ABSTRACT
Extracellular vesicles (EVs), which can be found in almost all body fluids, consist of a lipid bilayer enclosing proteins and nucleic acids from their cells of origin. EVs can transport their cargo to target cells and have therefore emerged as key players in intercellular communication. Their potential as either diagnostic and prognostic biomarkers or therapeutic drug delivery systems (DDSs) has generated considerable interest in recent years. However, conventional methods used to study EVs still have significant limitations including the time-consuming and low throughput techniques required, while at the same time the demand for better research tools is getting stronger and stronger. In the past few years, microfluidics-based technologies have gradually emerged and have come to play an essential role in the isolation, detection and analysis of EVs. Such technologies have several advantages, including low cost, low sample volumes, high throughput and precision. This review summarizes recent advances in microfluidics-based technologies, compares conventional and microfluidics-based technologies, and includes a brief survey of recent progress towards integrated “on-a-chip” systems. In addition, this review also discusses the potential clinical applications of “on-a-chip” systems, including both “liquid biopsies” for personalized medicine and DDS devices for precision medicine, and then anticipates the possible future participation of cloud-based portable disease diagnosis and monitoring systems, possibly with the participation of artificial intelligence (AI).
KEYWORDS: Extracellular vesicle, exosomes, biomarker, cell-free therapy, drug delivery system, DDS, microfluidics, on-a-chip, protein, RNA
Introduction
Extracellular vesicles (EVs) are found in the extracellular space as well as in almost all body fluids, including blood [1], urine [2], saliva [3] and cerebrospinal fluid [4]. EVs carry and protect cargoes, including proteins, nucleic acids and lipids, from their cells of origin [5,6] (Figure 1(a)). For this reason, the vast potential of EVs as biomarkers, vaccines and therapeutic drug delivery systems (DDSs) has attracted increasing attention in recent years [7–9]. However, conventional methods used to study EVs still have significant limitations; for example, they are often time-consuming and low throughput [10–12]. Recently, microfluidics technology has been applied to the study of EVs and has several advantages, including low cost, low sample volumes, high throughput and precision [13]. Initially, the microfluidic method was developed at the interface of physics and chemistry [14]. The emerging cross-disciplinary interface between microfluidics and biology is quite natural [15,16], given the need for such technology to study complicated biological systems. Microfluidics allows the development of “on-a-chip” systems for the isolation, detection and analysis of EVs on a single integrated circuit of only a few square centimetres. This technology has already been combined with smartphone technology [17], and we anticipate the advent of a cloud storage- and cloud computing-based future of disease diagnosis and monitoring, possibly with the participation of artificial intelligence (AI).
Figure 1.
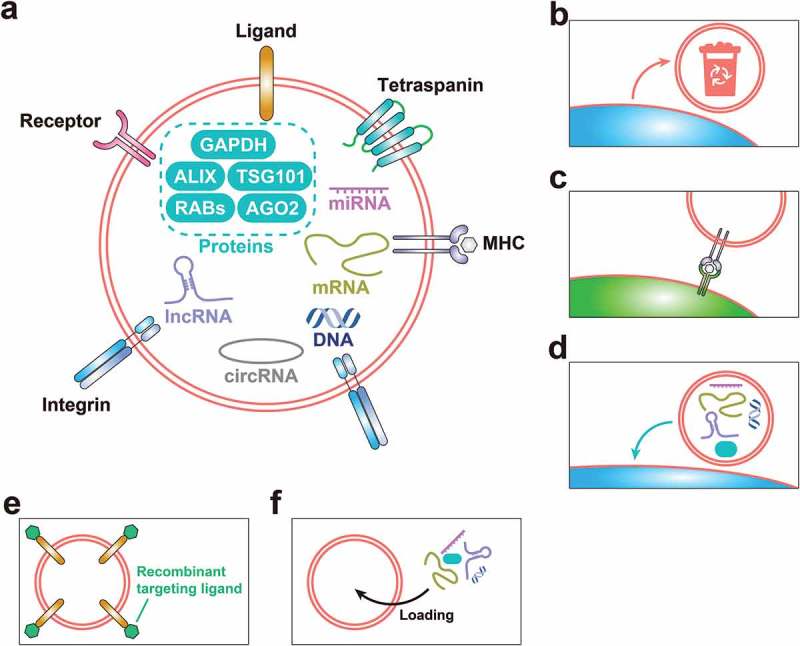
Current basic knowledge of EVs. (a) The constitution of EVs, involving membrane proteins (including ligands, receptors, tetraspanins, integrins and MHC – major histocompatibility complex – proteins), cargo proteins, nucleic acids (including DNA, messenger RNA (mRNA), microRNA (miRNA), long noncoding RNA (lncRNA) and circular RNA (circRNA)) and lipids, derived from their cells of origin. (b) Removing unwanted “trash” from the cell via the release of EVs. (c) The involvement of MHC-II-containing EVs in antigen presentation. (d) EV-mediated intracellular communication through transportation of proteins, nucleic acids and lipids from parent cells to distant cells. (e) EV surface protein modification, for example, based on C1C2 domain from the EV-surface-specific protein MFG-E8 (milk fat globule-epidermal growth factor 8 protein), or the N-terminus of lysosomal associated membrane protein 2b (Lamp2b), to customise targeting ability. (f) Cargo loading of EVs, based on cell engineering, which is particularly suitable for delivering nucleic acids, and specific delivery technology for proteins – for example, intracellular proteins labelled by a WW tag for the recognition by late-domain (L-domain) proteins and specifically loaded into EVs during their biogenesis.
This review focuses on comparisons between conventional and microfluidics-based technologies, and the impact of recent developments in microfluidic technologies on EV-related research. This includes a brief survey of recent progress towards integrated on-a-chip systems. Finally, we discuss the limitations of current systems and possible directions for future progress.
Extracellular vesicles
EVs are released from cells through two principal pathways: the “inward budding” pathway for the biogenesis of exosomes (Exos) from endosomes/multivesicular bodies (MVBs) via the endocytic pathway, and the “outward budding” pathway for the biogenesis of microvesicles (MVs) from the plasma membrane [18,19]. The endocytic pathway involves the formation of endosomes via a fundamental pathway for the entrance into the cell of large cargoes; the biogenesis of intraluminal vesicles, which are precursors of Exos, in endosomes for the transformation from early endosomes to MVBs, and the fusion between cytomembrane and MVB membrane to release Exos to the outside of the cell. Although initially dismissed as artefacts or “cell debris”, it is now clear that these nano-sized vesicles act as “correspondents” in intercellular communication [18,20,21]. EVs tend to express similar membrane proteins to their parent cells, which could give them potential targeting ability, and, in addition, could make them potential detection targets for use in liquid biopsy. For instance, EVs derived from endothelial cells have endothelial cell-specific surface markers including E-selectin (CD62E) and vascular endothelial cadherin (VE-cadherin) [22]. Glioblastomas and gliomas can express tumour-specific epidermal growth factor receptor variant III (EGFRvIII) on the surface of their EVs [23,24]. EVs containing the cancer-specific surface protein glypican-1 can be used for early detection of pancreatic cancer [25]. Dendritic cells, one type of antigen-presenting cells, release EVs containing surface proteins including intercellular adhesion molecule-1 (ICAM-1), which can be captured by leukocyte function-associated antigen-1 (LFA-1) on the T-cell surface via the high-affinity LFA-1/ICAM-1 interactions and thus activate T cells [26]. Some surface proteins can target specific tissues by targeting the extracellular matrix of target organ. For example, specific expression of integrins α6β4 and α6β1 on tumour-derived EVs (T-EVs) is associated with lung metastasis, and specific expression of integrin αvβ5 is associated with liver metastasis [27].
EV subtypes
EV subtypes differ in size and composition, and there is still a need for a complete set of improved protocols to isolate and distinguish these subtypes [28,29]. A conventional classification method, based on particle size, divides EVs into two subtypes: Exos (30–100 nm in diameter) and MVs (100–1000 nm in diameter) [18,30]. However, this classification method has been exposed as inadequate. For example, the biogenesis of some small vesicles (approximately 100 nm in diameter) is similar to that of MVs, formed by budding from the surface of the cytoplasmic membrane, but they can be isolated together with Exos [31]. Moreover, apoptotic bodies (800–5000 nm in diameter), which contain genomic DNA and have a variety of sizes, are difficult to distinguish from other EV subtypes [18,32].
Although the biogenesis of Exos and MVs is very different, the isolation method used in most EV-related research cannot distinguish their intracellular origin strictly following the definition of Exos and MVs. In most studies, small EVs are regarded as Exos [29,33] even though this is not the strict definition of Exos. Therefore, to avoid confusion, we make no distinction between Exos and MVs, consistent with most other EV-related reviews [32].
Physiological functions of EVs
Removing unwanted substances from a cell is one purpose for the release of EVs from a cell (Figure 1(b)). Initially, EVs were found to emerge during the maturation process of reticulocytes, in which the release of EVs containing transferrin receptors ensures the shutoff of iron import to prevent toxic iron overload [34–36]. However, subsequent researches, including investigation of the involvement of major histocompatibility complex II-containing EVs in antigen presentation (Figure 1(c)), revealed the participation of EVs in intracellular communication [37]. Recently, a growing body of evidence has shown that EVs can also transfer intracellular information, including proteins, nucleic acids and lipids, from parent cells to distant cells (Figure 1(d)) and can even participate in the re-programming/phenotype-alteration of the recipient cells [1,38–40].
Pathology related to EVs
In cancer progression, T-EVs modify the tumour-related microenvironment to make it more suitable for tumour cells by inducing angiogenesis and suppressing the reaction of immune cells [6]. EVs derived from malignant tumours at different stages are very different, so T-EVs can be used as biomarkers for the recognition and classification of tumours at very early stages [41]. EVs are also potential diagnostic biomarkers for infectious diseases because of the infection-related proteins and RNAs they carry [42]. However, a convenient yet accurate detection method is still needed to realise the potential of EVs in diagnostics.
EV-related cell-free therapy
EVs could be used therapeutically in two main ways: (1) by exploiting their natural properties and (2) by using them as drug delivery agents. Over the past few years, there has been much research into the wide range of potential applications of EVs in tissue engineering and regenerative medicine. For example, EVs from mesenchymal stem cells (MSCs) can promote tissue regeneration via the extracellular signal-regulated kinase pathway [43]. Moreover, MSC-derived EVs may induce angiogenesis in ischaemia-related disease [44]. The potential of human platelet-derived EVs in tissue regeneration is also emerging [18,45,46].
There is increasing exploration of nano-DDSs including liposomes and polymeric nanoparticles for delivery of therapeutic agents such as small molecule drugs and nucleic acids [47–49]. However, there are several disadvantages to the use of synthetic nanoparticles, such as low biocompatibility, immunoreaction and safety problems [50]. In comparison, EV-based DDSs have several advantages, for example, (1) low toxicity[51,52], (2) good stability in the circulation [52,53], (3) specific targeting ability [51,52], (4) customisation of targeting ability by modifying surface proteins, for example, based on the C1C2 domain from the EV-surface-specific protein MFG-E8 (milk fat globule-epidermal growth factor 8 protein), or the N-terminus of lysosomal-associated membrane protein 2b (Lamp2b) [52,54] (Figure 1(e)) and (5) straightforward cargo loading, especially for biomacromolecules, based on cell engineering, which is particularly suitable for delivering nucleic acids, as well as providing specific delivery technology for proteins – for example, intracellular proteins labelled by a WW tag for the recognition by late-domain (L-domain) proteins and specifically loaded into EVs during their biogenesis [9,52] (Figure 1(f)). As a result, we believe that EVs will open up a new frontier in the field of regenerative medicine.
Microfluidics-based EV isolation
Conventional methods
Conventional EV isolation techniques make use of (1) the specific size and density of EVs (differential centrifugation (DC), density gradient centrifugation (DGC), sucrose cushion centrifugation, ultrafiltration (UF) and chromatography), (2) specific EV surface markers (immunoaffinity capture, IAC) and (3) simple non-specific concentration (precipitation).
DC isolates EVs using increasing centrifugal force in three steps: (1) <1500×g (pelleting cell debris), (2) 10,000–20,000×g (pelleting large EVs) and (3) 100,000–200,000×g (pelleting small EVs) [55]. However, DC has limitations: (1) absolute separation is not possible using size alone [55,56]; (2) EVs may clump together [57,58]; (3) non-EV nanoparticles, including viruses [59] and protein complexes [60], may be co-isolated; (4) EVs may be damaged by ultracentrifugation [61]; and (5) the recovery rate is variable (2%–80%), resulting in poor comparability between studies [62,63].
DGC, an improvement on DC, uses a density gradient to isolate EVs according to their size and density [59]. EV isolation by DGC is free from protein contaminants but is also time-consuming and low throughput, limiting its practicability in clinical use [10–12]. A variation or simplified version of DGC, sucrose cushion centrifugation with a specific density rather than a continuous density gradient, is often used for morphological observation of EVs [64].
UF uses a membrane filter with pressure, either directly applied or generated by a centrifuge/ultracentrifuge [65]. UF can rapidly isolate EVs from body fluids [66,67], but the yield is low because of the loss of small size EVs, which have a similar size to protein aggregates, necessary to guarantee the purity [68].
Size exclusion chromatography (SEC) is a size-based high-yield isolation method, which has the advantage of simplicity, due to use of a single column, and the ability to isolate EVs from other soluble components [69]. However, this method can co-isolate other components, including viruses, protein aggregates, very large proteins (such as von Willebrand factor and chylomicrons) or low-density lipoproteins [70–73]. Other disadvantages include the requirement for specialised equipment, the time required and the loss of small EVs [69].
In principle, IAC of EVs uses a combination of specific monoclonal antibodies (mA) and suitable carriers such as plates or beads [74,75]. The IAC technology can be used on conventional systems including columns, plates and tubes, or with the help of more advanced microfluidic methods [76].
EV precipitation uses a synthetic polymer (available in commercial kits such as ExoQuickTM , System Biosciences (SBI), Palo Alto, CA, USA), which can rapidly precipitate both EVs and non-EV components, but the purity of precipitated EVs is very low [69], and the precipitation reagents influence the biological functions of EVs [77].
In summary, conventional methods usually require specific reagents/equipment, which are expensive and/or occupy a large amount of space. They are often cumbersome and associated with poor purity, potential effects on biological properties or low yield. These problems are present to a greater or lesser degree in all conventional methods, which cannot, therefore, entirely meet the needs of clinical usage. Microfluidic technologies have the potential to overcome these challenges.
Microfluidics-based isolation on-a-chip
Microfluidics-based EV isolation has several applications: (1) specific surface markers (microfluidics-based immunoaffinity capture, Mf-IAC) and (2) specific size and density (microfluidics-based membrane filtration (Mf-F), nanowire-based traps (NTs), nano-sized deterministic lateral displacement (nano-DLD), viscoelastic flow and acoustic isolation).
Mf-IAC uses “capture antibodies” or “capture beads” (beads coated with capture antibodies) targeting specific surface markers of EV subpopulations [78]. There are two main approaches: (1) inner surface modification with capture antibodies (Figure 2(e)) and (2) use of capture beads (Figure 2(f)). Chen et al. employed a surface-modified microfluidic channel with “herringbone grooves” to efficiently capture EVs [79]. Ashcroft et al. designed a mica surface with an antibody coat to efficiently isolate EVs from plasma [80]. Kanwar et al. efficiently captured circulating EVs using the “ExoChip” platform [76]. Moreover, with continual advances in nanotechnology such as nanoshearing effects [81] and nanostructured interfaces [82], the sensitivity and specificity of Mf-IAC devices has been substantially improved. For instance, Zhang et al. produced a Y-shaped micropost nanostructured interface to enhance the capture surface area [82], while Kang et al. designed a device that can generate mechanical whirling to increase the probability of binding between the antibody and the EVs [83]. Procedures combining Mf-IAC with capture beads consist of two steps: (1) binding between EVs in the sample and capture beads and (2) washing and separation of the capture beads. Dudani et al. developed an efficient Mf-IAC system by combining a rapid inertial solution exchange system with capture beads [84]. Meanwhile, Shao et al. developed a magnet-separating Mf-IAC system using magnetic capture beads and isolated EVs with a high yield [85]. Zhao et al. successfully developed a serpentine channel combined with magnetic capture beads that showed significant diagnostic potential [86]. The advantage of Mf-IAC with capture beads compared to Mf-IAC with inner surface modification is its relative convenience for follow-up analysis. Furthermore, we believe that the combination of a size-based method and an antibody-based method will provide a more advanced input–output ratio and enable isolation of EV subpopulations.
Figure 2.
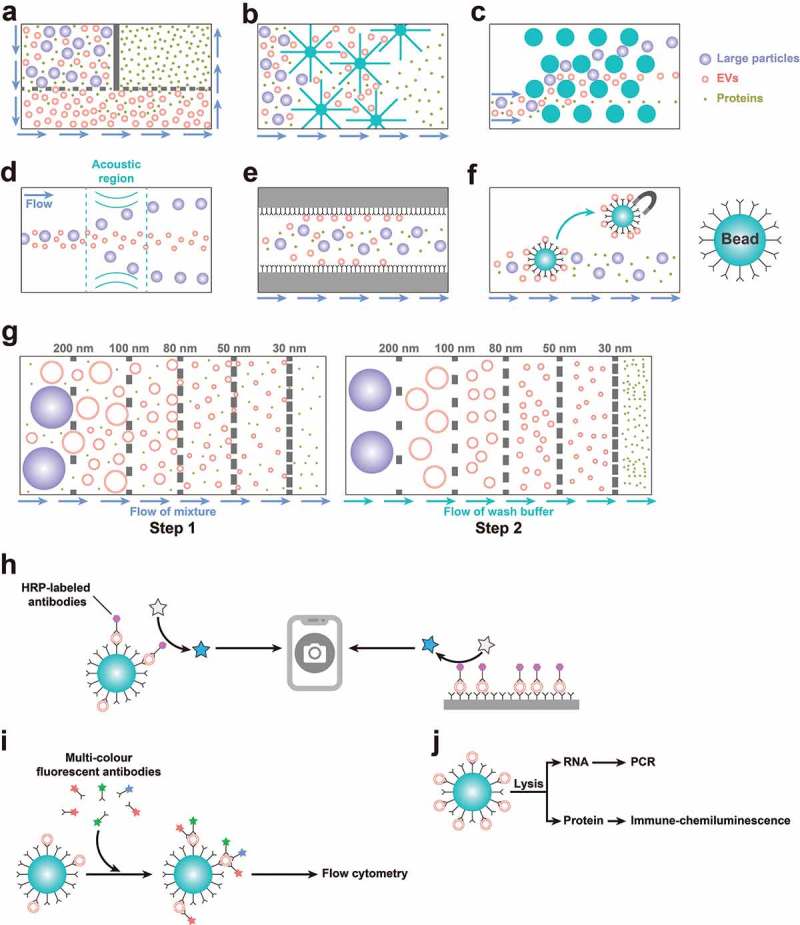
Current methods of on-chip isolation, detection and analysis. (a) A double-filtration Mf-F system for isolating small EVs. (b) An NT system with nanowire-on-a-micropillar structure to trap EVs. (c) A pillar-array-based microfluidic device or nano-DLD to isolate EVs by sorting particles in a continuous flow. (d) An acoustic isolation system to isolate EVs. (e) An Mf-IAC device using inner surface modification with capture antibodies. (f) An Mf-IAC device using capture beads. (g) ExoTIC: a modular platform with the ability to isolate EV subpopulations based on size. (h) A fluorescence imaging system integrated with Mf-IAC. (i) Multicolour FCM integrated with Mf-IAC. (j) Mf-IAC integrated with an “analyser-on-a-chip”.
Mf-IAC can isolate EVs from culture medium or body fluid. However, only EVs with a known surface antigen can be isolated. This might not be a problem when using EVs as therapeutic agents. However, when using them as diagnostic agents, it is better to capture all EVs, and not just a subpopulation with a specific antigen. Hence, an antigen-independent method would be better for developing an on-a-chip system for EV-based diagnosis.
Mf-F is based on the specific size of EVs. In their pioneering work, Davies et al. developed two kinds of Mf-F devices – pressure- and electrophoresis-driven – that separate cells, debris and small EVs via a nanoporous membrane with an adjustable pore size [87]. The limitation of pressure-driven Mf-F is that the pores become blocked after obtaining approximately 4 μL of filtrate. Using electrophoresis avoids this problem and increases the separation efficiency and purity [87]. Cho et al. isolated EVs using electrophoretic migration across a dialysis membrane with 30-nm pores [88].
Subsequently, double-filtration approaches were developed. For example, Liang et al. constructed a double-filtration Mf-F system (Figure 2(a)) comprising a filter with a pore size of 200 nm to remove cells and large impurities and a second filter with a pore size of 30 nm that allows proteins to pass through [89]. This system achieves high throughput and yield, in comparison with ultracentrifugation, for isolation of 30–200 nm EVs. Woo et al. constructed a different double-filtration Mf-F system (called Exodisc) using membranes with pore sizes of 20 and 600 nm. This method co-isolates small EVs and some large EVs with high throughput, high yield and low levels of protein contaminants [90]. Liu et al. demonstrated a size-based modularised Mf-F system called ExoTIC, which is a modular platform with the ability to isolate EV subpopulations based on size [91] (Figure 2(g)).
The NT system is a multiscale filtration system using nanowires (Figure 2(b)). Wang et al. built a porous silicon nanowire-on-a-micropillar structural NT system to trap EVs [92]. In principle, NT selectively traps specific sizes of particles, and the idea is similar to SEC.
Nano-DLD is a type of pillar-array-based microfluidic device that sorts particles in a continuous flow (Figure 2(c)). In 2004, Huang et al., for the first time, successfully constructed a high-throughput particle-separation system to separate microparticles (0.8, 0.9 and 1.0 μm in diameter), in which the path taken by the particles is determined by their size [93], but the separation of nanoparticles remained challenging for a long time. Recently, considering the electrostatic interactions between nanoparticles and pillars, the ionic strength of the buffer solution was modulated and improved. As a result, 51–1500 nm particles were successfully separated by a large-pore DLD device (with 2 μm gaps between pillars) [94]. The true nano-DLD system was first constructed by Wunsch et al. to separate colloids and EVs using a pillar-constituent array with gap sizes ranging from 25 to 235 nm [95].
The viscoelastic flow system is a continuous, size-based and label-free method of directly separating EVs from culture medium or body fluid using viscoelastic forces [96]. This method has a very high level of purity (>90%) and good yields [96]. Nevertheless, there has been little research in this area, so further studies are still needed.
An acoustic isolation system can also separate nano-sized particles (Figure 2(d)). For instance, differential acoustic radiation forces can separate small particles based on their mechanical properties including compressibility, diameter and density [97]. In addition, surface acoustic waves opened a new frontier in “separator-on-a-chip” technology, with a variety of advantages, including simple fabrication and integration with other microfluidic components, biocompatibility and contact-free manipulation of nanoparticles [98,99]. Lee et al. recently established a symmetric standing surface acoustic wave perpendicular to the direction of flow, which causes larger particles to move to the channel wall and then allows isolation of nanoscale vesicles (<200 nm)[100]. Wu et al. employed an acoustofluidics technique that consisted of two modules: a cell-removal module and an EV-isolation module [101]. This two-step device greatly simplifies the pre-processing of complex samples consisting of cells such as blood [101]. However, further studies in this area are still needed.
Non-destructive methods to isolate intact EVs from culture medium or complex body fluids are very important for subsequent detection and application. As outlined above, development of such methods is proceeding rapidly and it is now feasible to build a microfluid-based isolating device on a chip, providing convenience, miniaturisation and potential high expansibility. A brief summary is provided in Figure 3.
Figure 3.
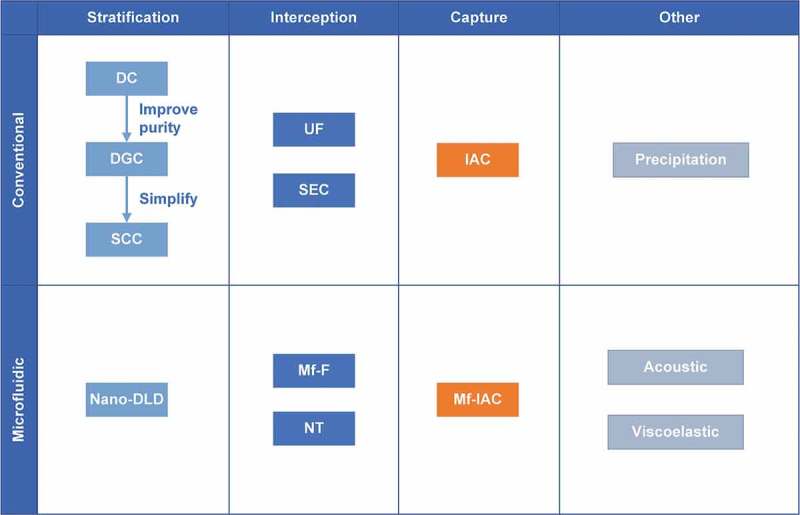
A brief summary of present isolation technology. The present isolation technologies are classified into conventional methods and microfluidics-based methods. There are three main guiding ideologies: stratification, interception and capture. For stratification, the idea is to sort EVs by some property (such as size or density). For interception, the idea is to set a cut-off or threshold value to influence the passage of specific particles. For capture, the idea is to proactively identify the specific particles with a particular characteristic (such as a specific surface marker). In addition, there are also some other technologies which need further study.
Microfluidics-based detection and analysis systems
The above on-a-chip isolation methods not only can be used as the preparatory work for downstream conventional detection and analysis but can also be integrated with a downstream microfluidics-based detection or analysis system. A detection system is defined as one that directly detects EVs without lysis, whereas an analysis system is defined as one that analyses the constituents of EVs. Compared to conventional methods, a microfluidics-based platform can provide high throughput, low consumption of reagents and microminiaturisation, as well as the potential for portability. Here, we describe microfluidics-based detection and analysis systems, then later discuss their integration with isolation systems.
Detection on-a-chip
Microfluidics-based EV detection methods include those based on (1) fluorescence (fluorescence imaging and fluorescence analysis via flow cytometry, FCM), (2) colourimetric techniques (colourimetric detection), (3) optical properties (surface plasmon resonance (SPR)), (4) magnetism (miniaturised nuclear magnetic resonance) and (5) electrical properties (electro-kinetic systems and electrical detection).
The ExoChip platform stains isolated EVs with a fluorescent carbocyanine dye, and the chip is analysed using a plate reader with a sensitivity of up to 0.5 pM [76]. Fang et al. also used a fluorescent dye but instead used a microscope-based imaging system for detection [102]. In an alternative approach, Ko et al. designed a smartphone-enabled portable on-a-chip optofluidic detection system, in which bead-captured EVs are recognised by a horseradish peroxidase-modified antibody. A smartphone-based fluorimeter can then detect the fluorescent signal generated by enzyme amplification [17]. Therefore, fluorescence-based detection systems have become increasingly portable.
Fluorescent FCM systems also have great potential. Dudani et al. detected capture beads using a fluorescent FCM assay [84]. Zhao et al. reported a multiplexed measurement system (detecting CA-125, EpCAM and CD24) via multicolour FCM imaging [86]. Another FCM-based array consisting of parallel nanochannels was successfully built by Friedrich et al. and accurately performs vesicle counting down to 170 fM [103]. The trend of fluorescent FCM systems seems to be from monochrome (single index) and mono-channel to polychrome (multiple indices) and multichannel (Figure 2(i)).
The results of colourimetric detection can be either visually observed or detected via absorbance. Vaidyanathan et al. used nanoshearing induced by alternating current electrohydrodynamics to quantify EVs both visually and by measuring absorbance, with a three-fold enhancement in sensitivity over other reported assays based on hydrodynamic flow[81]. In their Exodisc system, Woo et al. performed an on-disc enzyme-linked immunosorbent assay (ELISA) and measured the OD value at 450 nm after transferring the solution to a detection chamber [90]. To make the system more portable, Liang et al. used a smartphone camera to measure the results of a colourimetric ELISA [89]. Using colourimetric detection helps to make the system increasingly portable and simple.
SPR is gradually coming to the forefront in the field of label-free biosensor technology. Im et al. used transmission mode SPR to detect EVs with 670 aM sensitivity [104]. Furthermore, Sina et al. used SPR detection as part of a two-step isolation and detection strategy [105]. This system is gradually being integrated with the “separator-on-a-chip”, but further advances are still needed, such as a portable SPR system.
Miniaturised nuclear magnetic resonance (μNMR) is based on the superparamagnetic characteristics of magnetic nanoparticles. Shao et al. used μNMR to detect target-specific magnetic nanoparticles after on-chip isolation [106].
Akagi et al. introduced an electro-kinetic system to detect a marked positive shift in the distribution of zeta potential to detect the presence of EVs [107], while Fraikin et al. demonstrated a high-throughput electrical detection method for multicomponent mixtures of nanoparticles [108]. Zhou et al. designed an aptamer-based electrochemical biosensor to detect EVs, and when the EVs were bound to complementary aptamers, the released probing strands with redox moieties caused a decrease in the electrochemical signal [109].
Analysis on-a-chip
Microfluidics-based EV analysis includes (Figure 2(j)) (1) analysis of protein content (on-chip immuno-chemiluminescence) and (2) analysis of RNA content (on-chip quantitative PCR, qPCR). He et al. captured EpCAM(+) EVs and incubated them with lysis buffer to obtain the cargo proteins [110]. They then analysed total IGF-1R and p-IGF-1R using an immuno-chemiluminescence assay after on-chip mixing with specific antibodies [110]. Shao et al. performed on-chip RNA extraction, reverse transcription (RT) and qPCR to detect and quantify mRNAs [85]. Richards et al. used RT-qPCR to quantify the microRNA (miRNA) level in EVs [111]. In addition to quantifying the level of selected RNAs, Ko et al. employed a machine learning algorithm to analyse the RNA profile and further distinguish cancer, pre-cancer and healthy control tissues [112]. However, research into the field is still in the initial stages and needs further attention and study.
Efforts and progress towards integrated on-a-chip systems for EVs
There is increasing interest in point-of-care (POC) devices. Much effort has been put into the development of various in vitro diagnostic approaches such as POC devices, whose market volume is estimated to be 75.1 billion US dollars by the year 2020 [113]. The aim of POC devices is the realisation of rapid, highly sensitive, user-friendly and portable medical facilities [114,115]. Highly integrated microfluidic devices have shown great promise for making such POC devices a reality.
In recent years, researchers have designed integrated microfluidic systems for the isolation and detection of EVs, and the quantification of their cargoes. Compared to conventional protocols, these integrated devices have advantages including time saving, high throughput, high sensitivity and a low sample volume requirement. Currently, most of these POC candidates are based on Mf-IAC, and this indicates that immunoaffinity-based systems have better extensibility and are likely to be the “mainline” of future integrated on-a-chip systems or POC devices.
The first integrated microfluidic system, ExoChip, integrates immunocapture by a CD63 antibody and fluorescence quantification using a plate reader [76]. Although it used an old detection method, ExoChip showed the way to future integrated devices.
Later, FCM was incorporated into these systems. Dudani et al. employed fluorescent FCM to detect EVs after capture using an Mf-IAC system [84]. Moreover, Zhao et al. used multicolour FCM as a multiplexed measurement system (detecting CA-125, EpCAM and CD24) [86]. Meanwhile, Friedrich and colleagues created an FCM-based parallel nanochannel array and were able to accurately perform vesicle counting down to 170 fM [103].
The advent of a smartphone-based system will bring excellent portability and accessibility to POC devices. Ko et al. designed a smartphone-based on-a-chip system, in which the smartphone-based fluorimeter can detect a fluorescent signal generated by enzyme amplification [17]. In addition, colourimetric detection systems are also showing potential, especially with regard to portability and expandability. Liang et al. obtained results of a colourimetric ELISA using a smartphone camera [89], demonstrating the great potential of this approach in creating a portable POC device.
In addition, SPR is gradually showing potential in becoming a leader of various label-free biosensor technologies with multiplexing ability, high sensitivity and high-level integration as described by Im et al. [104]. Furthermore, Sina et al. integrated SPR detection into their two-step isolation and detection strategy [105].
In addition to the integration between isolation and detection, there is also growing attention being paid to the integration between isolation and analysis, which could reveal more in-depth information about EVs. He et al. captured EpCAM(+) EVs using magnetic Mf-IAC, and analysed the total IGF-1R and p-IGF-1R levels on-chip [110]. Shao et al. performed chip-based EV isolation, RNA extraction and RT-qPCR in one device to detect and quantify multiple target mRNAs [85]. Considering that the quantification of the level of selected mRNAs alone is far from enough, Ko et al. employed a machine learning algorithm to analyse the RNA profile and were able to further distinguish cancer, pre-cancer and healthy control tissues [112]. This aspect is very important, but the recent studies are not sufficient, so further research is needed. A brief summary of efforts on this area is shown in Figure 4.
Figure 4.

Recent efforts towards integrated on-a-chip systems for generation of EVs.
Discussion and outlook
In many cases, an inherently invasive and risky surgical biopsy is the “gold standard” of malignant tumour diagnosis. However, in some situations, the suspected region cannot be frequently sampled or is surgically inaccessible. By contrast, liquid biopsies, which test body fluids, are minimally invasive. The liquid biomarkers include cell-free nucleic acids (including DNAs, mRNAs and ncRNAs), circulating tumour cells and EVs [116]. These liquid biomarkers usually represent the microenvironment of malignant tumours, whereas surgically biopsied tissues are often heterogeneous, leading to ambiguous conclusions [116]. EVs, which contain the overwhelming majority of cell-free nucleic acids [52,117,118] and whose cargoes are specific to the cell of origin [52,117,118], show the greatest potential in non-invasive liquid biopsy.
In particular, T-EVs have the potential to be the most informative predictors in non-invasive tumour monitoring [119,120]. For example, Taylor et al. found and verified eight miRNAs of EVs that could identify and differentiate stages of tumours, and potentially be diagnostic/prognostic biomarkers of ovarian cancer [121]. However, many issues remain to be resolved, including the isolation of EVs from complex body fluids.
Before liquid biopsies can be developed, the devices for isolation, detection and analysis need to be high throughput, portable and intelligent. Fortunately, recent advances in microfluidic devices, especially on-a-chip EV isolation systems, have come a long way towards achieving these goals. Furthermore, there have been promising advances in technology used in the manipulation of cells and particles other than EVs, among which one of the most promising is label-free microfluidic manipulation technology or negative magnetophoresis-based technology [122]. These technologies also have the potential to advance the current state of liquid biopsies.
Over the past few years, many studies have reported the potential of EV-based cell-free therapy in the treatment of different diseases. In addition, clinical trials using therapeutic EVs have gradually begun to appear. To make EVs more versatile rather than limited to their native functions, there have been an increasing number of studies focused on molecular engineering of EVs. The traditional or conventional methods of using engineered EVs as a DDS have been comprehensively reviewed in our previous literature review [123]. Technically speaking, these strategies for EV engineering could be briefly classified into two levels: (1) engineering at the parent cell level – involving manipulation using genetic or metabolic methods; and (2) engineering at the EV level – including surface molecular modification methods and permeabilisation of the membrane.
For engineering at the parent cell level, after genetic modification of parent cells to generate EVs carrying proteins/peptides, microfluidics-based isolation technology, instead of a conventional isolating method, could be employed to isolate EVs. This is a relatively easy way to bring microfluidic technology into engineered EVs as a DDS.
For engineering at the EV level, much effort has been made to bring the advantages of microfluidics into the region of engineered EVs as a DDS, although the development is still at a very early stage. One potential avenue of exploration is the microfluidic extrusion method. In 2014, Jo et al. developed a microfluidic instrument to mechanically break down cells into EV-mimetics (EVMs), which have similar properties to naturally secreted EVs [124]. Then, EVMs were used to load siRNA via random packaging and re-assembly after the breakdown [125]. As research delves deeper, EVMs gradually show their potential as a DDS [126]. However, we believe that bringing the fundamental principles of conventional methods into microfluidic instruments has more potential for such applications. For example, the principles used in conventional methods, including the use of lipophilic/amphipathic molecules, electroporation and combination with liposomes [123], can be combined with the advantages of a microfluidic system such as accurate control of mass transfer [127]. Unfortunately, there have been barely any major breakthroughs in this direction. This field of study might offer potential for future in-depth studies.
The potential of integration of a smartphone [17] in such a microfluidic system cannot be underestimated. In the future, data from smartphone-based portable POC devices could be uploaded to a cloud drive. With the development of AI technology, we envision a situation in which data on a cloud drive could be pre-examined by AI. AI may recognise suspicious early signs, and, could then recommend appropriate experienced experts. Experienced physicians could use the data on the cloud server to further diagnose the issues, and then make an appointment for necessary auxiliary examinations and meetings with appropriate medical experts. After a definite diagnosis, doctors and scientists could have access to massive amounts of data to study diseases and to improve the diagnostic level of the medical community, as well as improving the pre-examination level of AI.
Funding Statement
This work was supported by the National Natural Science Foundation of China [81802226, 81871834, 81301589 and 81472066].
Author Contributions
Shi-Cong Tao planned and wrote the manuscript. Shang-Chun Guo helped with planning and writing. Helen Dawn gave basic advice and suggestions. All authors reviewed the manuscript.
Disclosure statement
The authors have declared that no competing interest exists.
References
- [1].Deregibus MC, Cantaluppi V, Calogero R, et al. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007October01;110(7):2440–15. PubMed PMID: 17536014; eng. [DOI] [PubMed] [Google Scholar]
- [2].van Balkom BW, Pisitkun T, Verhaar MC, et al. Exosomes and the kidney: prospects for diagnosis and therapy of renal diseases. Kidney Int. 2011December;80(11):1138–1145. PubMed PMID: 21881557; PubMed Central PMCID: PMCPMC3412193. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Michael A, Bajracharya SD, Yuen PS, et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010January;16(1):34–38. PubMed PMID: 19627513; PubMed Central PMCID: PMCPMC2844919. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Street JM, Barran PE, Mackay CL, et al. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med. 2012January05;10:5 PubMed PMID: 22221959; PubMed Central PMCID: PMCPMC3275480. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schorey JS, Bhatnagar S.. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008June;9(6):871–881. PubMed PMID: 18331451; PubMed Central PMCID: PMCPMC3636814. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Reviews Immunol. 2014March;14(3):195–208. PubMed PMID: 24566916; PubMed Central PMCID: PMCPMC4350779. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nawaz M, Camussi G, Valadi H, et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat Reviews Urol. 2014December;11(12):688–701. PubMed PMID: 25403245; eng. [DOI] [PubMed] [Google Scholar]
- [8].Lawson C, Vicencio JM, Yellon DM, et al. Microvesicles and exosomes: new players in metabolic and cardiovascular disease. J Endocrinol. 2016February;228(2):R57R71 PubMed PMID: 26743452; eng. [DOI] [PubMed] [Google Scholar]
- [9].Kooijmans SA, Vader P, van Dommelen SM, et al. Exosome mimetics: a novel class of drug delivery systems. Int J Nanomedicine. 2012;7:1525–1541. PubMed PMID: 22619510; PubMed Central PMCID: PMCPMC3356169. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tauro BJ, Greening DW, Mathias RA, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods (San Diego, CA). 2012February;56(2):293–304. PubMed PMID: 22285593; eng. [DOI] [PubMed] [Google Scholar]
- [11].Greening DW, Xu R, Ji H, et al. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol (Clifton, NJ). 2015;1295:179–209. PubMed PMID: 25820723; eng. [DOI] [PubMed] [Google Scholar]
- [12].Webber J, Clayton A. How pure are your vesicles? J Extracell Vesicles. 2013;2 PubMed PMID: 24009896; PubMed Central PMCID: PMCPMC3760653. eng DOI: 10.3402/jev.v2i0.19861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Salafi T, Zeming KK, Zhang Y. Advancements in microfluidics for nanoparticle separation. Lab Chip. 2016December20;17(1):11–33. PubMed PMID: 27830852; eng. [DOI] [PubMed] [Google Scholar]
- [14].Beebe DJ, Mensing GA, Walker GM. Physics and applications of microfluidics in biology. Annu Rev Biomed Eng. 2002;4:261–286. PubMed PMID: 12117759; eng. [DOI] [PubMed] [Google Scholar]
- [15].Paguirigan AL, Beebe DJ. Microfluidics meet cell biology: bridging the gap by validation and application of microscale techniques for cell biological assays. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology. 2008September;30(9):811–821. PubMed PMID: 18693260; PubMed Central PMCID: PMCPMC2814162. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Walker GM, Zeringue HC, Beebe DJ. Microenvironment design considerations for cellular scale studies. Lab Chip. 2004April;4(2):91–97. PubMed PMID: 15052346; eng. [DOI] [PubMed] [Google Scholar]
- [17].Ko J, Hemphill MA, Gabrieli D, et al. Smartphone-enabled optofluidic exosome diagnostic for concussion recovery. Sci Rep. 2016August08;6:31215 PubMed PMID: 27498963; PubMed Central PMCID: PMCPMC4976377. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tao SC, Guo SC, Zhang CQ. Platelet-derived extracellular vesicles: an emerging therapeutic approach. Int J Biol Sci. 2017;137:828–834. PubMed PMID: 28808416; PubMed Central PMCID: PMCPMC5555101. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Maas SL, Breakefield XO, Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 2017March;27(3):172–188. PubMed PMID: 27979573; PubMed Central PMCID: PMCPMC5318253. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen CY, Rao SS, Ren L, et al. Exosomal DMBT1 from human urine-derived stem cells facilitates diabetic wound repair by promoting angiogenesis. Theranostics. 2018;8(6):1607–1623. PubMed PMID: 29556344; PubMed Central PMCID: PMCPMC5858170. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hu Y, Rao SS, Wang ZX, et al. Exosomes from human umbilical cord blood accelerate cutaneous wound healing through miR-21-3p-mediated promotion of angiogenesis and fibroblast function. Theranostics. 2018;8(1):169–184. PubMed PMID: 29290800; PubMed Central PMCID: PMCPMC5743467. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dignat-George F, Boulanger CM. The many faces of endothelial microparticles. Arterioscler Thromb Vasc Biol. 2011January;31(1):27–33. . PubMed PMID: 21160065; eng. [DOI] [PubMed] [Google Scholar]
- [23].Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008December;10(12):1470–1476. PubMed PMID: 19011622; PubMed Central PMCID: PMCPMC3423894. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Al-Nedawi K, Meehan B, Micallef J, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008May;10(5):619–624. PubMed PMID: 18425114; eng. [DOI] [PubMed] [Google Scholar]
- [25].Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015July9;523(7559):177–182. PubMed PMID: 26106858; PubMed Central PMCID: PMCPMC4825698. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nolte-’t Hoen EN, Buschow SI, Anderton SM, et al. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood. 2009February26;113(9):1977–1981. . PubMed PMID: 19064723; eng. [DOI] [PubMed] [Google Scholar]
- [27].Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015November19;527(7578):329–335. PubMed PMID: 26524530; PubMed Central PMCID: PMCPMC4788391. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lai RC, Tan SS, Yeo RW, et al. MSC secretes at least 3 EV types each with a unique permutation of membrane lipid, protein and RNA. J Extracell Vesicles. 2016;5:29828 PubMed PMID: 26928672; PubMed Central PMCID: PMCPMC4770866. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016February23;113(8):E968E977 PubMed PMID: 26858453; PubMed Central PMCID: PMCPMC4776515. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gan X, Gould SJ. Identification of an inhibitory budding signal that blocks the release of HIV particles and exosome/microvesicle proteins. Mol Biol Cell. 2011March15;22(6):817–830. PubMed PMID: 21248205; PubMed Central PMCID: PMCPMC3057706. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nabhan JF, Hu R, Oh RS, et al. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci USA. 2012March13;109(11):4146–4151. PubMed PMID: 22315426; PubMed Central PMCID: PMCPMC3306724. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tkach M, Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016March10;164(6):1226–1232. PubMed PMID: 26967288; eng. [DOI] [PubMed] [Google Scholar]
- [33].Gould SJ, Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles. 2013;2 PubMed PMID: 24009890; PubMed Central PMCID: PMCPMC3760635. eng DOI: 10.3402/jev.v2i0.20389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983July;33(3):967–978. PubMed PMID: 6307529; eng. [DOI] [PubMed] [Google Scholar]
- [35].Johnstone RM, Adam M, Hammond JR, et al. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987July05;262(19):9412–9420. PubMed PMID: 3597417; eng. [PubMed] [Google Scholar]
- [36].Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983August;97(2):329–339. PubMed PMID: 6309857; PubMed Central PMCID: PMCPMC2112509. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996March01;183(3):1161–1172. PubMed PMID: 8642258; PubMed Central PMCID: PMCPMC2192324. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mause SF, Weber C. Microparticles: protagonists of a novel communication network for intercellular information exchange. Circ Res. 2010October29;107(9):1047–1057. PubMed PMID: 21030722; eng. [DOI] [PubMed] [Google Scholar]
- [39].Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007June;9(6):654–659. PubMed PMID: 17486113; eng. [DOI] [PubMed] [Google Scholar]
- [40].Ratajczak J, Wysoczynski M, Hayek F, et al. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia. 2006September;20(9):1487–1495. PubMed PMID: 16791265; eng. [DOI] [PubMed] [Google Scholar]
- [41].Choi DS, Lee J, Go G, et al. Circulating extracellular vesicles in cancer diagnosis and monitoring: an appraisal of clinical potential. Mol Diagn Ther. 2013October;17(5):265–271. PubMed PMID: 23729224; eng. [DOI] [PubMed] [Google Scholar]
- [42].Vlassov AV, Magdaleno S, Setterquist R, et al. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012July;1820(7):940–948. PubMed PMID: 22503788; eng. [DOI] [PubMed] [Google Scholar]
- [43].Zhou Y, Xu H, Xu W, et al. Exosomes released by human umbilical cord mesenchymal stem cells protect against cisplatin-induced renal oxidative stress and apoptosis in vivo and in vitro. Stem Cell Res Ther. 2013April25;4(2):34 PubMed PMID: 23618405; PubMed Central PMCID: PMCPMC3707035. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gatti S, Bruno S, Deregibus MC, et al. Microvesicles derived from human adult mesenchymal stem cells protect against ischaemia-reperfusion-induced acute and chronic kidney injury. Nephrology, Dialysis, Transplantation: Official Publication Eur Dial Transpl Assoc Eur Ren Assoc. 2011May;26(5):1474–1483. PubMed PMID: 21324974; eng. [DOI] [PubMed] [Google Scholar]
- [45].Tao SC, Yuan T, Rui BY, et al. Exosomes derived from human platelet-rich plasma prevent apoptosis induced by glucocorticoid-associated endoplasmic reticulum stress in rat osteonecrosis of the femoral head via the Akt/Bad/Bcl-2 signal pathway. Theranostics. 2017;7(3):733–750. PubMed PMID: 28255363; PubMed Central PMCID: PMCPMC5327646. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Guo SC, Tao SC, Yin WJ, et al. Exosomes derived from platelet-rich plasma promote the re-epithelization of chronic cutaneous wounds via activation of YAP in a diabetic rat model. Theranostics. 2017;7(1):81–96. PubMed PMID: 28042318; PubMed Central PMCID: PMCPMC5196887. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wilczewska AZ, Niemirowicz K, Markiewicz KH, et al. Nanoparticles as drug delivery systems. Pharmacol Rep. 2012;64(5):1020–1037. PubMed PMID: 23238461; eng. [DOI] [PubMed] [Google Scholar]
- [48].Zhang ZC, Tang C, Dong Y, et al. Targeting the long noncoding RNA MALAT1 blocks the pro-angiogenic effects of osteosarcoma and suppresses tumour growth. Int J Biol Sci. 2017;13(11):1398–1408. PubMed PMID: 29209144; PubMed Central PMCID: PMCPMC5715523. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tao SC, Rui BY, Wang QY, et al. Extracellular vesicle-mimetic nanovesicles transport LncRNA-H19 as competing endogenous RNA for the treatment of diabetic wounds. Drug Deliv. 2018November;25(1):241–255. PubMed PMID: 29334272; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Raemdonck K, Braeckmans K, Demeester J, et al. Merging the best of both worlds: hybrid lipid-enveloped matrix nanocomposites in drug delivery. Chem Soc Rev. 2014January07;43(1):444–472. PubMed PMID: 24100581; eng. [DOI] [PubMed] [Google Scholar]
- [51].Jang SC, Gho YS. Could bioengineered exosome-mimetic nanovesicles be an efficient strategy for the delivery of chemotherapeutics? Nanomedicine (London, England). 2014February;9(2):177–180. PubMed PMID: 24552557; eng. [DOI] [PubMed] [Google Scholar]
- [52].Tao SC, Guo SC, Zhang CQ. Modularized extracellular vesicles: the dawn of prospective personalized and precision medicine. Advanced Sci. Adv Sci (Weinh). 2018;5(2):1700449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Clayton A, Harris CL, Court J, et al. Antigen-presenting cell exosomes are protected from complement-mediated lysis by expression of CD55 and CD59. Eur J Immunol. 2003February;33(2):522–531. PubMed PMID: 12645951; eng. [DOI] [PubMed] [Google Scholar]
- [54].Ohno S, Takanashi M, Sudo K, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013January;21(1):185–191. PubMed PMID: 23032975; PubMed Central PMCID: PMCPMC3538304. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Momen-Heravi F, Balaj L, Alian S, et al. Current methods for the isolation of extracellular vesicles. Biol Chem. 2013October;394(10):1253–1262. PubMed PMID: 23770532; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Taylor DD, Shah S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods (San Diego, CA). 2015October01;87:3–10. [DOI] [PubMed] [Google Scholar]
- [57].Yuana Y, Boing AN, Grootemaat AE, et al. Handling and storage of human body fluids for analysis of extracellular vesicles. J Extracell Vesicles. 2015;4:29260 PubMed PMID: 26563735; PubMed Central PMCID: PMCPMC4643195. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Linares R, Tan S, Gounou C, et al. High-speed centrifugation induces aggregation of extracellular vesicles. J Extracell Vesicles. 2015;4:29509 PubMed PMID: 26700615; PubMed Central PMCID: PMCPMC4689953. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cantin R, Diou J, Belanger D, et al. Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. J Immunol Methods. 2008September30;338(1–2):21–30. PubMed PMID: 18675270; eng. [DOI] [PubMed] [Google Scholar]
- [60].Gyorgy B, Modos K, Pallinger E, et al. Detection and isolation of cell-derived microparticles are compromised by protein complexes resulting from shared biophysical parameters. Blood. 2011January27;117(4):e39e48 PubMed PMID: 21041717; eng. [DOI] [PubMed] [Google Scholar]
- [61].Ismail N, Wang Y, Dakhlallah D, et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood. 2013February07;121(6):984–995. PubMed PMID: 23144169; PubMed Central PMCID: PMCPMC3567345. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jayachandran M, Miller VM, Heit JA, et al. Methodology for isolation, identification and characterization of microvesicles in peripheral blood. J Immunol Methods. 2012January31;375(1–2):207–214. PubMed PMID: 22075275; PubMed Central PMCID: PMCPMC3253871. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Momen-Heravi F, Balaj L, Alian S, et al. Impact of biofluid viscosity on size and sedimentation efficiency of the isolated microvesicles. Front Physiol. 2012;3:162 PubMed PMID: 22661955; PubMed Central PMCID: PMCPMC3362089. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Thery C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current Protocols in Cell Biology. 2006April;Chapter 3:Unit 3.22 PubMed PMID: 18228490; eng DOI: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- [65].Coumans FAW, Brisson AR, Buzas EI, et al. Methodological guidelines to study extracellular vesicles. Circ Res. 2017May12;120(10):1632–1648. PubMed PMID: 28495994; eng. [DOI] [PubMed] [Google Scholar]
- [66].Merchant ML, Powell DW, Wilkey DW, et al. Microfiltration isolation of human urinary exosomes for characterization by MS. Proteomics Clinical Applications. 2010January;4(1):84–96. PubMed PMID: 21137018; eng. [DOI] [PubMed] [Google Scholar]
- [67].Grant R, Ansa-Addo E, Stratton D, et al. A filtration-based protocol to isolate human plasma membrane-derived vesicles and exosomes from blood plasma. J Immunol Methods. 2011August31;371(1–2):143–151. PubMed PMID: 21741384; eng. [DOI] [PubMed] [Google Scholar]
- [68].Lobb RJ, Becker M, Wen SW, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. 2015;4:27031 PubMed PMID: 26194179; PubMed Central PMCID: PMCPMC4507751. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Xu R, Greening DW, Zhu HJ, et al. Extracellular vesicle isolation and characterization: toward clinical application. J Clin Invest. 2016April1;126(4):1152–1162. PubMed PMID: 27035807; PubMed Central PMCID: PMCPMC4811150. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Sodar BW, Kittel A, Paloczi K, et al. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci Rep. 2016April18;6:24316 PubMed PMID: 27087061; PubMed Central PMCID: PMCPMC4834552. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Baranyai T, Herczeg K, Onodi Z, et al. Isolation of exosomes from blood plasma: qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PloS One. 2015;10(12):e0145686 PubMed PMID: 26690353; PubMed Central PMCID: PMCPMC4686892. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hong CS, Funk S, Muller L, et al. Isolation of biologically active and morphologically intact exosomes from plasma of patients with cancer. J Extracell Vesicles. 2016;5:29289 PubMed PMID: 27018366; PubMed Central PMCID: PMCPMC4808740. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Welton JL, Webber JP, Botos LA, et al. Ready-made chromatography columns for extracellular vesicle isolation from plasma. J Extracell Vesicles. 2015;4:27269 PubMed PMID: 25819214; PubMed Central PMCID: PMCPMC4376847. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Shih CL, Chong KY, Hsu SC, et al. Development of a magnetic bead-based method for the collection of circulating extracellular vesicles. N Biotechnol. 2016January25;33(1):116–122. PubMed PMID: 26409934; eng. [DOI] [PubMed] [Google Scholar]
- [75].Clayton A, Court J, Navabi H, et al. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J Immunol Methods. 2001January01;247(1–2):163–174. PubMed PMID: 11150547; eng. [DOI] [PubMed] [Google Scholar]
- [76].Kanwar SS, Dunlay CJ, Simeone DM, et al. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip. 2014June07;14(11):1891–1900. PubMed PMID: 24722878; PubMed Central PMCID: PMCPMC4134440. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Paolini L, Zendrini A, Di Noto G, et al. Residual matrix from different separation techniques impacts exosome biological activity. Sci Rep. 2016March24;6:23550 PubMed PMID: 27009329; PubMed Central PMCID: PMCPMC4806376. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Contreras-Naranjo JC, Wu HJ, Ugaz VM. Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine. Lab Chip. 2017August23 PubMed PMID: 28832692; eng DOI: 10.1039/c7lc00592j [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Chen C, Skog J, Hsu CH, et al. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip. 2010February21;10(4):505–511. PubMed PMID: 20126692; PubMed Central PMCID: PMCPMC3136803. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Ashcroft BA, De Sonneville J, Yuana Y, et al. Determination of the size distribution of blood microparticles directly in plasma using atomic force microscopy and microfluidics. Biomedical Microdevices. 2012August;14(4):641–649. PubMed PMID: 22391880; PubMed Central PMCID: PMCPMC3388260. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Vaidyanathan R, Naghibosadat M, Rauf S, et al. Detecting exosomes specifically: a multiplexed device based on alternating current electrohydrodynamic induced nanoshearing. Anal Chem. 2014November18;86(22):11125–11132. PubMed PMID: 25324037; eng. [DOI] [PubMed] [Google Scholar]
- [82].Zhang P, He M, Zeng Y. Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab Chip. 2016August02;16(16):3033–3042. PubMed PMID: 27045543; PubMed Central PMCID: PMCPMC4970962. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kang YT, Kim YJ, Bu J, et al. High-purity capture and release of circulating exosomes using an exosome-specific dual-patterned immunofiltration (ExoDIF) device. Nanoscale. 2017September21;9(36):13495–13505. PubMed PMID: 28862274; eng. [DOI] [PubMed] [Google Scholar]
- [84].Dudani JS, Gossett DR, Tse HT, et al. Rapid inertial solution exchange for enrichment and flow cytometric detection of microvesicles. Biomicrofluidics. 2015January;9(1):014112 PubMed PMID: 25713694; PubMed Central PMCID: PMCPMC4320146. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Shao H, Chung J, Lee K, et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun. 2015May11;6:6999 PubMed PMID: 25959588; PubMed Central PMCID: PMCPMC4430127. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Zhao Z, Yang Y, Zeng Y, et al. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip. 2016February07;16(3):489–496. PubMed PMID: 26645590; PubMed Central PMCID: PMCPMC4729647. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Rt D, Kim J, Sc J, et al. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip. 2012December21;12(24):5202–5210. PubMed PMID: 23111789; eng. [DOI] [PubMed] [Google Scholar]
- [88].Cho S, Jo W, Heo Y, et al. Isolation of extracellular vesicle from blood plasma using electrophoretic migration through porous membrane. Sensors and Actuators B: Chemical. 2016;233:289–297. [Google Scholar]
- [89].Liang LG, Kong MQ, Zhou S, et al. An integrated double-filtration microfluidic device for isolation, enrichment and quantification of urinary extracellular vesicles for detection of bladder cancer. Sci Rep. 2017April24;7:46224 PubMed PMID: 28436447; PubMed Central PMCID: PMCPMC5402302. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Woo HK, Sunkara V, Park J, et al. Exodisc for rapid, size-selective, and efficient isolation and analysis of nanoscale extracellular vesicles from biological samples. ACS Nano. 2017February28;11(2):1360–1370. PubMed PMID: 28068467; eng. [DOI] [PubMed] [Google Scholar]
- [91].Liu F, Vermesh O, Mani V, et al. The exosome total isolation chip. ACS Nano. 2017November01PubMed PMID: 29090896; eng DOI: 10.1021/acsnano.7b04878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Wang Z, Wu HJ, Fine D, et al. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab Chip. 2013August07;13(15):2879–2882. PubMed PMID: 23743667; PubMed Central PMCID: PMCPMC3740541. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Huang LR, Cox EC, Austin RH, et al. Continuous particle separation through deterministic lateral displacement. Science (New York, NY). 2004May14;304(5673):987–990. PubMed PMID: 15143275; eng. [DOI] [PubMed] [Google Scholar]
- [94].Zeming KK, Thakor NV, Zhang Y, et al. Real-time modulated nanoparticle separation with an ultra-large dynamic range. Lab Chip. 2016January07;16(1):75–85. PubMed PMID: 26575003; eng. [DOI] [PubMed] [Google Scholar]
- [95].Wunsch BH, Smith JT, Gifford SM, et al. Nanoscale lateral displacement arrays for the separation of exosomes and colloids down to 20 nm. Nat Nanotechnol. 2016November;11(11):936–940. PubMed PMID: 27479757; eng. [DOI] [PubMed] [Google Scholar]
- [96].Liu C, Guo J, Tian F, et al. Field-free isolation of exosomes from extracellular vesicles by microfluidic viscoelastic flows. ACS Nano. 2017July25;11(7):6968–6976. PubMed PMID: 28679045; eng. [DOI] [PubMed] [Google Scholar]
- [97].Bruus H. Acoustofluidics 7: the acoustic radiation force on small particles. Lab Chip. 2012March21;12(6):1014–1021. PubMed PMID: 22349937; eng. [DOI] [PubMed] [Google Scholar]
- [98].Ding X, Li P, Lin SC, et al. Surface acoustic wave microfluidics. Lab Chip. 2013September21;13(18):3626–3649. PubMed PMID: 23900527; PubMed Central PMCID: PMCPMC3992948. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Destgeer G, Sung HJ. Recent advances in microfluidic actuation and micro-object manipulation via surface acoustic waves. Lab Chip. 2015July07;15(13):2722–2738. . PubMed PMID: 26016538; eng. [DOI] [PubMed] [Google Scholar]
- [100].Lee K, Shao H, Weissleder R, et al. Acoustic purification of extracellular microvesicles. ACS Nano. 2015March24;9(3):2321–2327. PubMed PMID: 25672598; PubMed Central PMCID: PMCPMC4373978. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Wu M, Ouyang Y, Wang Z, et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proceedings of the National Academy of Sciences of the USA 2017September18PubMed PMID: 28923936; eng DOI: 10.1073/pnas.1709210114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Fang S, Tian H, Li X, et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PloS One. 2017;12(4):e0175050 PubMed PMID: 28369094; PubMed Central PMCID: PMCPMC5378374. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Friedrich R, Block S, Alizadehheidari M, et al. A nano flow cytometer for single lipid vesicle analysis. Lab Chip. 2017February28;17(5):830–841. PubMed PMID: 28128381; eng. [DOI] [PubMed] [Google Scholar]
- [104].Im H, Shao H, Park YI, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol. 2014May;32(5):490–495. PubMed PMID: 24752081; PubMed Central PMCID: PMCPMC4356947. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Sina AA, Vaidyanathan R, Dey S, et al. Real time and label free profiling of clinically relevant exosomes. Sci Rep. 2016July28;6:30460 PubMed PMID: 27464736; PubMed Central PMCID: PMCPMC4964344. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Shao H, Chung J, Balaj L, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012December;18(12):1835–1840. PubMed PMID: 23142818; PubMed Central PMCID: PMCPMC3518564. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Akagi T, Kato K, Kobayashi M, et al. On-chip immunoelectrophoresis of extracellular vesicles released from human breast cancer cells. PloS One. 2015;10(4):e0123603 PubMed PMID: 25928805; PubMed Central PMCID: PMCPMC4415775. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Fraikin JL, Teesalu T, McKenney CM, et al. A high-throughput label-free nanoparticle analyser. Nat Nanotechnol. 2011May;6(5):308–313. PubMed PMID: 21378975; eng. [DOI] [PubMed] [Google Scholar]
- [109].Zhou Q, Rahimian A, Son K, et al. Development of an aptasensor for electrochemical detection of exosomes. Methods (San Diego, CA). 2016March15;97:88–93. PubMed PMID: 26500145; eng. [DOI] [PubMed] [Google Scholar]
- [110].He M, Crow J, Roth M, et al. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip. 2014October07;14(19):3773–3780. PubMed PMID: 25099143; PubMed Central PMCID: PMCPMC4161194. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Richards KE, Go DB, Hill R. Surface acoustic wave lysis and ion-exchange membrane quantification of exosomal microRNA. Methods Mol Biol (Clifton, NJ). 2017;1580:59–70. PubMed PMID: 28439826; eng. [DOI] [PubMed] [Google Scholar]
- [112].Ko J, Bhagwat N, Yee SS, et al. Combining machine learning and nanofluidic technology to diagnose pancreatic cancer using exosomes. ACS Nano. 2017October17PubMed PMID: 29019651; eng DOI: 10.1021/acsnano.7b05503. [DOI] [PubMed] [Google Scholar]
- [113].Inan H, Poyraz M, Inci F, et al. Photonic crystals: emerging biosensors and their promise for point-of-care applications. Chem Soc Rev. 2017January23;46(2):366–388. PubMed PMID: 27841420; PubMed Central PMCID: PMCPMC5529146. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Lifson MA, Ozen MO, Inci F, et al. Advances in biosensing strategies for HIV-1 detection, diagnosis, and therapeutic monitoring. Adv Drug Deliv Rev. 2016August01;103:90–104. PubMed PMID: 27262924; PubMed Central PMCID: PMCPMC4943868. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Yager P, Domingo GJ, Gerdes J. Point-of-care diagnostics for global health. Annu Rev Biomed Eng. 2008;10:107–144. PubMed PMID: 18358075; eng. [DOI] [PubMed] [Google Scholar]
- [116].Marczak S, Richards K, Ramshani Z, et al. Simultaneous isolation and preconcentration of exosomes by ion concentration polarization. Electrophoresis. 2018February27PubMed PMID: 29484678; eng DOI: 10.1002/elps.201700491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Gallo A, Tandon M, Alevizos I, et al. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PloS One. 2012;7(3):e30679 PubMed PMID: 22427800; PubMed Central PMCID: PMCPMC3302865. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Whiteside TL. Tumor-derived exosomes and their role in cancer progression. Adv Clin Chem. 2016;74:103–141. PubMed PMID: 27117662; PubMed Central PMCID: PMCPMC5382933. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Boyiadzis M, Whiteside TL. Plasma-derived exosomes in acute myeloid leukemia for detection of minimal residual disease: are we ready? Expert Rev Mol Diagn. 2016June;16(6):623–629. PubMed PMID: 27043038; PubMed Central PMCID: PMCPMC5400097. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Whiteside TL. The potential of tumor-derived exosomes for noninvasive cancer monitoring. PubMed PMID: 26289602; PubMed Central PMCID: PMCPMC4813325. engExpert Rev Mol Diagn. 2015;1510:1293–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008July;110(1):13–21. . PubMed PMID: 18589210; eng. [DOI] [PubMed] [Google Scholar]
- [122].Zhao W, Cheng R, Miller JR, et al. Label-free microfluidic manipulation of particles and cells in magnetic liquids. Adv Funct Mater. 2016June14;26(22):3916–3932. PubMed PMID: 28663720; PubMed Central PMCID: PMCPMC5487005. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Tao SC, Guo SC, Zhang CQ. Modularized extracellular vesicles: the dawn of prospective personalized and precision medicine. Adv Sci (Weinheim, Baden-Wurttemberg, Germany). 2018February;5(2):1700449 PubMed PMID: 29619297; PubMed Central PMCID: PMCPMC5827100. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Jo W, Jeong D, Kim J, et al. Microfluidic fabrication of cell-derived nanovesicles as endogenous RNA carriers. Lab Chip. 2014April7;14(7):1261–1269. PubMed PMID: 24493004; eng. [DOI] [PubMed] [Google Scholar]
- [125].Lunavat TR, Jang SC, Nilsson L, et al. RNAi delivery by exosome-mimetic nanovesicles – implications for targeting c-Myc in cancer. Biomaterials. 2016September;102:231–238. PubMed PMID: 27344366; eng. [DOI] [PubMed] [Google Scholar]
- [126].Kim OY, Lee J, Gho YS. Extracellular vesicle mimetics: novel alternatives to extracellular vesicle-based theranostics, drug delivery, and vaccines. Semin Cell Dev Biol. 2017July;67:74–82. PubMed PMID: 27916566; eng. [DOI] [PubMed] [Google Scholar]
- [127].Kuo JS, Chiu DT. Controlling mass transport in microfluidic devices. Annual Review of Analytical Chemistry (Palo Alto, CA). 2011;4:275–296. PubMed PMID: 21456968; PubMed Central PMCID: PMCPMC5724977. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]


