ABSTRACT
In yeast, transcription of ribosomal DNA (rDNA) by RNA polymerase I (Pol I) is regulated by unique mechanisms acting at the level of the enzyme. Under stress situations such as starvation, Pol I hibernates through dimerization. When growth conditions are restored, dimer disassembly and Rrn3 binding drive enzyme activation and subsequent recruitment to rDNA.
KEYWORDS: ribosomal DNA, RNA polymerase I, transcriptional regulation, conformational state, enzyme activation, enzyme hibernation, cell growth
Introduction
Ribosomal DNA (rDNA) presents distinctive features. First, it is formed by several hundreds of tandem repeats of about 10 kb in yeast that cluster together to form the nucleolus [1]. Second, promoters contain two main regions termed upstream element (UE) and core element (CE) in yeast, with the latter overlapping with the transcription start site and the former extending to about 150 bp upstream of this site [2]. Third, in actively growing cells rDNA transcription represents 60% of the total transcriptional activity [3], which correlates with a high content of RNA polymerase occupancy on rDNA genes [4]. As a consequence, rDNA transcription is a major point for the regulation of cell growth and, thus, misregulation of the system is related with tumour development [5].
Initiation of rDNA transcription in yeast requires the sequential assembly of factors on different promoter elements (reviewed in [6,7]). The upstream activating factor (UAF) binds the UE [2], while the core factor (CF) binds the CE [8]. The two protein complexes interact with each other and are bridged through TATA-box binding protein (TBP) binding [9]. While the CF is sufficient for promoter recognition, UAF and TBP enhance rDNA transcription [10]. Promoter-attached initiation factors foster binding of RNA polymerase I (Pol I) in complex with the Rrn3 protein [11–13]. The complete assembly of enzyme and factors on the promoter constitutes the pre-initiation complex (PIC) that has been proposed to evolve from a closed to an open complex upon DNA melting, which in Pol I is independent from ATP hydrolysis [14–16]. As proposed by these authors, an initially transcribing complex (ITC) forms upon addition of the first RNA nucleotides in the presence of initiation factors, and subsequent enzyme dissociation from the PIC leads to the formation of an elongation complex (EC). Elongation is paused in case of an obstacle in the transcribed region, which is resolved by enzyme backtracking and subsequent RNA cleavage [17]. At the terminator, a similar situation likely leads to enzyme detachment, which may be mediated by external factors such as Nsi1 in yeast [18].
In spite of its central role in cell homeostasis, the study of the Pol I transcription system was neglected in favour of Pol II, responsible for the transcription of protein-coding genes. In recent years, however, a series of structural biology investigations have started to unveil the mechanisms regulating Pol I-mediated transcription. Two independent studies reported the crystal structure of Pol I in its inactive dimeric conformation [19,20]. More recently, electron cryomicroscopy (cryo-EM) analysis revealed the structures of Pol I in the free monomeric and Rrn3-bound states [21–24]. Additional cryo-EM studies reported the structures of Pol I engaged in elongation [22,25]. Finally, the structures of a minimal PIC comprising the CF, Rrn3 and Pol I in the absence and presence of melted DNA provided hints into the initiation process [14–16].
Pol I subunits, modules and structural domains
Pol I is formed by 14 subunits with a total mass of 590 kDa (Figure 1(A)). The enzyme conserves the overall crab-claw shape defined for other multi-subunit RNA polymerases [19,20], with a central cleft that binds downstream DNA [22,25]. Below the active site, located at the centre of the enzyme, a secondary pore opens in the floor of the cleft. In Pol II, the pore was suggested to constitute the entry site for NTPs [26] and shown to harbour extruded RNA during backtracking [27]. Next to the pore, the bridge helix crosses the cleft in the vicinity of the trigger loop. In Pol II, these structural elements participate in enzyme translocation along DNA during elongation [27].
Figure 1.
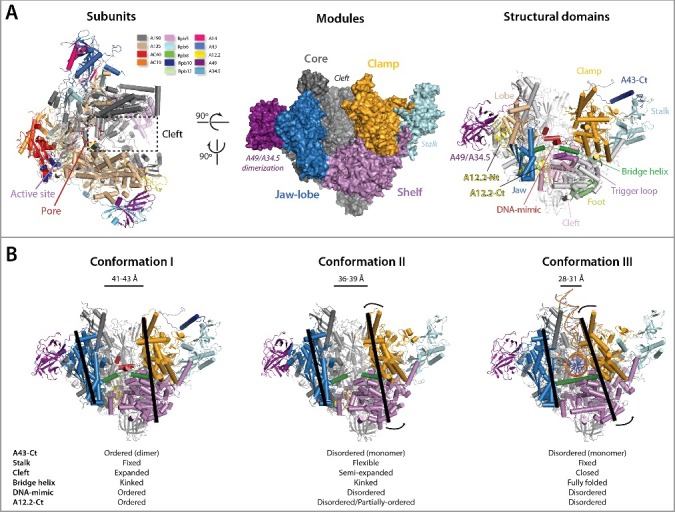
(A) Pol I subunits, modules and domains. On the left, the 14 subunits forming the complex are coloured according to the top legend. The middle panel shows the 4 modules defined for Pol II [26] as well as the stalk and the A49/A34.5 dimerization region. On the right, the most important structural domains are labelled. (B) The three major conformations of the Pol I enzyme as derived from recent structural studies, with defining features below. Black bars on the jaw-lobe and the rigid shelf-clamp-stalk are depicted to help visualising their relative movements.
In analogy to Pol II and bacterial RNA polymerase [26,28], four mobile modules can be defined in Pol I (Figure 1(A)). The core module harbours essential elements for RNA polymerization such as the active centre and the pore, as well as several subunits and regions involved in enzyme assembly, including subunits AC40, AC19, Rpb10 and Rpb12. The three remaining modules lie on both sides of the DNA-binding cleft. On one side is the lobe-jaw module, which in Pol I comprises the lobe domain in subunit A135, the jaw domain in A190, and the N-terminal and linker regions of A12.2. As the dimerization region of the A49/A34.5 heterodimer binds on the lobe, it can be included inside this module. The shelf and clamp modules locate on the opposite side of the cleft. The shelf includes the foot and cleft regions of A190, and subunits Rpb5 and Rpb6. The clamp essentially comprises domains of A190 with a minor contribution of A135. In contrast to Pol II where clamp swinging over the shelf was shown to play a role in transcriptional regulation [26], the clamp and shelf modules in Pol I constitute a rigid mobile unit [22]. Because the Pol I stalk, formed by the A43/A14 heterodimer, is tightly attached to both the shelf and the clamp, these two modules and the stalk can be regarded as a single unit that pivots against the core and jaw-lobe modules for cleft closure [20,25]. Two additional structural elements that are unique in Pol I are only ordered in the inactive homodimeric configuration. The A43 C-terminal tail (residues 251–326) including the connector element (residues 262–326) is essential for enzyme dimerization. The expander in subunit A190 (residues 1337–1439) including the DNA-mimicking loop (residues 1361–1399) blocks the binding site of nucleic acids inside the cleft.
Conformational states in Pol I
Structural studies allow definition of three major conformations of the Pol I enzyme (Figure 1(B)). Conformation I corresponds to the inactive state of the enzyme, as observed both in crystals and in solution [19,20,23]. This configuration is defined by an expanded DNA-binding cleft of about 42 Å in width and a bridge helix that is partially unfolded in its central region. In addition, the DNA-mimicking loop is well ordered inside the cleft. Moreover, the A12.2 C-terminal domain, involved in RNA cleavage [29], is located inside the pore. As mentioned, ordering of the A43 C-terminal tail enables essential contacts for enzyme dimerization. Conformation II corresponds to free monomeric Pol I, and it has also been observed in Rrn3-bound Pol I [21–24]. This configuration presents a semi-expanded cleft of about 38 Å in width, a partially-unfolded bridge helix and a disordered DNA-mimicking loop. This conformation has been compared with the ratcheted state in bacterial RNA polymerase and suggested to reflect the backtracked state of the Pol I enzyme [22]. In this conformation, the A12.2 C-terminal domain has been found either disordered or partially ordered inside the pore. The major difference between free monomeric and Rrn3-bound Pol I is the stalk, which appears flexible in the former while it is fixed upon Rrn3 interaction, with the exception of the A43 C-terminal tail [24]. Finally, conformation III corresponds to Pol I in the pre-initiation and elongation complexes, and is defined by a closed cleft of about 30 Å in width and a fully ordered bridge helix [14–16,22,25]. As expected by the presence of nucleic acids in the cleft, the DNA-mimicking loop is disordered. In addition, the 12.2 C-terminal domain is disordered, suggesting that in the presence of nucleic acids it may only access the pore during enzyme backtracking. Minor differences between reported structures in conformation III allowed definition of three structural states of the enzyme in the minimal PIC [15].
The transition between conformations I and II involves partial closure of the DNA-binding cleft by approach of the rigid shelf-clamp module towards the jaw-lobe (Figure 1(B)). The lobe and clamp approach, while the jaw and shelf slightly move away from each other. Within each structural module, minimal rearrangements of internal domains are observed. The bridge helix remains partially unfolded and the trigger loop is disordered. This transition, which implies disruption of Pol I homodimers into monomers and disordering of the DNA-mimicking loop, plays an important role in the regulation of rDNA transcription [24]. The transition between conformations II and III involves full closure of the cleft by further approach of the rigid shelf-clamp module towards the jaw-lobe (Figure 1(B)). It has been suggested that binding of nucleic acids in the cleft maintains conformation III, while their absence allows access of the A12.2 C-terminal domain inside the pore, which partially widens the cleft by opening of the clamp-shelf and jaw-lobe modules [22]. The transition between conformations II and III likely plays a central role in the progression from closed to open PIC, as well as during transcriptional pausing.
Pol I enzyme activation
In actively growing cells, most cellular Pol I is engaged in rDNA transcription. When detached from DNA, free monomeric Pol I cannot directly be recruited to rDNA promoters but requires prior binding to the activating factor Rrn3 [12,13]. This event allows direct downregulation of rRNA production by Rrn3 degradation [24,30]. In addition, rDNA transcription is also down-tuned by Pol I dimerization [24]. Upon nutrient addition, the transition from inactive homodimers to activated Pol I requires at least two events (Figure 2). First, enzyme dimers must dissociate to yield monomeric Pol I. Second, Pol I monomers need to bind Rrn3 in order to be recruited to promoter DNA. Rrn3 contacts the enzyme at both stalk subunits, A43 and A14, as well as A190 and the AC40/AC19 heterodimer [21,23,24]. Enzyme activation can, thus, be seen as a two-step process in the recovery from stress conditions such as nutrient deprivation. Enzyme inactivation applies to the opposite route.
Figure 2.

Principal mechanisms at the level of the Pol I enzyme regulating rDNA transcription. Dotted lines indicate flexible regions, while encircled “P” symbols represent described phosphorylations [39,41].
In bacteria, a holoenzyme was defined to designate the complex between the RNA polymerase core and the σ factor, which is sufficient for promoter binding and DNA melting [31]. In archaea and eukaryotes the situation is more sophisticated because, for promoter-specific transcription, general transcription factors must bind DNA prior to enzyme recruitment. Nevertheless, a holoenzyme was defined for Pol II when complexed to the Mediator [32]. While description of eukaryotic holoenzymes may be controversial, the transcription-competent Pol I-Rrn3 complex could be regarded as a holoenzyme that, in analogy to Pol II, requires contact with promoter-bound transcription factors to initiate rRNA synthesis. In agreement, Rrn3 occupies a similar area as the Mediator in Pol II.
Pol I hibernation by dimerization
In vitro Pol I dimerization was first observed by the end of last century [33] and later postulated to represent a regulatory mechanism [19]. We showed for the first time that Pol I dimerization occurs in vivo when cells are subjected to stress conditions that compromise processes downstream of rRNA synthesis [24]. Nutrient depletion but also the use of drugs blocking ribosome biogenesis or protein synthesis, all trigger the formation of Pol I dimers, which associates with reduced levels of the enzyme on rDNA promoters. In agreement with structural data, this process relies on the A43 C-terminal tail, as removal of the last 20 residues in this subunit hampers Pol I dimerization. Moreover, in starving conditions, this deletion strain presents higher levels of Pol I and Rrn3 associated to rDNA promoters than the wild-type strain, suggesting that Pol I dimerization downregulates rDNA transcription. In addition, it was also shown that Pol II or Pol III do not homodimerize under nutrient starvation, indicating that this mode of transcriptional inactivation is unique for Pol I.
Pol I dimerization can be seen as a hibernating mechanism under harsh environmental conditions (Figure 2). Pol I hibernation might protect the enzyme from degradation and, at the same time, allow fast reactivation when favourable growth conditions are restored. Interestingly, a similar mode of hibernation by dimerization has been observed for bacterial ribosomes [34,35]. Nevertheless, while the formation of ribosome homodimers, also termed disomes, relies on external factors that bind prior to dimerization, structural studies established that Pol I dimerization does not require binding of external factors [19,20]. Moreover, it was shown that Rrn3 addition is unable to disassemble Pol I dimers [23]. Therefore, control of the Pol I monomer-dimer transition relies on yet undescribed regulatory mechanisms. In addition to dimerization, hibernation implies cleft expansion and ordering of the DNA-mimicking loop inside the cleft. In Pol II and bacterial RNA polymerase, it was shown that certain RNAs and proteins can block the enzyme by binding inside the cleft [36–38]. The DNA-mimicking loop within the expander could have a protective function in the Pol I hibernating state, by hampering the binding of macromolecules that could compromise enzyme reactivation.
The role of phosphorylation
Phosphorylation might play an important role in Pol I activation and inactivation. It was shown that only unphosphorylated Rrn3 is able to bind Pol I in yeast, while the polymerase must be phosphorylated for this interaction to occur [39]. In agreement, the S145D phospho-mimetic mutation in yeast Rrn3 impairs the formation of the Pol I-Rrn3 complex and associates with reduced levels of both Pol I and Rrn3 on rDNA promoters [40]. In addition, a proteomic study in yeast revealed several phosphosites in Pol I-specific subunits A190, A34.5 and A43, but single mutations of specific residues did not affect Pol I-Rrn3 complex formation [41]. However, all A43 phosphosites identified in this report locate in regions connected with Pol I dimerization. In particular, Ser208 and Ser220 lie next to the Pol I dimer interface, while Ser262/263 and Ser285 belong to the A43 C-terminal tail, which is essential for dimerization. This suggests that, rather than a direct effect on Rrn3 interaction, phosphorylation of the A43 C-terminal region may regulate the Pol I monomer-dimer transition. Interestingly, Ser220 and Ser262/263 are fully exposed in the dimeric configuration, while Ser208 is in a flexible loop [19,20]. Therefore, phosphorylation of these residues may drive dimer disassembly, while Ser285 may play a role at a later stage. In a scenario of nutrient deprivation, dephosphorylation of the A43 C-terminal region would allow dimer formation (Figure 2). When nutrients are restored, phosphorylation of this region in Pol I dimers would increase the levels of free monomeric Pol I, while dephosphorylation of Rrn3 would allow the formation of Pol I-Rrn3 complexes to restore rDNA transcription. Identification of the kinases and phosphatases controlling this process will likely provide clues to understand how this transcription system is regulated.
Finally, phosphorylation may also play a role in the regulatory function of the expander. Ser1413, Ser1415 and Ser1417 in subunit A190, all belonging to this loop, were identified as phosphosites in the proteomic study [41]. Deletion of the DNA-mimicking loop within the expander exhibits a mild growth phenotype at 37 ºC [20] but the phosphosites lie outside the deleted region. Further investigations are, thus, required to address how phosphorylation may influence the role of the expander in Pol I transcription.
Concluding remarks
Structural and functional studies of the Pol I system have revealed unique mechanisms that regulate rDNA transcription. Importantly, the stalk has emerged as a central Pol I platform to direct the enzyme towards activation or hibernation, depending on external conditions. Future structural research will likely shed light on additional conformational changes and interactions regulating the system. In addition, additional studies on how phosphorylation controls enzyme activation by Rrn3 binding and enzyme inactivation by hibernation will deepen our understanding of this essential cell process and its relation to cell growth control.
Funding Statement
This work was supported by the Spanish Ministry of Economy and Competitiveness (MINECO) under Grant BFU2013-48374-P and by the Ramón Areces Foundation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgements
I thank Olivier Gadal and Srdja Drakulic for critically reading the manuscript and Marta Sanz-Murillo for helpful discussion during manuscript preparation.
References
- [1].Birch JL, Zomerdijk JC. Structure and function of ribosomal RNA gene chromatin. Biochem Soc Trans. 2008;36:619–624. doi: 10.1042/BST0360619. PMID: 18631128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Keys DA, Lee BS, Dodd JA, et al. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 1996;10:887–903. doi: 10.1101/gad.10.7.887. PMID: 8846924 [DOI] [PubMed] [Google Scholar]
- [3].Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/S0968-0004(99)01460-7. PMID: 10542411 [DOI] [PubMed] [Google Scholar]
- [4].Osheim YN, French SL, Sikes ML, et al. Electron microscope visualization of RNA transcription and processing in Saccharomyces cerevisiae by Miller chromatin spreading. Methods Mol Biol. 2009;464:55–69. doi: 10.1007/978-1-60327-461-6_4. PMID: 18951179 [DOI] [PubMed] [Google Scholar]
- [5].Bywater MJ, Pearson RB, McArthur GA, et al. Dysregulation of the basal RNA polymerase transcription apparatus in cancer. Nat Rev Cancer. 2013;13:299–314. doi: 10.1038/nrc3496. PMID: 23612459 [DOI] [PubMed] [Google Scholar]
- [6].Goodfellow SJ, Zomerdijk JC. Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Subcell Biochem. 2013;61:211–236. doi: 10.1007/978-94-007-4525-4_10. PMID: 23150253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schneider DA. RNA polymerase I activity is regulated at multiple steps in the transcription cycle: recent insights into factors that influence transcription elongation. Gene. 2012;493:176–184. doi: 10.1016/j.gene.2011.08.006. PMID: 21893173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Steffan JS, Keys DA, Dodd JA, et al. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 1996;10:2551–2563. doi: 10.1101/gad.10.20.2551. PMID: 8895657 [DOI] [PubMed] [Google Scholar]
- [9].Lin CW, Moorefield B, Payne J, et al. A novel 66-kilodalton protein complexes with Rrn6, Rrn7, and TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:6436–6443. doi: 10.1128/MCB.16.11.6436. PMID: 8887672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bedwell GJ, Appling FD, Anderson SJ, et al. Efficient transcription by RNA polymerase I using recombinant core factor. Gene. 2012;492:94–99. doi: 10.1016/j.gene.2011.10.049. PMID: 22093875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Aprikian P, Moorefield B, Reeder RH. New model for the yeast RNA polymerase I transcription cycle. Mol Cell Biol. 2001;21:4847–4855. doi: 10.1128/MCB.21.15.4847-4855.2001. PMID: 11438642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Keener J, Josaitis CA, Dodd JA, et al. Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J Biol Chem. 1998;273:33795–33802. doi: 10.1074/jbc.273.50.33795. PMID: 9837969 [DOI] [PubMed] [Google Scholar]
- [13].Yamamoto RT, Nogi Y, Dodd JA, et al. RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J. 1996;15:3964–3973. PMID: 8670901 [PMC free article] [PubMed] [Google Scholar]
- [14].Engel C, Gubbey T, Neyer S, et al. Structural basis of RNA polymerase I transcription initiation. Cell. 2017;169:120–131, e22. doi: 10.1016/j.cell.2017.03.003. PMID: 28340337 [DOI] [PubMed] [Google Scholar]
- [15].Han Y, Yan C, Nguyen THD, et al. Structural mechanism of ATP-independent transcription initiation by RNA polymerase I. Elife. 2017;6: PMID: 28623663; doi: 10.7554/eLife.27414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sadian Y, Tafur L, Kosinski J, et al. Structural insights into transcription initiation by yeast RNA polymerase I. EMBO J. 2017;36:2698–2709. doi: 10.15252/embj.201796958. PMID: 28739580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lisica A, Engel C, Jahnel M, et al. Mechanisms of backtrack recovery by RNA polymerases I and II. Proc Natl Acad Sci USA. 2016;113:2946–2951. doi: 10.1073/pnas.1517011113. PMID: 26929337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Reiter A, Hamperl S, Seitz H, et al. The Reb1-homologue Ydr026c/Nsi1 is required for efficient RNA polymerase I termination in yeast. EMBO J. 2012;31:3480–3493. doi: 10.1038/emboj.2012.185. PMID: 22805593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Engel C, Sainsbury S, Cheung AC, et al. RNA polymerase I structure and transcription regulation. Nature. 2013;502:650–655. doi: 10.1038/nature12712. PMID: 24153182 [DOI] [PubMed] [Google Scholar]
- [20].Fernandez-Tornero C, Moreno-Morcillo M, Rashid UJ, et al. Crystal structure of the 14-subunit RNA polymerase I. Nature. 2013;502:644–649. doi: 10.1038/nature12636. PMID: 24153184 [DOI] [PubMed] [Google Scholar]
- [21].Engel C, Plitzko J, Cramer P. RNA polymerase I-Rrn3 complex at 4.8 A resolution. Nat Commun. 2016;7:12129. doi: 10.1038/ncomms12129. PMID: 27418309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Neyer S, Kunz M, Geiss C, et al. Structure of RNA polymerase I transcribing ribosomal DNA genes. Nature. 2016; doi: 10.1038/nature20561. PMID: 27842382 [DOI] [PubMed] [Google Scholar]
- [23].Pilsl M, Crucifix C, Papai G, et al. Structure of the initiation-competent RNA polymerase I and its implication for transcription. Nat Commun. 2016;7:12126. doi: 10.1038/ncomms12126. PMID: 27418187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Torreira E, Louro JA, Pazos I, et al. The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription. Elife. 2017;6: doi: 10.7554/eLife.20832. PMID: 28262097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tafur L, Sadian Y, Hoffmann NA, et al. Molecular Structures of Transcribing RNA Polymerase I. Mol Cell. 2016;64:1135–1143. doi: 10.1016/j.molcel.2016.11.013. PMID: 27867008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. PMID: 11313498 [DOI] [PubMed] [Google Scholar]
- [27].Cheung AC, Cramer P. A movie of RNA polymerase II transcription. Cell. 2012;149:1431–1437. doi: 10.1016/j.cell.2012.06.006. PMID: 22726432 [DOI] [PubMed] [Google Scholar]
- [28].Tagami S, Sekine S, Kumarevel T, et al. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–982. doi: 10.1038/nature09573. PMID: 21124318 [DOI] [PubMed] [Google Scholar]
- [29].Kuhn CD, Geiger SR, Baumli S, et al. Functional architecture of RNA polymerase I. Cell. 2007;131:1260–1272. doi: 10.1016/j.cell.2007.10.051. PMID: 18160037 [DOI] [PubMed] [Google Scholar]
- [30].Philippi A, Steinbauer R, Reiter A, et al. TOR-dependent reduction in the expression level of Rrn3p lowers the activity of the yeast RNA Pol I machinery, but does not account for the strong inhibition of rRNA production. Nucleic Acids Res. 2010;38:5315–5326. doi: 10.1093/nar/gkq264. PMID: 20421203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gross CA, Chan C, Dombroski A, et al. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb Symp Quant Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. PMID: 10384278 [DOI] [PubMed] [Google Scholar]
- [32].Kim YJ, Bjorklund S, Li Y, et al. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. PMID: 8187178 [DOI] [PubMed] [Google Scholar]
- [33].Milkereit P, Schultz P, Tschochner H. Resolution of RNA polymerase I into dimers and monomers and their function in transcription. Biol Chem. 1997;378:1433–1443. doi: 10.1515/bchm.1997.378.12.1433. PMID: 9461342 [DOI] [PubMed] [Google Scholar]
- [34].Beckert B, Abdelshahid M, Schafer H, et al. Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J. 2017;36:2061–2072. doi: 10.15252/embj.201696189. PMID: 28468753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Khusainov I, Vicens Q, Bochler A, et al. Structure of the 70S ribosome from human pathogen Staphylococcus aureus. Nucleic Acids Res. 2017;45:1026. doi: 10.1093/nar/gkw1126. PMID: 28123039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chen J, Wassarman KM, Feng S, et al. 6S RNA Mimics B-Form DNA to regulate escherichia coli RNA polymerase. Mol Cell. 2017;68:388–397, e6. doi: 10.1016/j.molcel.2017.09.006. PMID: 28988932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kang JY, Olinares PD, Chen J, et al. Structural basis of transcription arrest by coliphage HK022 Nun in an Escherichia coli RNA polymerase elongation complex. Elife. 2017;6: doi: 10.7554/eLife.25478. PMID: 28318486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kettenberger H, Eisenfuhr A, Brueckner F, et al. Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs. Nat Struct Mol Biol. 2006;13:44–48. doi: 10.1038/nsmb1032. PMID: 16341226 [DOI] [PubMed] [Google Scholar]
- [39].Fath S, Milkereit P, Peyroche G, et al. Differential roles of phosphorylation in the formation of transcriptional active RNA polymerase I. Proc Natl Acad Sci USA. 2001;98:14334–14339. doi: 10.1073/pnas.231181398. PMID: 11717393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Blattner C, Jennebach S, Herzog F, et al. Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth. Genes Dev. 2011;25:2093–2105. doi: 10.1101/gad.17363311. PMID: 21940764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gerber J, Reiter A, Steinbauer R, et al. Site specific phosphorylation of yeast RNA polymerase I. Nucleic Acids Res. 2008;36:793–802. doi: 10.1093/nar/gkm1093. PMID: 18084032 [DOI] [PMC free article] [PubMed] [Google Scholar]


