Abstract
Background:
Eosinophilic esophagitis (EoE) is a chronic, inflammatory disease of the esophagus which currently requires repeated endoscopic biopsies for diagnosis and monitoring as no reliable non-invasive markers have been identified.
Objective:
To identify promising minimally-invasive EoE biomarkers and remaining gaps in biomarker validation.
Methods:
We performed a systematic review of EMBASE, Ovid Medline, PubMed, and Web of Science from inception to June 6, 2017. Studies were included if subjects met the 2007 consensus criteria for EoE diagnosis, a minimally-invasive biomarker was assessed, and the study included at least 1 control for comparison.
Results:
The search identified 2094 studies, with 234 reviewed at full text level, and 49 included in the analysis (20 adult, 19 pediatric, 7 pediatric and adult, and 3 not stated). The majority (26 of 49) were published after 2014. Thirty-five studies included normal controls, 9 analyzed atopic controls, and 29 compared samples from subjects with active and inactive EoE. Minimally-invasive biomarkers were obtained from peripheral blood (n=41 studies), sponge/string samples (3), oral/throat swab secretions (2), breath condensate (2), stool (2), and urine (2). The most commonly reported biomarkers were peripheral blood eosinophils (16), blood and string eosinophil granule proteins (14), and eosinophil surface or intracellular markers (12). EoE biomarkers distinguished active EoE from normal controls in 23 studies, atopic controls in 2 studies, and inactive EoE controls in 20 studies.
Conclusion:
Several promising minimally-invasive biomarkers for EoE have emerged; however, few are able to differentiate EoE from other atopic diseases.
Keywords: Eosinophilic esophagitis, biomarker, diagnosis, monitor, string test, esophageal brushings, noninvasive, minimally invasive
INTRODUCTION
Eosinophilic esophagitis (EoE) is a clinicopathologic diagnosis characterized by symptoms of esophageal dysfunction and eosinophilia of the esophageal epithelium. For many years, esophageal eosinophilia was considered a manifestation of gastroesophageal reflux disease (GERD)1; however, a retrospective case series by Attwood et al2 in 1993 described 12 adults with dysphagia, dense intraepithelial esophageal eosinophils in the absence of reflux. One year later, Straumann et al described 10 patients with acute recurrent dysphagia with discrete endoscopic findings and high concentrations of intraepithelial eosinophils, responsive to treatment with systemic steroid and antihistamine; further defining a new disease entity termed Idiopathic Eosinophilic Esophagitis.3 Finally, in 1995 Kelly et al4 demonstrated disease remission with institution of an elemental diet, suggesting that EoE is a food-driven disorder. Despite these descriptions, formalized diagnostic criteria for IEE were lacking until 2007, when the first consensus guidelines were developed for evaluation and management of the condition now termed eosinophilic esophagitis (EoE).5
The clinical presentation of EoE varies by age. Children suffer mainly from feeding difficulty and failure to thrive, with symptoms including vomiting and abdominal pain. In older children, complaints may include chest pain and dysphagia. This is in contrast to adolescents and adults, who present with symptoms of dysphagia, chest pain, and food impaction.6 The 2007 consensus definition of EoE requires at least one symptom of esophageal dysfunction along with at least 15 eosinophils per high-power field on esophageal biopsy. Other causes of esophageal eosinophilia – in particular GERD – must be excluded before the diagnosis can be established.5 Newer guidelines7 have been published further refining the diagnosis of EoE in which GERD and EoE may coexist and interact. We have chosen to use the 2007 consensus definition of EoE to ensure that all pertinent papers prior to the publication of these updated guidelines would be included in our review. Current treatment modalities are elimination diets (empiric, skin test-directed, or elemental), swallowed topical corticosteroids, and proton pump inhibitors. For patients who develop esophageal narrowing, esophageal dilation is used as treatment to alleviate symptoms. Controversy remains regarding the diagnostic and therapeutic long-term management given no evidence this is a pre-malignant condition, and few studies have investigated long-term outcomes associated with diet or topical corticosteroid therapy after symptom and histologic remission is achieved. Nevertheless, in the vast majority of patients, EoE is a chronic disease process, and if therapy is discontinued, inflammation recurs which can affect quality of life and result in complications (e.g. stricture formation).6,8–10 Current expert consensus recommends maintenance therapy for patients with evidence of chronic esophageal remodeling, a history of food impactions or severe symptoms, or rapid recurrence of symptoms while not on therapy.6
One of the challenges with EoE is discordance between symptoms and histopathologic features, making diagnosis and monitoring response to therapy challenging. For example, some patients with minimal symptoms may have significantly elevated eos/hpf on esophageal biopsy indicating ongoing inflammation and active disease. The current recommendations for initial diagnosis and disease monitoring involve serial endoscopic evaluations with biopsies. This invasive approach poses risk to patients especially in children younger than 3 years. In April 2017, the U.S. Food and Drug Administration (FDA) issued a new warning of possible negative effects on brain development in children below 3 years undergoing recurrent or lengthy procedure requiring sedation or general anesthesia.11 The negative effects on brain development associated with short duration anesthesia required for one upper endoscopy is unknown; however, patients with EoE typically undergo multiple procedures. In addition to the risks posed to patients, there are also significant health care costs associated with these procedures.12–14 Identifying a reliable, non-invasive or minimally invasive biomarker for diagnosing and monitoring could help reduce the need for risky, invasive procedures, potentially increasing safety and reducing health care expenditures.13
Several non-invasive biomarkers have been studied in patients with EoE but none have yet been incorporated into treatment guidelines or routine clinical practice. Recent efforts to identify EoE biomarkers have rapidly expanded;13,15 and while there are some published reviews on this subject, these publications do not employ systematic review methods in order to ensure all relevant studies are identified. Therefore, we undertook a systematic review to: (1) identify study design strengths and weaknesses that inform design of future EoE biomarker studies and (2) identify the most promising biomarkers so that attempts at reproduction and validation in other populations may propel the field forward.
METHODS
Eligibility criteria and literature search
This systematic review contains the elements of the 27-item checklist put forth in the Preferred Reporting Items for Systematic Reviews and Meta-analysis statement (PRISMA) statement.16 Articles were included that diagnosed EoE based on 2007 consensus definition which requires at least one symptom of esophageal dysfunction and at least 15 eosinophils/hpf on esophageal biopsy.5 The 2007 consensus definition was chosen so that articles prior to 2011 would not be excluded if they did not fulfill all of the 2011 EoE diagnosis criteria. In addition, articles were also required to study a non-invasive or minimally invasive biomarker. A non-invasive or minimally invasive test was defined as one that can be collected without an endoscope. Human case reports, case series, cross-sectional and cohort studies, and clinical trials were included. All non-human studies and studies not containing new clinical information were excluded. We also excluded studies investigating allergy testing (serum-specific IgE, prick skin testing, atopy patch testing), and radiologic modalities based on consensus of the authors.
We performed a systematic review of English-language and non-English-language articles using MEDLINE, PubMed, Web of Science and Embase (inception to June 6, 2017) with the assistance of an experienced medical librarian (LAM). The following search terms were used: eosinophil*, hypereosinophil*, serologic marker, peripheral blood, marker, biomarker, Cytosponge, string test, non-invasive, minimally invasive, semi-invasive, brush, and assay. The search strategy used for MEDLINE is detailed in Table 1. A similar search strategy was adapted by an experienced medical librarian for the other electronic databases. To identify additional relevant articles, bibliographies of included articles were searched. Published proceedings from 2013–2016 American College of Gastroenterology (ACG), Digestive Diseases Week (DDW), American Academy of Allergy, Asthma and Immunology (AAAAI), and United European Gastroenterology (UEG) annual meetings were searched online using the term eosinophilic esophagitis in portable document format (PDF). When a PDF was not available, meeting programs were searched. Once all studies had been reviewed and appropriate articles included, the bibliographies from all included articles were compared to our original search and duplicates removed. Content experts among the authors (BLW, MG, AS, ESD) were also queried regarding their knowledge of unpublished data or studies omitted from the list of eligible studies.
Table 1:
Search Strategy
| Search Terms | |
|---|---|
| 1 | (eosinophil*or hypereosinophil*) or exp eosinophil/or exp eosinophilia/ |
| 2 | 1 and (exp esophagitis/ or (esophag* or oesophag*) |
| 3 | serologic *marker* |
| 4 | peripheral blood |
| 5 | marker* |
| 6 | 3 or 4 or 5 |
| 7 | exp biomarkers/or (biomarker* or bio-marker* or cytosponge* or enterotest* or brush*or assay or (((sponge or string)adj2 (techn*1 or capsule*or sampl*)) or (((gel or gelatin)adj2 capsule*))) |
| 8 | 6 or 7 |
| 9 | (noninvasive* or non-invasive* or non-endoscop* or nonendoscop* or ((minim* or less)adj3 invasive*)) |
| 10 | (semi-invasive or semi invasive) [title, abstract, original title, name of substance word, subject heading word, protocol supplementary concept word, rare disease supplementary concept word, unique identifier] |
| 11 | 9 or 10 |
| 12 | 2 and (8 or 11) |
| 13 | remove duplicates from 12 |
Study selection
Two abstract reviewers worked independently to consider whether each of the abstracts identified would meet eligibility criteria. The reviewers were not blinded to the author, institution or journal of publication. If it was unclear whether the article met inclusion criteria based on the abstract or if the reviewers disagreed on whether to include or exclude the study, a full text review was performed. In order to include all possible relevant studies, we did not specifically exclude other causes of esophageal eosinophilia such as GERD when reviewing at the abstract level. Disagreement at full text inclusion levels was resolved by consensus.
Consensus was obtained upon discussion and agreement among 3 authors. Once all duplicates had been removed from the bibliography search of the included articles, the titles were reviewed by a single reviewer.
Data Collection
One author (MAR) independently extracted relevant data into a spreadsheet and these data were rechecked by a second reviewer (BTH). Any discrepancies were resolved through author consensus. Extracted data from each study included: author, year, age of participants, number of participants, information on study controls (normal, atopic, disease activity), the non-invasive collection method (e.g. blood, urine, sponge), a complete list of all biomarkers studied, and a list of all biomarkers where statistical significance was found when compared to normal controls, atopic controls, and disease responsiveness to treatment measures. Authors were contacted if data were missing. The method of author contacts were: (1) e-mail briefly explaining the study and asking specifically for data relevant to the review and (2) a 2nd email 1 week later if no response with a scaled-back request to share the most important missing data.
Assessment of methodological quality
To assess risk of bias, 2 independent reviewers followed instructions from the Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy.17 For each study, we used the recommended quality items derived from the QUADAS-2 tool by determining the spectrum of patients represented, the likelihood the reference standard was to identify the target condition, time between the index and reference tests, the allocation of reference standard on the study population with regard to whole group and independence of index test, whether interpretation of tests was independent, clinical data available when tests were interpreted and whether this would be available in practice.17,18
RESULTS:
Our initial search of electronic databases yielded 1454 articles after removal of duplicates. By searching bibliographies, reviewing meeting programs, and contacting content experts an additional 640 studies were identified after removal of duplicates. Of these 2094 articles, 234 met criteria for full-text review. Forty-nine studies19–67 were included after full-text review. The bibliographies of these 49 studies were searched and there were 614 non-duplicate references derived which were reviewed by a single reviewer at title level. Seven articles were identified for review at the abstract level. Four articles did not include minimally invasive biomarkers, 1 article was a duplicate that had already been included, 1 article was an abstract for an article we had already included, and 1 article was an abstract only that described a minimally invasive biomarker. We were unable to identify an associated full-length manuscript for these abstract data, and therefore elected to contact the authors to see if they could share their data (author did not respond). Figure 1 describes the flow of information through the different phases of the systematic review using the PRIMSA flow-sheet template.
Figure 1.
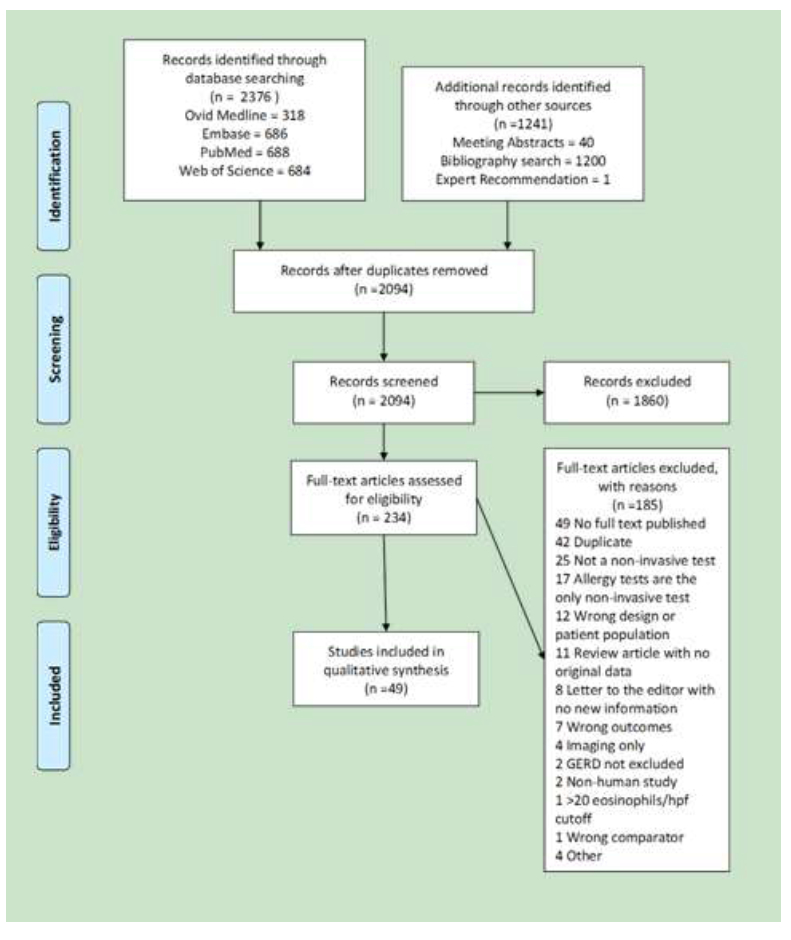
Legend: The PRISMA diagram details the search and selection process14
Table 2 details the study characteristics and biomarkers studied for each of the included articles. The search identified 20 adult, 19 pediatric, and 7 combined age studies, in addition to 3 studies which did not state the age of the participants. The majority of the publications were published after 2014 (26 of 49). Biomarkers were assessed from peripheral blood (41 studies), Cytosponge/esophageal string test (3), breath sampling (2), oropharyngeal swabs (2), stool (2), and urine (2). With regards to study controls, 35 studies included a normal control group, 9 included atopic controls, and 29 studies compared active to a “treated EoE” (trEoE) control group. There were 23 studies that noted significant differences in a variety of markers in subjects with EoE versus normal controls. Only increased urinary 3-bromotyrosine (3-BT)26 and decreased total IgE61 (EoE vs allergic rhinitis) demonstrated significant difference when compared to atopic controls.
Table 2:
Characteristics of All Included Studies
| Author | Year | Age group | Number of EoE patients | Number of Controls | Normal Control | Atopic Control | Disease Activity Control | Biomarker(s) Studied | Biomarker source |
|---|---|---|---|---|---|---|---|---|---|
| Benitez | 2015 | Pediatric | 33 | 35 | Yes | No | Yes | 16S RNA bacterial sequencing | Oral swab |
| Blanchard | 2011 | Not stated | 226 | 14 GERD, 14 Normal | Yes | No | Yes | Cytokine array | Blood |
| Botan | 2017 | Both | 31 | 10 | Yes | No | No | “activated” appearance of eosinophils | Blood |
| Bullock | 2007 | Pediatric | 12 | 8 Normal, 5 Atopic | Yes | Yes | Yes | AEC, CCR3 on eosinophils, CD4 expression of IL-5 | Blood |
| Clayton | 2014 | Adults (Except for 3 subjects age 15–17) | 15 | 41 Normal | Yes | No | No | IgG4 (total and food-specific) | Blood |
| Colson | 2014 | Pediatric | 59 | 0 | No | No | Yes | AEC | Blood |
| Conus | 2009 | Not stated | 11 (5mepolizumab, 6placebo) | 0 | No | No | Yes | AEC, IL-5Rα, CCR3 | Blood |
| Cunnion | 2016 | Pediatric | 27 | 24 Normal, 24 Atopic | Yes | Yes | No | 3-BT | Urine |
| Dellon | 2015 | Adult | 61 | 87 | Yes | No | Yes | IL-4, IL-5, IL-6, IL-9, IL-13, TGF-α, TGF-β, TNF-α, Eotaxin 1/2/3, TSLP, MBP, EDN | Blood |
| Dellon | 2016 | Adult | 61 | 87 | Yes | No | Yes | Periostin | Blood |
| Domenech Witek | 2017 | Adult | 19 | 0 | No | No | Yes | tIgE, AEC, ECP | Blood |
| Fuentebella | 2010 | Pediatric | 33 | 7 GERD, 8 Normal | Yes | No | No | Treg: CD4+CD25hi CD127lo | Blood |
| Fujiwara | 2002 | Adult | 1 | 0 | No | No | Yes | AEC, Eotaxin (total and free), ECP | Blood |
| Furuta | 2013 | Pediatric | 14 Active,8 Disease remission | 4 GERD, 15 Normal | Yes | No | Yes | MBP, EDN, ECP, EPX, CLC/Gal-10 | Esophageal String |
| Huang | 2010 | Pediatric | 35 Newly diagnosed 9 Treated | 8 GERD, 5 Ulcerative colitis, 5 Crohn’s disease, 8 Normal | Yes | No | Yes | 35 chemokine/cytokines including: bFGF or FGF-2; eotaxin 1/2/3; IL-1α; IL-1β; IL-1RA; IL-2; IL-4; IL-5; IL-6; IL-7; IL-8; IL-10; IL-12-p40; IL-12-p70; IL-13; IL-15; IL-17; IL-17F; ENA78; GCSF; GM-CSF, GRO-α; IFN-γ; IP10; leptin; MCP-3; MIG; MIP-1α; MIP-1β;NGF;PDGF-BB; RANTES; TGF-β;TNF-α; TNF-β, and VEGF | Blood |
| Johnsson | 2011 | Adult | 12 | 8 Ulcerative colitis, 10 Airway allergy 10 Normal | Yes | Yes | No | AEC, CD23, CD54, CRTH2, CD11c, CCR3, CD44, CD11b, CD18, CD58, CCL5(RANTES), CCL11 (eotaxin-1), CCL26, IL-2, IL-3, IL-4, IL-5, GM-CSF | Blood |
| Jyonouchi | 2013 | Pediatric | 10 Active 10 Disease Remission | 16 Normal | Yes | No | Yes | iNKTs | Blood |
| Katzka | 2015 | Adult | 13 Active 7 Disease Remission | 0 | No | No | Yes | Eos/hpf, EDN | Cytosponge |
| Kinoshita | 2012 | Adult | 18 | 18 EGID 30 Normal | Yes | No | No | IL-5, IL-13, IL-15, Eotaxin-3, TSLP | Blood |
| Knipping | 2014 | Pediatric | 91 | 45 | Yes | No | No | TSLP, TARC, KFLC, L-FLC | Blood |
| Konikoff | 2006 | Pediatric | 16 Active 16 Disease remission 1 Intermediate | 9 Normal5 EGID | Yes | Yes | Yes | AEC, IL-5, Eotaxins 1/2/3 EDN (blood/stool) |
Blood, Stool |
| Krupp | 2016 | Pediatric | 33 | 37 | Yes | No | No | IL-5, IL-9, Eotaxin, EGF, FGF-2 | Blood |
| Lanz | 2012 | Pediatric | 18 | 23 Gastritis 14 Normal | Yes | No | No | eNO | Breath |
| Leung | 2012 | Both | 14 | 0 | No | No | Yes | eNO | Breath |
| Lexmond | 2013 | Pediatric | 30 | 20 Reflux, 20 Normal | Yes | No | No | Urine LTE4, Serum LTC4 | Urine, Blood |
| Lingblom | 2014 | Adult | 21 | 15 | Yes | No | Yes | CD18, CD44, CD40, CCR3, CD23, CD54, FPR, CRTH2 | Blood |
| Lingblom | 2017 | Both | 53 | 51 | Yes | No | No | CD23, CD44, CD54, CRTH2, FoxP3, Galectin-10 | Blood |
| Lucendo | 2013 | Adult | 17 | 0 | No | No | Yes | AEC, tIgE, ECP | Blood |
| Min | 2016 | Both | 46 | 53 | Yes | No | Yes | AEC, Eotaxin-3, EDN, ECP, IL-5 | Blood |
| Morris | 2016 | Pediatric | 17 Active, 14 Disease Remission | 10 Atopic | No | Yes | Yes | AEC, Eosinophil progenitor | Blood |
| Nguyen | 2011 | Pediatric | 35 Newly Diagnosed EoE off therapy 7 Known EoE on therapy | 35 | Yes | Yes | Yes | PBMC transcript analysis of STAT1, STAT6, and CD66b, Surface CD66b on peripheral eosinophils | Blood |
| Patel | 2010 | Pediatric | 10 | 11 GERD,10 Normal | Yes | No | No | HLA-DR | Blood |
| Paterson | 2016 | Adult | 6 | 166 esophagitis 10 Candida 638 Normal | Yes | No | No | Eos/hpf | Cytosponge |
| Paz Zafra | 2012 | Both | 25 | 17 | Yes | No | No | AEC, tIgE, C5a, CD40 ligand, GCSF, GM-CSF, CXCL1, CCL1, CD54, IFN-c, IL-1a, IL-1b, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12 p70, IL-13, IL-16, IL-17, IL-17E, IL-23, IL-27, IL-32a, CXCL10, CXCL11, CCL2, MIF, CCL3, CCL4, Ser- pin E1, RANTES,CXCL12, and TNFa | Blood |
| Philpott | 2015 | Adult | 85 | 193 | Yes | No | No | AEC | Blood |
| Rawson | 2016 | Pediatric | 27 | 11 | Yes | No | Yes | TGF-β, PAI-1, PF4 | Blood |
| Rayapudi | 2014 | Not stated | 7 | 6 | Yes | No | No | iNKTs | Blood |
| Rodriguez-Sanchez | 2013 | Both | 22 Responders, 8 Nonresponders | 0 | No | No | Yes | AEC, ECP, tIgE | Blood |
| Saffari | 2017 | Adult | 8 | 21 | Yes | No | Yes | EPO activity | Throat swab |
| Sawant | 2015 | Pediatric | 7 | 8 Asthma8 Normal | Yes | Yes (asthma) | No | miR-21 | Blood |
| Schlag | 2014 | Adult | 15 | 0 | No | No | Yes | ECP, MC tryptase | Blood |
| Schlag | 2015 | Adult | 69 | 39 Atopic controls (with EoE) | No | Yes | Yes | AEC, ECP, CCL-17, CCL-18, CCL-26, MC tryptase | Blood |
| Soylu | 2016 | Adult | 7 (also with allergic rhinitis) | 60 Allergic rhinitis | No | Yes | No | tIgE | Blood |
| Straumann | 2005 | Adult | 8 | 4 Dyspepsia, 6 Normal | Yes | No | No | AEC; CD25, IL-4, IL-5, IL-13, IL-10 expression on eosinophils | Blood |
| Straumann | 2010 | Adult | 11 (5mepolizumab, 6placebo) | 0 | No | No | Yes | ECP, EDN, Eotaxin, and TNF-α, IL-5Rα on eosinophils | Blood |
| Subbarao | 2011 | Pediatric | 60 | 20 | Yes | No | Yes | IL-5 (Blood); EDN (Blood/stool) | Blood, stool |
| Upparahal li Venkatesh aiah | 2016 | Both | 2 | 0 | No | No | Yes | CD274 | Blood |
| von Arnim | 2011 | Adult | 23 | 20 GERD | Yes | No | No | AEC, elevated or normal tIgE | Blood |
| Wright | 2016 | Adult | 20 | 10 | Yes | No | Yes | sIgG4 (total and food-specific) | Blood |
Abbreviations Table 2:3-bromotyrosine (3-BT), absolute eosinophil count (AEC), chemokine receptor type 3 (CCR3), chemokine ligand 1 (CCL1), chemokine ligand 3 (CCL2), chemokine ligand 3 (CCL3), chemokine ligand 4 (CCL4), chemokine ligand 17 (CCL17), chemokine ligand 18 (CCL18), chemokine ligand 26 (CCL26), CXC chemokine ligand 1 (CXCL1), CXC chemokine ligand 10 (CXCL10), CXC chemokine ligand 11 (CXCL11), CXC chemokine ligand 12 (CXCL12), fibroblast growth factor basic (bFGF or FGF-2), IL-1 receptor antagonist (IL-1RA), IL-5 receptor alpha (IL-5Rα), eosinophil cationic protein (ECP),eosinophil derived neurotoxin (EDN), eosinophil peroxidase (EPX or EPO), eosinophil per high power field (eos/hpf), epidermal growth factor (EGF), epithelial cell-derived neutrophil-activating protein-78 (ENA78), exhaled nitric oxide (FeNO), formyl peptide receptor (FPR), granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), growth-related oncogene-alpha (GRO- α), interferon-gamma (IFN-g), invariant natural killer T cells (iNKTs), kappa free light chain (k-FLC), lambda free light chain (L-FLC), leukotriene E4 (LTE4), leukotriene C4 (LTC4), macrophage inflammatory protein 1 alpha (MIP-1a); macrophage inflammatory protein 1 beta (MIP-1b), major basic protein (MBP), MC (mast cell), nerve growth factor (NGF), peripheral blood mononuclear cell (PBMC), plasminogen activator inhibitor 1 (PAI-1), platelet derived growth factor-BB (PDGFBB), platelet factor 4 (PF4), regulated upon activation normal T cell expressed and secreted (RANTES), ribonucleic acid (RNA), thymic stromal lymphopoietin (TSLP), thymus-and activation-regulated chemokine (TARC), transforming growth factor-α (TGF-α), transforming growth factor- β (TGF-β), tumor necrosis factor-α , vascular endothelial growth factor (VEGF)
The most common EoE biomarker assessed by 16 studies was the peripheral blood absolute eosinophil count (AEC). Among these studies, 7 reported a significant difference in AEC between subjects with active versus trEoE (Table 3). Only 5 studies had an atopic control group for comparison (no significant differences noted). Four studies were noted that observed a change in AEC compared to normal controls.
Table 3:
Studies Assessing Absolute Eosinophil Count (AEC)
| Author | Year | Age group | Normal Control | Atopic Control | Disease Activity Control | Significant vs normal control | Significant vs atopic controls | Significant for responsiveness |
|---|---|---|---|---|---|---|---|---|
| Bullock | 2007 | Pediatric | Yes | Yes | Yes | Yes | No difference | Yes |
| Colson | 2014 | Pediatric | No | No | Yes | Not done | Not done | Yes |
| Conus | 2009 | Not stated |
No | No | Yes | Not done | Not done | Yes |
| Domenech Witek | 2017 | Adult | No | No | Yes | Not done | Not done | No difference |
| Fujiwara | 2002 | Adult | No | No | Yes | Not done | Not done | Yes (n=1, case report) |
| Johnsson | 2011 | Adult | Yes | Yes | No | No difference | Possible difference but compared across 4 groups | Not done |
| Konikoff | 2006 | Pediatric | Yes | Yes | Yes | Yes | No difference | Yes |
| Lucendo | 2013 | Adult | No | No | Yes | Not done | Not done | No difference |
| Min | 2016 | Both | Yes | No | Yes | Yes | Not done | Yes |
| Morris | 2016 | Pediatric | No | Yes | Yes | Not done | No difference | No difference |
| Paz Zafra | 2012 | Both | Yes | No | No | Yes | Not done | Not done |
| Philpott | 2015 | Adults | Yes | No | No | No | Not done | Not done |
| Rodriguez-Sanchez | 2013 | Both | No | No | Yes | Not done | Not done | No difference |
| Schlag | 2015 | Adult | No | Yes | Yes | Not done | No difference | Yes |
| Straumann | 2005 | Adult | Yes | No | No | Not done | Not done | Not done |
| von Arnim | 2011 | Adult | Yes | No | No | No actual | Not done | Not done |
| values (AEC elevated (y/n)) |
Table 4 summarizes the findings for 14 studies reporting granule proteins as biomarkers, including eosinophilic cationic protein (ECP) (9 studies), eosinophil-derived neurotoxin (EDN) (7 studies) eosinophil peroxidase (EPX) (2 studies), and major basic protein (MBP) (2 studies). Only two of these studies (one assessing EDN39 and another assessing ECP60) had an atopic control group for comparison, and neither study noted a significant difference between groups. Compared to normal controls, EDN was significantly increased in four studies, ECP in 2, and MBP in 1. Four studies identified significantly different ECP levels in samples from patients with active vs treated EoE. One additional study was a case report (n=1) which noted normalization of the ECP level following EoE treatment.31 There were 12 studies which analyzed a variety of eosinophil surface/intracellular markers. Four of these studies compared EoE to an atopic control group, but none identified any significant difference between groups (Table 5).
Table 4:
Studies Assessing Eosinophil Granular Proteins
| Author | Year | Age group | Normal Control | Atopic Control | Disease Activity Control | Granule Protein(s) studied | Marker(s) significant vs normal controls | Marker(s) significant vs atopic controls | Marker(s) significant for responsiveness |
|---|---|---|---|---|---|---|---|---|---|
| Dellon | 2015 | Adult | Yes | No | Yes | MBP, EDN | No difference | Not done | No difference |
| Domenech Witek | 2017 | Adult | No | No | Yes | ECP | Not done | Not done | ECP |
| Fujiwara | 2002 | Adult | No | No | Yes | ECP | Not done | Not Done | Yes (n=1, case report) |
| Furuta | 2013 | Pediatric | Yes | No | Yes | MBP, EDN, ECP, EPX (string) | MBP, EDN, ECP | Not done | MBP, EPX |
| Katzka | 2015 | Adult | No | No | Yes | EDN | Not done | Not done | No difference |
| Konikoff | 2006 | Pediatric | Yes | Yes | Yes | EDN (blood/ | EDN | No | No difference |
| stool) | (Blood) | difference | |||||||
| Lucendo | 2013 | Adult | No | No | Yes | ECP | Not done | Not done | No difference |
| Min | 2016 | Both | Yes | No | Yes | EDN, ECP | EDN, ECP | Not done | No difference |
| Rodriguez-Sanchez | 2013 | Both | No | No | Yes | ECP | Not done | Not done | No difference |
| Saffari | 2017 | Adult | Yes | No | Yes | EPX activity | No difference | Not done | No difference |
| Schlag | 2014 | Adult | No | No | Yes | ECP | Not done | Not done | ECP |
| Schlag | 2015 | Adult | No | Yes | Yes | ECP | Not done | No | ECP |
| difference | |||||||||
| Straumann | 2010 | Adult | No | No | Yes | ECP, EDN | Not done | Not done | ECP, EDN |
| Subbarao | 2011 | Pediatric | Yes | No | Yes | EDN (blood/stool) | EDN (Blood) | Not done | EDN (Blood) |
Table 5:
Studies Assessing Eosinophil Surface or Intracellular Markers
| Author | Year | Age Group | Normal Control | Atopic Control | Disease Activity Control | Cell Surface/Intracellul ar Marker Studied | Marker(s) significant vs normal controls | Marker(s) significan t vs atopic controls | Marker(s) significant for responsiveness |
|---|---|---|---|---|---|---|---|---|---|
| Bullock | 2007 | Pediatric | Yes | Yes | Yes | AEC, CCR3 on eosinophils, CD4 expression of IL-5 | CCR3 on eosinophil s, CD4 expression IL-5 | No difference | CCR3, CD4 expression IL-5 |
| Conus | 2009 | Not stated | No | No | Yes | AEC, IL-5Ra,CCR3 | Not done | Not done | Not significant for IL-5Ra |
| Furuta | 2013 | Pediatric | Yes | No | Yes | MBP, EDN, ECP, EPX, CLC/Gal-10 | CLC/Gal-10 | Not done | CLC/Gal-10 |
| Johnsson | 2011 | Adult | Yes | Yes | No | CD23, CD54,CRTH2, CD11c, CCR3, CD44, CD11b, CD18, CD58 | CD23, CD54,CRTH2 CD1c,CCR3,CD44 | Possible differenc e but compared across 4 groups | Not done |
| Lingblom | 2014 | Adult | Yes | No | Yes | CD18, CD44, CD40, CCR3, CD23, CD54, FPR, CRTH2 | CD44,CCR3,CD23,CD54 | Not done | CD18 |
| Lingblom | 2017 | Both | Yes | No | No | CD23, CD44,CD54, CRTH2, FoxP3, Galectin-10 | CD44,CRTH2, FoxP3, Galectin-10 | Not done | Not done |
| Morris | 2016 | Pediatric | No | Yes | Yes | AEC, Eosinophil progenitor | Not done | No difference | Eosinophil progenitor |
| Nguyen | 2011 | Pediatric | Yes | Yes | Yes | PBMC transcript analysis of STAT1, STAT6, and CD66b, Surface CD66b on peripheral eosinophils | CD66b, STAT 6, STAT 1 | Not reported | STAT1 (eosinophils), STAT6 (eosinophils/lymphocytes) |
| Patel | 2010 | Pediatric | Yes | No | No | HLA-DR | No difference | Not done | Not done |
| Straumann | 2005 | Adult | Yes | No | No | AEC; CD25, IL-4, IL-5, IL-13, IL-10 expression on eosinophils | Eosinophil expression of IL-5 and IL-13 | Not done | Not done |
| Straumann | 2010 | Adult | No | No | Yes | Eos expression IL5 alpha receptor | Not done | Not done | IL-5Ra |
| Upparahalli Venkateshaiah | 2016 | Both | No | No | Yes | CD274 | Not done | Not done | CD274 (case report, n=2) |
We identified 29 studies which assessed for potential biomarkers to monitor response to treatment. Only 3 of these studies were randomized clinical trials,25,60,63 which noted significant changes in AEC, ECP, chemokine ligand-26 (CCL-26), chemokine ligand-17 (CCL-17), and mast cell tryptase (MCT) in patients with active versus treated EoE (Table 6). Finally, we identified 3 studies that assess RNA (Benitez, Nguyen and Sawant) and note that these are distinct from all of the other studies which measured proteins (Table 2).
Table 6:
Studies Assessing Biomarker Response to Treatment
| Randomized | |||||
| Author | Year | Age group | Biomarker source | Biomarker(s) Studied | Biomarkers with significant difference in response to treatment |
| Conus | 2009 | Not stated | Blood | AEC, IL-5Ra, CCR3 | AEC |
| Schlag | 2015 | Adult | Blood | AEC, ECP, CCL-17, CCL-18, CCL-26, MC tryptase | AEC, ECP, CCL-26, CCL-17, Serum MCT |
| Straumann | 2010 | Adult | Blood | ECP, EDN, eotaxin, and TNF-a, IL-5Ra on eosinphils | ECP, EDN |
| Non-Randomized | |||||
| Benitez | 2015 | Pediatric | Oral swab | 16S RNA bacterial sequencing | No Difference |
| Blanchard | 2011 | Not stated | Blood | Cytokine array | No Difference |
| Bullock | 2007 | Pediatric | Blood | AEC, CCR3 on eosinophils, CD4 expression of IL-5 | AEC, CCR3, CD4 expression IL-5 |
| Colson | 2014 | Pediatric | Blood | AEC | AEC |
| Dellon | 2015 | Adult | Blood | IL-4, IL-5, Il-6, IL-9, Il-13, TGF-α, TGF-β, TNF-α, Eotaxin 1/2/3, TSLP, MBP, EDN | No Difference |
| Dellon | 2016 | Adult | Blood | Periostin | No Difference |
| Domenech Witek | 2017 | Adult | Blood | tIgE, AEC, ECP | ECP |
| Fujiwara | 2002 | Adult | Blood | AEC, Eotaxin (total and free), ECP | AEC, Eotaxin (total), ECP (n=1, case report) |
| Furuta | 2013 | Pediatric | Esophageal String | MBP, EDN, ECP, EPX, CLC/Gal-10 | MBP, EPX, CLC/Gal-10 |
| Huang | 2010 | Pediatric | Blood | 35 chemokine/cytokines including: bFGF or FGF-2; eotaxin 1/2/3; IL-1α; IL-1β; IL-1RA; IL-2; IL-4; IL-5; IL-6; IL-7; IL-8; IL-10; IL-12-p40; IL-12-p70; IL-13; IL-15; IL-17; IL-17F; ENA78; GCSF; GM-CSF, GRO-α; IFN-γ; IP10; leptin; MCP-3; MIG; MIP-1α; MIP-1β; NGF; PDGF-BB; RANTES; TGF-β;TNF-α; TNF-β, and VEGF | bFGF, IL-5 |
| Jyonouchi | 2013 | Pediatric | Blood | iNKTs | iNKTs |
| Katzka | 2015 | Adult | Cytosponge | Eosinophils, EDN | Eosinophils |
| Konikoff | 2006 | Pediatric | Blood, stool | AEC, IL-5, Eotaxins 1/2/3, EDN (blood and stool) | AEC |
| Leung | 2012 | Both | Breath | eNO | No Difference |
| Lingblom | 2014 | Adult | Blood | CD18, CD44, CD40, CCR3, CD23, CD54, PFR, CRTH2 | CD18 |
| Lucendo | 2013 | Adult | Blood | AEC, tIgE, ECP | No Difference |
| Min | 2016 | Both | Blood | AEC, Eotaxin-3, EDN, ECP, IL-5 | AEC |
| Morris | 2016 | Pediatric | Blood | AEC, Eosinophil progenitor | Eosinophil progenitor |
| Nguyen | 2011 | Pediatric | Blood | PBMC transcript analysis of STAT1, STAT6, and CD66b, Surface CD66b on peripheral eosinophils | STAT1 (eosinophils), STAT6 (eosinophils/lymphocytes) |
| Rawson | 2016 | Pediatric | Blood | TGF-β, PAI-1 | No Difference |
| Rodriguez-Sanchez | 2013 | Both | Blood | AEC, ECP, tIgE | No Difference |
| Saffari | 2017 | Adult | Throat swab | EPX activity | No Difference |
| Schlag | 2014 | Adult | Blood | ECP, tryptase | ECP, tryptase |
| Subbarao | 2011 | Pediatric | Blood, stool | IL-5 (Blood); EDN (Blood and stool) | EDN (Blood) |
| Upparahalli Venkateshaiah | 2016 | Both | Blood | CD274 | CD274 but case report (n=2) |
| Wright | 2016 | Adult | Blood | sIgG4 (total and foodspecific) | sIgG4 |
Quality Assessment:
In 22 of the 49 included studies, there was clear declaration that patient samples were obtained either by sampling consecutive patients or randomly selected, and therefore, at low risk of multiple biases. However, almost all of the included studies were derived from samples obtained from patients seen at specialty referral centers, resulting in possible selection biases and issues related to uncertain generalizability to other patient populations. Four studies were determined to have a high risk of bias based on the increased time frame between collection of the esophageal biopsies and measurement of the non-invasive biomarker while 16 studies did not clearly state what time frame separated the collection of biopsies and the measurement of the minimally invasive biomarker. Details of the quality assessments are displayed in Table 7.
Table 7:
QUADAS-2 for Eosinophilic Esophagitis Minimally Invasive Biomarker Studies
 |
DISCUSSION:
This study is the first to utilize a systematic approach to identify relevant articles, in contrast to other literature reviews for minimally invasive EoE biomarkers. Using a systematic approach, we identified 2094 potential articles, of which 49 met our inclusion criteria. Twenty-six of these articles have been published since 2014, a testament to the rapid pace at which the EoE biomarker field is moving; however, only 3 of these studies were randomized-controlled studies.
A key objective of this review was to identify methodological strengths and weaknesses of the identified studies. Many weaknesses were identified including specimen timing, retrospective design of many studies, absence of an atopic control group and selection of patients which would represent the general population--suggesting there is opportunity for improvement in the design of future studies. First, we strongly recommend that study protocols specify that biomarkers be measured at the same time as the reference standard (i.e. peak number of eosinophils in esophageal histology). This recommendation favors prospectively designed studies which can pre-specify sample collection timing. For example, blood, saliva, or urine biomarkers should ideally be collected immediately prior to esophageal biopsy to account for the possibility that the biopsy itself could impact the biomarker measurement. In addition, randomized-controlled studies (representing only 3 of 49 included studies) allow for comparison in both placebo and intervention groups and most importantly, reduce the risks posed by confounding factors. Second, EoE biomarker studies should incorporate a prospective design using random or consecutive selection to prevent selection bias in determining which patients are tested; a phenomenon that is difficult to avoid in retrospective studies. Third, the generalizability of samples derived primarily from highly specialized tertiary referral centers should be considered. Attempts to include community-based samples or design studies that utilize a population-based sampling frame may improve applicability of study findings. Finally, we identified only 9 studies that included atopic controls--only 2 of which identified biomarkers (increased urinary 3-BT and decreased total IgE) that distinguish EoE from other allergic disease states. Inclusion of atopic controls is critical given the fact that a large proportion of patients with EoE have allergic comorbidities associated with eosinophilia (i.e. asthma, allergic rhinitis). Consequently, biomarker comparison in EoE subjects and other atopic diseases enhances the ability to control for a more robust range of possible confounders.
The other objective of this study was to identify the most promising EoE biomarkers. Future plans include a meta-analysis of biomarkers which were investigated across multiple studies to determine whether pooled data can enhance power and provide more robust point estimate compared to estimates derived from smaller, individual studies. AEC, ECP, eotaxin, and chemokine receptor type 3 (CCR3) are of interest for meta-analysis based on the number of studies which measured these biomarkers along with selecting biomarkers representing different general categories of biomarkers (granule protein, chemokine, eosinophil cell surface protein). We note that almost all of the studies performed to date were investigating blood-based biomarkers (AEC more often than any other), which identifies a need to develop alternative sampling techniques. Given the risk of potential confounding due to other eosinophilic/atopic disorders, minimally-invasive sampling of the esophagus or a contiguous site may prove critical. Early findings from esophageal string test and Cytosponge are encouraging but represent only a small fraction of EoE biomarker studies. Additional controlled studies are also needed to validate mass spectroscopy assessment of brominated urinary tyrosine in EoE subjects. While the degree of 3-BT elevation may distinguish EoE subjects, this has also been used as a marker of pediatric asthma.68 Another potential approach might be to combine a biomarker (e.g. AEC) with symptom assessment in order to confer site specificity. This too has certain pitfalls particularly because subjects with EoE /or asthma may have subclinical or comorbid disease. Further efforts to build the evidence base around non-blood-based EoE biomarkers is an important focus of ongoing research efforts.
In summary, we identified 49 studies that examined minimally invasive EoE biomarkers, the majority of which were identified over the past 3 years. Blood-based biomarkers are the most frequently investigated however early findings from other non-invasive methods (esophageal string test and Cytosponge) seem promising. We identified timing of specimen collection, patient selection, and inclusion of an atopic control group as important study design considerations for future EoE biomarker studies. The absence of meta-analysis is the main limitation of this study; however, this is being actively pursued. Despite the increased interest in this area and the clear clinical need for minimally-invasive biomarkers, there is still not a minimally-invasive biomarker that has been incorporated into guideline recommendations or routine clinical practice. Fortunately, several promising biomarkers are under study which may reduce the need for repeated endoscopic biopsies.
Acknowledgments
Funding Source: NIH Grant R01 DK101856
Abbreviations:
- 3-BT
3-bromotyrosine
- AAAAI
American Academy of Allergy, Asthma and Immunology
- ACG
American College of Gastroenterology
- AEC
absolute eosinophil count
- CCL-26
chemokine ligand-26
- CCL-17
chemokine ligand-17
- CCR3
chemokine receptor type 3
- DDW
Digestive Diseases Week
- ECP
eosinophilic cationic protein
- EDN
eosinophil-derived neurotoxin
- EoE
eosinophilic esophagitis
- EPX
eosinophil peroxidase
- FDA
Food and Drug Administration
- GERD
Gastroesophageal reflux disease
- MBP
major basic protein
- MCT
mast cell tryptase
portable document format
- PRIMSA
Preferred Reporting Items for Systematic Reviews and Meta-analysis
- trEoE
treated EoE
- UEG
United European Gastroenterology
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: none
REFERENCES:
- 1.Winter HS, Madara JL, Stafford RJ, Grand RJ, Quinlan JE G H. Intraepithelial eosinophils: A new diagnostic criterion for reflux esophagitis. Gastroenterology. 1982;83:818–823. [PubMed] [Google Scholar]
- 2.Attwood SEA, Smyrk TC, Demeester TR, Jones JB. Esophageal eosinophilia with dysphagia - A distinct clinicopathologic syndrome. Dig Dis Sci. 1993;38(1):109–116. doi: 10.1007/BF01296781 [DOI] [PubMed] [Google Scholar]
- 3.Straumann A, Bernoulli R, Spichtin H, Vogtlin J. Idiopathic eosinophilic esophagitis: a frequently overlooked disease with typical clinical aspects and discrete endoscopic findings. Schweiz Med Wochenschr. 1994;124:1419–1429. [PubMed] [Google Scholar]
- 4.Kelly KJ, Lazenby AJ, Rowe PC, Yardley JH, Perman JA, Sampson HA. Eosinophilic esophagitis attributed to gastroesophageal reflux: Improvement with an amino acid-based formula. Gastroenterology. 1995;109(5):1503–1512. doi: 10.1016/0016-5085(95)90637-1 [DOI] [PubMed] [Google Scholar]
- 5.Furuta GT, Liacouras CA, Collins MH, et al. Eosinophilic Esophagitis in Children and Adults: A Systematic Review and Consensus Recommendations for Diagnosis and Treatment. Gastroenterology. 2007;133(4):1342–1363. doi: 10.1053/j.gastro.2007.08.017 [DOI] [PubMed] [Google Scholar]
- 6.Furuta GT, Katzka DA. Eosinophilic Esophagitis. N Engl J Med. 2015;373(17):1640–1648. doi: 10.1056/NEJMra1502863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lucendo AJ, Molina-Infante J, Arias Á, et al. Guidelines on eosinophilic esophagitis: evidence-based statements and recommendations for diagnosis and management in children and adults. United Eur Gastroenterol J. 2017;5(3):335–358. doi: 10.1177/2050640616689525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greuter T, Bussmann C, Safroneeva E, et al. Long-Term Treatment of Eosinophilic Esophagitis With Swallowed Topical Corticosteroids: Development and Evaluation of a Therapeutic Concept. Am J Gastroenterol. 2017;112(10):1527–1535. doi: 10.1038/ajg.2017.202 [DOI] [PubMed] [Google Scholar]
- 9.Straumann A, Conus S, Degen L, et al. Budesonide is effective in adolescent and adult patients with active eosinophilic esophagitis. Gastroenterology. 2010;139(5):1526–1537. doi: 10.1053/j.gastro.2010.07.048 [DOI] [PubMed] [Google Scholar]
- 10.Schaefer ET, Fitzgerald JF, Molleston JP, et al. Comparison of Oral Prednisone and Topical Fluticasone in the Treatment of Eosinophilic Esophagitis: A Randomized Trial in Children. Clin Gastroenterol Hepatol. 2008;6(2):165–173. doi: 10.1016/j.cgh.2007.11.008 [DOI] [PubMed] [Google Scholar]
- 11.FDA. FDA Drug Safety Communication: FDA approves label changes for use of general anesthetic and sedation drugs in young children. 2017. [Google Scholar]
- 12.Liacouras CA, Furuta GT, Hirano I, et al. Eosinophilic esophagitis: Updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128(1):3–20. doi: 10.1016/j.jaci.2011.02.040 [DOI] [PubMed] [Google Scholar]
- 13.Watts A, Alexander JA, Gupta SK. Eosinophilic esophagitis: Search for noninvasive techniques for long-term monitoring. Gastrointest Endosc. 2016;83(2):307–308. doi: 10.1016/j.gie.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen ET, Kappelman MD, Martin CF, Dellon ES. Health-Care Utilization, Costs, and the Burden of Disease Related to Eosinophilic Esophagitis in the United States. Am J Gastroenterol. 2015;110(5):626–632. doi: 10.1038/ajg.2014.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhardwaj N, Ghaffari G. Biomarkers for eosinophilic esophagitis: A review. Ann Allergy, Asthma Immunol. 2012;109(3):155–159. doi: 10.1016/j.anai.2012.06.014 [DOI] [PubMed] [Google Scholar]
- 16.Moher D, Liberati A, Tetzlaff J, Altman DG. Academia and Clinic Annals of Internal Medicine Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. Annu Intern Med. 2009;151(4):264–269. doi: 10.1371/journal.pmed1000097 [DOI] [PubMed] [Google Scholar]
- 17.Reitsma JB, Rutjes A, Whiting P, Vlassov V, Leeflang M, Deeks JJ. Assessing methodological quality. Cochrane Handb Syst Rev Diagnsotic Test Accuracy. 2009:0–27. doi: 10.3109/17549507.2010.492873 [DOI] [Google Scholar]
- 18.Whiting P, Rutjes AWS, Reitsma JB, Bossuyt PMM, Kleijnen J. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol. 2003;3:25. doi: 10.1186/1471-2288-3-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benitez AJ, Hoffmann C, Muir AB, et al. Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome. 2015;3:23. doi: 10.1186/s40168-015-0085-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanchard C, Stucke EM, Rodriguez-Jimenez B, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127(1):208–217.e7. doi: 10.1016/j.jaci.2010.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Botan V, dos Santos Borges TK, Rocha Alves ÉA, Claudino Pereira Couto S, Bender Kohnert Seidler H, Muniz-Junqueira MI. Enhanced activation of eosinophils in peripheral blood and implications for eosinophilic esophagitis diagnosis. J Gastroenterol Hepatol. 2017;32(7):1318–1327. doi: 10.1111/jgh.13710 [DOI] [PubMed] [Google Scholar]
- 22.Bullock JZ, Villanueva JM, Blanchard C, et al. Interplay of adaptive th2 immunity with eotaxin-3/c-C chemokine receptor 3 in eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2007;45(1):22–31. doi: 10.1097/MPG.0b013e318043c097 [DOI] [PubMed] [Google Scholar]
- 23.Clayton F, Fang JC, Gleich GJ, et al. Eosinophilic esophagitis in adults is associated with IgG4 and not mediated by IgE. Gastroenterology. 2014;147(3):602–609. doi: 10.1053/j.gastro.2014.05.036 [DOI] [PubMed] [Google Scholar]
- 24.Colson D, Kalach N, Soulaines P, et al. The Impact of Dietary Therapy on Clinical and Biologic Parameters of Pediatric Patients with Eosinophilic Esophagitis. J Allergy Clin Immunol Pract. 2014;2(5):587–593. doi: 10.1016/j.jaip.2014.05.012 [DOI] [PubMed] [Google Scholar]
- 25.Conus S, Straumann A, Simon HU. Anti-IL-5 (mepolizumab) therapy does not alter IL-5 receptor alpha levels in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2009;123(1):269. doi: 10.1016/j.jaci.2008.09.031 [DOI] [PubMed] [Google Scholar]
- 26.Cunnion KM, Willis LK, Minto HB, et al. Eosinophil Quantitated Urine Kinetic: A novel assay for assessment of eosinophilic esophagitis. Ann Allergy, Asthma Immunol. 2016;116(5):435–439. doi: 10.1016/j.anai.2016.02.011 [DOI] [PubMed] [Google Scholar]
- 27.Dellon ES, Rusin S, Gebhart JH, et al. Utility of a Noninvasive Serum Biomarker Panel for Diagnosis and Monitoring of Eosinophilic Esophagitis: A Prospective Study. Am J Gastroenterol. 2015;110(6):821–827. doi: 10.1038/ajg.2015.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dellon ES, Higgins LL, Beitia R, et al. Prospective assessment of serum periostin as a biomarker for diagnosis and monitoring of eosinophilic oesophagitis. Aliment Pharmacol Ther. 2016;44(2):189–197. doi: 10.1111/apt.13672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Domenech Witek J, Jover Cerda V, Gil Guillen V, Domenech Clar JB, Rodriguez Pacheco R. Assessing eosinophilic cationic protein as a biomarker for monitoring patients with eosinophilic esophagitis treated with specific exclusion diets. World Allergy Organ J. 2017;10(1):12. doi: 10.1186/s40413-017-0143-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuentebella J, Patel A, Nguyen T, et al. Increased Number of Regulatory T Cells in Children With Eosinophilic Esophagitis. J Pediatr Gastroenterol Nutr. 2010;51(3):1. doi: 10.1097/MPG.0b013e3181e0817b [DOI] [PubMed] [Google Scholar]
- 31.Fujiwara H, Morita A, Kobayashi H, et al. Infiltrating eosinophils and eotaxin: Their association with idiopathic eosinophilic esophagitis. Ann Allergy, Asthma Immunol. 2002;89(4):429–432. doi: 10.1016/S1081-1206(10)62047-9 [DOI] [PubMed] [Google Scholar]
- 32.Furuta GT, Kagalwalla AF, Lee JJ, et al. The oesophageal string test: a novel, minimally invasive method measures mucosal inflammation in eosinophilic oesophagitis. Gut. 2013;62(10):1395–1405. doi: 10.1136/gutjnl-2012-303171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang JJ, Joh JW, Fuentebella J, et al. Eotaxin and FGF enhance signaling through an extracellular signal-related kinase (ERK)-dependent pathway in the pathogenesis of Eosinophilic esophagitis. Allergy Asthma Clin Immunol. 2010;6(1):25. doi: 10.1186/1710-1492-6-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnsson M, Bove M, Bergquist H, et al. Distinctive blood eosinophilic phenotypes and cytokine patterns in eosinophilic esophagitis, inflammatory Bowel disease and airway allergy. J Innate Immun. 2011;3(6):594–604. doi: 10.1159/000331326 [DOI] [PubMed] [Google Scholar]
- 35.Jyonouchi S, Smith CL, Saretta F, et al. Invariant natural killer T cells in children with eosinophilic esophagitis. Clin Exp Allergy. 2014;44(1):58–68. doi: 10.1111/cea.12201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katzka DA, Geno DM, Ravi A, et al. Accuracy, safety, and tolerability of tissue collection by cytosponge vs endoscopy for evaluation of eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2015;13(1):77–83. doi: 10.1016/j.cgh.2014.06.026 [DOI] [PubMed] [Google Scholar]
- 37.Kinoshita Y, Furuta K, Ishimura N, Ishihara S. Elevated Plasma Cytokines in Japanese Patients with Eosinophilic Esophagitis and Gastroenteritis. Digestion. 2012;86:238–243. [DOI] [PubMed] [Google Scholar]
- 38.Knipping K, Colson D, Soulaines P, Redegeld F, Garssen J, Dupont C. Serum immunoglobulin free light chain levels are higher in girls than boys during eosinophilic oesophagitis. Acta Paediatr. 2014;103(7):766–774. doi: 10.1111/apa.12651 [DOI] [PubMed] [Google Scholar]
- 39.Konikoff MR, Blanchard C, Kirby C, et al. Potential of Blood Eosinophils, Eosinophil-Derived Neurotoxin, and Eotaxin-3 as Biomarkers of Eosinophilic Esophagitis. Clin Gastroenterol Hepatol. 2006;4(11):1328–1336. doi: 10.1016/j.cgh.2006.08.013 [DOI] [PubMed] [Google Scholar]
- 40.Krupp NL, Sehra S, Slaven JE, Kaplan MH, Gupta S, Tepper RS. Increased prevalence of airway reactivity in children with eosinophilic esophagitis. Pediatr Pulmonol. 2016;51(5):478–483. doi: 10.1002/ppul.23327 [DOI] [PubMed] [Google Scholar]
- 41.Lanz MJ, Guerrero RA, Gonzalez-Vallina R. Measurement of exhaled nitric oxide in the evaluation for eosinophilic esophagitis in children. Ann Allergy, Asthma Immunol. 2012;109(1):81–82. doi: 10.1016/j.anai.2012.05.009 [DOI] [PubMed] [Google Scholar]
- 42.Leung J, Nguyen-Traxler A, Lee EM, et al. Assessment of fractionated exhaled nitric oxide as a biomarker for the treatment of eosinophilic esophagitis. Allergy Asthma Proc. 2012;33(6):519–524. doi: 10.2500/aap.2012.33.3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lexmond WS, Pardo M, Rooney K, et al. Elevated levels of leukotriene C4 synthase mRNA distinguish a subpopulation of eosinophilic oesophagitis patients. Clin Exp Allergy. 2013;43(8):902–913. doi: 10.1111/cea.12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lingblom C, Käppi T, Bergquist H, et al. Differences in eosinophil molecular profiles between children and adults with eosinophilic esophagitis. Allergy Eur J Allergy Clin Immunol. 2017;72(9):1406–1414. doi: 10.1111/all.13140 [DOI] [PubMed] [Google Scholar]
- 45.Lingblom C, Bergquist H, Johnsson M, et al. Topical Corticosteroids Do Not Revert the Activated Phenotype of Eosinophils in Eosinophilic Esophagitis but Decrease Surface Levels of CD18 Resulting in Diminished Adherence to ICAM-1, ICAM-2, and Endothelial Cells. Inflammation. 2014;37(6):1932–1944. doi: 10.1007/s10753-014-9926-x [DOI] [PubMed] [Google Scholar]
- 46.Lucendo AJ, Arias Á, González-Cervera J, Mota-Huertas T, Yagüe-Compadre JL. Tolerance of a cow’s milk-based hydrolyzed formula in patients with eosinophilic esophagitis triggered by milk. Allergy Eur J Allergy Clin Immunol. 2013;68(8):1065–1072. doi: 10.1111/all.12200 [DOI] [PubMed] [Google Scholar]
- 47.Min SB, Nylund CM, Baker TP, et al. Longitudinal Evaluation of Noninvasive Biomarkers for Eosinophilic Esophagitis. J Clin Gastroenterol. 2016;0(0):1–9. doi: 10.1097/MCG.0000000000000621 [DOI] [PubMed] [Google Scholar]
- 48.Morris DW, Stucke EM, Martin LJ, et al. Eosinophil progenitor levels are increased in patients with active pediatric eosinophilic esophagitis. J Allergy Clin Immunol. 2016;138(3):915–918.e5. doi: 10.1016/j.jaci.2016.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen T, Gernez Y, Fuentebella J, et al. Immunophenotyping of peripheral eosinophils demonstrates activation in eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2011;53(1):40–47. doi: 10.1097/MPG.0b013e318212647a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel AJ, Fuentebella J, Gernez Y, et al. Increased HLA-DR expression on tissue eosinophils in eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2010;51(3):290–294. doi: 10.1097/MPG.0b013e3181e083e7 [DOI] [PubMed] [Google Scholar]
- 51.Paterson AL, Lao-Sirieix P, O’Donovan M, et al. Range of pathologies diagnosed using a minimally invasive capsule sponge to evaluate patients with reflux symptoms. Histopathology. 2017;70(2):203–210. doi: 10.1111/his.13039 [DOI] [PubMed] [Google Scholar]
- 52.Zafra MP, Cancelliere N, Rodríguez Del Río P, et al. Misregulation of suppressors of cytokine signaling in eosinophilic esophagitis. J Gastroenterol. 2013;48(8):910–920. doi: 10.1007/s00535-012-0723-8 [DOI] [PubMed] [Google Scholar]
- 53.Philpott HL, Nandurkar S, Thien F, et al. Seasonal recurrence of food bolus obstruction in eosinophilic esophagitis. Intern Med J. 2015;45(9):939–943. doi: 10.1111/imj.12790 [DOI] [PubMed] [Google Scholar]
- 54.Rawson R, Yang T, Newbury RO, et al. TGF-β1–induced PAI-1 contributes to a profibrotic network in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2016;138(3):791–800.e4. doi: 10.1016/j.jaci.2016.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rayapudi M, Rajavelu P, Zhu X, et al. Invariant natural killer T-cell neutralization is a possible novel therapy for human eosinophilic esophagitis. Clin Transl Immunol. 2014;3(1):e9. doi: 10.1038/cti.2013.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodríguez-Sánchez J, Gómez-Torrijos E, De-la-Santa-Belda E, et al. Effectiveness of serological markers of eosinophil activity in monitoring eosinophilic esophagitis. Rev Esp Enferm Dig. 2013;105:462–468. doi: 10.4321/S1130-01082013000800004 [DOI] [PubMed] [Google Scholar]
- 57.Saffari H, Baer K, Boynton KK, Gleich GJ, Peterson KA. Pharyngeal mucosa brushing does not correlate with disease activity in patients with eosinophilic esophagitis. J Allergy Clin Immunol Pract. 2017;5(5):1455–1456. doi: 10.1016/j.jaip.2017.03.012 [DOI] [PubMed] [Google Scholar]
- 58.Sawant D, Weiguio Y, Wright Z, et al. Serum MicroRNA-21 as a Biomarker for Allergic Inflammatory Disease in Children. MicroRNA. 2015;4(1):36–40. [DOI] [PubMed] [Google Scholar]
- 59.Schlag C, Pfefferkorn S, Brockow K, et al. Serum Eosinophil Cationic Protein is Superior to Mast Cell Tryptase as Marker for Response to Topical Corticosteroid Therapy in Eosinophilic Esophagitis. J Clin Gastroenterol. 2014;48(7):600–606. doi: 10.1097/01.mcg.0000436439.67768.8d [DOI] [PubMed] [Google Scholar]
- 60.Schlag C, Miehlke S, Heiseke A, et al. Peripheral blood eosinophils and other non-invasive biomarkers can monitor treatment response in eosinophilic oesophagitis. Aliment Pharmacol Ther. 2015;42(9):1122–1130. doi: 10.1111/apt.13386 [DOI] [PubMed] [Google Scholar]
- 61.Soylu A, Altintas A, Cakmak S, et al. The coexistence of eosinophilic esophagitis with allergic rhinitis. Eur Rev Med Pharmacol Sci. 2016;20(11):2315–2323. http://www.ncbi.nlm.nih.gov/pubmed/27338057. [PubMed] [Google Scholar]
- 62.Straumann A, Kristl J, Conus S, et al. Cytokine expression in healthy and inflamed mucosa: probing the role of eosinophils in the digestive tract. Inflamm Bowel Dis. 2005;11(8):720–726. doi: 10.1097/01.mib.0000172557.39767.53 [DOI] [PubMed] [Google Scholar]
- 63.Straumann A, Conus S, Grzonka P, et al. Anti-interleukin-5 antibody treatment (mepolizumab) in active eosinophilic oesophagitis: A randomised, placebo-controlled, double-blind trial. Gut. 2010:21–30. doi: 10.1136/gut.2009.178558 [DOI] [PubMed] [Google Scholar]
- 64.Wright BL, Kulis M, Guo R, et al. Food-specific IgG4 is associated with eosinophilic esophagitis. J Allergy Clin Immunol. 2016;138(4):1190–1192.e3. doi: 10.1016/j.jaci.2016.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Venkateshaiah SU, Manohar M, Verma AK, Blecker U, Mishra A. Possible noninvasive biomarker of eosinophilic esophagitis: Clinical and experimental evidence. Case Rep Gastroenterol. 2016;10(3):685–692. doi: 10.1159/000452654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Subbarao G, Rosenman MB, Ohnuki L, et al. Exploring Potential Non-Invasive Biomarkers in Eosinophilic Esophagitis: A Longitudinal Study in Children. J Pediatr Gastroenterol Nutr. 2011;53(6):1. doi: 10.1097/MPG.0b013e318228cee6 [DOI] [PubMed] [Google Scholar]
- 67.Von Arnim U, Wex T, Röhl FW, et al. Identification of clinical and laboratory markers for predicting eosinophilic esophagitis in adults. Digestion. 2011;84(4):323–327. doi: 10.1159/000331142 [DOI] [PubMed] [Google Scholar]
- 68.Wang YA, Yu X, Silverman PM, Harris RL, Edward H. NIH Public Access. 2010;385(1):22–29. doi: 10.1016/j.jmb.2008.10.054.The [DOI] [Google Scholar]


