Abstract
The human gut microbiome plays a key role in human health1, but 16S characterization lacks quantitative functional annotation2. The fecal metabolome provides a functional readout of microbial activity and could be used as an intermediate phenotype mediating these interactions3. In the first comprehensive description of the fecal metabolome, examining 1116 metabolites of 786 individuals from a population-based twin study (TwinsUK), the fecal metabolome was found to be only modestly influenced by host genetics (h2=17.9%). One replicated locus at the NAT2 gene was associated with fecal metabolic traits. The fecal metabolome largely reflects gut microbial composition explaining on average 67.7% (±18.8%) of its variance. It is strongly associated with visceral fat mass, illustrating potential mechanisms underlying the well-established microbial influence on abdominal obesity. Fecal metabolic profiling appears as a novel tool to explore links between microbiome composition, host phenotypes, and heritable complex traits.
There is growing evidence that the gut microbiome contributes to maintain homeostasis of host metabolism1. Disruption of this intricate system is associated with diseases such as obesity4,5 and insulin resistance6. Metabolomics and the gut microbiome are strongly related, with microbes producing many of the body’s chemicals, hormones and vitamins7. The gut microbiome has been reported to have an effect on circulating levels of several metabolites such as branched-chain amino acids potentially causing insulin resistance6. However, despite the advances of next generation sequencing platforms, which allow profiling of complex microbial communities using 16S sequencing, annotation is sparse. Moreover, the microbiome only codes microbial possibilities rather than their actual activity and cannot differentiate between alive and dead microbes8, nor determine the transcriptional activity of the genes within each bacterial genome2. Fecal metabolomics, however, reports specifically on the metabolic interplay between the host, diet and the gut microbiota3 and complements sequencing-based approaches with a functional readout of the microbiome. Here we provide the first comprehensive description of the fecal metabolome in a large population-based setting with the additional advantage of the twin model. We report (i) fecal metabolites associations with age, gender and obesity; (ii) host genetic influences; and (iii) uni- and multivariable dependencies with the gut microbiome.
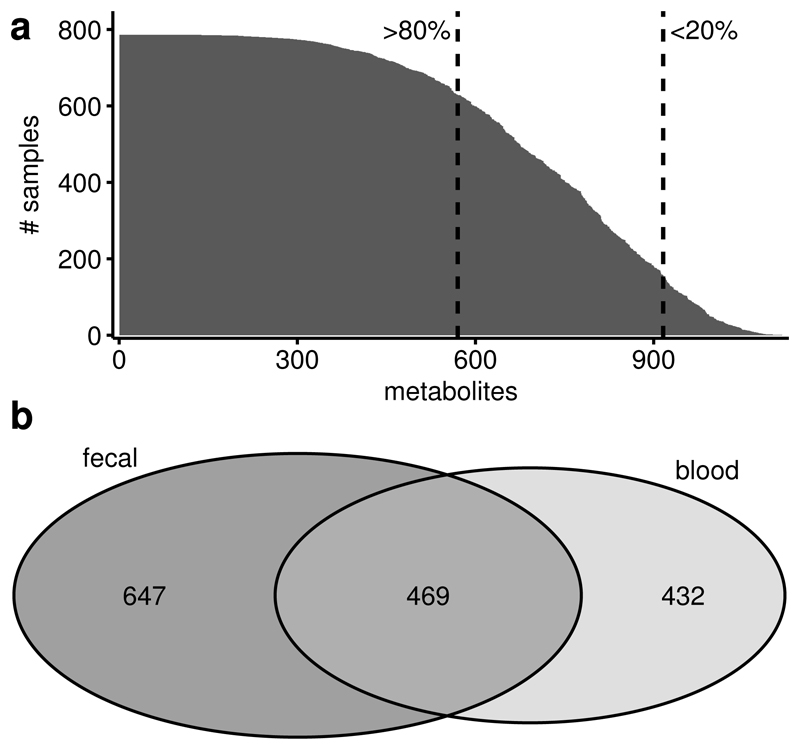
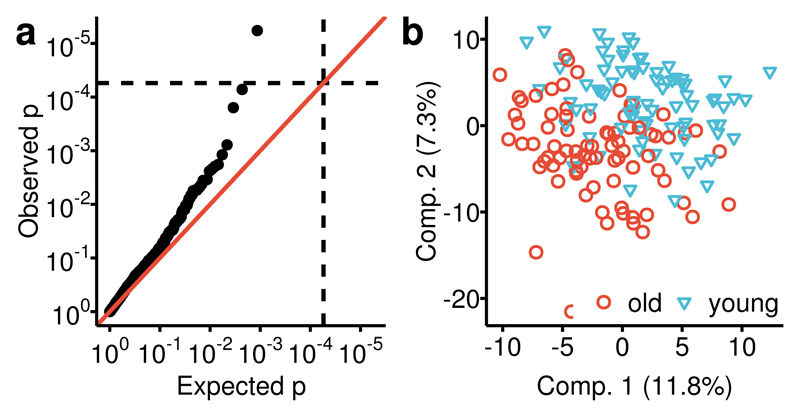
We analyzed fecal samples of 786 predominantly female twins of the TwinsUK cohort, aged 65.2 (±7.6), with an average BMI of 26.1 (±4.7) (Supplementary Table 1) and we replicated our genetics results in an independent sample of 230 individuals, aged 66.9 (±8.6) with an average BMI of 27.2 (±5.2). Untargeted metabolomics profiling of the participants’ fecal samples was conducted by Metabolon, Inc., using mass spectrometry, measuring a total of 1116 metabolites, 866 of them with known chemical identity. Among the metabolites identified, 570 were common and detected in at least 80% of the samples, while 345 were detected in at least 20% but in less than 80% of all samples (Fig 1a). The latter were analyzed as dichotomous traits (present/absent in a sample) and metabolites measured in less than 20% of the samples were discarded from further analysis. 647 of the 1116 measured metabolites were not detected in blood samples of the same individuals profiled on the same platform (Fig 1b). This suggests that the fecal metabolome provides complementary information to blood metabolomics. We did not find significant associations between 915 fecal metabolites and age after correcting for multiple testing. However, a multivariate partial least squares discriminant analysis incorporating all the common 570 metabolites could distinguish the oldest decile (>75 years) from the youngest decile (<56 years) of the study population (AUC=0.71, p=6.8×10-6, Fig 2b) and one metabolite, phytanate, was significantly different between the oldest and youngest deciles (p=5.0×10-3) (Fig 2a), suggesting non-linear associations between the fecal metabolome and age in line with previous reports on the effects of age on the gut microbiome9,10.
Figure 1. Number of measured fecal metabolites.
1116 metabolites were detected in 786 fecal samples. (a) 570 of those were detected in at least 80% of all samples and 345 were detected in less than 80% but more than 20% of all samples. The first were analyzed continuously, while we dichotomized the latter in present/absent. 210 metabolites that were present in less than 20% of the samples were excluded from further analysis. (b) 469 metabolites where observed in both, fecal and blood samples of the sample individuals, while 647 metabolites are unique to feces. 499 of these 647 metabolites were observed in at least 20% of the fecal samples.
Figure 2. Association of the fecal metabolome with age.
We compared the fecal metabolome between the oldest (>75 yrs., n=79) and youngest decile (<56 yrs., n=80) of the study population. (a) First, we investigated the age effect for all metabolites individually using logistic regression models and found one metabolite, phytanate, significantly different between the youngest and the oldest decile of our data. (b) Then, we fitted a multivariate PLS-DA model to distinguish the older (red) from the younger (blue) group. We estimated the area under the receiver operations curve (AUC) at 0.71 (p=6.8×10-6) in a 10-fold cross-validation.
BMI was associated with eight metabolites at an FDR (Benjamini-Hochberg) of 5%: five fecal lipids, including arachidonate (β [95% CI] = 0.13 [0.07:0.19], p=1.1×10-5), the hemoglobin metabolite bilirubin (β [95% CI] = 0.13 [0.06:0.19], p=8.9×10-5), and two unknown metabolites (Supplementary Table 2). We then looked for associations with visceral fat mass, a measure of abdominal obesity, correcting for BMI, and found a total of 102 statistically significant associations (FDR<5%, 13 passing Bonferroni correction), which together explain 28.4% of the observed total variance of visceral fat (p<2.2×10-16) (Supplementary Table 2). In contrast, we find only 8 metabolites associated with BMI, which is an imprecise measure of adiposity and measures overall mass without distinction between lean and fat mass11. However, the gut microbiome is known to play a major role in fatty acid metabolism and adiposity which may be better reflected by visceral fat measures12,13. Emerging evidence also suggests a role of the intestinal microbiota in visceral development by interacting with dietary components14. We have previously shown strong associations between visceral fat mass and the gut microbiome composition15,16. The much larger number of fecal metabolite associations with visceral fat than with BMI are consistent with these findings and highlight the strong influence of metabolic processes in the gut influencing abdominal adiposity.
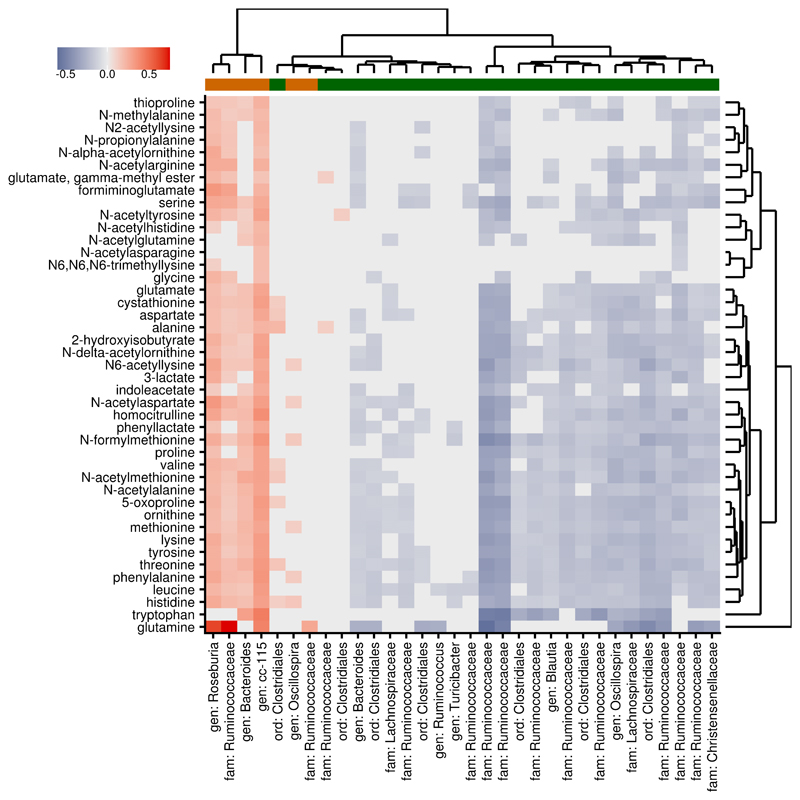
Visceral fat-associated metabolites were significantly enriched for amino acids (43 metabolites, enrichment p<2×10-4) but also included 14 fatty acids, including arachidonate (β=5.07 [2.55:7.59], p= 8.2×10-5), 8 nucleotides, 6 sugars and 6 vitamins. The strong association between the fecal metabolome and central obesity confirms hypotheses on the involvement of microbial amino acid metabolism in obesity and suggests new mechanisms, such as microbial vitamin B metabolism. We have previously found several microbe families associated with lower visceral fat mass15 and reduced weight gain in germ-free mice receiving human fecal transplants17. By analyzing the fecal metabolome, we found the abundance of the same families to be strongly associated with decreased abundance of amino-acids (see below), suggesting that their association with visceral fat may be mediated by the availability of amino acids (Fig 3). This may be due to increased utilization or decreased production of amino acids by these bacteria, or the result of more complex host-microbe interactions.
Figure 3. Associations of fecal metabolites with gut microbiome corespond to microbial effect on visceral fat.
Visceral fat mass was significantly associated with 43 fecal amino acids (all positively) (n=647) and 32 OTUs (n=540) (6 positively in orange, 26 negatively in green). Red tiles indicate positive associations between these metabolites and OTUs (β > 0) and blue tiles negative associations (β < 0); grey tiles indicate non-significant associations (FDR > 5%). Microbial associations with fecal metabolites correspond to their respective associations with visceral fat, indicating that the microbial metabolic profile is more closely related to the host phenotype than taxonomy.
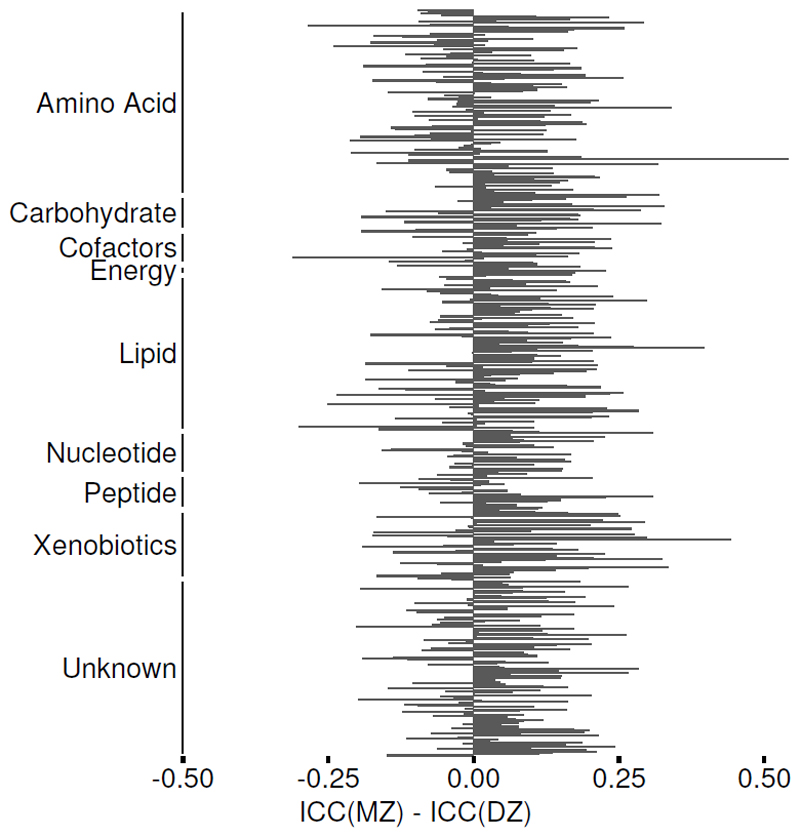
The gut microbiome is heritable17,18 and we found a heritable variance component for 210 OTUs, which explained on average 22.7% of the observed total variance. To test whether host genetic also influences the fecal metabolome we first estimated its heritability, taking advantage of the twin structure in our data (Fig 4) using structural equation modelling (148 MZ pairs, 155 DZ pairs). For 428 metabolites the best fitting model contained a heritable variance component (A), which explained on average 17.9% (±9.7%) of the metabolite variation. Long chain fatty acid-containing metabolites, such as 1-palmitoyl-2-arachidonoyl-GPC (H2=60.7% [95% CI 43.4:78.0]) and stearoylcarnitine (H2=54.3% [36.4:72.3]), were amongst the most heritable metabolites. For 279 metabolites, including the coffee-metabolite 5-acetylamino-6-amino-3-methyluracil (C=30.3% [20.0:40.6]), the best fitting model was the CE model, where the common environment component (C) explained on average 14.8% (±8.1%) of the variance. For the remaining 208 metabolites, the best fitting model was the E model where the entire variation of the metabolite is due to individual differences such as the microbiome or individual diet (Supplementary Table 2). We found a significantly stronger environmental effect on lipids than other metabolites (enrichment p-value < 2×10-4).
Figure 4. Intraclass correlation of fecal metabolites in MZ and DZ twins.
The intraclass correlation coefficients (ICCs) were calculated from variance components of a one-way analysis of variance separately for monozygotic (MZ, n=148 pairs) and dizygotic (DZ, n=155 pairs) twins for each metabolite. Positive values of their respective differences indicate more similar metabolic profiles between MZ than DZ twins.
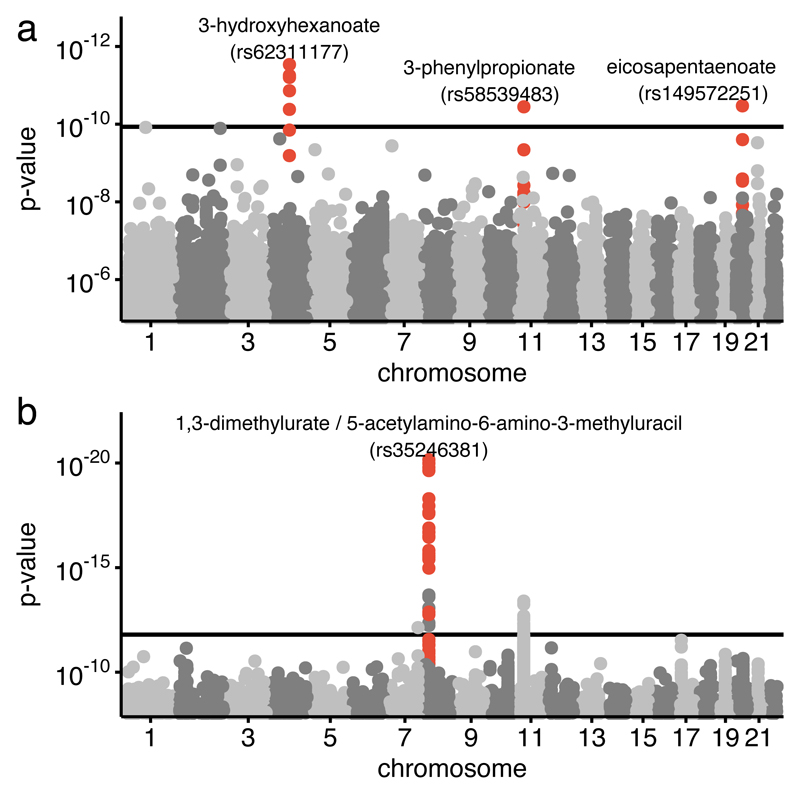
We subsequently conducted GWAS for the 428 metabolites with a heritable component (Supplementary Table 3) and identified three metabolites (the amino-acid 3-phenylpropionate and two lipids eicosapentaenoate and 3-hydroxyhexanoate) significantly associated with genetic loci after correcting for multiple testing (p<1.2×10-10=5×10-8/428) (Table 1, Fig 5a). We also tested for genetic associations of metabolites ratios, which can be better proxies for chemical reactions than single metabolites19. After correcting for 31,226 tested ratios we found the ratio of 5-acetylamino-6-amino-3-methyluracil and 1,3-dimethylurate was associated with a locus on chromosome 8 (rs35246381, p=7.0×10-21, p-gain=7.5×109) (Table 1, Fig 5b). We replicated our GWAS results in an independent sample of 230 individuals. Out of the 4 loci we tested, only the metabolite ratio of 5-acetylamino-6-amino-3-methyluracil and 1,3-dimethylurate was significantly associated in the replication cohort (p=3.6×10-10; meta-analysis p=3.3×10-36). The two metabolites 5-acetylamino-6-amino-3-methyluracil and 1,3-dimethylurate are products of caffeine metabolism20. The associated locus at the NAT2 gene codes for a N-acetyltransferase, which catalyzes the degradation of caffeine metabolites21 (Supplementary Figure 4). Associations of this locus with other caffeine-metabolites (1-methylxanthine, 4-acetamidobutanoate and 1-methylurate) have been previously observed in blood22 and urine23 and likely reflect efficiency of the degradation of caffeine. We then explored if there were any eQTLs or other functional variants in strong LD with the top SNP. Although we found three eQTLs (rs11996129, rs1112005, rs1799930) for NAT224, these are only in weak LD (r2<0.16) with rs3524638125 and the associations between these SNPs and the metabolite ratio is weaker than that of the top SNP (p=3.6×10-10 vs p=9.4×10-7). The tissues where the expression of NAT2 is highest, after liver, are in the jejunal and colonic mucosa, duodenum colon and small intestine26 (see URLs). This is consistent with polymorphisms in the NAT2 gene being associated with the concentration of caffeine derived metabolites in feces. We explored the relationship between caffeine and the fecal metabolites 5-acetylamino-6-amino-3-methyluracil and 1,3-dimethylurate and find that their ratio is positively correlated with both coffee intake and serum caffeine levels (Supplementary Figure 4). These genetic association data illustrate how part of the complex metabolism of caffeine takes place in the intestine before reaching the liver and that the links between the host genetic makeup and xenobiotic concentrations can be captured by fecal metabolites. In addition to caffeine, the NAT2 enzyme is also involved in metabolism of various xenobiotics and is therefore related to variance in drug response and toxicity27. There is work showing that the composition of the gut microbiome regulates xenobiotic enzymes, for instance the expression of NAT2 is 1.5 higher in germ free animals than in the large intestine of control animals28. Taken together with our results these data fit with a picture of xenobiotic metabolism being regulated jointly by host genetic variation and gut microbiome composition.
Table 1. Genetic associations of fecal metabolites.
Three metabolites and one metabolite ratio significantly associated with genetic loci in the discovery cohort (n=739). We report their respective heritabilities (H2), the associated variant along with its chromosomal position and the nearest gene, and the effect (± its standard error). P-values in the discovery and replication cohorts were calculated using the two-sided score test implemented in GEMMA and combined using fixed-effects inverse variance meta-analysis. The p-gain describes the strength of the association of the metabolite ratio relative to the associations of each individual metabolite.
| Discovery | Replication | Meta-Analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite | H2 | MAF | SNP | Gene | Effect | P | p-gain | Effect | P | Effect | P |
| 1,3-dimethylurate/ 5-acetylamino-6-amino-3-methyluracil |
40.2% | 24.7% | rs35246381 (8:1841502) |
NAT2 | -0.17±0.02 | 7.0×10-21 | 7.5×109 | -0.22±0.03 | 3.5×10-10 | -0.18±0.01 | 3.3×10-36 |
| 3-hydroxyhexanoate | 20.7% | 3.7% | rs62311177 (4:92962004) |
GRID2 | 0.41±0.06 | 2.9×10-12 | 0.07±0.09 | 0.429 | 0.32±0.05 | 3.0×10-11 | |
| eicosapentaenoate (EPA; 20:5n3) | 16.1% | 1.4% | rs149572251 (20:34322936) |
ITCH | 1.45±0.21 | 3.4×10-11 | -0.23±0.38 | 0.544 | 1.06±0.18 | 6.8×10-9 | |
| 3-phenylpropionate (hydrocinnamate) | 23.9% | 1.6% | rs58539483 (11:28830501) |
AC090833.1 | -1.31±0.19 | 3.6×10-11 | -0.39±0.22 | 0.076 | -0.92±0.14 | 1.5×10-10 | |
Figure 5. Host genetic influence on the fecal metabolome.
Genome-wide association studies were conducted for 428 heritable fecal metabolites and 31,226 fecal metabolite ratios (n=739). P-values were calculated using the score test implemented in GEMMA. (a) The Manhattan plot illustrates the genetic associations of fecal metabolites in the discovery sample. The horizontal line indicates the Bonferroni cutoff of 1.2×10-10. Three loci (red) pass the Bonferroni threshold. (b) The second Manhattan plot shows genetic associations with metabolite ratios in the discovery sample. The horizontal line indicates the Bonferroni cutoff of (p<1.6×10-12). Two loci pass the threshold, however only the association with 1,3-dimethylurate / 5-acetylamino-6-amino-3-methyluracil (p = 6.2×10-21, red) passed filtering by p-gain (p-gain > 8.9×105) and thus is considerably stronger than the association of each individual metabolite. Boxplots, QQ-plots, and regional association plots for each of the four loci are shown in Supplementary Figures 1-3.
We then investigated the extent to which the fecal metabolome reflect metabolic processes of the gut microbiome. We regressed metabolite levels against microbial diversity (quantified by the Shannon index), and found that 575 metabolites across all pathways showed a significant association with microbial diversity at a FDR of 5%, with 347 passing a Bonferroni correction. We estimated the proportion of variance in each metabolite explained by microbiome composition using the unweighted UniFrac beta-diversity metric, a measure of overall phylogenetic dissimilarity between individuals’ microbiota29. We found that gut microbial composition explained a significant proportion of the observed variance of 710 metabolites, on average 67.7% (±18.8%) of the observed variance, ranging from 22.1% for 1-linolenoylglycerol to 100% for several amino acids (Supplementary Table 2). Amongst others, the microbiome explained a significant proportion of the variance of the 8 BMI-related and 101 of the visceral fat-related metabolites. Xenobiotics showed the strongest associations with microbial composition (enrichment p-value <1×10-4), which explained the entire observed variance for some of them including the B-vitamins nicotinate and pantothenate.
To explore the associations of the fecal metabolome with gut microbes at a finer taxonomic resolution we regressed each metabolite against the 581 most abundant operational taxonomic units (OTUs), adjusting for potential confounding factors including Shannon diversity. We found 42,645 significant associations of 907 different metabolites with 579 different OTUs after adjusting for multiple testing (FDR<5%). We also calculated associations of fecal metabolites with collapsed taxonomical levels, ranging from genus to phylum level (Supplementary Table 4). 264 metabolites were only associated with microbes at the OTU level, with the remainder also associating with broader taxonomic groupings.
Lastly, to investigate the connectivity of the fecal metabolome with microbes, we calculated a Gaussian graphical model (GGM) combining 435 common metabolites with a known chemical identity with 241 OTUs with complete taxonomy assignment to at least genus level. The resulting model consists of 2553 independent associations, 1035 of them amongst metabolites, 946 amongst microbes and 572 connecting metabolites and microbes (Supplementary Table 5 and Figure 5). All but 9 variables form one connected component. We detect 19 clusters in the largest component, 9 of which contain both microbes and fecal metabolites and 10 consist of metabolites only. Xenobiotics have higher node degrees (p<3×10-4) and were more densely connected with OTUs (p<2.4×10-3). Our model demonstrates the high degree of interrelatedness between gut microbiome and fecal metabolome, despite the very different technologies used.
In conclusion, although state of the art metagenomic sequencing allows quantitative and functional annotation of species and microbial pathways30, 16S sequencing data has limitations including the lack of quantitative functional annotations. Fecal metabolomics provides a complimentary functional readout of microbial metabolism as well as its interaction with host and environmental factors. We have focused on the relationship between fecal metabolites and host and microbial genetics. Future studies should further investigate the influence of environmental factors, particularly nutrition and consider the influence of stool frequency/type on fecal metabolite measurements, as these are known to be associated with fecal microbiome composition31,32. The fecal metabolome can thus be used as intermediate phenotype that promotes microbial effects on the host and vice-versa. Using associations with obesity, we demonstrate that fecal metabolomics are a useful tool to complement future genomic and microbiome studies.
Online Methods
Study population
Study participants were 786 twins from the TwinsUK cohort. TwinsUK (a national twin registry) has been recruited since 1992 through media campaigns and is representative of the population of the UK in terms of life style33. The study population is predominantly female (93.4% females), with an average age of 65.2 (±7.6) and an average of BMI of 26.1 (±4.7). Ethical approval by St Thomas’ Hospital ethics committee; all participants provided informed written consent.
Results of the genome-wide association study were replicated in an independent set of 230 individuals (98.3% female) from the TwinsUK study, aged 66.9 (±8.6) and an average BMI of 27.2 (±5.2) (Supplementary Table 1).
Data collection
Sample collection, DNA extraction, and sequencing of the samples within this study has been described previously17,18. Briefly, the fecal samples were collected, refrigerated and kept in ice packs until they were frozen at -80°C (mostly within 24 hours from collection) before further processing. A number of participants (15%) sent their samples by post.
Metabolomics profiling
Metabolite concentrations were measured from fecal samples by Metabolon Inc., Durham, USA, using an untargeted LC/MS platform as previously described22,34 (see supplemental methods for details).
Sample preparation for global metabolomics
Samples were stored at –80°C until processed. Sample preparation was carried out as described previously34 at Metabolon, Inc. Lyophilized feces samples were extracted at a constant per mass basis. Briefly, recovery standards were added prior to the first step in the extraction process for quality control purposes. To remove protein, dissociate small molecules bound to protein or trapped in the precipitated protein matrix, and to recover chemically diverse metabolites, proteins were precipitated with methanol under vigorous shaking for 2 min (Glen Mills Genogrinder 2000) followed by centrifugation. The resulting extract was divided into five fractions: 1) acidic positive ion conditions, chromatographically optimized for more hydrophilic compounds; 2) acidic positive ion conditions, chromatographically optimized for more hydrophobic compounds; 3) basic negative ion optimized conditions using a separate dedicated C18 column; 4) negative ionization following elution from a HILIC column; 5) reserved for backup.
Three types of controls were analyzed in concert with the experimental samples: a pooled sample generated from a small portion of each experimental sample of interest served as a technical replicate throughout the platform run; extracted water samples served as process blanks; and a cocktail of standards spiked into every analyzed sample allowed instrument performance monitoring. Instrument variability was determined by calculating the median relative standard deviation (RSD) for the standards that were added to each sample prior to injection into the mass spectrometers (median RSDs were determined to be 5%; n = 31 standards). Overall process variability was determined by calculating the median RSD for all endogenous metabolites (i.e., non-instrument standards) present in 90% or more of the pooled technical replicate samples (median RSD = 12%, n = 832 metabolites). Experimental samples and controls were randomized across the platform run.
Mass spectrometry analysis
Extracts were subjected to UPLC-MS/MS35. The chromatography was standardized and, once the method was validated, no further changes were made. As part of Metabolon’s general practice, all columns were purchased from a single manufacturer’s lot at the outset of experiments. All solvents were similarly purchased in bulk from a single manufacturer’s lot in sufficient quantity to complete all related experiments. For each sample, vacuum-dried samples were dissolved in injection solvent containing eight or more injection standards at fixed concentrations, depending on the platform. The internal standards were used both to assure injection and chromatographic consistency. Instruments were tuned and calibrated for mass resolution and mass accuracy daily.
All methods utilized a Waters ACQUITY UPLC and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution. The sample extract was dried then reconstituted in solvents compatible to each of the four methods. Each reconstitution solvent contained a series of standards at fixed concentrations to ensure injection and chromatographic consistency. One aliquot was analyzed using acidic positive ion conditions, chromatographically optimized for more hydrophilic compounds. In this method, the extract was gradient eluted from a C18 column (Waters UPLC BEH C18-2.1x100 mm, 1.7 µm) using water and methanol, containing 0.05% perfluoropentanoic acid (PFPA) and 0.1% formic acid (FA). Another aliquot was also analyzed using acidic positive ion conditions; however, it was chromatographically optimized for more hydrophobic compounds. In this method, the extract was gradient eluted from the same aforementioned C18 column using methanol, acetonitrile, water, 0.05% PFPA and 0.01% FA and was operated at an overall higher organic content. Another aliquot was analyzed using basic negative ion optimized conditions using a separate dedicated C18 column. The basic extracts were gradient eluted from the column using methanol and water, however with 6.5mM ammonium bicarbonate at pH 8. The fourth aliquot was analyzed via negative ionization following elution from a HILIC column (Waters UPLC BEH Amide 2.1x150 mm, 1.7 µm) using a gradient consisting of water and acetonitrile with 10mM ammonium formate, pH 10.8. The MS analysis alternated between MS and data-dependent MSn scans using dynamic exclusion. The scan range varied slightly between methods but covered 80-1000 m/z.
Compound identification, quantification, and data curation
Metabolites were identified by automated comparison of the ion features in the experimental samples to a reference library of chemical standard entries that included retention time, molecular weight (m/z), preferred adducts, and in-source fragments as well as associated MS spectra and curated by visual inspection for quality control using software developed at Metabolon35,36. Identification of known chemical entities is based on comparison to metabolomic library entries of purified standards. Commercially available purified standard compounds have been acquired and registered into LIMS for distribution to the various UPLC-MS/MS platforms for determination of their detectable characteristics. Additional mass spectral entries have been created for structurally unnamed biochemicals, which have been identified by virtue of their recurrent nature (both chromatographic and mass spectral). These compounds have the potential to be identified by future acquisition of a matching purified standard or by classical structural analysis. Peaks were quantified using area-under-the-curve. Raw area counts for each metabolite in each sample were normalized to correct for variation resulting from instrument inter-day tuning differences by the median value for each run-day, therefore, setting the medians to 1.0 for each run. This preserved variation between samples but allowed metabolites of widely different raw peak areas to be compared on a similar graphical scale.
A total of 1116 different metabolites were measured in the 786 fecal samples, of which 210 were observed in less than 20% of the samples and thus excluded from further analysis due to lack of power. 345 metabolites were observed in more than 20% but less than 80% of the samples and were thus analyzed qualitatively as dichotomous traits (observed in a sample vs. not observed). The remaining 570 metabolites, which were observed in at least 80% of all samples, were scaled by run-day medians, log-transformed and scaled to uniform mean 0 and standard deviation 1 and analyzed quantitatively (Fig 1). Metabolite ratios were calculated from the run-day median normalized metabolite levels and subsequently log-transformed and scaled to a mean of 0 and standard deviation of 1.
We analyzed effects of sample storage time (i) in the fridge before samples were frozen and (ii) in the freezer before being further analyzed. To this end we regressed metabolite concentrations against storage times. After correcting for multiple testing, we found significant storage effects on 7 metabolites (FDR < 0.05) (Supplementary Figure 6). We, thus, corrected all further analyses for both storage time in the fridge and freezer, to avoid spurious results. Despite correcting all models for the storage time, we cannot ultimately eliminate a potential confounding effect due to storage time and future studies should investigate its influence on fecal metabolites.
Microbial sequencing
16S rRNA was extracted from fecal samples, PCR amplified, barcoded per sample and sequenced using the Illumina MiSeq, as previously described18. DNA was isolated from the samples using the PowerSoil kit. The V4 region of bacterial 16S rRNA gene sequences was PCR amplified using the 515F and 806R primers37. Reads were barcoded per sample and combined for multiplexed sequencing using the Illumina MiSeq to generate 250bp paired-end reads. Paired-end reads were merged with a minimum overlap of 200nt using fastq join within QIIME, which was also used to de-multiplex the sequence data.
Pre-processing of sequences and their clustering to operational taxonomic units followed the Sumaclust de novo approach as previously described for a subset of samples from38. In brief, 16S sequence reads for all samples were filtered to remove chimeric sequences produced during PCR, using USEARCH for chimera identification39. Remaining reads were then collapsed to operational taxonomic units (OTUs) using the Sumaclust algorithm for de novo clustering in QIIME 1.9.0, this method selected as de novo OTUs more accurately group reads at the 97% level40, and Sumaclust was the one of the best performing greedy clustering algorithms in previous comparisons38. Taxonomy was assigned to OTUs by alignment of representative sequences to the Greengenes 13_8 database with a 97% similarity threshold using UCLUST.
De novo clustering across all samples within the TwinsUK cohort produced ~300,000 OTUs after singleton removal; however, the majority of these were of low abundance and found in very few samples (table density 0.002). The OTU table was subset, discarding samples with fewer than 10,000 reads and OTUs that were not found in at least 25% of these samples. This resulted in a table of 581 OTUs (table density 0.547) (Supplementary Table 7). OTU counts were converted to relative abundances (over all reads in each sample), a pseudo count of 10-6 was added to account for zero counts, and the abundances were log transformed. The transformed abundances were then used as the response in models with sequencing run, sequencing depth, individual who extracted the DNA, individual who loaded the DNA and sample collection method as technical covariates. The residuals of these models were then used in downstream analysis of OTU abundances. The same normalization and control for technical effects was also carried out on taxonomic abundances collapsed at each taxonomic level. Collapsed taxonomies included counts from all OTUs. Shannon alpha diversity was also calculated from the complete OTU table. Each sample was rarefied to a depth of 10,000 reads 50 times. Diversity metrics were calculated for each sample in each table and the mean across all tables taken as the final measure. Beta diversity was calculated from all OTUs excluding singletons using the unweighted UniFrac algorithm29.
Visceral fat measurements
Measurements of whole body composition were performed using the DXA fan-beam technology (Hologic QDR; Hologic, Inc., Waltham, MA, USA) as previously described41. Briefly, subjects were positioned in a standardized manner, in a supine position with the clothes removed and wearing a gown. The DXA machine was calibrated on a daily basis using a spine phantom and on a weekly basis using a step phantom, as suggested by the manufacturer. The scans were analyzed using the QDR System Software Version 12.6.
Regions of interest were defined manually by the same operator following the SOP, which was derived from the manufacturer’s guidelines. The lower horizontal margins were placed above the pelvis, just above the iliac crest, while the upper horizontal margins were placed at the half of the distance between the acromions and the iliac crest. The vertical margins were adjusted just at the external borders of the body so that all the soft tissue was included.
This DXA-based measurement of visceral fat has been validated against VF measured by CT scan and shown to be reliable and reproducible42.
Statistical analysis
To assess the influence of age and gender on metabolite measurements, we regressed all metabolites against age and gender, correcting for family structure as random intercept using the R package lme443. Moreover, we calculated linear and logistic regression models, respectively, to assess the relationship of the fecal metabolome with obesity, measured as BMI and visceral fat mass (measured by double X-ray absorptiometry, see supplementary methods), respectively adjusting for age, sex, storage time and family as random intercept (see supplemental methods for formulas). Visceral fat measurements were available for 647 individuals. The following regression models were used:
-
a)
Associations of metabolites with age and gender
Metabolite ~ age + gender + storage time (fridge) + storage time (freezer) + (1 |family)
-
b)
Associations with BMI
BMI ~ Metabolite + age + sex + storage time (fridge) + storage time (freezer) + (1 |family)
-
c)
Associations with visceral fat mass
visceral fat ~ Metabolite + age + sex + BMI + height + height2 + storage time (fridge) + storage time (freezer) + (1 |family)
-
d)
Associations with microbial alpha diversity
Metabolite ~ microbiome diversity + age + sex + BMI + storage time (fridge) + storage time (freezer) + (1 |family)
-
e)
Associations with OTUs and taxa
Metabolite ~ microbe (OTU/taxa) + microbiome diversity + age + sex + BMI + storage time (fridge) + storage time (freezer) + (1 |family)
Partial Least Squares Discriminant Analysis
We used a partial least squares discriminant analysis (PLD-DA) to investigate global differences between the metabolic profiles of the youngest and oldest decile of our study population. To this end, all missing values were imputed using the mice package44 and metabolite levels were adjusted for storage times and family structure by linear mixed models. Residuals of these models were then used to train a PLSDA model as implemented in the mixOmics package45. The predictive performance was assessed using a 10-fold cross validation.
Heritability analysis
We used structural equation modelling to estimate the genetic (A), the common environment (C) and the unique environment (E) components of the total variance for each metabolite46. To this end we used the R package mets (version 1.1.1) to fit maximum-likelihood models, adjusting for age and sex and storage time. For each metabolite we fitted four models, estimating (1) A, C and E components (2) A and E components, (3) C and E components and (4) E component only. The best model was selected by minimizing the Akaike Information Criterion (AIC). In case of dichotomous metabolite abundances, a liability-threshold model was fitted using the bptwin function of the mets package. Additionally, intraclass correlation coefficients (ICCs) were calculated from variance components of a one-way analysis of variance for MZ and DZ twins individually, using the ICC package47.
Genome-wide association study
Genetic variation was measured using whole genome sequencing, as previously described (Nature genetics, in revision). In brief, samples were sequenced using the Illumina HiSeqX sequencer with 150 base paired reads. Reads were then mapped to hg38 genome using ISIS Analysis Software (v. 2.5.26.13; Illumina) and missing genotypes were filled in with reference homozygous calls48. Genomes with a ratio of heterozygous to homozygous variants higher than 2.5 were excluded leaving 739 individuals for further analysis. A cohort-based high confidence region of the genome was constructed by concatenating positions with greater than 90% “PASS” call rate using data from 3 sets of 100 randomly selected genomes. Variants outside of the high confidence region and duplicated variants were removed. We moreover excluded 273,355 variants with Hardy-Weinberg p<10-6, calculated from 420 unrelated individuals, leaving 8,208,502 biallelic SNPs and 1,408,051 InDels with minor allele frequency higher than 1% for further analysis.
We fitted linear mixed models to test for associations of heritable fecal metabolites with genetic variants, correcting for age, sex, storage time using GEMMA49 incorporating data from 739 individuals with fecal metabolomics and sequencing data. The twin structure of our data was taken into account by adjusting for the family relatedness using the sample kinship matrix. The score test implemented in GEMMA was used to assess significance of the associations. We considered metabolite-associations with a p-value lower than 1.2×10-10 significant, which corresponds to a genome-wide significance cut-off of 5.0×10-8, corrected for 428 tested metabolites. Additionally, we tested for genetic associations with all pairwise metabolite ratios of fecal metabolites with known chemical identity and a heritable variance component. We used the p-gain statistic to assess independence of the single metabolites19. The p-gain is defined as the minimal p-value of the associations of either of the single metabolites divided by the p-value of the metabolite ratio. A high p-gain statistic indicates that the ratio carries additional information compared to individual metabolites. We considered metabolite ratios with p < 1.6×10-12 (= 5×10-8 / 31,226 metabolite ratios) and p-gain > 3.1×105 (= 10 × 31,226 metabolite ratios) significant.
Four genome-wide significant associations were replicated in 230 individuals of the TwinsUK study, adjusting for the same confounding factors. Results of discovery and replication were combined using fixed-effects inverse variance meta-analysis.
Associations of the fecal metabolome with the gut microbiome
To assess associations of the fecal metabolome with the gut microbiome, we first regressed metabolite concentrations against the Shannon alpha diversity, adjusting for age, sex, BMI, storage time and family structure using 644 individuals with both fecal metabolomics and 16S sequencing data available.
We then estimated the proportions of variance of each metabolite explained by the microbiome by regressing the fecal metabolite concentration against the microbial beta-diversity. This technique is commonly used to estimate heritability from genetic kinship matrices50,51. To this end we calculated a restricted maximum likelihood model (REML), regressing the metabolite level against the microbial similarity, adjusting for age, sex, BMI and the storage time in fridge and freezer and technical covariates (sequencing run, sequencing depth, individual who extracted the DNA, individual who loaded the DNA and sample collection method) using the R package regress. The proportion of variance explained by microbial similarity (M2) and its standard error were calculated from the variance components using the R package gap52 and p-values were calculated from the ratio of M2 over its standard error.
Next, we aimed to identify microbes and taxonomical units that are associated with metabolite levels. To this end we regressed 581 inverse normalized operational taxonomic units (OTUs) against all 915 metabolites, adjusting for age, sex, BMI, sample storage times, family structure and the alpha diversity. Benjamini-Hochberg correction was applied to account for multiple testing. We further calculated associations at different taxonomical units, from genus to phylum level.
Lastly, to assess multivariate dependencies between the fecal metabolome and the microbiome, we inferred a graphical model combining 423 metabolites with known chemical identity that were observed in at least 80% of the samples with 241 OTUs that were assigned complete taxonomy at least to the genus level. Sparse graphical models were inferred using the GeneNet package53 and edges with false discovery rate < 0.05 were included in the model. We used the Fruchterman-Reingold algorithm54 to determine an unbiased graph layout and identified network modules by optimizing the modularity score as implemented in the igraph package55 (see supplemental methods for further details).
Pathway enrichment
We used pathway annotation as provided by Metabolon for pathway enrichment using the page algorithm. Enrichment p-values were estimated using permutation tests with 10,000 random permutations as implemented in the R-package piano56.
Graphical model inference
To assess multivariate dependencies between the fecal metabolome and the microbiome we inferred a graphical model combining 435 metabolites with known chemical identity that were observed in at least 80% of the samples with 241 OTUs that were annotated at least down to the genus level. To obtain a full data matrix we first imputed missing metabolite levels using mice44. A sparse graphical models was inferred using the GeneNet algorithm53 selecting edges with FDR < 0.05. We used the Fruchterman-Reingold algorithm54 to determine an unbiased graph layout and identified network modules by optimizing the modularity score as implemented in the igraph package55. For visualization purposes, we collapsed all metabolites of the same pathway to one node, which was connected to all neighbors of all its members. Microbes were collapsed by family. For both the full and the collapsed network, we calculated several centrality measures of nodes, including node degrees, as number of neighbors, clustering coefficients, as proportion of neighbors that are connected, and betweenness centralities, as proportion of shortest paths incorporating a node.
Supplementary Material
Acknowledgements
The study was funded by the Wellcome Trust; European Community’s Seventh Framework Programme (FP7/2007-2013). The study also receives support from the National Institute for Health Research (NIHR)- funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London. HLI collaborated with KCL to produce the metabolomics data from Metabolon Inc. CM is funded by the MRC AIM HY (MR/M016560/1) project grant.
We thank Julia Goodrich and Ruth Ley for their support in sequencing of the fecal samples.
Footnotes
Data availability
16S sequencing data used for this study is deposited in the European Nucleotide Archive (ERP015317). All other TwinsUK data are available upon request on the department website (http://www.twinsuk.ac.uk/data-access/accessmanagement/).
Author Contributions
Conceived and designed the experiments: AT, TDS, CM. Performed the experiments: RPM. Analyzed the data: JZ, MJ, TL, CM. Contributed reagents/materials/analysis tools: MM, GK, TL, AT, KS, CJS, JTB, AMV. Wrote the manuscript: JZ, MJ, RPM, AMV, TDS, CM. All authors revised the manuscript.
Competing financial interests
RPM is employee of Metabolon, Inc. TL and AT were employees of HLI, Inc. at the time this work was conducted. TDS is co-founder of MapMyGut Ltd. All other authors declare no competing financial interests.
References
- 1.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frias-Lopez J, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci U S A. 2008;105:3805–10. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcobal A, et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7:1933–43. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley R, Turnbaugh P, Klein S, Gordon J. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedersen HK, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–81. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 7.Clarke G, et al. Gut Microbiota: The Neglected Endocrine Organ. Mol Endocrinol. 2014;28:1221–1238. doi: 10.1210/me.2014-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cangelosi GA, Meschke JS. Dead or alive: molecular assessment of microbial viability. Appl Environ Microbiol. 2014;80:5884–91. doi: 10.1128/AEM.01763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Toole PW, Claesson MJ. Gut microbiota: Changes throughout the lifespan from infancy to elderly. Int Dairy J. 2010;20:281–291. [Google Scholar]
- 10.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romero-Corral A, et al. Accuracy of body mass index in diagnosing obesity in the adult general population. Int J Obes (Lond) 2008;32:959–66. doi: 10.1038/ijo.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arora T, Bäckhed F. The gut microbiota and metabolic disease: current understanding and future perspectives. J Intern Med. 2016;280:339–49. doi: 10.1111/joim.12508. [DOI] [PubMed] [Google Scholar]
- 13.Parséus A, et al. Microbiota-induced obesity requires farnesoid X receptor. Gut. 2017;66:429–437. doi: 10.1136/gutjnl-2015-310283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shoaie S, et al. Quantifying Diet-Induced Metabolic Changes of the Human Gut Microbiome. Cell Metab. 2015;22:320–31. doi: 10.1016/j.cmet.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Beaumont M, et al. Heritable components of the human fecal microbiome are associated with visceral fat. Genome Biol. 2016;17:189. doi: 10.1186/s13059-016-1052-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pallister T, et al. Untangling the relationship between diet and visceral fat mass through blood metabolomics and gut microbiome profiling. Int J Obes (Lond) 2017;41:1106–1113. doi: 10.1038/ijo.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodrich JK, et al. Human Genetics Shape the Gut Microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodrich JK, et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe. 2016;19:731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen A-K, et al. On the hypothesis-free testing of metabolite ratios in genome-wide and metabolome-wide association studies. BMC Bioinformatics. 2012;13:120. doi: 10.1186/1471-2105-13-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weimann A, Sabroe M, Poulsen HE. Measurement of caffeine and five of the major metabolites in urine by high-performance liquid chromatography/tandem mass spectrometry. J Mass Spectrom. 2005;40:307–16. doi: 10.1002/jms.785. [DOI] [PubMed] [Google Scholar]
- 21.Nyéki A, Buclin T, Biollaz J, Decosterd LA. NAT2 and CYP1A2 phenotyping with caffeine: head-to-head comparison of AFMU vs. AAMU in the urine metabolite ratios. Br J Clin Pharmacol. 2003;55:62–7. doi: 10.1046/j.1365-2125.2003.01730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin S-Y, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–50. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raffler J, et al. Genome-Wide Association Study with Targeted and Non-targeted NMR Metabolomics Identifies 15 Novel Loci of Urinary Human Metabolic Individuality. PLoS Genet. 2015;11:e1005487. doi: 10.1371/journal.pgen.1005487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.GTEx Consortium et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zerbino DR, et al. Ensembl 2018. Nucleic Acids Res. 2017;46:754–761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bastian F, et al. Data Integration in the Life Sciences. Springer; Berlin Heidelberg: 2008. Bgee: Integrating and Comparing Heterogeneous Transcriptome Data Among Species; pp. 124–131. [DOI] [Google Scholar]
- 27.McDonagh EM, et al. PharmGKB summary: very important pharmacogene information for N-acetyltransferase 2. Pharmacogenet Genomics. 2014;24:409–25. doi: 10.1097/FPC.0000000000000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meinl W, Sczesny S, Brigelius-Flohé R, Blaut M, Glatt H. Impact of gut microbiota on intestinal and hepatic levels of phase 2 xenobiotic-metabolizing enzymes in the rat. Drug Metab Dispos. 2009;37:1179–86. doi: 10.1124/dmd.108.025916. [DOI] [PubMed] [Google Scholar]
- 29.Lozupone C, Knight R. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keegan KP, Glass EM, Meyer F. MG-RAST, a Metagenomics Service for Analysis of Microbial Community Structure and Function. Methods Mol Biol. 2016;1399:207–33. doi: 10.1007/978-1-4939-3369-3_13. [DOI] [PubMed] [Google Scholar]
- 31.Vandeputte D, et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65:57–62. doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tigchelaar EF, et al. Gut microbiota composition associated with stool consistency. Gut. 2016;65:540–2. doi: 10.1136/gutjnl-2015-310328. [DOI] [PubMed] [Google Scholar]
- 33.Moayyeri A, Hammond CJ, Valdes AM, Spector TD. Cohort Profile: TwinsUK and Healthy Ageing Twin Study. Int J Epidemiol. 2013;42:76–85. doi: 10.1093/ije/dyr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans A, et al. High Resolution Mass Spectrometry Improves Data Quantity and Quality as Compared to Unit Mass Resolution Mass Spectrometry in High-Throughput Profiling Metabolomics. J Postgenomics Drug Biomark Dev. 2014;4:S24–S36. [Google Scholar]
- 35.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, Nontargeted Ultrahigh Performance Liquid Chromatography/Electrospray Ionization Tandem Mass Spectrometry Platform for the Identification and Relative Quantification of the Small-Molecule Complement of Biological Systems. Anal Chem. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 36.Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform. 2010;2:9. doi: 10.1186/1758-2946-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl):4516–22. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson MA, Bell JT, Spector TD, Steves CJ. A heritability-based comparison of methods used to cluster 16S rRNA gene sequences into operational taxonomic units. PeerJ. 2016;4:e2341. doi: 10.7717/peerj.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westcott SL, Schloss PD. De novo clustering methods outperform reference-based methods for assigning 16S rRNA gene sequences to operational taxonomic units. PeerJ. 2015;3:e1487. doi: 10.7717/peerj.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menni C, et al. Metabolomic profiling to dissect the role of visceral fat in cardiometabolic health. Obesity. 2016;24:1380–1388. doi: 10.1002/oby.21488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaul S, et al. Dual-energy X-ray absorptiometry for quantification of visceral fat. Obesity (Silver Spring) 2012;20:1313–8. doi: 10.1038/oby.2011.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bates D, Mächler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. J Stat Softw. 2015;67:51. [Google Scholar]
- 44.van Buuren S, Groothuis-Oudshoorn K. mice : Multivariate Imputation by Chained Equations in R. J Stat Softw. 2011;45:1–67. [Google Scholar]
- 45.Dejean S, et al. mixOmics: Omics Data Integration Project. 2013.
- 46.Neale M, Cardon L. Methodology for genetic studies of twins and families. Springer; Netherlands: 1994. [Google Scholar]
- 47.Wolak ME, Fairbairn DJ, Paulsen YR. Guidelines for estimating repeatability. Methods Ecol Evol. 2012;3:129–137. [Google Scholar]
- 48.Telenti A, et al. Deep sequencing of 10,000 human genomes. Proc Natl Acad Sci U S A. 2016;113:11901–11906. doi: 10.1073/pnas.1613365113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–4. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Speed D, Hemani G, Johnson MR, Balding DJ. Improved Heritability Estimation from Genome-wide SNPs. Am J Hum Genet. 2012;91:1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao JH. gap: Genetic Analysis Package. J Stat Softw. 2007;23:1–18. [Google Scholar]
- 53.Schaefer J, Opgen-Rhein R, Strimmer K. GeneNet: Modeling and Inferring Gene Networks. 2014 [Google Scholar]
- 54.Fruchterman TMJ, Reingold EM. Graph Drawing by Force-directed Placement. Software-Practice Exp. 1991;21:1129–1164. [Google Scholar]
- 55.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal Complex Sy. 2006:1695. [Google Scholar]
- 56.Väremo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41:4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.