ABSTRACT
Zinc-finger and homeodomain transcription factors have been shown in vitro to bind to recognition motifs containing a methylated CpG. However, accessing these motifs in vivo might be seriously impeded by the inclusion of DNA in nucleosomes and by the condensed structure adopted by chromatin formed on methylated DNA. Here, we discuss how oxidation of 5-methylcytosine into 5-hydroxymethylcytosine could provide the initial destabilizing clue for such transcription factors to get access to nucleosomal DNA and read epigenetic information.
KEYWORDS: DNA methylation, transcription factors, ten eleven translocation, 5-methylcytosine, 5-hydroxymethylcytosine chromatin
Introduction
“Epicytosine”, also known as 5-methyl-deoxycytidine (5mC), was discovered in mammalian DNA in the mid 20th century, well before being recognized as an epigenetic mark capable of influencing transcription [1]. Although the biological impact of the presence of 5mC in DNA is probably multifaceted, our general understanding of how 5mC can alter transcription relies mainly on two non-exclusive concepts: (i) 5mC can affect the stability of DNA interaction with sequence-specific DNA binding proteins like transcription factors (TFs), and (ii) the presence of 5mC in CpG dinucleotides allows interaction with methyl-DNA binding domain (MBD) proteins such as MeCP2, which in turn recruit protein complexes favouring chromatin condensation [2–4]. Accordingly, enhancers (one of the main TF binding platforms together with promoters), are genomic regions showing cell-type specific DNA methylation profiles and undergoing methylation/demethylation processes. These dynamic modifications pertain to adaptative changes in the transcriptional program of cells including rewiring of transcription factor networks [5–9]. Seminal work from A. Rao's and N. Heintz's laboratories on 5mC oxidation into 5-hydroxymethylcytosine (5hmC) by Ten Eleven Translocation (TET) dioxygenases paved the way to investigations on active DNA demethylation events and their link to transcription regulation [10,11]. TET proteins were then further shown to be implicated in iterative oxidation of 5hmC into 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) [12,13]. Both 5fC and 5caC are recognized and eliminated by the base excision repair (BER) machinery, including the T:G mismatch DNA glycosylase TDG which interacts with and processes 5fC and 5caC [14,15]. Accordingly, data from Y. Zhang's group showed that, upon depletion of TDG in mouse embryonic stem cells (mESCs), a significant fraction of 5hmC-marked enhancers become enriched in 5fC and 5caC [16]. Whereas 5fC and 5caC are labile entities in DNA due to removal by the BER system, 5hmC is readily detectable in genomic DNA and appears to be quite stable in certain genomic regions according to kinetic studies, suggesting that it may not just be a mere intermediate of demethylation but could also bear signalling potential [17–19]. Interestingly, the observation that TET enzymes can act in a non-processive manner suggests that each demethylation intermediate could serve specific functions and allow their recognition by selective readers, including transcription factors [20].
Transcription factor families rule 5mC sensitivity of TF binding to DNA in vitro
Although initial studies on the influence of 5mC on TF binding to DNA were devoted to single motifs and single TFs (for review see Tate and Bird [21]), recent technological advances like protein and DNA microarrays, SELEX (systematic evolution of ligands by exponential enrichment) coupled to deep sequencing, and mass spectrometry (MS) analysis of pull-down assays have allowed to run in vitro investigations at a larger scale [22–25]. Despite the fact that each of these techniques has its own drawbacks (for discussion see Zhu et al. [26]), they generated valuable information on how TFs can sense DNA methylation. Table 1 compiles the DNA binding preferences of given TFs and shows a striking dichotomy between families of TFs. Indeed, TFs from the basic Helix-Loop-Helix (bHLH) and the basic-Zipper (bZIP) families tend to prefer motifs with unmethylated CpGs, whereas Zinc-finger and Homeodomain (HD) TFs clearly prefer 5mCpGs. This striking difference between different classes of TF was recently confirmed in a large scale study interrogating the preference of 542 full-length TFs by high throughput (HT) SELEX [27]. However, the preference of bZIP and bHLH TFs for unmethylated CpGs does not systematically apply. The bZIP factor CREB1, for instance, binds preferentially to the unmethylated consensus motif TGACGTCA whereas CEBPα and CEBPβ prefer TGAmCGTCA [28,29]. In addition, a same bZIP factor can prefer either methylated or unmethylated CpGs in their motifs, depending on their sequence. ARNT2, for instance, prefers AAAmCGCTTCCC, whereas binding to the canonical sequence CACGTG requires an unmethylated CpG [25,27]. Contrary to bZIP and bHLH, Zinc-finger and HD TFs show a clear in vitro preference for 5mC (see Table 1). Such a behaviour can be inferred from studies using various technologies, leaving no doubt that these proteins indeed interact better with methylated sequences. Interestingly, KLF4 recognition of 5mCpG involves an arginine/glutamate pair of residues contacting the methyl group of 5mC, and such a pair is also implicated in 5mC interaction with KAISO, another Zinc-finger protein [30,31]. KAISO binds both mCpG and TpG containing sequences, with the Thymine methyl group being engaged in similar interactions than its 5mC counterpart [31–33]. Since methylation of cytosines in the context of CpAs has been evidenced in ES cells and 5mCpAs are efficiently oxidized by TET enzymes, binding of TFs to 5mCpAs could indeed occur in vivo and be regulated through active demethylation [34,35]. Thymine mimicry could also explain the impact of cytosine methylation on the binding of HD proteins [23]. Indeed, a number of HD TFs, including HOX proteins and the HOX partners two amino acids tale extension (TALE)-HD proteins, bind sites containing a TpG dinucleotide which are also efficiently bound when the TpG is replaced by a 5mCpG [23]. In particular, the TALE-HD protein PBX1 interacts with both TGATTG and 5mCGATTG but poorly with a CGATTG sequence [23]. PBX1 form heterodimers with the TALE-HD protein MEIS1 and we showed that enhancers bound by these TFs are particularly enriched in 5hmC, suggesting that these factors could also bind 5hmCpGs [18,19]. Interestingly, analysis of the structure of MEIS1 dimers bound to a TGACAG site (RCSB number 4XRM – Fig. 1A) shows that HD residues I50 and R54 from the DNA recognition helix establish contacts with the methyl group of the thymine whereas residue N47 does not contact the adjacent cytosine but a water molecule. When superimposed with the recognition helix from HOXB13 bound to a 5mCpG containing TTACGA motif (RCSB number 5EGO), N47 is predicted to be engaged in an interaction with the methyl group, suggesting that cytosine modifications at this position could stabilize MEIS1 binding. We recently investigated the ability of 5mC and 5hmC to modulate in vitro DNA binding of TALE-HDs in pull-down assays using nuclear extracts of neural progenitor cells (NPCs) differentiated from embryonal carcinoma cells (EECs) [19]. Consistent with our structure-based hypothesis, the presence of 5mC and 5hmC at position 4 of the consensus MEIS1 site TGACAG could equally increase binding of MEIS1 (Fig. 1B). Nonetheless, binding of MEIS1 to the TGATTTACGA probe (known to bind PBX1/HOXA9 heterodimers) [36] was significantly more enhanced by 5mCpG than by 5hmCpG (Fig. 1B). Apart from favouring contacts between residues of the DNA recognition helix and the cytosine bases, 5mC and 5hmC could indirectly impact on the stability of TALE-HD and/or TALE-HD/HOX dimers by influencing DNA shape. The N-terminal extension of the homeodomain in TALE-HD/HOX heterodimers forms a loop enriched in basic residues that sense minor groove width and stabilize binding to DNA (for review see Merabet and Mann) [37,38]. Remarkably, insertion of arginine 5 (R5) in the minor groove has been directly correlated to the presence of a CpG which was found to influence minor groove width [37]. Importantly, cytosine methylation further decreases minor groove width as appreciated by the shape-sensitive clivage of DNA by DNAse I. [39] To examine whether 5mC could influence the binding of HD N-terminal basic residues, we interrogated 5EGO and its unmethylated counterpart 5EG0 crystal structures. Results indicate that indeed the minor groove is narrowed by methylation of the CpG and that the HOXB13 residue R5 adopts different configurations and is engaged in 5 H-bonds in 5EGO versus only 2 in 5EG0 (Fig. 1C). This observation supports the idea that 5mCpGs confer optimal characteristics to DNA for its recognition by HD proteins.
Table 1.
DNA binding preference of transcription factors.
| Transcription factor | Family | Binding site/probe | Technology | Preference | Reference |
|---|---|---|---|---|---|
| MAX | bHLH | CACGTG | Gel-shift | C, 5caC | [48] |
| MAX | bHLH | TGACGCGCGCG | Pull-down, MS/MS | C | [22] |
| MAX | bHLH | CACGTG | SELEX | C | [27] |
| Tcf3/Ascl1 | bHLH | CGCAGGTG | Gel-shift | 5caC | [49] |
| Tcf4 | bHLH | ACACGTG | DNA array | 5hmC | [50] |
| USF-1 | bHLH | ACACGTG | DNA array | C | [50] |
| ARNT2 | bHLH | AAACGCTTCC | Protein microarray | 5mC | [25] |
| CREB1 | bZIP | TGACGTCA | DNA array | C | [28] |
| CREB1 | bZIP | (TGAT)GCAA | DNA array | 5mC, 5hmC | [28] |
| CREB1 | bZIP | TGACGCGCGCG | Pull-down, MS/MS | C | [22] |
| JUN/FOS | bZIP | TGACTCG | Gel-shift | 5mC | [51] |
| JUN | bZIP | TGACGCGCGCG | Pull-down, MS/MS | C | [22] |
| FOS | bZIP | TGACGCGCGCG | Pull-down, MS/MS | C | [22] |
| CEBPB/ATF4 | bZIP | CGATGCAA | DNA array and gel-shift | 5mC | [24] |
| CEBPB | bZIP | ATTGCGCAA | DNA array and gel-shift | 5mC | [24] |
| ATF4 | bZIP | ATTGCGCAA | DNA array and gel-shift | C | [24] |
| CEBPA/B | bZIP | TGACGTCA | Gel-shift | 5mC | [29] |
| ELF3/5 | ETS | ACCCGGAAGT | SELEX | C | [27] |
| HOXB9 | HD | TGACGCGCGCG | Pull-down, MS/MS | 5mC | [22] |
| HDX | HD | TGACGCGCGCG | Pull-down, MS/MS | 5hmC | [22] |
| ZHX1/2 | HD/Zinc-finger | TGACGCGCGCG | Pull-down, MS/MS | 5hmC | [22] |
| HOXA5 | HD | AAACGCTTCC | Protein microarray | 5mC | [25] |
| HOXB13 | HD | CTCGTAAAA | SELEX | 5mC | [27] |
| CDX1/2 | HD | GTCGTAAAA | SELEX | 5mC | [27] |
| PBX1 | TALE-HD | TGACGCGCGCG | Pull-down, MS/MS | 5mC | [22] |
| MEIS1 | TALE-HD | TGACGCGCGCG | Pull-down, MS/MS | 5mC | [22] |
| MEIS1 | TALE-HD | TGACAG | Pull-down | 5mC, 5hmC | [19] |
| MEIS1 | TALE-HD | TGATTTACG | Pull-down | 5mC, 5hmC | [19] |
| HOX/PBX | HD | TGATTTACG | EpiSELEX-Seq | 5mC | [23] |
| OCT4 | POU domain | ATGCGCAT | SELEX | 5mC | [27] |
| KLF4 | Zinc-finger | TGACGCGCGCG | Pull-down, MS/MS | 5mC | [22] |
| KLF4 | Zinc-finger | CCCGCC | Protein microarray | 5mC | [25] |
| SALL2 | Zinc-finger | TGACGCGCGCG | Pull-down, MS/MS | 5mC | [22] |
| SALL4 | Zinc-finger | CG rich long probe | Pull-down and gel-shift | 5mC/5hmC | [47] |
| GATA4 | Zinc-finger | AAACGCTTCC | Protein microarray | 5mC | [25] |
| P53 | TGACGCGCGCG | Pull-down, MS/MS | 5fC | [22] | |
| P53 | RRRCACGYYY | EpiSELEX-Seq | 5mC | [23] |
Figure 1.
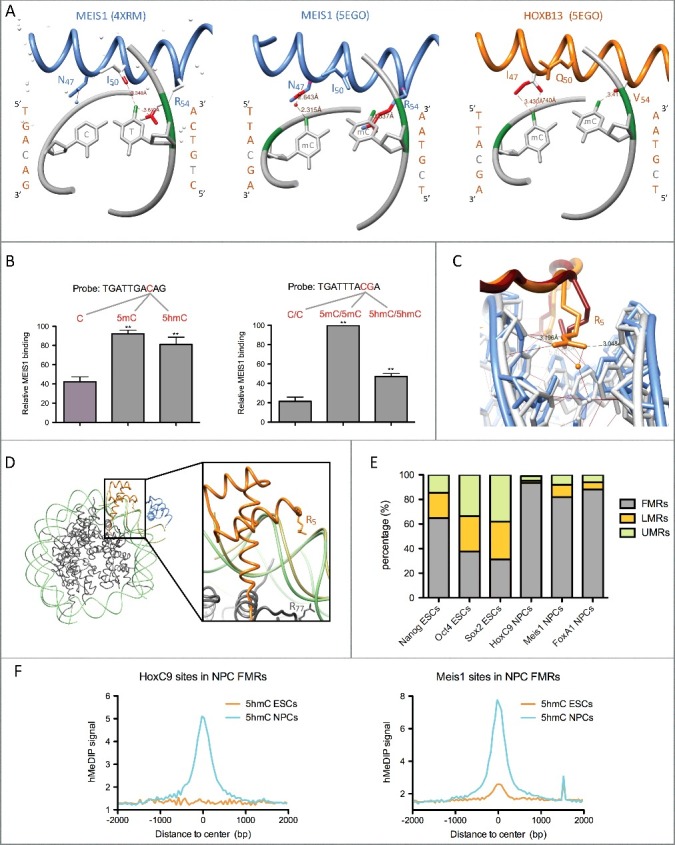
Homeodomain transcription factor interaction with methylated and hydroxymethylated DNA. (A) HD recognition helix contacts with CpA (4XRM) or 5mCpGs (5EGO). Contacts between the T or 5mC methyl groups (coloured in green) and HD residues are coloured in red. Contacts shown in the middle panel are predicted by the superimposition of the MEIS1 recognition helix to the one of HOXB13 bound to methylated DNA (5EGO), whereas the ones shown in the left and right panels are those observed in the 4XRM and 5EGO crystal structures. All structures were visualized with the UCSF Chimera 1.4.1 software. (B) In vitro pull-down assays showing binding preference of MEIS1 from NPC nuclear extracts for 5mC and 5hmC in two different DNA probes. Experimental data are from Mahé et al. [19] (C) Insertion of residue R5 within the minor groove in crystals of HOXB13 bound either to an unmethylated (5EG0, HOXB13 in red and DNA in blue) or to a methylated (5EGO, HOXB13 in orange and DNA in gray) TTACGA motif. (D) Superimposition of the crystal structures of HOXB13 (orange)/MEIS1 (blue) heterodimer bound to methylated DNA (kaki – 5EGO) and a nucleosome formed on methylated DNA (histones in dark gray and DNA in light green – 5B2J). The right panel shows an enlargement of the superimposed structures with the widened minor groove of the nucleosomal DNA likely unable to stabilize R5 insertion. (E) Distribution of TF binding sites in fully- (FMRs), low- (LMRs) and un-methylated regions (UMRs) of the mouse genome from ESCs and NPCs according to Stadler et al. [9] Mesi1 and FoxA1 binding sites are from Mahé et al. [19] NCBI GEO datasets for TF binding sites are as follows: GSM766061 (HoxC9), GSM1436068 (Nanog), GSM1436067 (Oct4), and GSM1436070 (Sox2). (F) Average ESC (GSM978374) and NPC (GSM978376) 5hmC profiles (hMeDIP-seq) centered on HoxC9 and Meis1 binding sites overlapping NPC FMRs.
In vivo constraints to TF interaction with cytosine modifications
Despite effects of cytosine methylation on DNA binding by TFs can be appreciated in vitro, their ability to access modified cytosines in vivo is complicated by the fact that DNA associates with histones to form chromatin. Indeed, insertion of a DNA sequence into a nucleosome can decrease binding site accessibility. This is the case for PBX1 that can be enriched in pull-down experiments with methylated naked DNA but not when this DNA is wrapped around histones, whereas the opposite is observed for MeCP2, raising the possibility that TALE-HDs may not efficiently bind to methylated nucleosomal DNA in cells [40]. It is established that, in nucleosomes, DNA is bent such as narrow minor groove sections facing the octamer favour insertion of histone arginines whereas minor grooves facing out of the nucleosome are enlarged above the limit required for arginine insertion [41]. Hence, as suggested by superimposition of the 5EGO structure with a nucleosome structure (5B2J), the recognition helix of a nucleosome-bound HD could access to the modified cytosines in the major groove but binding stabilization by arginines from the N-terminal part of the HD could be impeded by the enlargement of the minor groove (Fig. 1D). Another level of potential in vivo interference with TF binding to 5mCpGs could be attributed to MBDs. Indeed, MeCP2 could compete with homeodomains for 5mCpG binding. FRAP experiments have shown that wtHOXC13 and MECP2 have quite similar residency times in living cells (i.e. t1/2 recovery after photobleaching: 40 sec and 29 sec respectively) [42,43]. However, when R5 is mutated in HOXC13, t1/2 drops to 9 sec, twice as low as the one of a triple mutant (I47A, Q50A, N51A) in the recognition helix of HOXC13 (17.5 sec) [42]. Collectively, these observations suggest that R5 is a major determinant of HD stability in living cells and that impeding R5 function by inclusion of a binding site in a nucleosome could allow MeCP2 to outcompete HDs. In addition to a putative competition between MBDs and HDs for engaging interactions with 5mCpGs, there is evidence for a role of MeCP2 in condensing chromatin, either directly, as shown by atomic force microscopy for a tetranucleosome, or indirectly though recruiting repressor complexes containing histone deacetylases and H3K9 methyltransferases, leading to condensed nucleosomal arrays which are likely to decrease engagement of TFs [3,4,43]. Hence, although HD binding to naked DNA is clearly stabilized by 5mCpGs, for it to happen in vivo would require mechanisms that likely impact MeCP2/DNA interaction as well as nucleosome stability. In this context, oxidation of 5mC into 5hmC could initiate a reconfiguration of the chromatin structure leading to a more relaxed state more amenable to HD binding. Indeed, at physiological salt concentrations, MeCP2 affinity for symmetrically hydroxymethylated CpGs is 20 fold lower than that for fully methylated CpGs, and nucleosomes formed on an hydroxymethylated template tend to be less stable in vitro and likely in vivo [44–46]. Consistent with these observations, we evidenced that the presence of 5hmC in chromatin increases DNA accessibility [19]. In an apparent contradiction with the observation that TALE-HDs bind 5hmC-enriched regions, we show in Fig. 1E that, in in vitro-differentiated NPCs, the distribution of HD transcription factors is largely biased towards fully-methylated regions (FMRs), compared to pluripotency TFs which significantly occupy low (LMRs) or unmethylated (UMRs) regions. However, this partition of the genome is based on whole-genome bisulfite sequencing (WGBS), a technique which cannot discriminate between 5mC and 5hmC. In addition, our recent data indicate that binding of the TALE-HD protein MEIS1 to its target enhancers in ECC-derived NPCs is preceded by 5mC conversion into 5hmC [18,19]. To extend the relationship between DNA hydroxymethylation and HD binding to chromatin, we interrogated HoxC9 ChIP-seq and hMeDIP-seq data obtained in ESC-derived NPCs (Fig. 1F). Similarly to Meis1 sites, HoxC9 sites are highly hydroxymethylated in NPCs. Collectively, these observations indicate that HDs bind to chromatin sites that have experienced conversion of 5mC to 5hmC, a step that likely reverses the negative influence of a tight nucleosome structure on HD/DNA interaction.
Whether this requirement for 5mC to 5hmC conversion also applies to zinc-finger TFs is an interesting question. A recent study describing the relationship between the recruitment of the Zinc-finger protein SALL4 and the activity of TET enzymes in ESCs suggests that it might indeed be the case [47]. SALL4 binds to DNA through 7 Zinc-finger modules grouped into 3 clusters and each cluster has been tested in vitro for binding to CpG-, 5mCpG- and 5hmCpG-containing probes. Results indicated that, these modules bind preferentially to 5mCpGs and 5hmCpGs, although one of the modules preferred 5hmC over 5mC [47]. In ESCs, the recruitment of SALL4 to chromatin was shown to be dependent on TET-mediated oxidation of 5mC [47]. Hence, despite being described as 5mCpG-binding proteins, the biological activity of TFs from the ZF and HD families is likely to depend in vivo on an initial conversion of 5mC to 5hmC by TET proteins leading to an increase in DNA accessibility within chromatin.
Funding Statement
E.A.M. was funded by the French Ministry of Research and the Fondation ARC pour la Recherche sur le Cancer. G.S. aknowledges the University of Rennes 1 and the Centre National de la Recherche Scientifique for funding.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem. 1948;175(1):315–332. PMID:18873306 [PubMed] [Google Scholar]
- [2].Lewis JD, Meehan RR, Henzel WJ, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69(6):905–914. 10.1016/0092-8674(92)90610-O. PMID:1606614 [DOI] [PubMed] [Google Scholar]
- [3].Jones PL, Veenstra GJ, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19(2):187–191. 10.1038/561. PMID:9620779 [DOI] [PubMed] [Google Scholar]
- [4].Fuks F, Hurd PJ, Wolf D, et al. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278(6):4035–4040. 10.1074/jbc.M210256200. PMID:12427740 [DOI] [PubMed] [Google Scholar]
- [5].Martinowich K, Hattori D, Wu H, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302(5646):890–893. 10.1126/science.1090842. PMID:14593184 [DOI] [PubMed] [Google Scholar]
- [6].Mohn F, Weber M, Rebhan M, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30(6):755–766. 10.1016/j.molcel.2008.05.007. PMID:18514006 [DOI] [PubMed] [Google Scholar]
- [7].Schmidl C, Klug M, Boeld TJ, et al. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res. 2009;19(7):1165–1174. 10.1101/gr.091470.109. PMID:19494038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sérandour AA, Avner S, Percevault F, et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 2011;21(4):555–565. 10.1101/gr.111534.110. PMID:21233399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stadler MB, Murr R, Burger L, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480(7378):490–495. PMID:22170606 [DOI] [PubMed] [Google Scholar]
- [10].Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. 10.1126/science.1170116. PMID:19372391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. 10.1126/science.1169786. PMID:19372393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. 10.1126/science.1210944. PMID:21817016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. 10.1126/science.1210597. PMID:21778364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286(41):35334–35338. 10.1074/jbc.C111.284620. PMID:21862836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang L, Lu X, Lu J, et al. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol. 2012;8(4):328–330. 10.1038/nchembio.914. PMID:22327402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shen L, Wu H, Diep D, et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153(3):692–706. 10.1016/j.cell.2013.04.002. PMID:23602152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fu Y, He C. Nucleic acid modifications with epigenetic significance. Curr Opin Chem Biol. 2012;16(5–6):516–524. 10.1016/j.cbpa.2012.10.002. PMID:23092881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sérandour AA, Avner S, Oger F, et al. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res. 2012;40(17):8255–8265. 10.1093/nar/gks595. PMID:22730288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mahé EA, Madigou T, Sérandour AA, et al. Cytosine modifications modulate the chromatin architecture of transcriptional enhancers. Genome Res. 2017;27(6):947–958. 10.1101/gr.211466.116. PMID:28396520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tamanaha E, Guan S, Marks K, et al. Distributive Processing by the Iron(II)/α-Ketoglutarate-Dependent Catalytic Domains of the TET Enzymes Is Consistent with Epigenetic Roles for Oxidized 5-Methylcytosine Bases. J Am Chem Soc. 2016;138(30):9345–9348. 10.1021/jacs.6b03243. PMID:27362828 [DOI] [PubMed] [Google Scholar]
- [21].Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3(2):226–231. 10.1016/0959-437X(93)90027-M. PMID:8504247 [DOI] [PubMed] [Google Scholar]
- [22].Spruijt CG, Gnerlich F, Smits AH, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152(5):1146–1159. 10.1016/j.cell.2013.02.004. PMID:23434322 [DOI] [PubMed] [Google Scholar]
- [23].Kribelbauer JF, Laptenko O, Chen S, et al. Quantitative Analysis of the DNA Methylation Sensitivity of Transcription Factor Complexes. Cell Rep. 2017;19(11):2383–2395. 10.1016/j.celrep.2017.05.069. PMID:28614722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mann IK, Chatterjee R, Zhao J, et al. CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Res. 2013;23(6):988–997. 10.1101/gr.146654.112. PMID:23590861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hu S, Wan J, Su Y, et al. DNA methylation presents distinct binding sites for human transcription factors. Elife. 2013;2:e00726. 10.7554/eLife.00726. PMID:24015356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhu H, Wang G, Qian J. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet. 2016;17(9):551–565. 10.1038/nrg.2016.83. PMID:27479905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yin Y, Morgunova E, Jolma A, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356(6337). doi: 10.1126/science.aaj2239 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Syed KS, He X, Tillo D, et al. 5-Methylcytosine (5mC) and 5-Hydroxymethylcytosine (5hmC) Enhance the DNA Binding of CREB1 to the C/EBP Half-Site Tetranucleotide GCAA. Biochemistry. 2016;55(49):6940–6948. 10.1021/acs.biochem.6b00796. PMID:27951657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rishi V, Bhattacharya P, Chatterjee R, et al. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc Natl Acad Sci U S A. 2010;107(47):20311–20316. 10.1073/pnas.1008688107. PMID:21059933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu Y, Olanrewaju YO, Zheng Y, et al. Structural basis for Klf4 recognition of methylated DNA. Nucleic Acids Res. 2014;42(8):4859–4867. 10.1093/nar/gku134. PMID:24520114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Buck-Koehntop BA, Stanfield RL, Ekiert DC, et al. Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso. Proc Natl Acad Sci U S A. 2012;109(38):15229–15234. 10.1073/pnas.1213726109. PMID:22949637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Prokhortchouk A, Hendrich B, Jørgensen H, et al. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15(13):1613–1618. 10.1101/gad.198501. PMID:11445535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Daniel JM, Spring CM, Crawford HC, et al. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 2002;30(13):2911–2919. 10.1093/nar/gkf398. PMID:12087177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. 10.1038/nature08514. PMID:19829295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Papin C, Ibrahim A, Gras SL, et al. Combinatorial DNA methylation codes at repetitive elements. Genome Res. 2017;27(6):934–946. 10.1101/gr.213983.116. PMID:28348165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].LaRonde-LeBlanc NA, Wolberger C. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 2003;17(16):2060–2072. 10.1101/gad.1103303. PMID:12923056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Joshi R, Passner JM, Rohs R, et al. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131(3):530–43. 10.1016/j.cell.2007.09.024. PMID:17981120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Merabet S, Mann RS. To be specific or not: the critical relation between Hox and TALE proteins. Trends Genet. 2016;32(6):334–347. 10.1016/j.tig.2016.03.004. PMID:27066866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lazarovici A, Zhou T, Shafer A, et al. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc Natl Acad Sci USA. 2013;110(16):6376–6381. 10.1073/pnas.1216822110. PMID:23576721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bartke T, Vermeulen M, Xhemalce B, et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143(3):470–484. 10.1016/j.cell.2010.10.012. PMID:21029866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rohs R, West SM, Sosinsky A, et al. The role of DNA shape in protein-DNA recognition. Nature. 2009;461(7268):1248–1253. 10.1038/nature08473. PMID:19865164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Marchetti L, Comelli L, D'Innocenzo B, et al. Homeotic proteins participate in the function of human-DNA replication origins. Nucleic Acids Res. 2010;38(22):8105–8119. 10.1093/nar/gkq688. PMID:20693533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ghosh RP, Horowitz-Scherer RA, Nikitina T, et al. MeCP2 binds cooperatively to its substrate and competes with histone H1 for chromatin binding sites. Mol Cell Biol. 2010;30(19):4656–4670. 10.1128/MCB.00379-10. PMID:20679481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Khrapunov S, Warren C, Cheng H, et al. Unusual characteristics of the DNA binding domain of epigenetic regulatory protein MeCP2 determine its binding specificity. Biochemistry. 2014;53(21):3379–3391. 10.1021/bi500424z. PMID:24828757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mendonca A, Chang EH, Liu W, et al. Hydroxymethylation of DNA influences nucleosomal conformation and stability in vitro. Biochim Biophys Acta. 2014;1839(11):1323–1329. PMID:25263161 [DOI] [PubMed] [Google Scholar]
- [46].Teif VB, Beshnova DA, Vainshtein Y, et al. Nucleosome repositioning links DNA (de)methylation and differential CTCF binding during stem cell development. Genome Res. 2014;24(8):1285–1295. 10.1101/gr.164418.113. PMID:24812327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Xiong J, Zhang Z, Chen J, et al. Cooperative Action between SALL4A and TET Proteins in Stepwise Oxidation of 5-Methylcytosine. Mol Cell. 2016;64(5):913–925. 10.1016/j.molcel.2016.10.013. PMID:27840027 [DOI] [PubMed] [Google Scholar]
- [48].Wang D, Hashimoto H, Zhang X, et al. MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma. Nucleic Acids Res. 2017;45(5):2396–2407. 10.1093/nar/gkw1184. PMID:27903915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Golla JP, Zhao J, Mann IK, et al. Carboxylation of cytosine (5caC) in the CG dinucleotide in the E-box motif (CGCAG|GTG) increases binding of the Tcf3|Ascl1 helix-loop-helix heterodimer 10-fold. Biochem Biophys Res Commun. 2014;449(2):248–255. 10.1016/j.bbrc.2014.05.018. PMID:24835951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Khund-Sayeed S, He X, Holzberg T, et al. 5-Hydroxymethylcytosine in E-box motifs ACAT|GTG and ACAC|GTG increases DNA-binding of the B-HLH transcription factor TCF4. Integr Biol (Camb). 2016;8(9):936–945. 10.1039/C6IB00079G. PMID:27485769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Gustems M, Woellmer A, Rothbauer U, et al. c-Jun/c-Fos heterodimers regulate cellular genes via a newly identified class of methylated DNA sequence motifs. Nucleic Acids Res. 2014;42(5):3059–3072. 10.1093/nar/gkt1323. PMID:24371273 [DOI] [PMC free article] [PubMed] [Google Scholar]


