Abstract
Background/Aims
There are limited data from India on genetic variants influencing late age-related macular degeneration (AMD). We have previously reported associations from a population-based study in India (the India age-related eye disease study (INDEYE)) of early AMD and single nucleotide polymorphisms (SNPs) in ARMS2/HTRA1 and no association with CFH, C2 or CFB. Late AMD cases were too few for meaningful analyses. We aimed to investigate SNPs for late AMD through case enrichment and extend the loci for early AMD.
Methods
Fundus images of late AMD hospital cases were independently graded by the modified Wisconsin AMD grading scheme. In total 510 cases with late AMD (14 geographic atrophy and 496 neovascular AMD (nvAMD)), 1876 with early AMD and 1176 with no signs of AMD underwent genotyping for selected SNPs. We investigated genotype and per-allele additive associations (OR and 95% CIs) with nvAMD or early AMD. Bonferroni adjusted P values are presented.
Results
We found associations with nvAMD for CFHY402H variant (rs1061170) (OR=1.99, 95% CI 1.67 to 2.37, P=10−6), ARMS2 (rs10490924) (OR=2.94, 95% CI 2.45 to 3.52, P=10−9), C2 (rs547154) (OR=0.67, 95% CI 0.53 to 0.85, P=0.01), ABCA1 (rs1883025) (OR=0.77, 95% CI 0.65 to 0.92, P=0.04) and an SNP near VEGFA (rs4711751) (OR=0.64, 95% CI 0.54 to 0.77, P=10−3). We found no associations of TLR3 (rs3775291), CFD (rs3826945), FRK (rs1999930) or LIPC (rs10468017) or APOE ε4 alleles with nvAMD or early AMD, nor between early AMD and rs1883025 or rs4711751.
Conclusions
The major genetic determinants of nvAMD risk in India are similar to those in other ancestries, while findings for early AMD suggest potential differences in the pathophysiology of AMD development.
Keywords: retina, genetics, macula, epidemiology
Introduction
Genetic risk variants for late age-related macular degeneration (AMD) have been identified and further confirmed in genome-wide association studies (GWAS), the majority of which in studies of European ancestry.1 There is less information on late AMD genetic risk in India, with most data coming from one patient/control cohort.2–4 We have previously reported genetic results from a large population-based study of people aged 60 and over in India (the India age-related eye disease study (INDEYE)) for early AMD with variants in complement factor H (CFH), factor B (CFB), component 2 (C2) and ARMS2/HTRA1.5 Late AMD cases were too few for meaningful analyses. In the present paper we present results for late AMD based on an enriched sample and for other genetic loci with early AMD.
Materials and methods
INDEYE was conducted between 2005 and 2007 in two locations in south (Tamil Nadu) and north (Haryana) India. The study methods including sampling and recruitment, blood collection, ophthalmological examination and AMD grading, along with results on the prevalence of early and late AMD, have been published.6 In the present study, we recruited additional cases of late AMD between 2009 and 2011 from the hospitals that participated in the INDEYE study (All India Institute of Medical Sciences, Delhi, and Aravind Eye Hospital, Pondicherry, Tamil Nadu) and additionally from Aravind Eye Hospital, Madurai, Tamil Nadu. We aimed to achieve 600 late AMD cases plus two population controls per case to detect the twofold per-allele association of Y402H CFH (rs1061170) reported in a meta-analysis of primarily European ancestry7 at 90% power and alpha <0.001. Initial eligibility criteria were age 60 years and over, Indian descent and a diagnosis of late AMD by retinal ophthalmologists. Controls were participants in the INDEYE study with no signs of early or late AMD in either eye.
In both INDEYE and clinic participants, informed written consent was obtained prior to enrolment. If the participant was illiterate, the information sheet was read out aloud in the presence of a local witness, and a thumb impression of the participant signified assent. The study complied with the Declaration of Helsinki.
Full details of the method of ascertainment of AMD in the population study have previously been published.6 In brief two 35° stereo fundus photographs of each eye were taken and graded at Queens University Belfast (QUB) using the modified Wisconsin Age-Related Maculopathy Grading System.8 Each eye was classified into four mutually exclusive grades: grade 1: soft distinct drusen (≥63 µm) only or pigmentary irregularities only; grade 2: soft indistinct (≥125 µm) or reticular drusen only or soft distinct drusen (≥63 µm) with pigmentary irregularities; grade 3: soft indistinct (≥125 µm) or reticular drusen with pigmentary irregularities; grade 4: either neovascular AMD (nvAMD; presence of any of the following: serous or haemorrhagic retinal or retinal pigment epithelial detachment, subretinal neovascular membrane, periretinal fibrous scar) or geographic atrophy (GA; well-demarcated area of retinal pigment atrophy with visible choroidal vessels). Fundus images of cases recruited from hospital clinics were sent to QUB (colour photographs, optical coherence tomography (OCT)) and graded as above. In all graded images, GA and nvAMD present in the same eye were categorised as nvAMD. Images that showed no signs of any features of early or late AMD were categorised as having no AMD.
DNA extraction and genotyping
Genomic DNA was extracted from peripheral blood leucocytes using Qiagen kits. Single nucleotide polymorphisms (SNPs) were genotyped using TaqMan assays in an ABI 7900 real-time PCR. We limited our study to genes in biological pathways relevant to AMD pathogenesis, including complement activation (CFH, CFB, CFD) and deposition (Toll-like receptors (TLR 3, 4, 7)), lipid metabolism (ABCA1, APOE, CETP, LIPC), or the degradation of the extracellular matrix (TIMP3).9 We investigated two SNPs on chromosome 6, previously reported to be associated with late AMD10 (LOC107986598 rs4711751 located near VEGFA and FRK rs1999930 near COL10A1). We included SNPs in ARMS2/HTRA1 due to their demonstrated importance in many studies11 and recent evidence for an ARMS2 role in surface complement regulation.12 We tested for departures from Hardy-Weinberg equilibrium (HWE) in controls and excluded any SNPs with a P value ≤0.05. We used logistic regression in Stata V.14 to examine associations of (1) genotype and (2) per-allele additive models adjusted for age, sex and centre. We present additionally Bonferroni-adjusted P values for the number of independent SNPs tested. We created APOE alleles from the SNPs rs429358 (T/C) and rs7412 (C/T), resulting in three alleles: ε2 (TT), ε3 (TC) and ε4 (CC). Analyses of APOE alleles used ε3 as the reference group.
Results
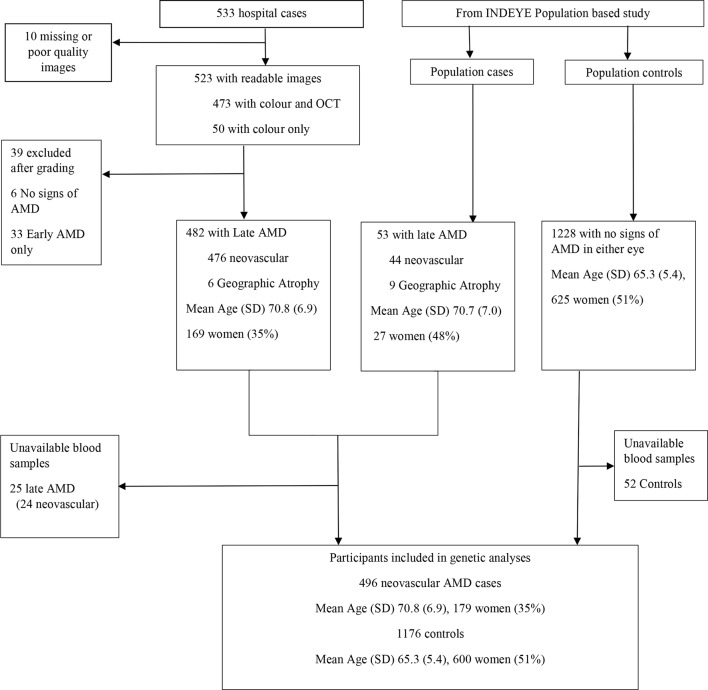
The prevalence of early and late AMD in the INDEYE population study has been published.6 There were 1986 cases of early AMD (1686 grade 1, 289 grade 2, 11 grade 3), 53 of late AMD (44 nvAMD, 9 GA) and 1228 population controls with no signs of AMD in either eye. Hospital retinal clinics recruited 533 cases based on ophthalmologists’ diagnoses. After exclusion of participants without confirmed late AMD or missing blood samples (figure 1), 496 nvAMD cases, 1876 early AMD and 1176 controls were available for analysis. We did not investigate GA because of a small number (n=14). The mean age in years (SD) was 65.3 (5.4) in population controls, in early AMD 67.0 (6.1) and in nvAMD 70.7 (6.9). The number and proportion of women were 600 (51%), 915 (49%) and 179 (36%), respectively. Two SNPs (rs4986790 TLR4/TLR7, rs9621532 TIMP3) failed HWE. HWE and minor allele frequencies (MAFs) for the remaining SNPs are shown in table 1. We also present MAFs for European and Indian ancestries from the 1000 genome study (https://www.ncbi.nlm.nih.gov/snp, accessed 5 December 2016). The control frequencies of APOE alleles were ε3 (0.73), ε2 (0.09) and ε4 (0.18).
Figure 1.
Flow chart of hospital case recruitment and population cases and controls. AMD, age-related macular degeneration; OCT, optical coherence tomography.
Table 1.
SNPs, MAF and test for HWE and corresponding reported MAF in the 1000 genomes project in South Asian and European populations
| Chromosome | Gene | SNP | Major/minor alleles | HWE* | MAF† | MAF EUR‡ | MAF SAS§ |
| 1 | Y402H | rs1061170 | T/C | 0.6854 | 0.323 | 0.362 | 0.287 |
| 4 | TLR3 | rs3775291 | C/T | 0.9347 | 0.235 | 0.324 | 0.263 |
| 6 | C2 | rs547154 | C/A | 0.3813 | 0.187 | 0.089 | 0.156 |
| 6 | SKIV2L | rs438999 | A/G | 0.6932 | 0.183 | 0.089 | 0.148 |
| 6 | LOC107986598¶ | rs4711751 | T/C | 1 | 0.423 | 0.487 | 0.330 |
| 6 | FRK | rs1999930 | C/T | 0.3957 | 0.075 | 0.281 | 0.052 |
| 9 | ABCA1 | rs1883025 | C/T | 0.0797 | 0.432 | 0.240 | 0.413 |
| 10 | ARMS2 | rs10490923 | G/A | 0.7299 | 0.149 | 0.130 | 0.148 |
| 10 | ARMS2 | rs10490924 | G/T | 0.1953 | 0.319 | 0.195 | 0.343 |
| 10 | HTRA1 | rs2672598 | T/C** | 0.2969 | 0.524 | 0.499 | 0.464 |
| 15 | LIPC | rs10468017 | C/T | 0.9195 | 0.176 | 0.283 | 0.184 |
| 16 | CETP | rs3764261 | C/A | 0.0737 | 0.295 | 0.292 | 0.321 |
| 19 | APOE | rs429358 | T/C | 1 | 0.097 | 0.155 | 0.087 |
| 19 | APOE | rs7412 | C/T | 0.5232 | 0.050 | 0.063 | 0.044 |
| 19 | CFD | rs3826945 | T/C | 0.8424 | 0.344 | 0.313 | 0.334 |
*P value for tests for departure from Hardy-Weinberg equilibrium (HWE) in controls.
†Minor allele frequency (MAF) in controls.
‡MAF from 1000 genome study for European ancestry available at https://www.ncbi.nlm.nih.gov/snp.
§MAF from 1000 genome study for South Asian ancestry available at https://www.ncbi.nlm.nih.gov/snp.
¶SNP located near VEGFA.
**Minor allele considered as C for comparison with other studies.
SNP, single nucleotide polymorphisms.
We found additive associations with nvAMD for Y402H (rs1061170), HTRA1 (rs2672598), ARMS2 (rs10490924, rs10490923), CFB (rs438999, rs547154), ABCA1 (rs1883025) and SNP (rs4711751 close to VEGFA) (table 2). We found no associations with TLR3 (rs3775291), CFD (rs3826945), FRK (rs1999930) or LIPC (rs10468017). There was no association between APOE ε4 and nvAMD (OR=0.72, 95% CI 0.52 to 1.01).
Table 2.
Association of neovascular age-related macular degeneration with SNPs
| Gene | SNP | Major/minor alleles | 1 vs 0 copy of minor allele | 2 vs 0 copies of minor allele | Additive per minor allele | |||||
| OR* | 95% CI | OR* | 95% CI | OR* | 95% CI | P | P† | |||
| Y402H | rs1061170 | T/C | 1.72 | 1.31 to 2.28 | 4.13 | 2.91 to 5.87 | 1.99 | 1.67 to 2.37 | 10−7 | 10−6 |
| TLR3 | rs3775291 | C/T | 1.18 | 0.92 to 1.51 | 0.88 | 0.51 to 1.53 | 1.06 | 0.87 to 1.30 | 0.545 | |
| C2 | rs547154 | C/A | 0.62 | 0.47 to 0. 82 | 0. 64 | 0.29 to 1.43 | 0.67 | 0.53 to 0. 85 | 0.001 | 0.01 |
| SKIV2L | rs438999 | A/G | 0.63 | 0.47 to 0.83 | 0.50 | 0.21 to 1.22 | 0.65 | 0.50 to 0.83 | 0.001 | 0.01 |
| LOC107986598 | rs4711751 | T/C | 0.35 | 0.27 to 0.46 | 0.65 | 0.45 to 0. 94 | 0.64 | 0.54 to 0. 77 | 10−4 | 10−3 |
| FRK | rs1999930 | C/T | 0.93 | 0.64 to 1.34 | 5.94 | 1.17 to 30.10 | 1.05 | 0.74 to 1.49 | 0.777 | |
| ABCA1 | rs1883025 | C/T | 0.81 | 0.61 to 1.07 | 0.58 | 0.41 to 0.83 | 0.77 | 0.65 to 0.92 | 0.003 | 0.04 |
| ARMS2 | rs10490923 | G/A | 0.49 | 0.35 to 0.67 | 0.85 | 0.33 to 2.17 | 0.57 | 0.43 to 0.75 | 10−3 | 0.04 |
| ARMS2 | rs10490924 | G/T | 1.86 | 1.37 to 2.51 | 8.73 | 6.11 to 12.48 | 2.94 | 2.45 to 3.52 | 10−10 | 10−9 |
| HTRA1 | rs2672598 | T/C | 1.53 | 1.01 to 2.32 | 5.42 | 3.58 to 8.21 | 2.67 | 2.19 to 3.25 | 10−9 | 10−8 |
| LIPC | rs10468017 | C/T | 1.13 | 0.87 to 1.47 | 1.12 | 0.56 to 2.23 | 1.11 | 0.89 to 1.37 | 0.370 | |
| CETP | rs3764261 | C/A | 1.27 | 0.98 to 1.64 | 1.26 | 0.82 to 1.91 | 1.17 | 0.98 to 1.41 | 0.087 | |
| APOE | rs429358 | T/C | 0.82 | 0.60 to 1.14 | NC‡ | NC‡ | ||||
| APOE | rs7412 | C/T | 0.87 | 0.58 to 1.32 | NC‡ | NC‡ | ||||
| CFD | rs3826945 | T/C | 1.02 | 0.79 to 1.31 | 1.05 | 0.70 to 1.58 | 1.02 | 0.85 to 1.23 | 0.820 | |
*Adjusted for age, sex and centre.
†Bonferroni-adjusted P value for 13 per-allele tests.
‡Not calculated, no cases with 2 copies of minor allele.
SNP, single nucleotide polymorphisms.
We combined grades 2 and 3 of early AMD due to the small numbers of grade 3. Subsequently we combined all grades of early AMD (1–3) because our preliminary analyses revealed no differences in genetic associations for these early stages. There were no associations with early AMD and any of the SNPs (table 3) or with APOE ε4 (OR=0.88, 95% CI 0.73 to 1.01).
Table 3.
Association of early age-related macular degeneration with SNPs
| Gene | SNP | 1 vs 0 copy of minor allele | 2 vs 0 copies of minor allele | Additive per allele | ||||||
| OR* | 95% CI | P | OR* | 95% CI | P | OR* | 95% CI | P | ||
| TLR3 | rs3775291 | 1.01 | 0.86 to 1.20 | 0.868 | 1.13 | 0.83 to 1.54 | 0.425 | 1.04 | 0.92 to 1.16 | 0.520 |
| LOC107986598 | rs4711751 | 0.95 | 0.78 to 1.14 | 0.558 | 0.91 | 0.69 to 1.20 | 0.502 | 0.95 | 0.84 to 1.08 | 0.452 |
| FRK | rs1999930 | 0.83 | 0.66 to 1.05 | 0.117 | 1.63 | 0.39 to 6.77 | 0.498 | 0.88 | 0.69 to 1.12 | 0.285 |
| ABCA1 | rs1883025 | 0.96 | 0.79 to 1.17 | 0.698 | 0.96 | 0.77 to 1.19 | 0.726 | 0.98 | 0.88 to 1.09 | 0.699 |
| LIPC | rs10468017 | 0.97 | 0.82 to 1.14 | 0.677 | 1.08 | 0.67 to 1.75 | 0.744 | 0.99 | 0.85 to 1.15 | 0.913 |
| CETP | rs3764261 | 1.08 | 0.91 to 1.28 | 0.365 | 1.10 | 0.90 to 1.33 | 0.339 | 1.06 | 0.96 to 1.17 | 0.228 |
| APOE | rs429358 | 0.93 | 0.77 to 1.12 | 0.454 | 0.80 | 0.35 to 1.84 | 0.594 | 0.93 | 0.77 to 1.10 | 0.380 |
| APOE | rs7412 | 0.97 | 0.74 to 1.27 | 0.826 | 1.38 | 0.42 to 4.55 | 0.594 | 1.00 | 0.79 to 1.27 | 0.994 |
| CFD | rs3826945 | 1.02 | 0.88 to 1.20 | 0.758 | 1.10 | 0.87 to 1.39 | 0.399 | 1.04 | 0.94 to 1.15 | 0.416 |
*Adjusted for age, sex and centre.
SNP, single nucleotide polymorphism.
Discussion
CFH and ARMS2/HTRA1 have been identified in numerous studies in European1 11 and East Asian ancestries13 as the most important genes for late AMD risk, with effect sizes around 2.5 and 3 per allele, respectively,1 7 11 and the top two variants at GWAS significance.1 Our effect sizes of 2 for the C allele of Y402H variant of CFH (rs1061170) and 3 for ARMS2 T allele (rs10490924) are consistent with these findings and add to the limited evidence for India.2 3 The MAF of rs1061170 is lower in East Asian (<0.10) compared with European ancestry (0.3),7 and higher for rs10490924 (0.4), almost twice that in European ancestry.9 Our MAFs for rs1061170 (0.32) and rs10490924 (0.32) concur with those for South Asians in the 1000 genome study (table 1) and other sources in India.2 3 14 It appears that rs1061170 allele frequencies in Indian ancestry are closer to European than East Asian and intermediate between European and East Asian for rs10490924.
We found associations with SNPs in other genes established predominantly in European ancestry, including C2, SKIV2L and ABCA1 and in an SNP (rs4711751) in an uncharacterised gene LOC107986598 close to VEGFA.1 We found a reduced risk with the T allele of ABCA1 (rs1883025) but not with CETP or LIPC. A meta-analysis of European ancestry studies found APOE ε4 haplotype was associated with a 30% lower risk of nvAMD15; we observed a similar effect but with wide CIs.
We found no association with early AMD and any of the variants reported in table 3. We have previously reported results for early AMD and found no association with Y402H (rs1061170), C2 (rs547154) and SKIVL (rs43899).5 ARMS2/HTRA1 variants (rs10490924 and rs2672598) were associated with early AMD; the OR per allele was 1.22 (95% CI 1.13 to 1.33, P<0.0001) and 1.12 (95% CI 1.02 to 1.23, P=0.02), respectively.5 A GWAS meta-analysis of 4089 early AMD cases, the majority of European ancestry, found associations between SNPs in CFH and ARMS2/HTRA1, but with smaller effect sizes than those reported for late AMD.16 Analyses by Asian ancestry found no association with any CFH SNP, whereas ARMS2 (rs10490924) was associated with an OR of 1.18 (95% CI 1.07 to 1.13), similar to our study, compared with 1.43 (95% CI 1.34 to 1.54) for European ancestry. The lower prevalence of early AMD in Asia17 and India6 may, in part, be explained by the apparently lesser role of genetic variants compared with studies in European ancestry, but caution is warranted due to the paucity of genetic studies of early AMD in Indian and East Asian ancestries.
Limitations
Although we did not attain the 600 planned cases, we confirmed the per-allele twofold risk of rs1061170 and nvAMD hypothesised for the sample size estimates. We had low power to investigate variants with low MAFs (compared with European ancestry) such as FRK and LIPC, or to identify smaller effects. The majority of late AMD cases were of nvAMD phenotype, similar to studies in East Asia,18 and we could not investigate genetic associations with GA. It is possible we misclassified population cases of late AMD. We had confirmatory OCTs in 89% of clinical late AMD cases, but the population-based study used colour images only.
Conclusions
Our findings suggest the major genetic determinants of nvAMD risk in India are similar to those in other populations, while findings for early AMD suggest potential differences in the pathophysiology of AMD development.
Footnotes
Contributors: AEF had full access to all the data in the study and takes full responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: AEF, RDR, UC, LS, DN. Acquisition, analysis or interpretation of data: all authors. Drafting of the manuscript: AEF, RDR. Critical revision of the manuscript for important intellectual content: all authors. Statistical analysis: AEF. Obtained funding: AEF, UC, RDR, DN, LS. Administrative, technical or material support: RDR, UC, PS, AEF. Study supervision: RDR, AEF.
Funding: Wellcome Trust UK Grants G073300 and G082571.
Competing interests: None declared.
Ethics approval: Ethics approval was received from the Indian Council for Medical Research, the Research Ethics Committees of All India Institute of Medical Sciences (AIIMS) Delhi, Aravind Eye Hospital Pondicherry (Tamil Nadu), Aravind Eye Hospital Madurai (Tamil Nadu), the London School of Hygiene & Tropical Medicine, and Queen’s University Belfast.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Fritsche LG, Igl W, Bailey JN, et al. . A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet 2016;48:134–43. 10.1038/ng.3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaur I, Hussain A, Hussain N, et al. . Analysis of CFH, TLR4, and APOE polymorphism in India suggests the Tyr402His variant of CFH to be a global marker for age-related macular degeneration. Invest Ophthalmol Vis Sci 2006;47:3729–35. 10.1167/iovs.05-1430 [DOI] [PubMed] [Google Scholar]
- 3. Kaur I, Katta S, Hussain A, et al. . Variants in the 10q26 gene cluster (LOC387715 and HTRA1) exhibit enhanced risk of age-related macular degeneration along with CFH in Indian patients. Invest Ophthalmol Vis Sci 2008;49:1771–6. 10.1167/iovs.07-0560 [DOI] [PubMed] [Google Scholar]
- 4. Kaur I, Katta S, Reddy RK, et al. . The involvement of complement factor B and complement component C2 in an Indian cohort with age-related macular degeneration. Invest Ophthalmol Vis Sci 2010;51:59–63. 10.1167/iovs.09-4135 [DOI] [PubMed] [Google Scholar]
- 5. Sundaresan P, Vashist P, Ravindran RD, et al. . Polymorphisms in ARMS2/HTRA1 and complement genes and age-related macular degeneration in India: findings from the INDEYE study. Invest Ophthalmol Vis Sci 2012;53:7492–7. 10.1167/iovs.12-10073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krishnan T, Ravindran RD, Murthy GV, et al. . Prevalence of early and late age-related macular degeneration in India: the INDEYE study. Invest Ophthalmol Vis Sci 2010;51:701–7. 10.1167/iovs.09-4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sofat R, Casas JP, Webster AR, et al. . Complement factor H genetic variant and age-related macular degeneration: effect size, modifiers and relationship to disease subtype. Int J Epidemiol 2012;41:250–62. 10.1093/ije/dyr204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bird AC, Bressler NM, Bressler SB, et al. . An international classification and grading system for age-related maculopathy and age-related macular degeneration. The international ARM epidemiological study group. Surv Ophthalmol 1995;39:367–74. [DOI] [PubMed] [Google Scholar]
- 9. Fritsche LG, Fariss RN, Stambolian D, et al. . Age-related macular degeneration: genetics and biology coming together. Annu Rev Genomics Hum Genet 2014;15:151–71. 10.1146/annurev-genom-090413-025610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu Y, Bhangale TR, Fagerness J, et al. . Common variants near FRK/COL10A1 and VEGFA are associated with advanced age-related macular degeneration. Hum Mol Genet 2011;20:3699–709. 10.1093/hmg/ddr270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fritsche LG, Chen W, Schu M, et al. . Seven new loci associated with age-related macular degeneration. Nat Genet 2013;45:433–9. 10.1038/ng.2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Micklisch S, Lin Y, Jacob S, et al. . Age-related macular degeneration associated polymorphism rs10490924 in ARMS2 results in deficiency of a complement activator. J Neuroinflammation 2017;14:4 10.1186/s12974-016-0776-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng CY, Yamashiro K, Chen LJ, et al. . New loci and coding variants confer risk for age-related macular degeneration in East Asians. Nat Commun 2015;6:6063 10.1038/ncomms7063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pemberton TJ, Mehta NU, Witonsky D, et al. . Prevalence of common disease-associated variants in Asian Indians. BMC Genet 2008;9:13–20. 10.1186/1471-2156-9-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McKay GJ, Patterson CC, Chakravarthy U, et al. . Evidence of association of APOE with age-related macular degeneration: a pooled analysis of 15 studies. Hum Mutat 2011;32:1407–16. 10.1002/humu.21577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holliday EG, Smith AV, Cornes BK, et al. . Insights into the genetic architecture of early stage age-related macular degeneration: a genome-wide association study meta-analysis. PLoS One 2013;8:e53830 10.1371/journal.pone.0053830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wong WL, Su X, Li X, et al. . Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health 2014;2:e106–e116. 10.1016/S2214-109X(13)70145-1 [DOI] [PubMed] [Google Scholar]
- 18. Kawasaki R, Yasuda M, Song SJ, et al. . The prevalence of age-related macular degeneration in Asians: a systematic review and meta-analysis. Ophthalmology 2010;117:921–7. 10.1016/j.ophtha.2009.10.007 [DOI] [PubMed] [Google Scholar]