beam expander. We routinely align within 10 microns. We use 200×200 micron Quake style valves.
Protocols for SU8:
The root channels are 110 microns high in SU8. GM-1075 SU8 photoresist (Gersteltec, Switzerland) is spin coated at 1530 RPM for 100 seconds resulting in 110 microns, baked at 40°C for 30 min, baked at 120°C for 2 minutes, and exposed to UV for 15 minutes. Post bake is 95°C for 30 minutes. 25-micron valves are made with Microchem SU8–3025 using the standard procedure.
Protocols for AZ-40XT:
The mold for rounded fluid layer channels is made with AZ-40XT. We first spin coat HMDS at 3000 RPM for 30 seconds, bake at 110°C for 1 min, spin coat AZ-40XT at 3000 RPM for 30 seconds, bake at 110°C for 5 min, expose ~1J/cm2, and post exposure bake for 2 min at 110°C. We develop in 327 MIF for five minutes and rinse in water. Reflow for several minutes is done at 130°C and inspected under the microscope to observer parabolic channels. The height of the channel in the center is roughly 26 microns by using a profilometer.
Silane treatment:
Wafers were treated in a vacuum desiccator with Trichloro (1H, 1H, 2H, 2H-perfluorooctyl) silane (Sigma) for 1 hour. The treatment needs only be applied once.
PDMS device protocol:
We weight approximately 60 g of 5:1 RTV 615 and 21 g of 20:1 RTV 615. We use a Thinky ARE-250 mixer 2000 RPM mixing and 2 min at 2000 RPM degassing. We make an aluminum foil dish for the wafer. We pour the 5:1 PDMS onto the fluid layer and degas one last time to remove bubbles in the photoresist for approximately 10 min. We then spin coat the 20:1 PDMS at 2000 RPM for 30 seconds. The 5:1 layer is baked for 1 hour at 80°C and the spin-coated layer is baked for 40 minutes at 80°C. The thick layer is cooled off and cut into pieces and holes are punched (0.5mm puncher for the flow inlets and 1.25mm for the root inlets). Scotch tape is then applied to remove particles, and the pieces are aligned by hand under the stereomicroscope. An additional 2 hours of bonding is done at 80°C in the oven. The pieces are then cut out and the final holes for the control valves are punched out (0.5 mm puncher). Scotch tape is applied. Glass slides are prepared by sonication briefly in isopropanol. The devices are plasma bonded using 1 min medium power in a Harrick plasma cleaner (pdc-02). After sealing, the devices they are baked at 100°C for 1 hour on a hotplate and then placed back in the oven overnight at 80°C and then are ready to go and solidly bonded.
Valve electronics:
We use an adaptation of Rafael Gomez circuit board design to control 16 Festo valves, but we use an Arduino to control the valves. The control software is programmed in Arduino and operated by serial communication of commands that control the valve states. The Auto-It scripting language can be used to make automated sequences of programs by interfacing with the serial communication window. The program changes the sequences of media applied and which roots or group of roots the media is directed to.
Growing plants into chip and chip usage
The vRootChip was mounted into a custom-made aluminum sealable holder of a size of the 96-well plate that fitted the microscope inset. The valve layer was first filled with sterile water, then the flow layer was filled with growth medium, all inlets were closed using the 3M tape, pressure of 0.2 bar applied, and we waited as long as all bubbles were removed from the system. Then the tape was removed, seedlings in the pipette tips were transferred into root inlets, the holder was sealed and placed under a blue-red LED lamp to support photosynthesis of the plants overnight to 24 hours, the flow was sustained by ~0.1 bar. Then the entire setup was transferred to the microscope and connected to pressure control and medium, and experiments were performed. Good chips were reused by flowing with 70% EtOH which enabled pulling out the shrunk roots and then cleaned with sterile water.
Statistics and Reproducibility and data availability statement
Number of experiments with similar results from which the illustrative microscopy images or individual measurements were selected: Fig. 1C: more than 3×; Fig. 2A: more than 3×; Fig. 2B: 2×; Fig. 3b: more than 3×; Fig. 4C: 2×; Fig. 4D: more than 3×.
Supplementary Material
Acknowledgments:
The authors thank Robert Hauschild for the help with image analysis. MF was supported by the Austrian Science Fund (FWF) [M 2128-B21], and in the final stages of the project by the Czech Science Foundation [18–10116Y]. MA was supported by the Austrian Science Fund (FWF) [M2379-B28]. Additional funding for MG was received from Ministry of Education of the Czech Republic/MŠMT proj. NPUI - LO1417. MEXT/JSPS KAKENHI JP16H01462 to NU, JP17H06350 to SH, JP26440140 to KT, and JP16H01237 to KUT. KUT is supported by GBMF Grant No. 3035. SH is a JST PRESTO Investigator, and KUT is an HHMI-GBMF Investigator. JF acknowledges the ERC Advanced Grants (ETAP).
Footnotes
Competing interests: the authors declare no competing interests.
All source data for the results in the manuscript are provided in the Supplementary Data 1 file.
References
- 1.Leyser O Auxin Signaling. Plant Physiol. 176, 465–479 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray WM, Kepinski S, Rouse D, Leyser O & Estelle M Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Evans ML, Ishikawa H & Estelle MA Responses of Arabidopsis Roots to Auxin Studied with High Temporal Resolution - Comparison of Wild-Type and Auxin-Response Mutants. Planta 194, 215–222 (1994). [Google Scholar]
- 4.Knight TA On the Direction of the Radicle and Germen during the Vegetation of Seeds. Phil. Trans. R. Soc. Lond. 96, 99–108 (1806). [Google Scholar]
- 5.Darwin C The Power of Movement in Plants. (John Murray, London, 1880). [Google Scholar]
- 6.Nemec B Ueber die art der wahrnehmung des schwek- raftreizes bei den pflanzen. Ber Dtsch Bot Ges 18, 241–245 (1900). [Google Scholar]
- 7.Adamowski M & Friml J PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27, 20–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Went F & Thimann K Phytohormones. (1937). doi: 10.1017/CBO9781107415324.004 [DOI]
- 9.Tan X et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–5 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Fendrych M, Leung J & Friml J TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. Elife 5, 1–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchida N et al. Chemical hijacking of auxin signaling with an engineered auxin–TIR1 pair. Nat. Chem. Biol. (2018). doi: 10.1038/nchembio.2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leyser HMO, Pickett FB, Dharmasiri S & Estelle M Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10, 403–413 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Ruegger M et al. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12, 198–207 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheitz K, Lüthen H & Schenck D Rapid auxin-induced root growth inhibition requires the TIR and AFB auxin receptors. Planta 238, 1171–1176 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Monshausen GB, Miller ND, Murphy AS & Gilroy S Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high-resolution imaging with automated computer vision-based analysis. Plant J. 65, 309–18 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Shih HW, Depew CL, Miller ND & Monshausen GB The cyclic nucleotide-gated channel CNGC14 regulates root gravitropism in arabidopsis thaliana. Curr. Biol. 25, 3119–3125 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Grossmann G et al. The RootChip: an integrated microfluidic chip for plant science. Plant Cell 23, 4234–4240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Wangenheim D et al. Live tracking of moving samples in confocal microscopy for vertically grown roots. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Band LRR et al. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc. Natl. Acad. Sci. U. S. A. 109, 4668–4673 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunoud G et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103–6 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Hoyle NP & Ish-Horowicz D Transcript processing and export kinetics are rate-limiting steps in expressing vertebrate segmentation clock genes. Proc. Natl. Acad. Sci. U. S. A. 110, E4316–24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClure BA, Hagen G, Brown CS, Gee MA & Guilfoyle TJ Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell 1, 229–39 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno-Risueno MA et al. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306–11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MACDONALD IR & ELLIS RJ Does Cycloheximide inhibit Protein Synthesis specifically in Plant Tissues? Nature 222, 791–792 (1969). [Google Scholar]
- 25.Okushima Y et al. Functional Genomic Analysis of the AUXIN RESPONSE FACTOR Gene Family Members in Arabidopsis thaliana. Plant Cell 17, 444–463 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swarup R et al. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7, 1057–1065 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Bennett MJ et al. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273, 948–50 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Delbarre A, Muller P, Imhoff V & Guern J Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198, 532–541 (1996). [DOI] [PubMed] [Google Scholar]
- 29.Dindas J et al. AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+signaling. Nat. Commun. 9, 1174 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dharmasiri N et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9, 109–119 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Hayashi K et al. Rational design of an auxin antagonist of the SCF(TIR1) auxin receptor complex. ACS Chem. Biol. 7, 590–8 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Swarup R et al. Structure-Function Analysis of the Presumptive Arabidopsis Auxin Permease AUX1. Plant Cell 16, 3069–3083 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grossmann G et al. Time-lapse Fluorescence Imaging of Arabidopsis Root Growth with Rapid Manipulation of The Root Environment Using The RootChip. J. Vis. Exp. e4290–e4290 (2012). doi: 10.3791/4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L, Krens SFG, Fendrych M & Friml J Real-time Analysis of Auxin Response, Cell Wall pH and Elongation in Arabidopsis thaliana Hypocotyls. Bio-Protocol 7, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thévenaz P, Ruttimann UE, U M, Thevenaz P, Ruttimann UE & Unser M A Pyramid Approach to Subpixel Registration Based on Intensity. IEEE Trans. Image Process. 7, 27–41 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Schneider C. a, Rasband WS & Eliceiri KW NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


