Abstract
It is striking that the mechanism by which birds sense geomagnetic fields during the biannual migration seasons is not entirely understood. A protein believed to be responsible for avian magnetoreception is the flavoprotein cryptochrome (CRY), which fulfills many of the criteria for a magnetic field sensor. Some experiments, however, indicate that magnetoreception in birds may be disturbed by extremely weak radio frequency fields, an effect that likely cannot be described by an isolated CRY protein. An explanation can possibly be delivered if CRY binds to another protein inside a cell that would possess certain biochemical properties, and it is, therefore, important to identify possible intracellular CRY interaction partners. The goal of this study is to investigate a possible interaction between CRY4 and the iron-sulfur-containing assembly protein (ISCA1) from Erithacus rubecula (European robin), which has recently been proposed to be relevant for magnetic field sensing. The interaction between the proteins is established through classical molecular dynamics simulations for several possible protein-docking modes. The analysis of these simulations concludes that the ISCA1 complex and CRY4 are capable of binding; however, the peculiarities of this binding argue strongly against ISCA1 as relevant for magnetoreception.
Introduction
Avian magnetoreception, the phenomenon of birds being able to sense magnetic fields and to particularly employ the Earth’s magnetic field for navigation during migration, has been observed experimentally for almost half a century (1, 2, 3, 4, 5), yet the biophysical mechanism whereby this occurs remains unclear. The mechanism underlying the magnetic compass sense in animals could possibly be described through spin chemistry that influences biochemical processes inside an animal in reaction to the Earth’s weak magnetic field of ∼50 μT (5, 6, 7, 8, 9, 10). The studies have strongly indicated that the so-called radical-pair mechanism could explain the phenomenon of the magnetic compass sense (5, 6, 7, 11, 12). The mechanism postulates that the spin states (singlet and triplet) of a radical pair are both precursors to specific spin-selective reactions, and the reaction yields of these reactions can be affected by the direction and intensity of the geomagnetic field (5, 8, 10, 12, 13, 14, 15). Such a magnetic-field-dependent process in geomagnetic fields is, however, only possible under certain conditions (5, 13, 15, 16, 17, 18, 19, 20, 21, 22, 23). First, a radical pair is expected to respond to changes of geomagnetic field efficiently once at least one of its radicals has hyperfine couplings weaker or comparable to the Earth’s magnetic field (5, 8, 9, 14, 15, 21, 22, 24). Second, the separation (i.e., edge-to-edge) distance between the radicals in the radical pair is expected to be greater than 1.5 nm, whereas a larger separation distance might be more favorable and could readily be facilitated through electron transfer (5, 25, 26, 27).
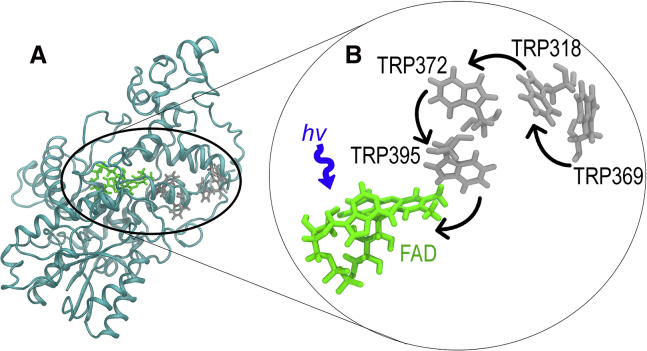
The most promising biological molecules currently found in birds that fulfill the many requirements for a potential magnetosensor are the photoreceptor protein cryptochromes (CRYs) (5, 8, 9, 10, 11, 12, 21, 28, 29, 30, 31, 32, 33, 34, 35, 36). CRYs can potentially act as a magnetic compass sensor through a radical-pair mechanism involving the flavin adenine dinucleotide (FAD) cofactor and a chain of tryptophan (TRP) residues (this chain can be a triad or tetrad of tryptophans, depending on the organism) (25, 28, 34, 37, 38, 39, 40). When FAD is excited by blue light, a sequential electron transfer is initiated; an electron helicopters along the chain of tryptophan residues to produce a radical pair involving FAD and the terminal tryptophan residue (5, 8, 12, 15, 21, 22, 25, 26, 38, 39, 40, 41, 42, 43), as illustrated in Fig. 1 for CRY4 found in the European robin (Erithacus rubecula) (36). Another hypothesis of cryptochrome-based magnetoreception involves the dark-state reoxidation by oxygen. Some evidence in favor of the reoxidation hypothesis has become available recently (44, 45, 46, 47, 48, 49, 50, 51) and delivers a possible explanation of how to alleviate the problem of fast spin relaxation, e.g., in the superoxide radical (22, 52). There is, however, not enough experimental evidence to refute either hypothesis yet.
Figure 1.
(A) CRY4 from Erithacus rubecula, visualized with the FAD cofactor in green and the tryptophan tetrad residues in gray. (B) A closer look at the electron-transfer pathway (shown with arrows) in CRY4 involving FAD and the tetrad of tryptophan residues is shown. To see this figure in color, go online.
Although CRY is a viable magnetoreceptor candidate, it cannot explain all experimental observations related to avian magnetoreception if it is assumed to be functioning in isolation (5, 10, 27). For example, CRY alone cannot explain the disturbance of the avian magnetic compass sense when exposed to weak radio fields (5, 53, 54). However, because the cellular environment is crowded with many different molecules and proteins, an interaction partner for CRY may be able to solve some of the existing inconsistencies (5, 10, 27, 55). For example, such an interaction partner may shed light on radiofrequency disturbances of the compass sense if that interaction partner is able to participate in an electron transfer with a radical in CRY. The FAD/TRP radical pair in cryptochrome does not have the optimal spin relaxation (21, 22, 56), but if the tryptophan radical was substituted with another one from an external protein with more favorable properties, the resulting radical pair could be much more sensitive to the external perturbing fields.
This study deals with a possible interaction partner for CRY and addresses the viability of its interaction with CRY. This interaction partner is an iron-sulfur-containing assembly protein (ISCA1) complex, originally proposed by Qin et al. (55). ISCA1 makes for an exciting possible interaction partner because it binds the iron sulfur clusters (Fe2S2) internally, which could potentially contribute to local magnetic properties at the ISCA1-CRY interface because of the magnetic iron atoms, and could also be involved in an electron transfer with CRY, as Fe2S2 clusters are known to participate in electron-transfer reactions in other biological systems (57, 58, 59, 60). Qin et al. proposed (55) the ISCA1 complex as part of a rod-like protein polymer structure; however, this structure has been shown to be questionable by Friis et al. after an independent reconstruction (27). Although the study revealed that the rod-like protein polymer structure of ISCA1 monomers is unlikely to bind CRY proteins in a robust manner, it is, however, still possible that CRY binds to isolated ISCA1 segments, which called for a separate investigation, performed here. This study focuses specifically on the inter- and intraprotein interactions of one CRY4 from E. rubecula (36) with the disk-like ISCA1 complex structure shown in Fig. 2, consisting of four ISCA1 monomers from the same organism. The three-dimensional building block of the disk-like ISCA1 complex allows for a larger number of possible binding configurations with CRY4. This work thus delivers a consistent investigation of binding of two proteins from the same organism; CRY4 from E. rubecula has a different structure than the Drosophila melanogaster cryptochrome that was studied previously (27). Considering two proteins from the same organism is crucial for revealing whether ISCA1 can possible serve as an important interaction partner for magnetoreception, as the European robin is known to be magnetosensitive, whereas this sense is more speculative among fruit flies (61). In humans, ISCA1 is known to interact with the IOP1/NARFL protein complex, which is important for iron-sulfur-cluster biogenesis, but it is not known if it has any other functions or interaction partners (62).
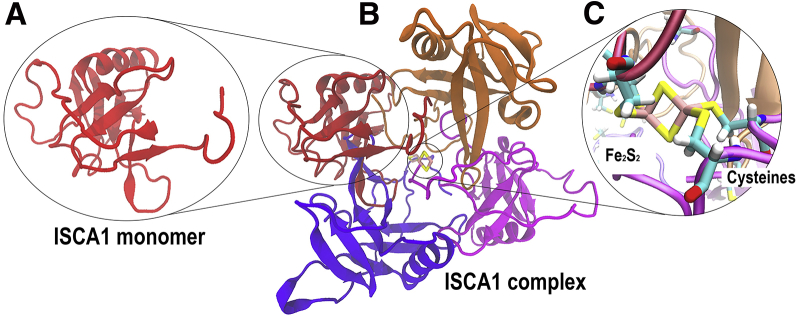
Figure 2.
(A) The single ISCA1 protein is the core building block of the studied ISCA1 protein complex. (B) The arrangement of four ISCA1 proteins in a disk-like complex is shown from one side. (C) An Fe2S2 cluster binds two ISCA1 monomers in the middle of the ISCA1 complex through the covalent bonds with four cysteine residues. The two remaining ISCA1 monomers are cross-linked in the middle of the ISCA1 complex in the same way by another Fe2S2 cluster (data not shown). To see this figure in color, go online.
The performed analysis aims to consider different possible spatial-binding modes between the ISCA1 complex and CRY4 to reveal if any are favorable, as well as whether the intraprotein distances of these configurations would potentially permit any enhancements of the chemical magnetoreception mechanism in E. rubecula CRY4. The interaction between the ISCA1 complex and CRY4 was studied using classical molecular dynamics (MD), which was able to reveal all dynamical traits of the ISCA1-CRY4 interaction and, in particular, allowed us to judge the stability of intraprotein bindings.
Methods
This section describes the computational protocol employed in this investigation. First, we outline the procedure for the ISCA1 complex construction for E. rubecula; then we describe the methodology used to generate different docking configurations between ISCA1 and CRY4; and finally, we provide the computational details of the classical MD simulations employing the program NAMD (63), whereas the analysis of the dynamical traits was accomplished with VMD (64).
Construction of the ISCA1 complex
The binding specificity of CRY4 to the ISCA1 complex was studied by constructing the two proteins’ molecular structures. The equilibrated structure of CRY4 from E. rubecula was taken from a previous investigation (36), whereas the ISCA1 protein complex structure was prepared using homology modeling (65), to a large degree following an earlier protocol (27). This protocol involved crystal packing and manual insertion of the Fe2S2 cluster inside the protein complex. This results in four monomers forming the ISCA1 complex, cross-linked pairwise diagonally by a Fe2S2 and creating a somewhat locked, disk-like structure as shown in Fig. 2. The system then underwent an extended equilibration procedure. The key steps of the ICSA1 molecular assembly protocol are described in greater detail below.
ISCA1 monomer constructed from its amino acid sequence
Because there is currently no crystal structure of ISCA1 from E. rubecula available, its structure was modeled using the program Phyre2 (65). The primary amino acid sequence of ISCA1 in E. rubecula was established by Dr. H. Wu and the team of AG biochemistry of neurosensory processes and signal transduction, Oldenburg University, and can be found in Fig. S1. The sequence was used to do a search on the Phyre2 server, which then produced a homology model, shown in Fig. 2 A. Homology modeling is a method that uses the primary amino acid sequence of a protein to produce an atomistic three-dimensional-structure model for that protein. It does so by finding structures of proteins with similar amino acid sequences for which crystalline structures exist and then creating a three-dimensional model of the desired protein based on these homologous proteins. One ISCA1 complex includes four segments, covalently interconnected through one Fe2S2 cluster as shown in Fig. 2. All of these segments were obtained through homology modeling, and the Fe2S2 cluster was added according to the chosen template molecule.
Docking CRY4 to ISCA1 disk-like complex
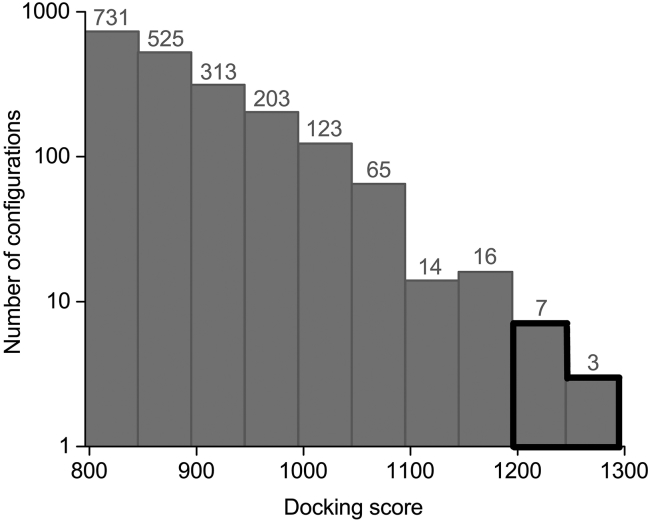
The structures of the possible CRY4-ISCA1 complexes were generated using ZDOCK (66), in which shape complementarity was taken into consideration. The algorithm considers one protein static (ISCA1 complex), whereas the other one (CRY4) was rotated and translated to find the configuration that resulted in the best fitting score. ZDOCK produced 2000 CRY4-ISCA1 configurations, among which 10 with the highest score were chosen for further analysis, as those are the configurations present in the top 20% of the scoring range, as shown in Fig. 3. Table 1 summarizes the corresponding score values of the chosen configurations.
Figure 3.
The histogram of the docking scores between ISCA1 and CRY as found by ZDOCK. The number of configurations with a docking score in the interval covered by the blue column is depicted above the column. The number of docking configurations is shown on a logarithmic scale, and the histograms corresponding to the 10 structures with the highest docking scores are outlined.
Table 1.
Docking Scores Obtained Using ZDOCK for the Top 10 Configurations when Docking CRY4 to the ISCA1 Disk-like Complex
| CRY4-ISCA1 Configuration | Docking Score (Arbitrary Units) |
|---|---|
| 1 | 1290.830 |
| 2 | 1274.191 |
| 3 | 1242.322 |
| 4 | 1220.775 |
| 5 | 1215.370 |
| 6 | 1193.980 |
| 7 | 1199.473 |
| 8 | 1192.894 |
| 9 | 1228.422 |
| 10 | 1191.583 |
Structure equilibration
Once constructed, the 10 most favorable CRY4-ISCA1 complexes were studied dynamically. A summary of the protocol used for equilibrating the isolated ISCA1 complex as well as the 10 CRY4-ISCA1 complexes is shown in Table 2. Equilibration is a necessary step in MD simulations, as the computationally constructed atomic models of the proteins are likely different from a stable configuration and, therefore, cannot be used to judge the protein’s dynamics at normal conditions. By equilibrating each configuration, we then produce more viable positions for the atoms within the configurations in a stable form.
Table 2.
The Equilibration Process for Both the Isolated ISCA1 Disk-like Complex and the 10 Studied ISCA1-CRY4 Complexes
| Structure | Area Where harmonic Constraints Were Applied | Simulation Duration (ns) | Statistical Ensemble |
|---|---|---|---|
| Isolated ISCA1 complex | ISCA1 complex protein backbone | 1.0 | NPT |
| 0.5 | NPT | ||
| 15.0 | NPT | ||
| CRY4-ISCA1 complexes | protein backbone | 2.0 | NPT |
| 15.0 | NVT | ||
| 200.0 | NVT |
For all simulations, the CHARMM36 force field for proteins, with CMAP corrections, was used (67, 68, 69, 70), as well as an additional force field for the FAD complex in CRY4 (36) and the iron sulfur clusters (27). Furthermore, periodic boundary conditions were imposed, the particle mesh Ewald summation method was used, and van der Waals energies were calculated using a cutoff distance of 12 Å. Simulations were run at a temperature of 300 K, controlled through the Langevin thermostat (67) and using a damping coefficient of 5 ps. Langevin piston pressure control (63) was applied in equilibration simulations to keep the pressure at 1 atm, with a period of 200 fs and a decay of 50 fs. Further information on the equilibration and simulation protocol is provided in Table 2.
Equilibration of the isolated ISCA1 complex
Because ISCA1 segments were constructed from a homology model and the Fe2S2 clusters were manually placed inside, the resulting ISCA1 disk-like complex was equilibrated before docking it to CRY4. To equilibrate the system, the ISCA1 complex was placed in a water box of 102.8 × 100.8 × 92.9 Å with a salt concentration of 0.05 mol/L NaCl, resulting in a total of 100,606 atoms. The chosen value of the salt concentration is considered standard in many MD simulations (57, 58, 60, 71, 72) and has, therefore, also been used here, because no specific experimental guidelines on a different value are presently available. After 10,000 NAMD minimization steps, harmonic constraints were introduced in the system and gradually released to achieve an equilibrium structure (see Table 2). These constraints serve to preserve the folding motifs in the proteins that could otherwise be severely damaged if all atoms were allowed to move at once from the start of the simulation. Firstly, the water, along with the Fe2S2 clusters that had been placed at the center of the complex, and the attached cysteine residues were equilibrated. Then the side chains of the proteins were equilibrated, and finally the entire complex, to ensure full stability of the system.
Equilibration of the CRY4-ISCA1 complexes
For each of the different CRY4-ISCA1 configurations, a similar simulation protocol was employed (although some configurations required extra equilibration steps). After the ISCA1 complexes were docked with CRY4, the CRY4-ISCA1 complexes were then resolvated in a new water box with a salt concentration of 0.05 mol/L NaCl. Each configuration required a water box of different size to provide the desired water padding distance, with dimensions ranging from 104 to 144 Å and a different number of total atoms due to these difference, each system containing ∼180,000–230,000 atoms (see Table 3). The first step in all equilibrations was a minimization of 10,000 NAMD steps, followed by an equilibration of the water molecules and side chains of the proteins (see Table 2), followed by equilibration of the entire complex for a total of 15 ns. After all the equilibration steps were complete, the integration time step was increased from 1 to 1.5 fs for the production simulation, which was carried out for 200 ns under temperature control employing the NVT statistical ensemble.
Table 3.
Dimensions of the Simulation Boxes and the Total Number of Atoms in the 10 Considered CRY4-ISCA1 Configurations
| Configuration | x (Å) | y (Å) | z (Å) | N Atoms |
|---|---|---|---|---|
| 1 | 145.6 | 119.1 | 135.4 | 136,240 |
| 2 | 140.3 | 140.8 | 101.4 | 201,898 |
| 3 | 133.5 | 118.8 | 142.9 | 223,452 |
| 4 | 144.7 | 140.4 | 110.4 | 221,023 |
| 5 | 118.0 | 132.3 | 112.9 | 184,314 |
| 6 | 133.2 | 124.9 | 103.1 | 179,339 |
| 7 | 129.8 | 131.7 | 104.5 | 174,618 |
| 8 | 133.7 | 107.3 | 142.9 | 193,626 |
| 9 | 109.6 | 129.3 | 141.8 | 189,664 |
| 10 | 150.8 | 149.7 | 103.1 | 215,715 |
Results
This section presents the results of MD simulations for the 10 different CRY4-ISCA1 configurations. First, the root mean-square deviation (RMSD) of the complexes is analyzed, providing insight into the stability of the CRY4-ISCA1 binding; then the interaction energy between CRY4 and ISCA1 is studied to judge the favorability of the binding; and finally, the hydrogen bonding networks are evaluated to provide a deeper insight into the binding between the two proteins components. The different methods for analysis complement each other to provide a conclusive description of how well the macromolecular configurations are equilibrated, whether the binding between CRY4 and ISCA1 is likely to occur, and how strong the binding could be. The ability of ISCA1 to bind to CRY4 is not, however, the only criteria for ISCA1 to be a viable interaction partner for CRY4 to boost its magnetoreceptive properties. The other factor that is relevant here is the possibility of an electron transfer between ISCA1 and CRY4, which can be concluded based on the distance analysis between charge transfer sites in the proteins.
CRY4-ISCA1 complexes
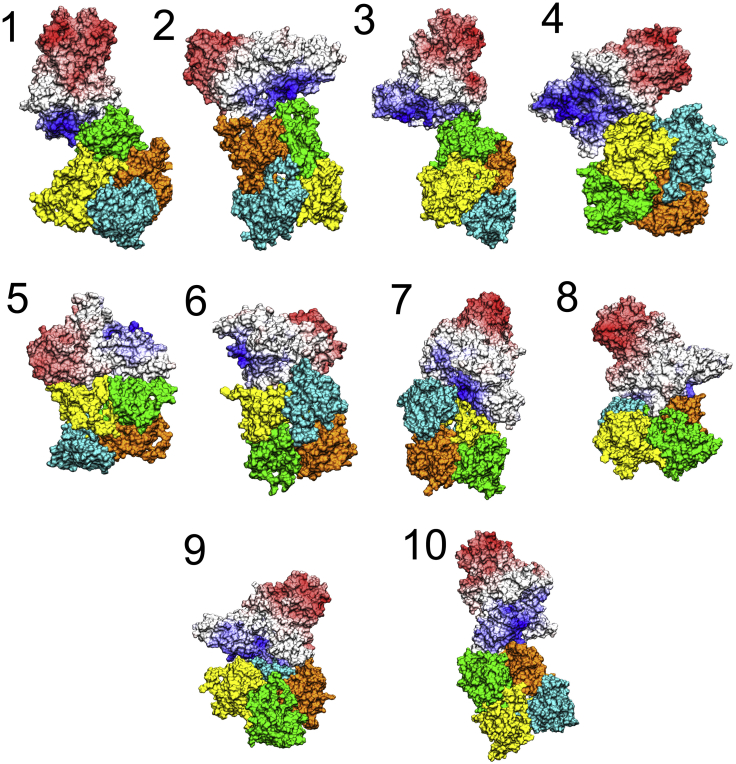
The program ZDOCK (66) allowed for the obtaining of several CRY4-ISCA1 configurations. Ten of these configurations were equilibrated and chosen for a further in-depth binding analysis. These 10 configurations were selected because of their high docking scores, as listed in Table 1. Only one of the configurations, namely configuration 5, is similar to the docking motif described by Qin et al. (55), in which the interaction of CRY and ISCA1 happened between two helices in each protein. This is a consequence of the smaller disk-like ISCA1 structure, which allows for more spatial freedom in the binding than the large rod-like configuration previously studied (27, 55). The 10 structural representations of the CRY4-ISCA1 configurations are shown in Fig. 4, in which cryptochrome is colored by a blue-to-red gradient representing the surface potential such that blue corresponds to a positive charge and red represents a negative charge. The four ISCA1 monomers are colored green, orange, cyan, and yellow.
Figure 4.
Surface representation of the 10 studied CRY4-ISCA1 complex configurations. The surface of CRY4 is shown in red/white/blue, whereas the surface of the four segments of the ISCA1 complex are each colored differently: ISC0 = green, ISC1 = orange, ISC2 = cyan, and ISC3 = yellow. The labels indicate configuration number (see Table 1). To see this figure in color, go online.
Stability of the CRY4-ISCA1 complexes
The RMSD characterizes the average displacement of a group of atoms at a given time instance relative to some reference configuration. RMSD is virtually always defined as
| (1) |
where Natoms is the number of displaceable atoms considered and ri(t) is the position of an atom i at the time instance t, while ri(tref) is its position at a reference time instance tref.
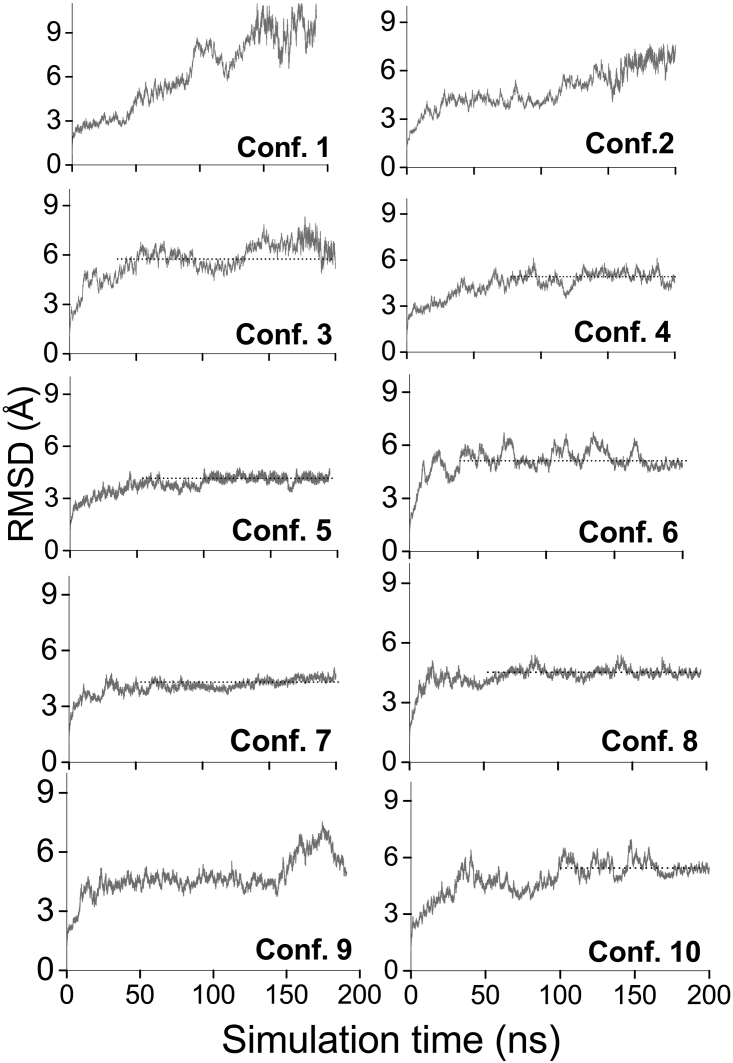
Fig. 5 shows a plot of the RMSD time evolution computed for each of the CRY4-ISCA1 complexes during the 200 ns of production simulations. The RMSD was calculated for all the 10 CRY4-ISCA1 configurations to describe how the structure of the proteins changed compared to the initial structure from after the equilibration (see Table 2). The dependencies in Fig. 5 reflect how well the structures are stabilized, because the more horizontal and linear they become, the less the atoms are drifting apart from their converged positions. The 10 considered CRY4-ISCA1 configurations require different simulation times for the RMSD of the proteins to flatten out. During the studied 200-ns-long simulation, configuration 1 never reached a stabilized RMSD value, implying that in this configuration, the ISCA1 complex and the CRY4 proteins may begin drifting away from each other, i.e., do not bind. Configuration 2 seemed to have reached a flat RMSD after 25 ns; however, it notably increases after ∼120 ns and keeps increasing. This RMSD increase in configuration 2 shows that the configuration experiences some perturbations in CRY4-ISCA1 binding and could also be unstable. However, the reasoning for the perturbation is unclear and can be elaborated by using other analysis methods, as will be described in the following sections.
Figure 5.
RMSD of the 10 CRY4-ISCA1 configurations (see Fig. 4) simulated in a water box during the 200 ns production simulation. For all calculations, the reference time is the beginning of the simulation, tref = 0 ns. In all the plots, the RMSD dependencies were computed for all the atoms in the entire protein complexes relative to the configuration after the completed equilibration simulations.
Configurations 3–8 and 10 show steady equilibration. The simulation times at which these stabilized structures emerge are different, however. Configuration 5 is the most peculiar one, as it features a stabilized configuration after a much shorter simulation time than the other configurations and has small fluctuations in RMSD, which also featured in configuration 7 and configuration 8. Another noticeable peculiarity of configuration 5 is that the RMSD value stabilizes at a lower value than in the case of the other five configurations. This could possibly indicate that configuration 5 was equilibrated better during the equilibration simulation, because on average, its atoms do not need to move far from the reference positions. The RMSD plot for configuration 9 reveals that during the last 50 ns of simulation, the system sought to find a new equilibrium and was apparently populating a metastable state in the beginning of the simulation.
Interaction energy analysis
The interaction energy between the two proteins is calculated by summing all contributions from the pairwise interactions arising from the van der Waals and electrostatic terms for atoms within the cutoff distance of the potentials. In the calculations, the van der Waals interaction is described by the Lennard-Jones potential, given as
| (2) |
where Emin is the energy corresponding to an equilibrium distance between two specific atoms, Rmin is the equilibrium distance of the corresponding van der Waals interaction between these atoms, and rij is the separation distance between them. Important to mention here is that atoms with indices i and j belong to two different proteins, ISCA1 and CRY4, respectively. The electrostatic potential is found from the Coulomb potential, which reads as follows:
| (3) |
where qi and qj are the electric charges of the atoms involved in the electrostatic interaction and C is the Coulomb constant. The long-range tails of the electrostatic energy are accounted for through the particle mesh Ewald method, as described elsewhere (73).
The interaction energy between two macromolecules reflects how energetically favorable their binding is. If the value is positive, the two molecules repel each other, whereas the more negative the value is, the more attraction they experience. The interaction energy between ISCA1 and CRY4, in each of the 10 considered configurations, was calculated for the production simulations, and the results are presented in Figs. 6 and 7 alongside the probability density of the interaction energy, defined as
| (4) |
Here, σ is the standard deviation of the interaction energy, σ2 is its variance, and is the average interaction energy.
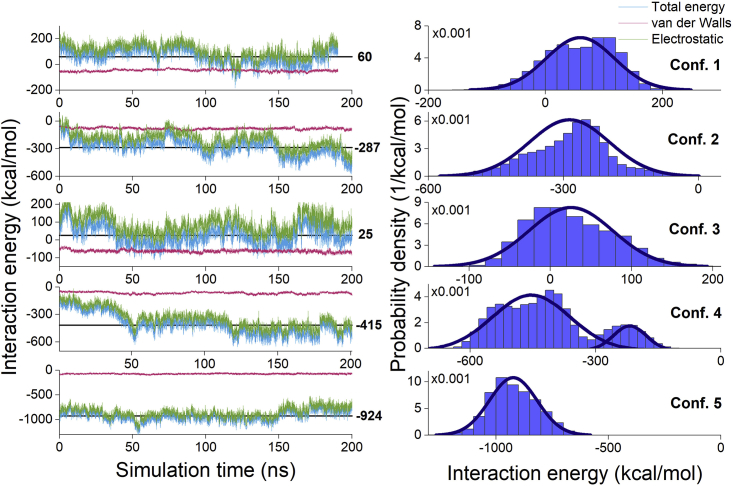
Figure 6.
Interaction energy between the ISCA1 complex and CRY4 for configurations 1–5. The plots in the left panel show the time evolution of the interaction energy as well as its van der Waals and Coulomb components; horizontal lines show the average values, which are indicated next to them. The plots to the right show the probability density of the interaction energy, which is expected to follow Eq. 4, as marked by the blue curve. To see this figure in color, go online.
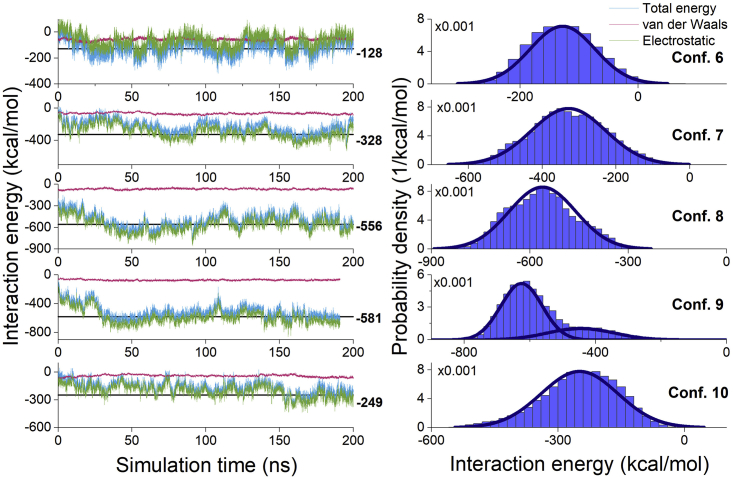
Figure 7.
Interaction energy between the ISCA1 complex and CRY4 for configurations 6–10. The details are the same as in Fig. 6. To see this figure in color, go online.
The probability density of the CRY4-ISCA1 interaction energy, as computed from the simulations, was fitted to the Gaussian distribution in Eq. 4, and the resulting fits are shown in Figs. 6 and 7 with blue lines. The probability densities of the interaction energy for configurations 5, 6, 7, 8, and 10 seem to fit the normal distribution reasonably well, whereas the distributions for configurations 1, 2, 3, 4, and 9 feature much larger distortions. Configurations 1 and 3 have positive interaction energies on average (see Figs. 6 and 7), and so they are not expected to bind, meaning the lack of a normal distribution of the interaction energy in these cases is not surprising, as the ISCA1 and CRY4 proteins are expected to be slowly drifting apart. Configurations 4 and 9 have probability densities of interaction energy that cannot be described by a single Gaussian distribution. These specific CRY4-ISCA1 configurations show a changing interaction energy between ISCA1 and CRY4, which is reflected in a double-peaked distribution of the probability density (see Figs. 6 and 7). Significant decreases in the interaction energy are observed at around 50 ns for configuration 4 and at around 30 ns for configuration 9, suggesting that one or more hydrogen bonds between CRY4 and ISCA1 has been broken at this point. In configuration 2, a small drop in interaction energy can be seen at 150 ns, which corresponds to a change in the RMSD, shown in Fig. 5. This indicates that configuration 2 has yet to reach its true equilibrium.
It is worth noting that the major contribution to the interaction energy between two proteins stems from the electrostatic energy arising between ISCA1 and CRY4, which is represented by the green line on the plots of the time evolution of the interaction energy. The red line on the same plots represents the van der Waals contribution to the interaction energy and is consistently around −50 kcal/mol for all the six configurations, whereas the electrostatic part can be as low as −1000 kcal/mol in the case of configuration 5.
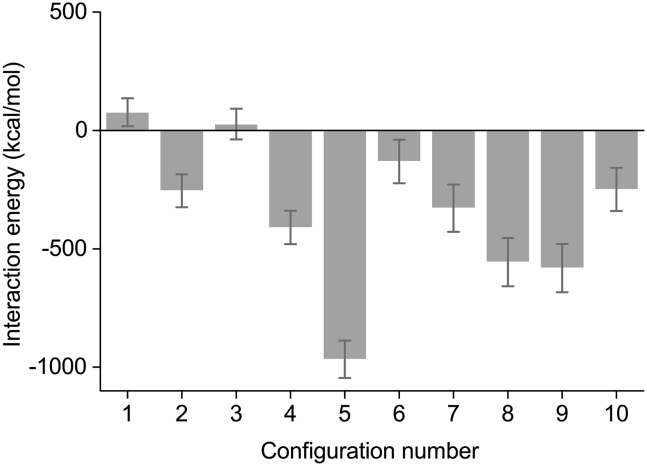
Fig. 8 provides a summary of the time-averaged interaction energies for all 10 configurations. It demonstrates that configurations 1 and 3 have, on average, a positive interaction energy and therefore should not bind on a long timescale, whereas the other studied configurations have varying negative values for the total interaction energy, with configuration 5 experiencing the strongest binding. These findings agree with the earlier analysis of the RMSD dependencies in Fig. 5, as configuration 1 was not able to stay in an equilibrium state and configuration 5 was the most stable according to the RMSD plot. Configuration 5 has an interaction-energy value close to the one found by Friis et al. for the most stable configuration of CRY and ISCA1 from D. melanogaster (fruit fly); Friis et al. found the most stable binding configuration to have an interaction energy of around −800 kcal/mol (27), whereas our configuration 5 has an interaction energy of −967 kcal/mol. The similarity between the findings for these two studies of CRY-ISCA1 interaction in two different species indicates that the binding energy of the two proteins could also be similar in other species and organisms.
Figure 8.
Summary of the average interaction energies for the 10 studied CRY4-ISCA1 configurations. The error bars indicate the standard deviations.
Hydrogen bonding network analysis
A hydrogen bond is typically caused by electrostatic attraction between an electropositive and an electronegative atom, for example, between a hydrogen and an oxygen. The hydrogen bonds between ISCA1 and CRY4 are what holds the two together, as can be concluded from the fact that the largest portion of the total interaction energy, calculated between the ISCA1 complex and CRY4 (see Figs. 6 and 7), is made up by the electrostatics. The hydrogen bond occupancy is the percentage of the simulation time for which a specific hydrogen bond is observed in a simulation. The hydrogen bonds between ISCA1 and CRY4 were calculated for each of the 10 CRY4-ISCA1 configurations studied, allowing for the bonds with a donor-acceptor distance of less than 3 Å and an angle cutoff of 20°. Table S1 lists all bonds with over 20% occupancy for the 10 CRY4-ISCA1 configurations.
There are, on average, approximately twice as many hydrogen bonds with occupancy over 20% in configuration 5, 8, and 9 compared to the other configurations, which is in agreement with the RMSD and interaction energy analysis that also demonstrate that these three configurations have a more favorable binding motif than the other seven. Another observation is that there are very few significant hydrogen bonds in configurations 1 and 3, the configurations that, according to their interaction energy, are unlikely to stay bound for a long time. The number of significant hydrogen bonds is, however, not the only factor that determines proteins binding affinity, as the strength of the hydrogen bonds also matters and will be discussed further.
A few of the most notable hydrogen bonds are visualized in Fig. 9, and further analysis can be found in Fig. S2. Amino acid residues participating in hydrogen bonds are labeled. Some of the irregular patterns in the RMSD and the interaction-energy time-evolution dependencies can be explained by looking at the formation and breakage of these hydrogen bonds over time. Fig. 9 shows an example of hydrogen bonds that break at some point in the simulation. The separation of the hydrogen bonds over time in the CRY4-ISCA1 configuration 1, seen in Fig. 9, is in agreement with the increasing RMSD of the proteins and the positive average interaction energy, all of which indicate that configuration 1 is an unfavorable binding motif even though it was suggested to have a very high docking score (see Table 1). Even within the CRY4-ISCA1 configurations that have a negative interaction energy, not all display signs of being stable during the performed 200 ns simulations.
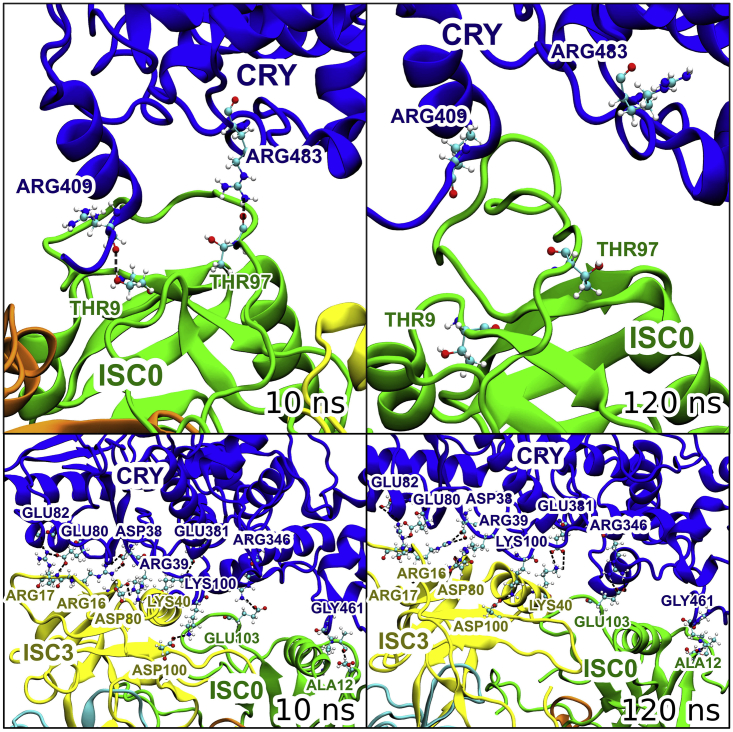
Figure 9.
Top: Visualizations of some of the hydrogen bonds in configuration 1. The bottom shows visualizations of some of the hydrogen bonds in configuration 5. The visualization is zoomed in on the hydrogen bonds discussed, and therefore not all ISCA1 monomers in the complex can be seen. The figure shows the hydrogen bonds in the beginning of the simulation (left) and a snapshot configuration at the binding site at a time instance of 120 ns (right). The colors denote visualized segments; the four ISCA1 monomers in the ISCA1 complex are labeled ISC0, ISC1, ISC2, and ISC3. ISC0 = green, ISC1 = orange, ISC2 = cyan, ISC3 = yellow, and CRY4 = blue. The labels specify the amino acid residues participating in the closest visualized hydrogen bond, with the colors of the labels matching the color of the amino acid the label represents. The hydrogen bonds themselves are visualized as a dotted line between the participating atoms. To see this figure in color, go online.
Configuration 5 has the most hydrogen bonds between charged amino acid residues, which is likely the reason for this configuration being the strongest binding configuration of ISCA1 and CRY4. The hydrogen bonds in configuration 5 were compared to those in the most stable configuration found by Friis et al. (27) (see Fig. 9), which also had a similar interaction energy, but because the study by Friis et al. was done for D. melanogaster, the specific amino acids involved in the hydrogen bonds are somewhat different.
Intraprotein distance analysis
The distances between important components of CRY4 and ISCA1 were calculated and analyzed. In particular, the distances between the FAD cofactor in CRY4 and the two Fe2S2 clusters in the ISCA1 complex were studied, as well as the distance between the terminal tryptophan residue (see Fig. 1) and the two Fe2S2 clusters (see Table 4 for a summary of these distances). These distances are of interest because small distances between redox centers of the ISCA1 complex and CRY4 could possibly help explain some of the experimental observations on noise disturbance of the magnetic compass in migratory birds (5, 53, 54), as ISCA1 and CRY4 could act jointly through an intraprotein electron transfer. However, for an efficient electron transfer to occur, the active sites of CRY4, FAD, and the terminal tryptophan (TRP369) must be close to one of the iron sulfur clusters in the ISCA1 complex. It is necessary that these charge-transfer sites are close enough to each other because the rate of the electron transfer process depends on the distance between the sites, as follows from the Marcus theory of electron transfer (8, 25, 26, 74):
| (5) |
where k is the electron transfer rate constant, HDA is the electronic coupling between the initial and final states of the donor and acceptor states participating in the electron transfer, λ is the reorganization energy, ΔG is the total Gibbs free-energy change for the electron transfer reaction, kB is Boltzmann’s constant, and T is the temperature. By treating the donor and acceptor as two bound states separated by a barrier, |HDA|2 can be parametrized as a function that decreases exponentially with the distance between the molecules involved in the electron transfer reaction (8, 25, 26), i.e.,
| (6) |
Table 4.
The Distances between CRY4 and ISCA1 Segments Calculated for Each CRY4-ISCA1 Complex as an Average over Their Values for the Entire Production Simulation
| Configuration | FAD-FES1 Distance (Å) | FAD-FES2 Distance (Å) | TRP-FES1 Distance (Å) | TRP-FES2 Distance (Å) |
|---|---|---|---|---|
| 1 | 49.3 (± 2.2) | 56.4 (± 3.1) | 38.8 (± 1.4) | 44.2 (± 1.0) |
| 2 | 45.1 (± 1.1) | 46.2 (± 1.2) | 54.7 (± 2.0) | 67.4 (± 1.8) |
| 3 | 51.8 (± 0.9) | 55.6 (± 1.1) | 40.2 (± 0.8) | 46.4 (± 1.1) |
| 4 | 53.5 (± 1.5) | 38.0 (± 1.7) | 44.1 (± 1.6) | 27.3 (± 1.8) |
| 5 | 40.1 (± 1.4) | 51.1 (± 0.9) | 34.1 (± 1.5) | 51.3 (± 1.3) |
| 6 | 47.2 (± 1.5) | 47.2 (± 1.5) | 66.6 (± 2.2) | 63.1 (± 1.5) |
| 7 | 34.9 (± 1.2) | 45.3 (± 0.8) | 63.90 (± 1.0) | 43.0 (± 0.8) |
| 8 | 22.8 (± 1.2) | 45.4 (± 1.2) | 18.6 (± 1.0) | 45.1 (± 1.1) |
| 9 | 24.3 (± 1.7) | 45.3 (± 0.9) | 29.6 (± 1.4) | 39.4 (± 1.4) |
| 10 | 54.8 (± 1.4) | 58.3 (± 2.0) | 41.4 (± 1.9) | 49.6 (± 1.4) |
FES1 and FES2 denote the two iron sulfur clusters in each CRY4-ISCA1 configuration. TRP refers to the terminal tryptophan in CRY (TRP369); see Fig. 1. The values in the brackets show the error bars as decided by the standard deviation, in which the average values correspond to the distance mean value.
Here, V0 is the coupling parameter and β is the parameter controlling the effective distance of wavefunction overlap. Because |HDA|2 decreases with R, so does the electron transfer rate k. For example, in the case of the chain of tryptophan residues in CRY of Arabidopsis thaliana (plant), it has been shown that a 0.7 Å increase in R would cause a fivefold decrease in the rate constant (25, 28).
Table 4 presents the distances between the protein compartments for the 10 CRY4-ISCA1 configurations. It can be concluded that overall, the distances between the charge transfer sites are significantly larger than those that are needed for an efficient one-step electron transfer (25), which indicates that the Fe2S2 would likely have a minimal influence, if any, on the electron transfer between FAD and tryptophan residues. Note that the distances found in this investigation are similar to those by Friis et al. (27), showing that different organisms, despite their differences in CRY residues, exhibit the same trends in terms of the interaction between CRY and ISCA1.
It would be expected that at least one of the stable configurations should feature a distance between the active site in CRY4 and ISCA1 that is comparable to those between the residues in the tryptophan chain to justify the cooperativity of CRY4 and the ISCA1 complex that was hypothesized by Qin et al. (55). The typical edge-to-edge distance between the tryptophan residues and FAD complex in CRY4 is ∼2–4 Å, which is the distance in which electron transfer can realistically happen with a biologically significant rate constant. This is, however, not the case, as one would require the distances between TRP369 and Fe2S2 to be around the same length (≤10 Å) (25, 28), but the measurement of distances generally shows results in the region of ≥20 Å, except in configuration 8, in which a distance of 18.6 Å is measured between FES1 and tryptophan, which is still much too large to support efficient direct electron transfer. It could also be theorized that instead of the direct electron-transfer route that follows from Fe2S2 via the terminal tryptophan, the electron transfer could be assisted by additional compounds in the CRY4-ISCA1 complex, such as tyrosine, cysteine, or tryptophan amino acids. However, none of the studied CRY4-ISCA1 complexes revealed a path involving these amino acids between Fe2S2 and the TRP369 suitable for an electron transfer.
Conclusion
Finding an intracellular interaction partner for CRY would mean a big step forward in understanding the mechanism of avian magnetoreception. The experiments of Qin et al. allowed for the proposal of a magnetoreceptor complex, which invited further investigation of the protein ISCA1 as a potential interaction partner for cryptochrome, which is here done in a migratory bird, the European robin (E. rubecula), known to navigate using the geomagnetic field. The aim of this study is to investigate and judge the interaction between CRY4 and the ISCA1 complex to see if it is plausible for the ISCA1 complex to be an interaction partner of CRY4 and if the CRY4-ISCA1 complex could potentially be involved in magnetoreception.
The disk-like ISCA1 complex was docked to CRY4 in several possible configurations. The stability, interaction energy, and hydrogen bonds of these configurations were analyzed. The interaction energies revealed that one of the binding configurations was much more stable than the others, whereas two of the 10 configurations revealed by the static docking analysis did not bind at all once dynamical investigation was performed. Trends that support this conclusion were found in the stability of the structures and in the hydrogen bonds holding CRY4 and the ISCA1 complex together. The finding of one very stable configuration indicates that a binding between CRY4 and the ISCA1 complex is indeed possible.
With the binding of CRY4 to ISCA1 shown to be possible, the questions are whether the electron transfer between CRY4 and ISCA1 will occur and, more generally, whether ISCA1 could influence the electron dynamics in CRY through its bound Fe2S2 cluster. The distances between the active sites of CRY (FAD and terminal tryptophan) and the nearest Fe2S2 clusters in the ISCA1-complex were measured for all the 10 configurations but turned out to be significantly larger than those expected for an efficient and realistic electron transfer (25).
The study of electron transfer from the Fe2S2 cluster demands a separate investigation, and likewise, the spin relaxation of this intraprotein component would be interesting to study once it gains a radical character. The close proximity of Fe2S2 to the FAD and TRP369 components of CRY4 may also be important in light of a recent suggestion of boosted sensitivity of a radical-pair compass magnetoreceptor through the so-called scavenging reaction, which may involve Fe2S2 clusters if these are placed appropriately (52, 75). The specificity of such a scavenging reaction is beyond the scope of this investigation and could be studied separately; however, the large distance between Fe2S2 clusters of the ISCA1 complex and the FAD/TRP369 cofactor suggests that this process is unlikely to happen.
In conclusion, although the findings of our study indicate that CRY4 and the ISCA1 complex can bind to each other, they do so very weakly due to the low number of found low-energy binding configurations, and the large distance between the radical pair in CRY and the Fe2S2 cluster in the ISCA1 complex eliminates the possibility of ISCA1 being involved in a joined electron transfer. This makes the ISCA1-CRY4 binding likely insignificant with respect to magnetoreception and ISCA1 an unlikely interaction partner for CRY4.
Author Contributions
I.A.S. designed and supervised the study. S.M.K. and I.F. performed the calculations. S.M.K., I.F., and I.A.S. performed the analysis. All authors contributed to writing the manuscript and have given approval to the final version of the manuscript.
Acknowledgments
The authors thank Haijia Wu for providing the ISCA1 amino acid sequence in E. rubecula.
Financial support by the Lundbeck Foundation, the Danish Councils for Independent Research, and the Russian Science Foundation (grant no. 17-72-20201) is greatly acknowledged. Computational resources for the simulations were provided by the DeiC National HPC Center, University of Southern Denmark.
Editor: Margaret Cheung.
Footnotes
Two figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30777-X.
Supporting Material
References
- 1.Merkel F., Wiltschko W. Magnetismus und Richtungsfinden zugunruhiger Rotkehlchen, Erithacus rubecula. Vogelwarte. 1965;23:71–77. [Google Scholar]
- 2.Wiltschko W., Wiltschko R. Magnetic compass of European robins. Science. 1972;176:62–64. doi: 10.1126/science.176.4030.62. [DOI] [PubMed] [Google Scholar]
- 3.Wiltschko R., Wiltschko W. Avian navigation: from historical to modern concepts. Anim. Behav. 2003;65:257–272. [Google Scholar]
- 4.Mouritsen H., Ritz T. Magnetoreception and its use in bird navigation. Curr. Opin. Neurobiol. 2005;15:406–414. doi: 10.1016/j.conb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Hore P.J., Mouritsen H. The radical-pair mechanism of magnetoreception. Annu. Rev. Biophys. 2016;45:299–344. doi: 10.1146/annurev-biophys-032116-094545. [DOI] [PubMed] [Google Scholar]
- 6.Brocklehurst B. Magnetic fields and radical reactions: recent developments and their role in nature. Chem. Soc. Rev. 2002;31:301–311. doi: 10.1039/b107250c. [DOI] [PubMed] [Google Scholar]
- 7.Rodgers C.T., Hore P.J. Chemical magnetoreception in birds: the radical pair mechanism. Proc. Natl. Acad. Sci. USA. 2009;106:353–360. doi: 10.1073/pnas.0711968106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solov’yov I.A., Chandler D.E., Schulten K. Magnetic field effects in Arabidopsis thaliana cryptochrome-1. Biophys. J. 2007;92:2711–2726. doi: 10.1529/biophysj.106.097139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritz T., Adem S., Schulten K. A model for photoreceptor-based magnetoreception in birds. Biophys. J. 2000;78:707–718. doi: 10.1016/S0006-3495(00)76629-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedersen J.B., Nielsen C., Solov’yov I.A. Multiscale description of avian migration: from chemical compass to behaviour modeling. Sci. Rep. 2016;6:36709. doi: 10.1038/srep36709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill E., Ritz T. Can disordered radical pair systems provide a basis for a magnetic compass in animals? J. R. Soc. Interface. 2010;7(Suppl 2):S265–S271. doi: 10.1098/rsif.2009.0378.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maeda K., Henbest K.B., Hore P.J. Chemical compass model of avian magnetoreception. Nature. 2008;453:387–390. doi: 10.1038/nature06834. [DOI] [PubMed] [Google Scholar]
- 13.Maeda K., Robinson A.J., Hore P.J. Magnetically sensitive light-induced reactions in cryptochrome are consistent with its proposed role as a magnetoreceptor. Proc. Natl. Acad. Sci. USA. 2012;109:4774–4779. doi: 10.1073/pnas.1118959109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barnes F.S., Greenebaum B. The effects of weak magnetic fields on radical pairs. Bioelectromagnetics. 2015;36:45–54. doi: 10.1002/bem.21883. [DOI] [PubMed] [Google Scholar]
- 15.Solov’yov I.A., Ritz T., Hore P.J. Quantum Effects in Biology. Cambridge University Press; 2014. Chapter A chemical compass for bird navigation; pp. 218–236. [Google Scholar]
- 16.Efimova O., Hore P.J. Role of exchange and dipolar interactions in the radical pair model of the avian magnetic compass. Biophys. J. 2008;94:1565–1574. doi: 10.1529/biophysj.107.119362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timmel C.R., Till U., Hore P.J. Effects of weak magnetic fields on free radical recombination reactions. Mol. Phys. 1998;95:71–89. [Google Scholar]
- 18.Cintolesi F., Ritz T., Hore P.J. Anisotropic recombination of an immobilized photoinduced radical pair in a 50-μT magnetic field: a model avian photomagnetoreceptor. Chem. Phys. 2003;294:707–718. [Google Scholar]
- 19.Hiscock H.G., Worster S., Hore P.J. The quantum needle of the avian magnetic compass. Proc. Natl. Acad. Sci. USA. 2016;113:4634–4639. doi: 10.1073/pnas.1600341113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kattnig D.R., Evans E.W., Hore P.J. Chemical amplification of magnetic field effects relevant to avian magnetoreception. Nat. Chem. 2016;8:384–391. doi: 10.1038/nchem.2447. [DOI] [PubMed] [Google Scholar]
- 21.Kattnig D.R., Solov’yov I.A., Hore P.J. Electron spin relaxation in cryptochrome-based magnetoreception. Phys. Chem. Chem. Phys. 2016;18:12443–12456. doi: 10.1039/c5cp06731f. [DOI] [PubMed] [Google Scholar]
- 22.Kattnig D.R., Sowa J.K., Hore P.J. Electron spin relaxation can enhance the performance of a cryptochrome-based magnetic compass sensor. New J. Phys. 2016;18:063007. [Google Scholar]
- 23.Nielsen C., Kattnig D.R., Solov’yov I.A. Ascorbic acid may not be involved in cryptochrome-based magnetoreception. J. R. Soc. Interface. 2017;14:20170657. doi: 10.1098/rsif.2017.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poonia V.S., Saha D., Ganguly S. State transitions and decoherence in the avian compass. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2015;91:052709. doi: 10.1103/PhysRevE.91.052709. [DOI] [PubMed] [Google Scholar]
- 25.Sjulstok E., Olsen J.M., Solov’yov I.A. Quantifying electron transfer reactions in biological systems: what interactions play the major role? Sci. Rep. 2015;5:18446. doi: 10.1038/srep18446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solov’yov I.A., Domratcheva T., Schulten K. Separation of photo-induced radical pair in cryptochrome to a functionally critical distance. Sci. Rep. 2014;4:3845. doi: 10.1038/srep03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friis I., Sjulstok E., Solov’yov I.A. Computational reconstruction reveals a candidate magnetic biocompass to be likely irrelevant for magnetoreception. Sci. Rep. 2017;7:13908. doi: 10.1038/s41598-017-13258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lüdemann G., Solov’yov I.A., Elstner M. Solvent driving force ensures fast formation of a persistent and well-separated radical pair in plant cryptochrome. J. Am. Chem. Soc. 2015;137:1147–1156. doi: 10.1021/ja510550g. [DOI] [PubMed] [Google Scholar]
- 29.Gegear R.J., Casselman A., Reppert S.M. Cryptochrome mediates light-dependent magnetosensitivity in Drosophila. Nature. 2008;454:1014–1018. doi: 10.1038/nature07183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niessner C., Denzau S., Wiltschko R. Avian ultraviolet/violet cones identified as probable magnetoreceptors. PLoS One. 2011;6:e20091. doi: 10.1371/journal.pone.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mouritsen H., Janssen-Bienhold U., Weiler R. Cryptochromes and neuronal-activity markers colocalize in the retina of migratory birds during magnetic orientation. Proc. Natl. Acad. Sci. USA. 2004;101:14294–14299. doi: 10.1073/pnas.0405968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmad M., Galland P., Wiltschko W. Magnetic intensity affects cryptochrome-dependent responses in Arabidopsis thaliana. Planta. 2007;225:615–624. doi: 10.1007/s00425-006-0383-0. [DOI] [PubMed] [Google Scholar]
- 33.Solov’yov I.A., Schulten K. Reaction kinetics and mechanism of magnetic field effects in cryptochrome. J. Phys. Chem. B. 2012;116:1089–1099. doi: 10.1021/jp209508y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans E.W., Dodson C.A., Timmel C.R. Magnetic field effects in flavoproteins and related systems. Interface Focus. 2013;3:20130037. doi: 10.1098/rsfs.2013.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solov’yov I.A., Mouritsen H., Schulten K. Acuity of a cryptochrome and vision-based magnetoreception system in birds. Biophys. J. 2010;99:40–49. doi: 10.1016/j.bpj.2010.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Günther A., Einwich A., Mouritsen H. Double-cone localization and seasonal expression pattern suggest a role in magnetoreception for European robin cryptochrome 4. Curr. Biol. 2018;28:211–223.e4. doi: 10.1016/j.cub.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Sheppard D.M., Li J., Mackenzie S.R. Millitesla magnetic field effects on the photocycle of an animal cryptochrome. Sci. Rep. 2017;7:42228. doi: 10.1038/srep42228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nohr D., Franz S., Schleicher E. Extended electron-transfer in animal cryptochromes mediated by a tetrad of aromatic amino acids. Biophys. J. 2016;111:301–311. doi: 10.1016/j.bpj.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller P., Yamamoto J., Brettel K. Discovery and functional analysis of a 4th electron-transferring tryptophan conserved exclusively in animal cryptochromes and (6-4) photolyases. Chem. Commun. (Camb.) 2015;51:15502–15505. doi: 10.1039/c5cc06276d. [DOI] [PubMed] [Google Scholar]
- 40.Müller P., Bouly J.P. Searching for the mechanism of signalling by plant photoreceptor cryptochrome. FEBS Lett. 2015;589:189–192. doi: 10.1016/j.febslet.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Immeln D., Weigel A., Pérez Lustres J.L. Primary events in the blue light sensor plant cryptochrome: intraprotein electron and proton transfer revealed by femtosecond spectroscopy. J. Am. Chem. Soc. 2012;134:12536–12546. doi: 10.1021/ja302121z. [DOI] [PubMed] [Google Scholar]
- 42.Kottke T., Batschauer A., Heberle J. Blue-light-induced changes in Arabidopsis cryptochrome 1 probed by FTIR difference spectroscopy. Biochemistry. 2006;45:2472–2479. doi: 10.1021/bi051964b. [DOI] [PubMed] [Google Scholar]
- 43.Langenbacher T., Immeln D., Kottke T. Microsecond light-induced proton transfer to flavin in the blue light sensor plant cryptochrome. J. Am. Chem. Soc. 2009;131:14274–14280. doi: 10.1021/ja901628y. [DOI] [PubMed] [Google Scholar]
- 44.Wiltschko R., Ahmad M., Wiltschko W. Light-dependent magnetoreception in birds: the crucial step occurs in the dark. J. R. Soc. Interface. 2016;13:20151010. doi: 10.1098/rsif.2015.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiltschko R., Gehring D., Wiltschko W. Magnetoreception in birds: II. Behavioural experiments concerning the cryptochrome cycle. J. Exp. Biol. 2014;217:4225–4228. doi: 10.1242/jeb.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiltschko R., Stapput K., Thalau P., Wiltschko W. Directional orientation of birds by the magnetic field under different light conditions. J. R. Soc. Interface. 2009;7:S163–S177. doi: 10.1098/rsif.2009.0367.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nießner C., Denzau S., Wiltschko R. Magnetoreception: activated cryptochrome 1a concurs with magnetic orientation in birds. J. R. Soc. Interface. 2013;10:20130638. doi: 10.1098/rsif.2013.0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritz T., Wiltschko R., Wiltschko W. Magnetic compass of birds is based on a molecule with optimal directional sensitivity. Biophys. J. 2009;96:3451–3457. doi: 10.1016/j.bpj.2008.11.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nießner C., Denzau S., Wiltschko R. Magnetoreception in birds: I. Immunohistochemical studies concerning the cryptochrome cycle. J. Exp. Biol. 2014;217:4221–4224. doi: 10.1242/jeb.110965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller P., Ahmad M. Light-activated cryptochrome reacts with molecular oxygen to form a flavin-superoxide radical pair consistent with magnetoreception. J. Biol. Chem. 2011;286:21033–21040. doi: 10.1074/jbc.M111.228940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Solov’yov I.A., Schulten K. Magnetoreception through cryptochrome may involve superoxide. Biophys. J. 2009;96:4804–4813. doi: 10.1016/j.bpj.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kattnig D.R. Radical-pair-based magnetoreception amplified by radical scavenging: resilience to spin relaxation. J. Phys. Chem. B. 2017;121:10215–10227. doi: 10.1021/acs.jpcb.7b07672. [DOI] [PubMed] [Google Scholar]
- 53.Wiltschko R., Thalau P., Wiltschko W. Magnetoreception in birds: the effect of radio-frequency fields. J. R. Soc. Interface. 2015;12:20141103. doi: 10.1098/rsif.2014.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Engels S., Schneider N.L., Mouritsen H. Anthropogenic electromagnetic noise disrupts magnetic compass orientation in a migratory bird. Nature. 2014;509:353–356. doi: 10.1038/nature13290. [DOI] [PubMed] [Google Scholar]
- 55.Qin S., Yin H., Xie C. A magnetic protein biocompass. Nat. Mater. 2016;15:217–226. doi: 10.1038/nmat4484. [DOI] [PubMed] [Google Scholar]
- 56.Worster S., Kattnig D.R., Hore P.J. Spin relaxation of radicals in cryptochrome and its role in avian magnetoreception. J. Chem. Phys. 2016;145:035104. doi: 10.1063/1.4958624. [DOI] [PubMed] [Google Scholar]
- 57.Husen P., Solov’yov I.A. Mutations at the Qo site of the cytochrome bc1 complex strongly affect oxygen binding. J. Phys. Chem. B. 2017;121:3308–3317. doi: 10.1021/acs.jpcb.6b08226. [DOI] [PubMed] [Google Scholar]
- 58.Barragan A.M., Schulten K., Solov’yov I.A. Mechanism of the primary charge transfer reaction in the cytochrome bc1 complex. J. Phys. Chem. B. 2016;120:11369–11380. doi: 10.1021/acs.jpcb.6b07394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salo A.B., Husen P., Solov’yov I.A. Charge transfer at the Qo-site of the cytochrome bc1 complex leads to superoxide production. J. Phys. Chem. B. 2017;121:1771–1782. doi: 10.1021/acs.jpcb.6b10403. [DOI] [PubMed] [Google Scholar]
- 60.Husen P., Solov’yov I.A. Spontaneous binding of molecular oxygen at the Qo-site of the bc1 complex could stimulate superoxide formation. J. Am. Chem. Soc. 2016;138:12150–12158. doi: 10.1021/jacs.6b04849. [DOI] [PubMed] [Google Scholar]
- 61.Dodson C.A., Hore P.J., Wallace M.I. A radical sense of direction: signalling and mechanism in cryptochrome magnetoreception. Trends Biochem. Sci. 2013;38:435–446. doi: 10.1016/j.tibs.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 62.Song D., Tu Z., Lee F.S. Human ISCA1 interacts with IOP1/NARFL and functions in both cytosolic and mitochondrial iron-sulfur protein biogenesis. J. Biol. Chem. 2009;284:35297–35307. doi: 10.1074/jbc.M109.040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 65.Kelley L.A., Mezulis S., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen R., Weng Z. A novel shape complementarity scoring function for protein-protein docking. Proteins. 2003;51:397–408. doi: 10.1002/prot.10334. [DOI] [PubMed] [Google Scholar]
- 67.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Best R.B., Zhu X., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mackerell A.D., Jr., Feig M., Brooks C.L., III Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 70.MacKerell A.D., Jr., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 71.Sachs J.N., Petrache H.I., Woolf T.B. Molecular dynamics simulations of ionic concentration gradients across model bilayers. J. Chem. Phys. 2003;118:1957–1969. [Google Scholar]
- 72.Almond A., Sheehan J.K., Brass A. Molecular dynamics simulations of the two disaccharides of hyaluronan in aqueous solution. Glycobiology. 1997;7:597–604. doi: 10.1093/glycob/7.5.597. [DOI] [PubMed] [Google Scholar]
- 73.Darden T., York D., Pedersen L. Particle mesh Ewald: An N log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 74.Marcus R., Sutin N. Electron transfer in chemistry and biology. Biochim. Biophys. Acta. 1985;811:265–322. [Google Scholar]
- 75.Kattnig D.R., Hore P.J. The sensitivity of a radical pair compass magnetoreceptor can be significantly amplified by radical scavengers. Sci. Rep. 2017;7:11640. doi: 10.1038/s41598-017-09914-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.