Abstract
Background
An Immunochip study recently identified the association of a number of new genetic loci with Behcet’s disease (BD).
Objective
To confirm the association between new genetic loci reported in an Immunochip study and BD in a Han Chinese population.
Methods
A two-stage association study was carried out in 1238 patients with BD and 1458 healthy controls. Twenty-two candidate single nucleotide polymorphisms (SNPs) were selected for genotyping by iPLEXGold genotyping or TaqMan SNP assays and a meta-analysis was performed for significantly associated markers.
Results
The results showed that four SNPs (LACC1/rs9316059, CEBPB-PTPN1/rs913678, ADO-EGR2/rs224127 and RIPK2/rs10094579) were associated with BD in an allelic association test (rs9316059 T allele: pc=4.95×10−8, OR=0.687; rs913678 C allele: pc=3.01×10−4, OR=1.297; rs224127 A allele: pc=3.77×10−4, OR=1.274; rs10094579 A allele: pc=6.93×10−4, OR=1.302). For four SNPs tested by meta-analysis, the association with BD was strengthened and all exceeded genome-wide significance (rs9316059: p=2.96×10−16; rs913678: p=2.09×10−16; rs224127: p=5.28×10−13; rs10094579: p=9.21×10−11).
Conclusions
Our findings confirmed the association of four loci (LACC1, CEBPB-PTPN1, ADO-EGR2 and RIPK2) in Chinese Han patients with BD.
Keywords: Genetics, Immunology, Inflammation
Introduction
BD is a chronic systemic vasculitis that mainly presents with recurrent uveitis, oral ulcers, genital ulcers and multiple skin lesions.1 BD is more common among countries along the ‘silk route’ from the Mediterranean, Middle East, China and Japan, but is rare in the USA and Europe.2 Although the aetiology and pathogenesis of BD remain unclear, it is currently thought that both genetic and environmental factors contribute to disease occurrence and development. In addition to HLA-B*51 which has been shown to have the strongest association with BD,3–7 a series of genome-wide association studies in different populations have identified a number of non-human leucocyte antigen susceptibility loci for BD, including IL23R-IL12RB2, IL10, STAT4, CCR1-CCR3, KLRC4, ERAP1, TNFAIP3, IL12A and FUT2.3–11 These findings have increased our understanding of immunogenetic factors involved in the disease. However, these identified genetic risk loci do not fully explain the genetic aetiology of BD, and other genetic factors remain to be identified.
Recently, Takeuchi et al 12 conducted a genetic association study using the Immunochip genotyping array in a Turkish cohort (1900 patients with BD and 1779 controls) and further confirmed the associations of BD with HLA-B*51, IL23R-IL12RB2, IL10, CCR1, KLRC4, ERAP1, IL12A and FUT2. More significantly, the same study identified six new BD risk loci (IL1A-IL1B, IRF8, CEBPB-PTPN1, ADO-EGR2, RIPK2 and LACC1) with genome-wide significance (p<5×10−8) and a number of new loci with a suggestive disease association (p<5×10−5). Takeuchi et al also performed a replication study12 and confirmed the association for some, but not all, loci in Iranian and Japanese cohorts. These findings indicated that the genetic background of BD may differ among different ethnic groups. Since the novel loci mentioned above have not yet been tested in other populations, we decided to perform a replication study to assess whether the findings of the Takeuchi study could be confirmed in Chinese Han patients with BD.
Materials and methods
Subjects
A total of 1238 patients with BD and 1458 healthy controls were included in the present study. All the patients were Han Chinese and recruited from the Department of Ophthalmology of the First Affiliated Hospital of Chongqing Medical University (Chongqing, China) from June 2008 to December 2016. BD was strictly diagnosed based on the criteria of the International Study Group for BD13 and all patients had uveitis. All control subjects were matched for age, sex, ethnicity (Han Chinese) and geographic origin with patients with BD. The present study was conducted in accordance with the tenets of the Declaration of Helsinki. Informed consent was obtained from all participating individuals.
SNPs selection
We selected 27 candidate SNPs in 20 susceptibility loci from the BD Immunochip association study.12 Criteria used were as follows: we selected the lead SNPs in the potential susceptibility loci with a p value less than 5×10−5. In order to increase the reliability of our results, the other susceptibility SNPs that were identified by meta-analysis and imputation were also included in this study.
Some SNPs were excluded from this study for the following reasons. (1) The SNPs in the loci that have been reported previously by our team (IL10, IL23R-IL12RB2, CCR1, ERAP1, KLRC4, IRF8, FOXP1).14–18 (2) The SNPs that were not polymorphic in Han Chinese. (3) The SNPs that were in high linkage disequilibrium (LD) with the lead SNPs in Chinese Han (r2>0.8, Han Chinese in Beijing, HCB) (table 1).
Table 1.
The potential susceptibility loci identified in the ‘Immunochip’ study for Behcet’s disease
| Number | SNP | Nearest gene(s) | Chromosome | Reported p values* |
| 1 | rs17753641† | IL12A | 3 | 8.11E−10 |
| 2 | rs17810546† | IL12A | 3 | 1.01E−07 |
| 3 | rs601338‡ | FUT2 | 19 | 6.51E−09 |
| 4 | rs1047781‡ | FUT2 | 19 | 6.50E−04 |
| 5 | rs3783550 | IL1A–IL1B | 2 | 1.29E−08 |
| 6 | rs913678 | CEBPB–PTPN1 | 20 | 1.10E−09 |
| 7 | rs7075773§ | ADO-EGR2 | 10 | 1.69E−09 |
| 8 | rs1509966‡ | ADO-EGR2 | 10 | 1.47E−06 |
| 9 | rs224127‡ | ADO-EGR2 | 10 | 1.56E−06 |
| 10 | rs9316059‡ | LACC1 | 13 | 1.16E−05 |
| 11 | rs10176241 | THADA | 2 | 3.05E−05 |
| 12 | rs79891766† | LONRF2 | 2 | 3.60E−05 |
| 13 | rs116379815† | RBM6 | 3 | 4.17E−07 |
| 14 | rs11248047 | CPLX1 | 4 | 1.27E−07 |
| 15 | rs13190001† | C5orf56 | 5 | 1.19E−05 |
| 16 | rs17705333 | INHBA | 7 | 1.82E−05 |
| 17 | rs9656588 | IKZF1 | 7 | 5.28E−06 |
| 18 | rs10094579 | RIPK2 | 8 | 6.03E−07 |
| 19 | rs2230801‡ | RIPK2 | 8 | 9.60E−06 |
| 20 | rs911603 | TNFSF8 | 9 | 1.17E−05 |
| 21 | rs28734985 | IPMK-UBE2D1 | 10 | 4.10E−05 |
| 22 | rs1698386§† | IPMK-UBE2D1 | 10 | 1.36E−05 |
| 23 | rs10896027 | MAP3K11-RELA | 11 | 2.58E−05 |
| 24 | rs58950470§ | MAP3K11-RELA | 11 | 6.25E−07 |
| 25 | rs4906762 | ATP10A | 15 | 3.81E−05 |
| 26 | rs3844576 | SOCS1-TNP2 | 16 | 3.09E−06 |
| 27 | rs1793978 | CKM-KLC3 | 19 | 2.70E−05 |
*The association of SNPs reported by the Immunochip study in Turkish patients.
†Some SNPs (six in total) were excluded since they were not polymorphic in Han Chinese (rs17753641, rs17810546, rs79891766, rs116379815, rs13190001 and rs1698386). The SNPrs2647935, with p<5×10−5, was selected as an alternative SNP in IL12A.
‡The SNPs identified by meta-analysis.
§Imputed SNPs that with more significant association than the lead SNPs in loci and that is not in high linkage disequilibrium with lead SNPs in Chinese Han population (r2 <0.8, Han Chinese Beijing).
SNP, single nucleotide polymorphism.
Genotyping
Genotyping of the selected 22 SNPs was performed in 478 BD cases and 662 controls drawn from a Chinese Han population in a first-stage study, and another independent cohort including 760 BD cases and 796 controls was examined in a second-stage confirmation study. Genotyping of 21 SNPs was performed using the MassARRAY platform (Sequenom, California, USA) and iPLEX Gold Assay. The SNPs rs1047781was genotyped by TaqMan SNP Genotyping Assay (Applied Biosystems, Foster City, California, USA) and the probe fluorescence signal was detected using the 7500 Real-Time PCR System (Applied Biosystems, USA). All SNPs tested had a high call rate (≥95% in all individual) and conformed to Hardy-Weinberg equilibrium (HWE) in the normal controls (p for HWE ≥0.05).
Statistical analysis
The χ2 test was applied for the evaluation of the HWE. Genotype and allele frequencies were compared between patients with BD and normal controls by the χ2 test or Fisher’s exact test using SPSS V.17.0. The Bonferroni correction method was applied for the correction of p values for multiple comparisons. HWE was tested by the SHEsis website. Meta-analysis in multiple populations was performed using STATA software V.12.0. The p value of heterogeneity and I2 were calculated to evaluate heterogeneity between populations. Phet<0.05 and I2 >0.5 were considered to be significant. Statistical power was estimated from effect size in the original Turkish data sets from the ‘Takeuchi’ study,12 allele frequency and sample size in the Chinese Han population. Statistical power analysis was performed using PS Power and Sample Size Calculations software (V.3.1.2; Department of Biostatistics, Vanderbilt University, Nashville, Tennessee, USA).
Results
Clinical characteristics of patients with BDs
All 1238 patients with BD had uveitis, of which 26% patients had hypopyon. The most frequent type of uveitis was panuveitis (93.2%), followed by posterior uveitis (4.9%) and anterior uveitis (1.9%). Oral ulcers (94.4%) were the most frequent extraocular manifestation, followed by skin lesions (75.2%) and genital ulcers (55.4%). The distribution of age and gender and clinical features of the enrolled patients with BD and the healthy controls in this study are shown in table 2.
Table 2.
Clinical features of patients with ocular Behcet’s disease (BD) and controls enrolled in the study
| Clinical features | Number | Percentage (%) |
| Patients with BD | 1238 | |
| Age (years), mean±SD | 33.9±9.1 | |
| Male | 1001 | 80.9 |
| Female | 237 | 19.1 |
| Uveitis | 1238 | 100 |
| Oral ulcer | 1169 | 94.4 |
| Genital ulcer | 686 | 55.4 |
| Arthritis | 223 | 18 |
| Skin lesions | 931 | 75.2 |
| Positive pathergy test | 47 | 3.8 |
| Controls | 1458 | |
| Age (years), mean±SD | 35.3±10.2 | |
| Male | 1191 | 81.7 |
| Female | 267 | 18.3 |
Association test of examined SNPs in the first phase
In the first-stage study, 22 SNPs were genotyped in 478 patients with BD and 662 normal controls. Significant higher frequencies of the CEBPB -PTPN1/rs913678 C allele (pc=1.01×10−2, OR=1.382), RIPK2/rs10094579 A allele (pc=3.16×10−2, OR=1.368) and ADO-EGR2/rs224127 A allele (pc=3.36×10−2, OR=1.318) were observed in patients with BD. In addition, the frequency of the T allele for LACC1/rs9316059 was significantly lower in patients with BD (pc=1.53×10−5; OR=0.613) (table 3). However, there was no association between the remaining SNPs and BD in this Chinese Han cohort (online supplementary table 1).
Table 3.
Main effects of tested SNPs on BD risk
| Nearest gene(s) | SNP | Stage | Genotype/allele | Case | freq | Control | freq | P values | Pc values | OR (95% CI) | Statistical power |
| CEBPB- PTPN1 | rs913678 | Stage1 | CC | 251 | 0.526 | 280 | 0.423 | 5.69E−04 | 3.75E−02 | 1.515 (1.196 to 1.920) | 0.998 |
| CT | 186 | 0.390 | 303 | 0.458 | 2.26E−02 | NS | 0.757 (0.596 to 0.962) | ||||
| TT | 40 | 0.084 | 79 | 0.119 | 0.054 | – | 0.675 (0.453 to 1.008) | ||||
| C | 688 | 0.721 | 863 | 0.652 | 4.59E−04 | 1.01E−02 | 1.382 (1.153 to 1.656) | ||||
| Stage2 | CC | 405 | 0.534 | 379 | 0.476 | 2.35E−02 | NS | 1.259 (1.031 to 1.536) | |||
| CT | 297 | 0.391 | 337 | 0.423 | 0.198 | – | 0.876 (0.715 to 1.072) | ||||
| TT | 57 | 0.075 | 80 | 0.101 | 0.077 | – | 0.727 (0.509 to 1.037) | ||||
| C | 1107 | 0.729 | 1095 | 0.688 | 1.11E−02 | 4.43E−02 | 1.222 (1.047 to 1.428) | ||||
| Combined | CC | 656 | 0.531 | 659 | 0.452 | 4.60E−05 | 3.04E−03 | 1.371 (1.178 to 1.596) | |||
| CT | 483 | 0.391 | 640 | 0.439 | 1.15E−02 | NS | 0.820 (0.703 to 0.956) | ||||
| TT | 97 | 0.078 | 159 | 0.109 | 7.01E−03 | NS | 0.696 (0.534 to 0.907) | ||||
| C | 1795 | 0.726 | 1958 | 0.671 | 1.37E−05 | 3.01E−04 | 1.297 (1.154 to 1.459) | ||||
| LACC1 | rs9316059 | Stage1 | TT | 17 | 0.036 | 56 | 0.085 | 7.87E−04 | 5.19E−02 | 0.397 (0.228 to 0.692) | 0.970 |
| TA | 170 | 0.357 | 292 | 0.445 | 2.95E−03 | NS | 0.693 (0.543 to 0.883) | ||||
| AA | 289 | 0.607 | 308 | 0.470 | 4.68E−06 | 3.09E−04 | 1.746 (1.374 to 2.219) | ||||
| T | 204 | 0.214 | 404 | 0.308 | 6.96E−07 | 1.53E−05 | 0.613 (0.505 to 0.744) | ||||
| Stage2 | TT | 46 | 0.061 | 62 | 0.078 | 0.176 | – | 0.762 (0.513 to 1.131) | |||
| TA | 260 | 0.342 | 334 | 0.420 | 1.55E−03 | 1.86E−02 | 0.718 (0.584 to 0.882) | ||||
| AA | 454 | 0.597 | 399 | 0.502 | 1.56E−04 | 1.87E−03 | 1.473 (1.205 to 1.808) | ||||
| T | 352 | 0.232 | 458 | 0.288 | 3.35E−04 | 1.34E−03 | 0.745 (0.634 to 0.875) | ||||
| Combined | TT | 63 | 0.051 | 118 | 0.081 | 1.76E−03 | NS | 0.607 (0.442 to 0.832) | |||
| TA | 430 | 0.348 | 626 | 0.431 | 9.95E−06 | 6.56E−04 | 0.703 (0.601 to 0.822) | ||||
| AA | 743 | 0.601 | 707 | 0.487 | 3.57E−09 | 2.36E−07 | 1.586 (1.360 to 1.849) | ||||
| T | 556 | 0.225 | 862 | 0.297 | 2.25E−09 | 4.95E−08 | 0.687 (0.607 to 0.777) | ||||
| RIPK2 | rs10094579 | Stage1 | AA | 29 | 0.061 | 30 | 0.045 | 0.246 | – | 1.336 (0.806 to 2.302) | 0.996 |
| CA | 205 | 0.431 | 228 | 0.345 | 3.52E−03 | NS | 1.433 (1.125 to 1.826) | ||||
| CC | 242 | 0.508 | 402 | 0.609 | 7.27E−04 | 4.80E−02 | 0.664 (0.523 to 0.842) | ||||
| A | 263 | 0.276 | 288 | 0.218 | 1.44E−03 | 3.16E−02 | 1.368 (1.128 to 1.659) | ||||
| Stage2 | AA | 58 | 0.077 | 47 | 0.059 | 0.157 | – | 1.332 (0.895 to 1.984) | |||
| CA | 294 | 0.393 | 273 | 0.344 | 4.95E−02 | NS | 1.231 (1.000 to 1.514) | ||||
| CC | 397 | 0.530 | 473 | 0.596 | 8.56E−03 | NS | 0.763 (0.624 to 0.934) | ||||
| A | 410 | 0.274 | 367 | 0.231 | 6.84E−03 | 2.74E−02 | 1.252 (1.064 to 1.473) | ||||
| Combined | AA | 87 | 0.071 | 77 | 0.053 | 0.053 | – | 1.366 (0.995 to 1.875) | |||
| CA | 499 | 0.407 | 501 | 0.345 | 8.58E−04 | NS | 1.306 (1.116 to 0.528) | ||||
| CC | 639 | 0.522 | 875 | 0.602 | 2.79E−05 | 1.84E−03 | 0.720 (0.618 to 0.840) | ||||
| A | 673 | 0.275 | 655 | 0.225 | 3.15E−05 | 6.93E−04 | 1.302 (1.149 to 1474) | ||||
| ADO- EGR2 | rs224127 | Stage1 | GG | 67 | 0.140 | 121 | 0.185 | 4.40E−02 | NS | 0.717 (0.518 to 0.992) | 0.985 |
| GA | 217 | 0.454 | 324 | 0.496 | 0.161 | – | 0.844 (0.666 to 1.070) | ||||
| AA | 194 | 0.406 | 208 | 0.319 | 2.44E−03 | NS | 1.461 (1.143 to 1.869) | ||||
| A | 605 | 0.636 | 740 | 0.567 | 1.53E−03 | 3.36E−02 | 1.318 (1.111 to 1.564) | ||||
| Stage2 | GG | 90 | 0.118 | 135 | 0.170 | 3.73E−03 | 4.48E−02 | 0.655 (0.491 to 0.873) | |||
| GA | 366 | 0.482 | 378 | 0.477 | 0.847 | – | 1.020 (0.836 to 1.245) | ||||
| AA | 304 | 0.400 | 280 | 0.353 | 5.64E−02 | – | 1.221 (0.994 to 1.500) | ||||
| A | 974 | 0.641 | 938 | 0.591 | 4.70E−03 | 1.88E−02 | 1.232 (1.066 to 1.425) | ||||
| Combined | GG | 157 | 0.127 | 256 | 0.177 | 3.25E−04 | 2.14E−02 | 0.675 (0.544 to 0.837) | |||
| GA | 583 | 0.471 | 702 | 0.485 | 0.452 | – | 0.943 (0.810 to 1.098) | ||||
| AA | 498 | 0.402 | 488 | 0.337 | 5.20E−04 | 3.43E−02 | 1.321 (1.129 to 1.546) | ||||
| A | 1579 | 0.638 | 1678 | 0.580 | 1.71E−05 | 3.77E−04 | 1.274 (1.141 to 1.422) |
Statistical power was estimated from effect size in the original Turkish data sets, allele frequency and sample size in the Chinese Han population.
BD, Behcet’s disease; pc value, the Bonferroni corrected p value; NS, not significant; SNP, single nucleotide polymorphism.
bjophthalmol-2017-311753supp001.pdf (223KB, pdf)
Association test of examined SNPs in the second phase and combined study
To further confirm the outcome of the first-stage study, we enrolled a separate set of 760 patients with BD and 796 healthy individuals for a second-stage test. We only tested the SNPs that showed a significant association in the first phase. The frequency of the T allele for LACC1/rs9316059 in patients with BD was confirmed to be significantly lower (pc=1.34×10−3, OR=0.745). In addition, the result again demonstrated significantly higher frequencies of the CEBPB-PTPN1/rs913678 C allele (pc=4.43×10−2, OR=1.222), RIPK2/rs10094579 A allele (pc=2.74×10−2, OR=1.252) and ADO-EGR2/rs224127 A allele (pc=1.88×10−2, OR=1.232) in patients with BD (table 3).
Combination of the data from the first-stage and second-stage study showed that four SNPs (rs913678, rs9316059, rs10094579 and rs224127) were significantly associated with BD (rs913678 C allele: p=1.37×10−5, pc=3.01× 10−4, OR=1.297; rs9316059 T allele: p=2.25×10−9, pc=4.95×10−8, OR=0.687; rs10094579 A allele: p=3.15×10−5, pc=6.93×10−4, OR=1.302; rs224127 A allele: p=1.71×10−5, pc=3.77×10−4, OR=1.274) (table 3).
Stratified analysis for rs913678, rs9316059, rs10094579 and rs224127
We also analysed whether rs913678, rs9316059, rs10094579 and rs224127 showed an association with the main clinical features of BD. The results did not show any significant association between the four tested SNPs and groups of patients with BD divided according to their clinical features (online supplementary table 2).
Meta-analysis
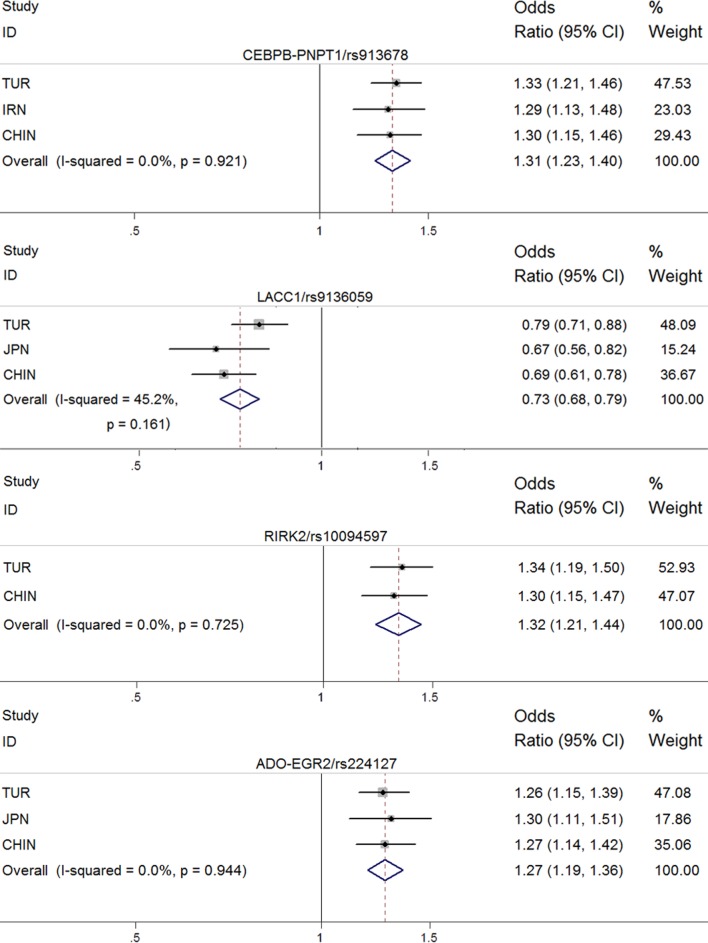
To further investigate the risk conferred by the SNPs (rs913678, rs9316059, rs10094579 and rs224127) associated with BD, we performed a meta-analysis of the genetic polymorphisms for which data were available from the Takeuchi association study and our study data sets. The results showed that the disease association of tested SNPs were reinforced after meta-analysis and all exceeded genome-wide significance (rs913678: p=2.09×10−16; rs9316059: p=2.96×10−16; rs10094579: p=9.21×10−11; rs224127: p=5.28×10−13) (table 4).
Table 4.
Meta-analysis of multiple populations for the markers replicated in the Han Chinese cohorts
| Marker (loci) | Risk allele | Population | OR | 95% CI | P values | I2 | Phet values |
| rs913678 (CEBPB-PTPN1) | C | Turkish | 1.33 | 1.21 to 1.46 | 1.10E−09 | 0 | 0.92 |
| Iranian | 1.29 | 1.13 to 1.48 | 1.59E−04 | ||||
| Han Chinese | 1.30 | 1.15 to 1.46 | 1.37E−05 | ||||
| Meta-analysis | 1.31 | 1.23 to 1.40 | 2.09E−-16 | ||||
| rs9316059 (LACC1) | T | Turkish | 0.79 | 0.71 to 0.88 | 1.16E−05 | 0.45 | 0.16 |
| Japanese | 0.67 | 0.56 to 0.82 | 5.41E−05 | ||||
| Han Chinese | 0.69 | 0.61 to 0.78 | 2.25E−09 | ||||
| Meta-analysis | 0.73 | 0.68 to 0.79 | 2.96E−16 | ||||
| rs10094579 (RIPK2) | A | Turkish | 1.34 | 1.19 to 1.50 | 6.03E−07 | 0 | 0.73 |
| Han Chinese | 1.30 | 1.15 to 1.47 | 3.15E−05 | ||||
| Meta-analysis | 1.32 | 1.21 to 1.44 | 9.21E−-11 | ||||
| rs224127 (ADO-EGR2) | A | Turkish | 1.26 | 1.15 to 1.39 | 1.56E−06 | 0 | 0.94 |
| Japanese | 1.30 | 1.11 to 1.51 | 1.10E−03 | ||||
| Han Chinese | 1.27 | 1.14 to 1.42 | 1.71E−05 | ||||
| Meta-analysis | 1.27 | 1.19 to 1.36 | 5.28E−-13 |
Meta-analysis was performed for populations in which association for the variant exceeded the replication threshold.
I2, inconsistency index; phet, p for heterogeneity.
Discussion
In the present study, we performed a replication study in a Han Chinese BD cohort for 22 candidate SNPs identified with an association p value <5× 10−5 with BD in a recent Immunochip study.12 The results showed that four SNPs (rs913678 in CEBPB - PTPN1, rs9316059 in LACC1, rs10094579 in RIPK2, rs224127 in ADO-EGR2) contribute to the genetic susceptibility to BD in a Chinese Han population.
LACC1/rs9316059, the most significantly associated SNP with BD in our study, displayed genome-wide significant association (table 3). The Immunochip study was performed in Turkish patients, and the findings concerning LACC1 rs9316059 was also confirmed in a Japanese cohort.12 In the same study, the association with SNP rs2121033 in LACC1 was identified by meta-analysis in three populations (Turkish, Iranian and Japanese). These two SNPs are in strong LD with each other (r2=0.891, in HCB). This study in combination with ours indicates that the protective LACC1 locus is a commonly associated gene for BD in all the populations tested including Chinese Han, Turkish, Iranian and Japanese. In addition, we identified the susceptibility SNP rs10094579 in RIPK2 in our cohort (table 3). To our knowledge, this is the first report showing that rs10094579 in RIPK2 confers risk to BD. This SNP only showed a suggestive disease association with a Turkish BD cohort but was not confirmed both in an Iranian and Japanese BD cohort.12 Moreover, the association we found for the other two SNPs (CEBPB-PTPN1/rs913678 and ADO-EGR2/rs224127) are in agreement with data in an Iranian population for CEBPB-PTPN1/rs913678 and a Japanese population for ADO-EGR2/rs224127 (table 3). In a meta-analysis of populations (table 4 and figure 1), we show that ADO-EGR2/rs224127, CEBPB-PTPN1/rs913678 and PIPK2/s10094579 all exceeded genome-wide significance. Based on these findings, we propose that LACC1, CEBPB–PTPN1, RIPK2 and ADO-EGR2 constitute BD susceptibility genes in Chinese Han, together with other established loci such as IL10, IL23R–IL12RB2, CCR1, IRF8, KLRC4, STAT4, ERAP1, TNFAIP3, TNFSF4, UBAC2, IL-37, IL-18RAP, GAS6, PROS1, CD6, CD11c, ATG5, TRAF5, TRAF3IP2, JAK1, MIF, PDGFRL, CD40, CIITA, NOD1, NOS3, REL and TLR2.14–34
Figure 1.
Forest plots for four SNPs associated with BD in the Chinese Han population compared with other populations. For the four SNPs, the meta-analysis refer to the C, T, A and A alleles, respectively. The broken vertical line shows the no effect point (OR 1). BD, Behcet’s disease; CHIN, HanChinese; JPN, Japanese; SNP, single nucleotide polymorphism; TUR, Turkish.
Of note, we also did not find evidence supporting the disease association with two other reported loci (IL1A-IL1B and FUT2), which both showed a genome-wide association (p<5×10−8) in the original ‘Immunochip’ report.12 IL1A-IL1B/rs3783550 that was identified in Turkish patients could also not be confirmed in Iranian as well as Japanese patients with BD. We could also not confirm the association with the FUT2/rs1047781 (T) allele, which is an ancestry-specific FUT2 non-secretor mutation (in Japanese and Han Chinese) with a significant association with BD in Japanese.12 The fact that the two SNPs mentioned above show a lack of association in Chinese Han is probably due to the different genetic background between Chinese Han and Japanese and Turkish populations, since our sample size was large enough to find a possible existing association (Power>0.8) (online supplementary table 1). Populations between different continents show between 16% and 19% genetic differences. Even within a continent, populations may differ genetically, whereby Japanese and Chinese, differ by 6.78%.35 Similarly, we did not confirm an association with the other two SNPs (rs1509966 and rs7075773) in ADO-EGR2 and BD susceptibility. Although they had a relatively high statistic power of 0.641 and 0.738, respectively, we could not exclude a false-negative disease association of rs1509966 and rs7075773 in our study. In addition, the difference of the associations for three SNPs in ADO-EGR2 among the nationalities may be partly explained by the population genetic heterogeneity. Likewise, RIPK2/rs2230801, another SNP identified in Turkish patients and confirmed in Japanese patients, did not show a significant association with BD in our study. Given that rs2230801is a rare variant in China (minor allele frequency<0.05), the statistical power to confirm this finding was low (0.23) (online supplementary table 1).
A recent study reported that the LACC1/rs3764147 (p.Ile254Val) is in high LD with rs9316059 (r2=0.892, HCB) and leads to impaired protein function.36 Furthermore, Lacc1−/− mice produce decreased IL-1β in response to lipopolysaccharide treatment, consistent with a role for IL-1β in BD pathogenesis.36 The minor allele of the other three SNPs increase the risk for BD. The CEBPB-PTPN1/rs913678 C allele is associated with decreased gene expression, and Cebpb−/− mice show increased susceptibility to pathogens.37 38 The RIPK2 kinase transduces signalling downstream of the intracellular peptidoglycan sensors NOD1 and NOD2 to promote a productive inflammatory response.39 However, excessive NOD2 signalling has been associated with numerous diseases, including inflammatory bowel disease (IBD), sarcoidosis and inflammatory arthritis.40–42 Interestingly, ADO-EGR2 has also been identified as a risk for Vogt-Koyanagi-Harada (VKH) syndrome by a previous genome-wide association study of our team.43 ADO and EGR2 were all expressed in the iris, whereas EGR2 was also expressed in ciliary body and choroid.43 VKH syndrome and BD are two of the most common types of uveitis in Chinese Han, and the fact that they share common susceptibility loci suggests that ADO-EGR2 may be a common genetic locus for uveitis, which may provide a theoretical basis for prevention and treatment of other type of uveitis.
The four novel BD susceptibility loci that we could confirm in Chinese patients have also been reported to be associated with other immune disorders. ADO-EGR2, LACC1 and CEBPB-PTPN1 are shared by BD and IBD.41 44–46 RIPK2, ADO-EGR2 and LACC1 have been shown to be associated with leprosy.47–50 These observations suggest that these diseases, whether being autoinflammatory (BD and IBD) or infectious (leprosy), may share molecular pathways although their exact role (protective or susceptibility) may differ markedly. The C allele of rs913678 in CEBPB-PTPN1 confers risk of BD but was protective for IBD and ulcerative colitis (UC).41 46 The minor allele of LACC1/rs9316059 confers protection for BD but is in high LD with a common coding variant, rs3764147 (r2=0.892, HCB), which increases risk for IBD, Crohn’s disease (CD) and leprosy.41 49 A similar discrepancy is also seen in RIPK2, where the minor allele of rs10094579 conferred risk of BD but is in high LD with rs7015630 (r2=0.976, HCB) that was protective for CD and leprosy.41 49 These discordant observations suggests that these genes which are involved in various signalling pathways may play opposite roles in BD as compared with IBD and leprosy. Further functional investigations may help to elucidate the molecular mechanisms underlying BD development and increase our understanding on the impact of these loci in the pathogenesis of autoinflammatory and infectious diseases.
Our study has several limitations. We only replicated the lead SNPs from the loci identified by a previous Immunochip study12 and we cannot exclude that other suggestive SNPs may show an association with BD in Chinese Han. In addition, there were four genes that did not show informative results. These included LONRF2, RBM6 and C5orf56 since the only SNP included was not polymorphic, and IL12A, where the two SNPs included, had one that was not polymorphic and data analysis of the other SNP IL12A/rs2647935 showed a statistical power that was too low to obtain a meaningful conclusion. Further, fine mapping for these gene regions is needed in Chinese Han to definitely show whether this is not a false-negative association. It should also be noted that all the patients with BD in the present study suffered from uveitis (100%), whereas the Turkish, Iranian and Japanese patients from the ‘Immunochip’ association study showed a lower uveitis incidence (39.4%, 56.3%, 86.9%, respectively),12 which indicates that there may be a selection bias towards ocular BD in our study. One should also be aware of the fact that most of our patients with BD are male (80.9%), which is in agreement with the previous reports from countries along the ancient Silk Road,51–53 whereas studies from Europe or the USA often show an almost equal gender distribution.54–56
In conclusion, our study not only confirms the association of LACC1/rs9316059, CEBPB-PTPN1/rs913678 and ADO-EGR2/rs224127 with BD but also identifies a novel RIPK2/rs10094579 polymorphism that affects BD susceptibility in Chinese Han. Our findings are an addition to the growing body of data from different ethnic populations, thereby gradually revealing the genetic risk landscape of BD. Further investigations on how these gene polymorphisms exactly affect BD are needed and may provide future targets for its treatment.
Acknowledgments
The authors thank the patients and controls for their participation in this study.
Footnotes
PW, LD and SH contributed equally.
Contributors: PW, LD and SH are joint first authors and anlaysed the data. PW and PY designed the study. PW, LY and JH collected the data. PY made clinical diagnoses. GS, LY, JH, JD, QC and CZ collected the samples. LY, JH, JD, QC and GY extracted the blood DNA. PW drafted the manuscript. PY, SH, GS and AK helped revise the manuscript. All authors have read and approved the final manuscript.
Funding: This work was supported by Major Research Development Program of China (2016YFC0904000), Natural Science Foundation Major International (Regional) Joint Research Project (81720108009), Natural Science Foundation Major International (Regional) Joint Research Project (81320108009), National Natural Science Foundation Project (81522013), Chongqing Outstanding Youth Grant (cstc2014jcyjjq10005), Chongqing Key Laboratory of Ophthalmology (CSTC2008CA5003), Chongqing Science and Technology Platform and Base Construction Program (cstc2014pt-sy10002) and Research fund for Traditional Chinese Medicine of Chongqing Health and FamilyPlanning Commission (ZY201401013). Thanks to all donors enrolled in the present study.
Competing interests: None declared.
Patient consent: Parental/guardian consent obtained.
Ethics approval: This study was approved by the ethics committee of the First Affiliated Hospital of Chongqing Medical University, Chongqing, China (Permit Number 2009–201008).
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Yang P, Fang W, Meng Q, et al. Clinical features of chinese patients with Behçet’s disease. Ophthalmology 2008;115:312–8. 10.1016/j.ophtha.2007.04.056 [DOI] [PubMed] [Google Scholar]
- 2. Skef W, Hamilton MJ, Arayssi T. Gastrointestinal Behçet’s disease: a review. World J Gastroenterol 2015;21:3801–12. 10.3748/wjg.v21.i13.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gul A, Ohno S. HLA-B*51 and Behçet Disease. Ocul Immunol Inflamm 2012;20:37–43. 10.3109/09273948.2011.634978 [DOI] [PubMed] [Google Scholar]
- 4. Remmers EF, Cosan F, Kirino Y, et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behçet’s disease. Nat Genet 2010;42:698–702. 10.1038/ng.625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mizuki N, Meguro A, Ota M, et al. Genome-wide association studies identify IL23R-IL12RB2 and IL10 as Behçet’s disease susceptibility loci. Nat Genet 2010;42:703–6. 10.1038/ng.624 [DOI] [PubMed] [Google Scholar]
- 6. Kirino Y, Bertsias G, Ishigatsubo Y, et al. Genome-wide association analysis identifies new susceptibility loci for Behçet’s disease and epistasis between HLA-B*51 and ERAP1. Nat Genet 2013;45:202–7. 10.1038/ng.2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ortiz-Fernández L, Carmona FD, Montes-Cano MA, et al. Genetic analysis with the immunochip platform in Behçet disease. Identification of residues associated in the HLA class I region and new susceptibility loci. PLoS One 2016;11:e0161305 10.1371/journal.pone.0161305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fei Y, Webb R, Cobb BL, et al. Identification of novel genetic susceptibility loci for Behçet’s disease using a genome-wide association study. Arthritis Res Ther 2009;11:R66 10.1186/ar2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hou S, Yang Z, Du L, et al. Identification of a susceptibility locus in STAT4 for Behçet’s disease in Han Chinese in a genome-wide association study. Arthritis Rheum 2012;64:4104–13. 10.1002/art.37708 [DOI] [PubMed] [Google Scholar]
- 10. Xavier JM, Shahram F, Sousa I, et al. FUT2: filling the gap between genes and environment in Behçet’s disease? Ann Rheum Dis 2015;74:618–24. 10.1136/annrheumdis-2013-204475 [DOI] [PubMed] [Google Scholar]
- 11. Kappen JH, Medina-Gomez C, van Hagen PM, et al. Genome-wide association study in an admixed case series reveals IL12A as a new candidate in Behçet disease. PLoS One 2015;10:e0119085 10.1371/journal.pone.0119085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takeuchi M, Mizuki N, Meguro A, et al. Dense genotyping of immune-related loci implicates host responses to microbial exposure in Behçet’s disease susceptibility. Nat Genet 2017;49:438–43. 10.1038/ng.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Criteria for diagnosis of Behçet’s disease. International Study Group for Behçet’s Disease. Lancet 1990;335:1078–80. [PubMed] [Google Scholar]
- 14. Yu H, Zheng M, Zhang L, et al. Identification of susceptibility SNPs in IL10 and IL23R-IL12RB2 for Behçet’s disease in Han Chinese. J Allergy Clin Immunol 2017;139:621–7. 10.1016/j.jaci.2016.05.024 [DOI] [PubMed] [Google Scholar]
- 15. Hou S, Xiao X, Li F, et al. Two-stage association study in Chinese Han identifies two independent associations in CCR1/CCR3 locus as candidate for Behçet’s disease susceptibility. Hum Genet 2012;131:1841–50. 10.1007/s00439-012-1200-4 [DOI] [PubMed] [Google Scholar]
- 16. Zhang L, Yu H, Zheng M, et al. Association of ERAP1 gene polymorphisms with Behçet’s disease in Han Chinese. Invest Ophthalmol Vis Sci 2015;56:6029–35. 10.1167/iovs.15-17544 [DOI] [PubMed] [Google Scholar]
- 17. Yang Y, Tan H, Deng B, et al. Genetic polymorphisms of C-type lectin receptors in Behcet’s disease in a Chinese Han population. Sci Rep 2017;7:5348 10.1038/s41598-017-05877-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang Y, Wang H, Yu H, et al. Two Genetic Variations in the IRF8 region are associated with Behçet’s disease in Han Chinese. Sci Rep 2016;6:19651 10.1038/srep19651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li H, Liu Q, Hou S, et al. TNFAIP3 gene polymorphisms confer risk for Behcet’s disease in a Chinese Han population. Hum Genet 2013;132:293–300. 10.1007/s00439-012-1250-7 [DOI] [PubMed] [Google Scholar]
- 20. Hou S, Shu Q, Jiang Z, et al. Replication study confirms the association between UBAC2 and Behçet’s disease in two independent Chinese sets of patients and controls. Arthritis Res Ther 2012;14:R70 10.1186/ar3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan H, Deng B, Yu H, et al. Genetic analysis of innate immunity in Behcet’s disease identifies an association with IL-37 and IL-18RAP. Sci Rep 2016;6:35802 10.1038/srep35802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qin J, Li L, Zhang D, et al. Analysis of receptor tyrosine kinase genetics identifies two novel risk loci in GAS6 and PROS1 in Behçet’s disease. Sci Rep 2016;6:26662 10.1038/srep26662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng M, Zhang L, Yu H, et al. Genetic polymorphisms of cell adhesion molecules in Behcet’s disease in a Chinese Han population. Sci Rep 2016;6:24974 10.1038/srep24974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng M, Yu H, Zhang L, et al. Association of ATG5 gene polymorphisms with Behçet’s disease and ATG10 gene polymorphisms with VKH syndrome in a Chinese Han population. Invest Ophthalmol Vis Sci 2015;56:8280–7. 10.1167/iovs.15-18035 [DOI] [PubMed] [Google Scholar]
- 25. Xiang Q, Chen L, Hou S, et al. TRAF5 and TRAF3IP2 gene polymorphisms are associated with Behçet’s disease and Vogt-Koyanagi-Harada syndrome: a case-control study. PLoS One 2014;9:e84214 10.1371/journal.pone.0084214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hou S, Qi J, Zhang Q, et al. Genetic variants in the JAK1 gene confer higher risk of Behcet’s disease with ocular involvement in Han Chinese. Hum Genet 2013;132:1049–58. 10.1007/s00439-013-1312-5 [DOI] [PubMed] [Google Scholar]
- 27. Zheng X, Wang D, Hou S, et al. Association of macrophage migration inhibitory factor gene polymorphisms with Behçet’s disease in a Han Chinese population. Ophthalmology 2012;119:2514–8. 10.1016/j.ophtha.2012.06.039 [DOI] [PubMed] [Google Scholar]
- 28. Hou S, Xiao X, Zhou Y, et al. Genetic variant on PDGFRL associated with Behçet disease in Chinese Han populations. Hum Mutat 2013;34:74–8. 10.1002/humu.22208 [DOI] [PubMed] [Google Scholar]
- 29. Chen F, Hou S, Jiang Z, et al. CD40 gene polymorphisms confer risk of Behcet’s disease but not of Vogt-Koyanagi-Harada syndrome in a Han Chinese population. Rheumatology 2012;51:47–51. 10.1093/rheumatology/ker345 [DOI] [PubMed] [Google Scholar]
- 30. Li L, Yu H, Jiang Y, et al. Genetic Variations of NLR family genes in Behcet’s Disease. Sci Rep 2016;6:20098 10.1038/srep20098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou Y, Yu H, Hou S, et al. Association of a NOS3 gene polymorphism with Behçet’s disease but not with Vogt-Koyanagi-Harada syndrome in Han Chinese. Mol Vis 2016;22:311–8. [PMC free article] [PubMed] [Google Scholar]
- 32. Chen F, Xu L, Zhao T, et al. Genetic variation in the REL gene increases risk of Behcet’s disease in a Chinese Han population but that of PRKCQ does not. PLoS One 2016;11:e0147350 10.1371/journal.pone.0147350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fang J, Hu R, Hou S, et al. Association of TLR2 gene polymorphisms with ocular Behcet’s disease in a Chinese Han population. Invest Ophthalmol Vis Sci 2013;54:8384–92. 10.1167/iovs.13-12878 [DOI] [PubMed] [Google Scholar]
- 34. Lu S, Song S, Hou S, et al. Association of TNFSF4 Polymorphisms with Vogt-Koyanagi-Harada and Behcet’s Disease in Han Chinese. Sci Rep 2016;6:37257 10.1038/srep37257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller RD, Phillips MS, Jo I, et al. SNP Consortium Allele Frequency Project. High-density single-nucleotide polymorphism maps of the human genome. Genomics 2005;86:117–26. 10.1016/j.ygeno.2005.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cader MZ, Boroviak K, Zhang Q, et al. C13orf31 (FAMIN) is a central regulator of immunometabolic function. Nat Immunol 2016;17:1046–56. 10.1038/ni.3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Screpanti I, Romani L, Musiani P, et al. Lymphoproliferative disorder and imbalanced T-helper response in C/EBP beta-deficient mice. EMBO J 1995;14:1932–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tanaka T, Akira S, Yoshida K, et al. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell 1995;80:353–61. 10.1016/0092-8674(95)90418-2 [DOI] [PubMed] [Google Scholar]
- 39. Nachbur U, Stafford CA, Bankovacki A, et al. A RIPK2 inhibitor delays NOD signalling events yet prevents inflammatory cytokine production. Nat Commun 2015;6:6442 10.1038/ncomms7442 [DOI] [PubMed] [Google Scholar]
- 40. Henckaerts L, Vermeire S. NOD2/CARD15 disease associations other than Crohn’s disease. Inflamm Bowel Dis 2007;13:235–41. 10.1002/ibd.20066 [DOI] [PubMed] [Google Scholar]
- 41. Jostins L, Ripke S, Weersma RK, et al. International IBD Genetics Consortium (IIBDGC). Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001;411:599–603. 10.1038/35079107 [DOI] [PubMed] [Google Scholar]
- 43. Hou S, Du L, Lei B, et al. Genome-wide association analysis of Vogt-Koyanagi-Harada syndrome identifies two new susceptibility loci at 1p31.2 and 10q21.3. Nat Genet 2014;46:1007–11. 10.1038/ng.3061 [DOI] [PubMed] [Google Scholar]
- 44. Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet 2010;42:1118–25. 10.1038/ng.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu JZ, van Sommeren S, Huang H, et al. International Multiple Sclerosis Genetics Consortium International IBD Genetics Consortium. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. 10.1038/ng.3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Orlando G, Law PJ, Palin K, et al. Variation at 2q35 (PNKD and TMBIM1) influences colorectal cancer risk and identifies a pleiotropic effect with inflammatory bowel disease. Hum Mol Genet 2016;25:2349–59. 10.1093/hmg/ddw087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tsoi LC, Spain SL, Knight J, et al. Collaborative Association Study of Psoriasis (CASP) Genetic Analysis of Psoriasis Consortium Psoriasis Association Genetics Extension Wellcome Trust Case Control Consortium 2. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet 2012;44:1341–8. 10.1038/ng.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang FR, Huang W, Chen SM, et al. Genomewide association study of leprosy. N Engl J Med 2009;361:2609–18. 10.1056/NEJMoa0903753 [DOI] [PubMed] [Google Scholar]
- 49. Liu H, Irwanto A, Fu X, et al. Discovery of six new susceptibility loci and analysis of pleiotropic effects in leprosy. Nat Genet 2015;47:267–71. 10.1038/ng.3212 [DOI] [PubMed] [Google Scholar]
- 50. Sales-Marques C, Salomão H, Fava VM, et al. NOD2 and CCDC122-LACC1 genes are associated with leprosy susceptibility in Brazilians. Hum Genet 2014;133:1525–32. 10.1007/s00439-014-1502-9 [DOI] [PubMed] [Google Scholar]
- 51. Yoshida A, Kawashima H, Motoyama Y, et al. Comparison of patients with Behçet’s disease in the 1980s and 1990s. Ophthalmology 2004;111:810–5. 10.1016/j.ophtha.2003.07.018 [DOI] [PubMed] [Google Scholar]
- 52. Tugal-Tutkun I, Onal S, Altan-Yaycioglu R, et al. Uveitis in Behçet disease: an analysis of 880 patients. Am J Ophthalmol 2004;138:373–80. 10.1016/j.ajo.2004.03.022 [DOI] [PubMed] [Google Scholar]
- 53. Keino H, Okada AA. Behçet’s disease: global epidemiology of an Old Silk Road disease. Br J Ophthalmol 2007;91:1573–4. 10.1136/bjo.2007.124875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kaçmaz RO, Kempen JH, Newcomb C, et al. Systemic Immunosuppressive Therapy for Eye Diseases Cohort Study Group. Ocular inflammation in Behçet disease: incidence of ocular complications and of loss of visual acuity. Am J Ophthalmol 2008;146:828–36. 10.1016/j.ajo.2008.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Krause I, Yankevich A, Fraser A, et al. Prevalence and clinical aspects of Behcet’s disease in the north of Israel. Clin Rheumatol 2007;26:555–60. 10.1007/s10067-006-0349-4 [DOI] [PubMed] [Google Scholar]
- 56. Salvarani C, Pipitone N, Catanoso MG, et al. Epidemiology and clinical course of Behçet’s disease in the Reggio Emilia area of Northern Italy: a seventeen-year population-based study. Arthritis Rheum 2007;57:171–8. 10.1002/art.22500 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bjophthalmol-2017-311753supp001.pdf (223KB, pdf)