Abstract
Dynamin 1-like proteins (DNM1-L) are mechanochemical GTPases that induce membrane fission in mitochondria and peroxisomes. Their mechanism depends on conformational changes driven by nucleotide and lipid cycling. Here we show the crystal structure of a mitochondrial fission dynamin (CmDnm1) from the algae Cyanidioschyzon merolae. Contrary to other eukaryotic dynamin structures, CmDnm1 is in a hinge 1 closed conformation with the GTPase domain compacted against the stalk. Within the crystal, CmDnm1 packs as a diamond shaped tetramer that is consistent with an inactive off-membrane state. Cross-linking, photoinduced electron transfer (PET) assays, and electron microscopy verify these structures. In vitro, CmDnm1 forms concentration dependent rings and protein-lipid tubes reminiscent of DNM1-L and classical dynamin with hinge 1 open. Our data provides a mechanism for filament collapse and membrane release that may extend to other dynamin family members. Additionally, hinge 1 closing may represent a key conformational change that contributes to membrane fission.
Mitochondrial membranes are maintained by an interplay of fission and fusion events mediated by different classes of dynamin-like proteins (DLPs). During membrane fission, the DLP DNM1-L controls the distribution of mitochondria in the cell 1 by locating to future division sites and driving membrane constriction and abscission 2. Human DNM1-L is an 80 kDa protein that comprises three core structural domains, the GTPase domain (G-domain), the bundle-signalling element (BSE), and the stalk. The stalk tip is augmented with a ~100 amino acid flexible lipid-binding domain called the B-insert for which no structural data currently exists. Hinge regions interconnect the BSE and stalk (hinge 1), and the G-domain and BSE (hinge 2). These regions are described in studies of BDLP1 where hinge 1 mediates a 135° rotation between the trunk and neck (equivalent to the stalk and BSE) from a closed to an open state, and hinge 2 mediates a 75° rotation between the neck and G-domain from a closed to open state (see Fig. 1a for a key to nomenclature) 3. Flexibility around hinge 2 has emerged as a conserved feature amongst many DLPs. In DNM1-L, comparison of GMPPCP and apo crystal structures shows a ~70° rotation of the BSE relative to the G-domain 4,5. Human Dynamin 1 undergoes a similar rotation when transitioning from the GMPPCP to GDP.AlF4- state 6. Arabidopsis DRP1A hinge 2 mediates a complex 95° and 45° twist observed between different crystal forms 7, whilst MxA hinge 2 undergoes a 110° rotation 8. Apart from BDLP1, common to all these structures was a requirement for the stalk to be truncated to facilitate crystallisation. Critically, this means that in these structures the hinge 1 position and conformation of the stalk relative to the BSE is unknown.
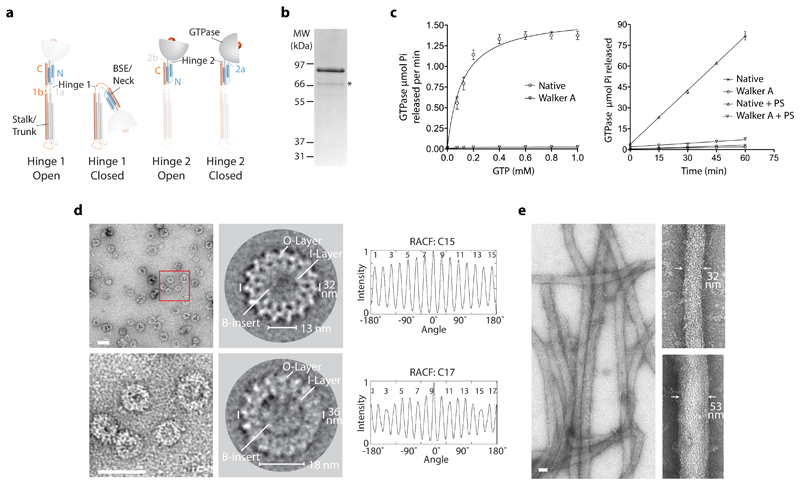
Figure 1. CmDnm1 catalytic activity and self-assembly.
a, Schematic showing nomenclature for hinge 1 and hinge 2 mediated conformations based on BDLP1 3. b, Native CmDnm1 purifies as a full-length band at 86 kDa and a minor proteolysis product at ~70 kDa as denoted by *. MW = molecular weight. c, (Left) Basal hydrolysis rate at variable GTP concentration. The Walker A K39A P-loop mutant shows negligible activity. (Right) Time course comparing GTP turnover in the presence and absence of PS liposomes. CmDnm1 shows significant assembly stimulated turnover. Error bars show the standard deviation based on n=3 independent replicates. d, (Top and bottom left) At 8 µM, CmDnm1 forms rings of variable diameter. Red box defines zoomed area. (Middle) Class average analysis of selected ring sizes derived from 6806 particles. Rings are comprised of an outer layer (O-layer) and inner layer (I-layer). Disordered densities may be observed for the B-insert within the central lumen. (Right) Rotation autocorrelation function for the selected rings show C15 and C17 symmetry. e, The addition of PS liposomes induces the formation of protein-lipid tubes of variable diameter between 30-75 nm. Scale bar for d and e = 50 nm.
For hinge 1, large scale rotation between the BSE and stalk to the closed conformation, as observed for BDLP1, has not yet been reported in eukaryotic systems. However, flexibility around hinge 1 has been observed through FRET experiments in the DLP MxA 9, and for DNM1-L where a 17.5° rotation was observed between symmetry mates within the crystal asymmetric unit 10. Early evidence for a BDLP1 closed state has been described for s-Mgm1 11 and speculatively for Mitofusin 1 through an Asp189 tether 12. Current data showing how a hinge 1 closed state may exist in eukaryotes is therefore restricted to DLPs involved in membrane fusion. The full-length crystal structure of DNM1-L 10 was captured in the apo state with the BSE and stalk in an open conformation, and hinge 2 between the G-domain and BSE in a closed conformation. Dynamin 1 and 3 were also observed in a similar DNM1-L apo conformation. For all these structures, stalk mutations that inhibit self-assembly were essential to facilitate crystallisation and structure determination. It is possible that these mutations or crystal lattice packing requirements may influence hinge 1 movement and have so far inhibited any alternative ground state conformations between the BSE and stalk to be captured.
DNM1-L is thought to sever mitochondria by assembling a helical filament that encompasses and constricts the outer membrane. The architecture of this helical filament depends on the DNM1-L subunit structure and its mode of self-assembly. DNM1-L subunits crystallize as a dimer with the stalks forming the interface 2 criss-cross motif 10. The same conserved dimerization mechanism has been observed in MxA 13,14 and classical Dynamin 1 and 3 15,16. In both DNM1-L and the classical dynamins, the stalks form a helical filament via self-assembly between interface 1 and 3 located towards the top and bottom of the stalk criss-cross, respectively. A relatively low-resolution reconstruction of the DNM1-L filament bound to lipid and GMPPCP is currently available 17. The lack of a high-resolution structure of the DNM1-L filament impedes the fitting of crystal structures and deducing subunit conformations. The reconstructed filament diameter is 129 nm and comprises a 2-start helix. The filament model utilises a contact, termed interface 4, between DNM1-L stalks to generate the 2-start helical arrangement 10. The binding of GTP and its non-hydrolysable analogues likely promotes G-dimerization between subunits on neighbouring rungs of the helical filament 5,10. The conformation of the DNM1-L subunit within the filament when bound to GTP and G-dimerised can be described with some certainty based on partial DNM1-L and Dynamin 1 crystal structures and relatively low-resolution EM reconstructions 4,6,18. Here, the data supports a model where hinge 1 and hinge 2 are in an open conformation. This is equivalent to Dynamin 1 under similar conditions 6. A question remains as to the conformation of DNM1-L and Dynamin 1 protein-lipid tubes in the apo state. It is unclear whether hinge 2 exists in an open conformation primed or weakly engaged in G-dimer formation, or is predominantly in a closed state reminiscent of the MxA polymer 19.
The mechanism for constriction and ultimately membrane fission is poorly understood for DNM1-L. However, it likely shares significant similarity with Dynamin 1 membrane fission for which two principle mechanisms are debated 20. Firstly, in the disassembly model, superconstriction of the membrane by a G-dimerised dynamin helix in the GDP and phosphate bound transition state is sufficient to provoke hemi-fission intermediates. Subsequent GDP or phosphate release induces filament disassembly and membrane destabilisation so that membrane fission results. Alternatively, the constriction/ratchet model couples nucleotide cycling to conformational changes that induce filament sliding and radial constriction. Ultimately, it is helix torsion that triggers membrane fission. The proposed conformational change, or power stroke, occurs as GTP is hydrolysed to the GDP and Pi bound transition state and drives hinge 2 closure. Transition to the GDP state induces G-dimer dissociation, nucleotide release, and the renewal of the cycle. During the power stroke, it is possible that a significant conformational change around hinge 1 is also triggered but has yet to be described given the absence of native full-length structures in the GDP bound state.
Given its relative simplicity, the ancient red algae Cyanidioschyzon merolae has emerged as a tractable model system for studying mitochondrial dynamics. Here, a single round mitochondrion in each algal cell undergoes orderly division driven by the DNM1-L homologue CmDnm1 21. Light and electron microscopy (EM) studies show that CmDnm1 is recruited from 10-20 cytoplasmic patches to the midpoint of the constricted mitochondrion-dividing ring late in the division cycle for final severance. Like DMN1-L in humans, CmDnm1 is critical for peroxisomal division 22,23. Both CmDnm1 and DNM1-L therefore represent key regulators and drivers of mitochondrial and peroxisome membrane fission in their respective hosts. Here we show the crystal structure of native CmDnm1 in the apo state at 7 Å resolution. The structure distinguishes itself from previous eukaryotic dynamin family structures, specifically those with G-dimer, BSE and stalk domains, as it requires no assembly inhibiting mutations or domain truncations for crystallisation. CmDnm1 is observed in a hinge 1 closed state reminiscent of BDLP1 in the GDP bound and apo state. This closed state is confirmed by PET and cross-linking assays. CmDnm1 also forms a novel tetrameric assembly that is suggestive of an inactive off-membrane storage state. Our results have broad mechanistic implication for how membrane fission, and possibly membrane fusion, are coordinated by members of the dynamin family.
Results
Purification and catalytic activity of CmDnm1
The gene encoding CmDnm1 from Cyanidioschyzon merolae (UniProt accession number Q84Y91) was cloned and expressed as an N-terminal maltose-binding protein (MBP) fusion in Escherichia coli. After initial purification using affinity chromatography, CmDnm1 and the MBP moiety were separated by TEV cleavage and size-exclusion chromatography (SEC) to yield purified CmDnm1 with mass 86 kDa (Fig. 1b and Supplementary Data Set 1). Full-length CmDnm1 purifies with a minor proteolysis product at ~70 kDa. Mass spectrometry analysis suggests that it is the B-insert that is cleaved. Working at a concentration of 5 µM, basal GTPase activity was measured with Km = 113 µM and kcat = 0.7 min-1 (Fig. 1c), which is comparable to Dynamin 1 24 but represents a lower maximum turnover rate when compared with yeast DNM1-L 25. The addition of phosphatidylserine (PS) liposomes induced a 27-fold increase in GTPase activity, which is indicative of G-dimerization and assembly stimulated turnover 18. In comparison, the GTPase defective CmDnm1 K39A P-loop mutant (Walker A) showed negligible activity both in the presence and absence of lipid.
CmDnm1 self-assembly is concentration dependent
Given the presence of assembly stimulated turnover and the propensity of DLPs to self-assemble, negative stain EM was used to visualise the oligomeric state of CmDnm1. At a low concentration (1 µM), CmDnm1 was typically observed as single particles ~7-12 nm in size (Supplementary Fig. 1). At 2-3 µM concentration, CmDnm1 initiates self-assembly and was observed as short crescent shaped filaments. Increasing concentration to 8 µM induced self-assembly into ordered rings (Supplementary Fig. 1 and Fig. 1d). Class averages reveal detailed ultrastructure where rings comprise concentric inner and outer layers. This architecture is compatible with DNM1-L and Dynamin 1 filaments in cross-section where the G-domain and BSE form the outer layer, and the stalks the inner layer 6,17. Rings predominantly range in rotational symmetry between C15, C16 and C17 symmetry (Fig. 1d). C13, C14 and C18 symmetry rings are also present but with low frequency. Within the lumen of the inner ring disordered densities were observed consistent with the lipid-binding B-insert. For smaller ring diameters, B-insert densities fill substantial parts of the inner lumen. At high concentration, the addition of PS liposomes induced formation of extensive tubular networks (Fig. 1e) reminiscent of those previously observed for DNM1-L 17. Tube diameter varies between ~30-75 nm suggesting high levels of constriction are achievable in the absence of nucleotide. The addition of nucleotide including GMPPCP and GDP both in the presence or absence of lipid yielded similar results to the apo state (Supplementary Fig. 2).
The crystal structure of CmDnm1
Crystallisation trials with CmDnm1 in the apo state yielded hexagonal crystals with P6(2)22 space group that diffracted to 7.0 Å resolution (Table 1). Strategies to improve resolution either inhibited crystallisation or yielded equivalent crystals (see Methods). The structure was solved by molecular replacement (MR) using two fragments as search models comprising the DNM1-L G-domain with BSE, and the stalk 10. A CmDnm1 homology model based on the DNM1-L MR solution was generated and refined using Phenix 26 (Table 1).
Table 1. Data collection and refinement statistics.
| CmDnm1 native (6FGZ) |
|
|---|---|
| Data collection | |
| Space group | P6(2)22 |
| Cell dimensions | |
| a, b, c (Å) | 188.2, 188.2, 163.6 |
| α, β, γ(˚) | 90.0, 90.0, 120.0 |
| Resolution (Å) | 40.84-7.00 (7.38)a |
| Rmerge | 0.14 (0.746) |
| I/σ(I) | 19.5 (3.3) |
| Completeness (%) | 99.5 (99.5) |
| Redundancy | 18.8 (20.8) |
| Refinement | |
| Resolution (Å) | 40.84-7.0 |
| No. reflections | 2950 |
| Rwork / Rfree | 34.4/41.1 |
| No. atoms | |
| Protein | 2611 |
| Ligand/ion | 0 |
| Water | 0 |
| B factors | |
| Protein | 390.0 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.022 |
| Bond angles (˚) | 2.13 |
Values in parentheses are for highest-resolution shell.
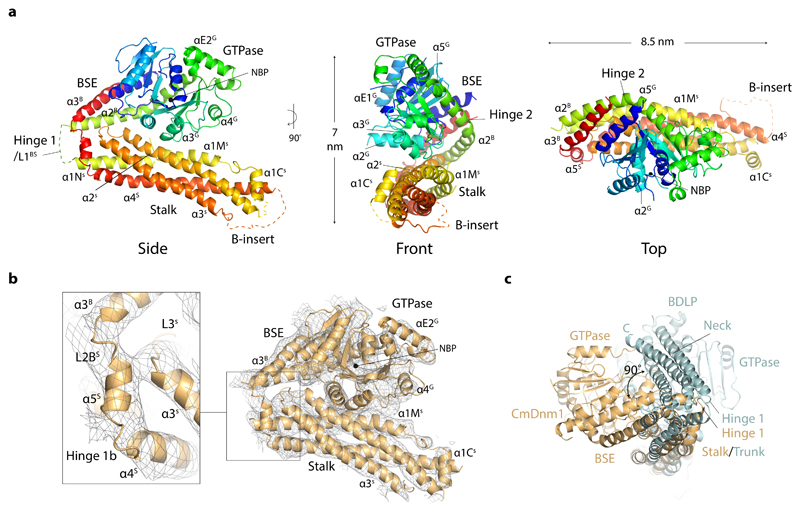
The asymmetric unit is a monomer and comprises GTPase, BSE and stalk domains equivalent to those of DNM1-L (Fig. 2a). This is expected given the 41 % sequence identity between CmDnm1 and DNM1-L (Supplementary Fig. 3). Significant electron density was not observed for the B-insert, which is likely a consequence of high flexibility in this motif 10,17. Hinge 2 located between the G-domain and BSE is in a closed position as it is for all other full-length eukaryotic classical dynamin 15,16,27, DNM1-L 10 and MxA 9,13 apo crystal structures. However, hinge 1 is also observed in a closed conformation so that the G-domain and BSE are compacted against the stalk. Well resolved electron density showing the position of the hinge 1b strand 3 facilitates the connection between the stalk and BSE to be modelled (Fig. 2b). This hinge 1 conformation is reminiscent of BDLP1 in the apo and GDP bound state where hinge 1 is closed, and the G-domain and neck form extensive contacts along the trunk 28. Superposition of CmDnm1 stalk and BDLP1 trunk (PDB 2J69) show that these domains share the same overall fold. However, the structures deviate as the CmDnm1 BSE is rotated 90 ° relative to the BDLP1 neck around the trunk/stalk long axis so that the plane of hinge 1 closure is different (Fig. 2c). Whilst the CmDnm1 GTPase domain contacts helices α2S and α1MS (Fig. 2a), the BDLP1 GTPase domain contacts helices 15 and 20 28, which constitute neighbouring faces on their respective stalk and trunk domains. As CmDnm1 hinge 2 is in a closed position, it is the G-domain side that contacts the stalk via the helix α3G C-terminus, whilst for BDLP1, hinge 2 is in an open conformation so that the GTPase base now contacts the trunk via helices 7 and 8.
Figure 2. The crystal structure of the CmDnm1 monomer.
a, Cartoon schematic showing the CmDnm1 monomer in a hinge 1 closed conformation. Blue N-terminus through to the red C-terminus. b, Electron density contoured at 1σ showing map quality and CmDnm1 model fit. No significant electron density is observed for the flexible B-insert. Zoom panel shows hinge 1b detail with associated electron density. c, End view superposition of CmDnm1 and BDLP1 (PDB 2J69) aligned by their respective stalk and trunk domains (C-α RMSD = 11.2 Å). The CmDnm1 G-domain and BSE is offset 90° relative to the BDLP1 G-domain and neck. The plane of hinge 1 closure therefore differs between CmDnm1 and BDLP1.
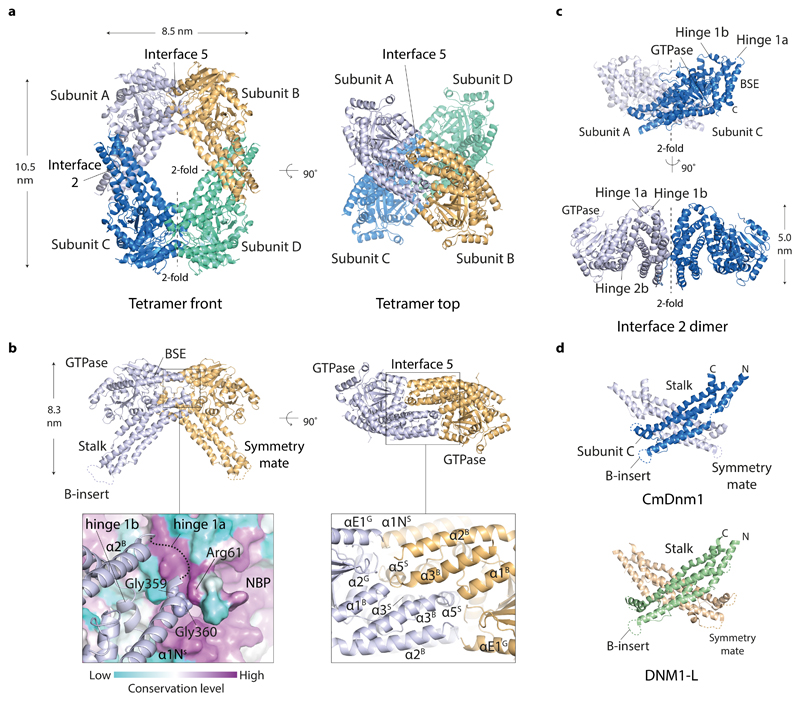
Within the crystal, the CmDnm1 subunit packs as a diamond shaped tetramer where the crystallographic 2-fold axes generate a pair of oligomerisation interfaces, termed interface 2 and interface 5 (Fig. 3a). This nomenclature aims to be consistent with previously identified oligomerisation interfaces 1-4 10,15,27. Interface 5 is novel and is formed by the back-to-back dovetailing of two CmDnm1 subunits where the convex outward face of each hinge 1 packs intimately against the backside of the symmetry mate GTPase domain (Fig. 3b). The buried surface area comprises 8 % of the monomer surface (2359 Å2 from a total of 28,931 Å2). Within interface 5, the hinge 1a loop at the hinge tip (Figs. 2a and 3b) neighbours the highly conserved Arg61, which is located on the side of the nucleotide binding pocket and is proximal to the catalytically crucial Switch 1 Thr60 (Supplementary Fig. 3). This arrangement suggests a mechanism for how nucleotide state may directly modulate hinge 1 conformation and consequently oligomerisation state. Hinge 1b helix α5S is entirely buried within interface 5 (Fig. 3b) and contacts both the C-terminal end of stalk helix α1NS and α2G located on the bottom of the GTPase domain within the symmetry mate.
Figure 3. The crystal structure of the CmDnm1 tetramer.
a, CmDnm1 packs as a diamond-shaped tetramer generated by crystallographic 2-fold axes around interface 2 and interface 5. Note how interface 5 dimers are offset by a ~ 90° screw or twist around the tetramer long axis. b, Cartoon schematic showing the arrangement of the interface 5 dimer. (Left) Front view of the interface 5 dimer. Zoom panel shows surface colouring based on residue conservation level calculated against 150 CmDnm1 homologues. Hinge 1a nestles in a highly conserved groove at the backside of the nucleotide binding pocket and contacts Arg61 located next to the Switch 1 Thr60. NBP = nucleotide binding pocket. (Right) Top view of interface 5 dimer. Zoom panel shows key helices contributing to the interface. c, Cartoon schematic showing the highly conserved interface 2 dimer. d, CmDnm1 and DNM1-L (PDB 4BEJ) form equivalent interface 2 stalk dimers.
Interface 2 constitutes the same criss-cross motif that has been observed previously in the DNM1-L 10, Dynamin 1 15,27 and MxA 13 interface 2 stalk dimer (Fig. 3c and 3d). The CmDnm1 and DNM1-L stalk dimer may be superimposed with Cα RMSD = 1.9 Å.
Validation of CmDnm1 hinge 1 closed conformation in solution
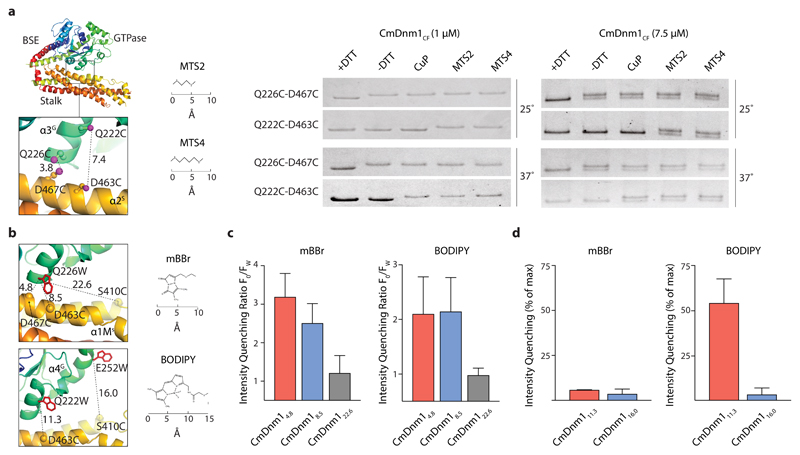
In order to validate the CmDnm1 model and the hinge 1 closed conformation observed in the crystal, cross-linking and PET assays were undertaken. A similar suite of assays has previously been used to observe conformational changes in Dynamin 1 29. Focusing initially on confirming the hinge 1 closed conformation, the cysteine residues at positions 70, 270, 461, 476, 494 and 506 were converted to alanine so as to create a cysteine-free CmDnm1 mutant, termed CmDnm1CF. A cysteine pair was then generated by introducing Q226C and D467C mutations into CmDnm1CF, termed CmDnm1CF-Q226C-D467C. These residues are located at a contact point between helix α3G located on the side of the GTPase domain and helix α2S on the stalk (Fig. 4a). The CmDnm1 model predicts the sulphur atoms between Q226C and D467C to be 3.8 Å apart. For human DNM1-L (PDB 4BEJ), where hinge 1 is in an open conformation, the equivalent Glu202 and Glu437 are 62 Å apart. At 25°C and at low concentration (1 µM) when CmDnm1 polymerisation is not favoured (Supplementary Fig. 1), CmDnm1CF-Q226C-D467C yielded a single band in the presence of DTT as confirmed by SDS-PAGE. When DTT was removed, a band shift was observed as disulphide bond formation between Q226C and D467C reduced migration rate during SDS-PAGE (Fig. 4a and Supplementary Data Set 1). Given disulphide bonds are 2.05 Å in length, this data suggests that Q226C and D467C are in close contact with each other. The addition of the oxidising agent Cu(II)-phenanthroline (CuP), or the cross-linkers MTS2 with 5.2 Å span or MTS4 with 7.8 Å span, yielded the equivalent band shift. The subsequent addition of DTT abolished all observed band shifts due to reversible disulphide bond reduction (Supplementary Fig. 4). Raising reaction temperature from 25°C to 37°C has no additional effect. CmDnm1CF-Q226C-D467C cross-linked at 1 µM and subsequently concentrated to 10 µM does not self-assemble into rings or tubulate PS liposomes (Supplementary Fig. 5). CmDnm1 in the closed conformation does not therefore support polymerisation. An alternative cysteine pair was then generated by introducing Q222C and D463C mutations into CmDnm1CF, termed CmDnm1CF-Q222C-D463C. The position of Q222C and D463C relate to CmDnm1CF-Q226C-D467C by a shift of 1 helical turn so that the sulphur atom pair is now 7.4 Å apart in the model (Fig. 4a). At 25°C, the band shift was only observed in the presence of MTS2 and MTS4 cross-linkers, whilst at 37°C, a shift was also observed in the presence of CuP (Fig. 4a and Supplementary Data Set 1). This data shows that the cysteines at positions 222 and 463 are sufficiently close for MTS2 to facilitate cross-linking, and that by increasing thermal motion in the presence of CuP, direct disulphide bond formation may be induced. Again, band shifts were reversible with the addition of DTT (Supplementary Fig. 4). The activity of the mutant constructs CmDnm1CF, CmDnm1CF-Q226C-D467C, and CmDnm1CF-Q222C-D463C was verified by their ability to self-assemble and form protein-lipid tubes (Supplementary Fig. 6a). Selected mutants were also tested for their ability to hydrolyse GTP in the presence and absence of lipids (Supplementary Fig. 6b). CmDnm1CF and CmDnm1CF-226C-467C showed similar levels of lipid stimulated GTP turnover when compared with native CmDnm1. Taken together, these cross-linking results are consistent with the CmDnm1 structure both in the amino acid register for the cysteine mutations, and in the overall conformation of the monomer with hinge 1 closed.
Figure 4. Biochemical validation of CmDnm1 hinge 1 closed conformation in solution.
a, CmDnm1CF-Q226C-D467C and CmDnm1CF-Q222C-D463 cysteine cross-linking (S-S distance = 3.8 Å and 7.4 Å, respectively) with copper phenanthroline (CuP) and MTS cross linkers. At low concentration (1 μM), cross-linking induces slower migration rates by SDS-PAGE and consequently a band shift. At high concentrations (7.5 µM), self-assembly reduces the efficiency of cross-linking, presumably as a consequence of hinge 1 opening, so that a band doublet is observed. The experiment was repeated n=3 times with similar results. b, (Top left) Cartoon schematic showing the fluorescent conjugate pairs CmDnm1mBBr/4.8, CmDnm1mBBr/8.5, and CmDnm1mBBr/22.6 relative to PET data shown in c. Distances are shown in Ångstrom. (Bottom left) The fluorescent conjugate pairs CmDnm111.3 and CmDnm116.0 relative to PET data shown in d. c, PET data showing the emission intensity quenching ratio (F0/Fw) for mBBr and BODIPY in variable distance fluorescent conjugate pairs. Measurements performed at CmDnm1 concentration of 1 µM. CmDnm14.8 and CmDnm18.5 show significant intensity quenching whilst CmDnm122.6 is unquenched. Error bars show the standard deviation based on n=3 independent replicates. d, Only BODIPY quenches at a Cα-Cα distance = 11.3 Å, whilst the smaller mBBr moiety remains unquenched. Error bars show the standard deviation based on n=3 independent replicates.
The same set of cross-linking experiments was then repeated but at a concentration where CmDnm1 was at the transition point for polymerisation into rings (7.5 µM) (Supplementary Fig. 1). Whilst the overall pattern of band shifting remained the same as at low concentration, it differed as the efficiency of disulphide linkage or cross-linking was reduced so that a mixed population of shifted and non-shifted was now observed (Fig. 4a and Supplementary Data Set 1). This data supports a model where polymerisation releases GTPase domain contact from the stalk and allows hinge 1 to open.
To confirm the cross-linking results, PET experiments were undertaken where a tryptophan quenching moiety and a fluorophore were introduced at specific sites and fluorescence emission measured. Fluorescence varies based on the distance between the tryptophan and fluorophore with efficient PET quenching typically occurring at < 1 nm 30. Using CmDnm1CF, Q226W was mutated as a quenching moiety, and D467C for the attachment of the thiol-reactive fluorescent conjugate monobromobimane (mBBr). This fluorescent conjugate pair has a predicted distance of 4.8 Å based on the CmDnm1 model between main chain Cα atoms and is therefore termed CmDnm1mBBr/4.8 (Fig. 4b). Additional conjugate pairs were then generated by moving the position of the fluorophore to D463C and S410C measured to be either 8.5 Å or 22.6 Å away from Q226W, respectively. The resulting constructs were termed CmDnm1mBBr/8.5 and CmDnm1mBBr/22.6. All conjugate pairs were competent to self-assemble (Supplementary Fig. 6c). Fluorescent intensities (FW) were calculated as an intensity quenching ratio (IQR) 31 against equivalent mBBr constructs that lacked the tryptophan at position 226 (F0). Comparing IQR showed that quenching declines with increasing distance between conjugate pairs with CmDnm1mBBr/4.8 and CmDnm1mBBr/8.5 showing a significant decrease in fluorescent output, whilst CmDnm1mBBr/22.6 showed an IQR close to F0 (Fig. 4c). Increasing the size of the fluorescent moiety by swopping mBBr for a thiol-reactive iodoacetamide derivative of BODIPY-FL (BODIPY) (Fig. 4b) now yielded comparable IQRs between CmDnm1BODIPY/4.8 and CmDnm1BODIPY/8.5, whilst CmDnm1BODIPY/22.6 IQR was close to maximum fluorescence.
In order to confirm that the observed PET was not specific to the Q226W quenching moiety, two novel conjugate pairs were generated comprising Q222W and D463C with a main chain Cα distance of 11.3 Å, and E252W and S410C with a main chain Cα distance of 16.0 Å (Fig. 4b). These constructs were termed CmDnm111.3 and CmDnm116.0, and were competent to self-assemble (Supplementary Fig. 6c). To each conjugate pair, either mBBr or BODIPY was attached for comparison. For CmDnm111.3 and CmDnm116.0 attached to mBBr negligible quenching was observed due to the limited fluorophore length. However, when fluorophore length was increased by attaching BODIPY, CmDnm111.3 became quenched whilst CmDnm116.0 remained out of range (Fig. 4d). Collectively, these PET experiments support the cross-linking data and are consistent with a model where the CmDnm1 GTPase domain is in contact with the stalk. CmDnm1 is therefore in a hinge 1 closed conformation, contrary to the DNM1-L apo crystal structure (PDB 4BEJ) where hinge 1 is extended. Repetition of these PET experiments but with the addition of GMPPCP or GDP yielded results equivalent to the apo state (data not shown). This suggests that at low CmDnm1 concentration (< 1 µM), nucleotide alone is not sufficient to trigger hinge 1 opening in vitro.
Validation of the CmDnm1 tetramer in solution
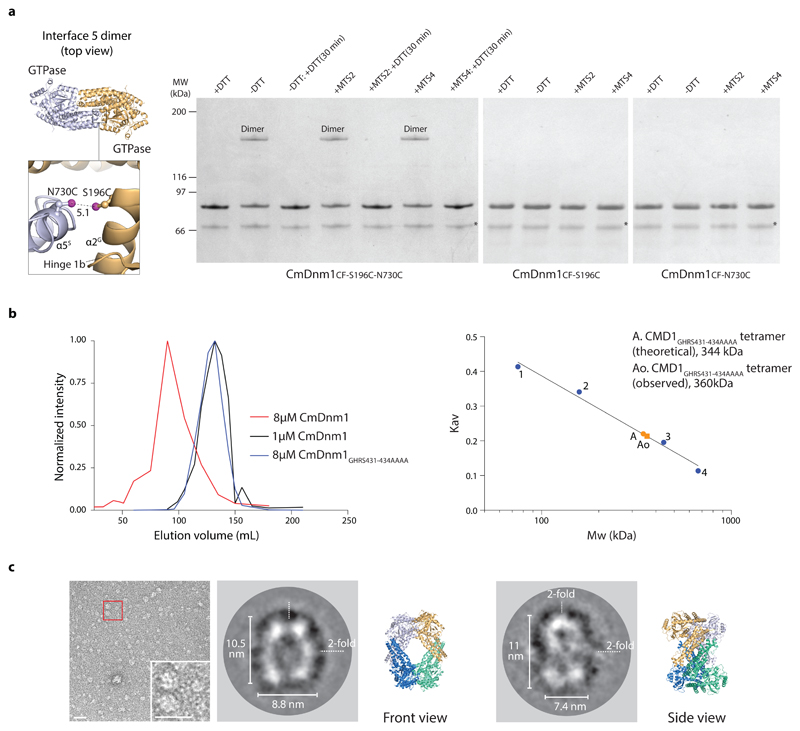
Crystal packing generates interface 2 and interface 5, and the formation of a diamond shaped tetramer. Interface 2 is a conserved feature of many eukaryotic dynamins, including human DNM1-L, with a fundamental role in self-assembly. The formation of CmDnm1 rings (Fig. 1d) reminiscent of Dynamin1 filaments in cross section 6, and CmDnm1 protein-lipid tubes (Fig. 1e) similar to DNM1-L protein-lipid tubes 17, strongly suggests that self-assembly via interface 2 is conserved in CmDnm1. In order to validate interface 5, a cross-linking experiment was set up where the mutations S196C and N730C were introduced into CmDnm1CF, to generate CmDnm1CF-S196C-N730C. These residues are located at the base of the GTPase domain, and the outside face of hinge 1b, respectively, and constitute a buried contact point within interface 5 (Fig. 5a). In silico mutation of S196C and N730C based on the CmDnm1 structure, positions the sulphur atoms within the cysteine pair to be 5.1 Å apart. Given the proximity of S196C and N730C to each other and their buried location within interface 5, the structure predicts that disulphide bond formation will yield a dimer only. In the presence of DTT, CmDnm1CF-S196C-N730C exists as a monomer as observed by SDS-PAGE (Fig. 5a and Supplementary Data Set 1). However, removal of DTT induced the appearance of an additional band on the gel at a position consistent with the theoretical 172 kDa dimer. No higher order oligomeric species was observed even with the addition of MTS2 and MTS4 cross-linker. Subsequent addition of DTT reversed dimerization so that only the monomer remained. To ensure the dimerization was specific between S196C and N730C, single cysteine mutations were introduced into CmDnm1CF to generate either CmDnm1CF-S196C or CmDnm1CF-N730C. Removal of DTT or addition of MTS2 and MTS4 cross-linker with either CmDnm1CF-S196C or CmDnm1CF-N730C failed to induce dimerization (Fig. 5a and Supplementary Data Set 1). CmDnm1CF-S196C-N730C, CmDnm1CF-S196C and CmDnm1CF-N730C are functional as shown by their ability to polymerise and tubulate liposomes (Supplementary Fig. 6d). These cross-linking experiments are consistent with interface 5 and the CmDnm1 tetramer existing in solution. In addition, SEC shows that at 1 µM concentration CmDnm1 elutes with an observed molecular weight of 360 kDa, which is close to the theoretical tetramer of 344 kDa (Fig. 5b). This is in contrast to SEC at 8 µM concentration when CmDnm1 is polymerised and elutes in the void volume. Note that the self-assembly limiting stalk mutant GHRS431-434AAAA 15 also elutes as a tetramer at 8 µM concentration (Fig. 5b). This is in contrast to Dynamin 1 15, DNM1-L 10 and MxA 13 which elute as a dimer with the equivalent mutation.
Figure 5. Validation of the CmDnm1 tetramer in solution.
a, CmDnm1CF-S196C-N730C cross-linking (S-S distance = 5.1 Å) forms a dimer in non-reducing conditions, or with MTS cross linkers. No higher order oxidation oligomers are observed. Cross-linking is not observed in the single cysteine mutants CmDnm1CF-S196C and CmDnm1CF-N730C. Cross-linking is reversible upon incubation with DTT after 30 mins reaction time. The experiment was repeated n=3 times with similar results. MW = molecular weight. b, SEC analysis of CmDnm1 oligomeric state based on concentration. At 8.0 µM, CmDnm1 is polymerised and elutes in the void volume. At 1.0 µM, CmDnm1 elutes as a tetramer (Kav = 0.25). At 8.0 µM, the self-assembly limiting stalk mutant GHRS431-434AAAA elutes as a tetramer. Calibration data includes conalbumin 75 kDa (1), aldolase 158 kDa (2), ferritin 440 kDa (3), thyroglobulin 669 kDa (4). c, Negative stain EM showing CmDnm1 single particles at low concentration (1.0 µM) (left). Class averages derived from 7740 particles typically show diamond or oval shaped particles (middle), or figure-of-8 shapes (right) that are consistent with front or side views of the CmDnm1 tetramer structure.
Negative stain EM class average analysis of the CmDnm1 single particles observed at 1 µM concentration (Supplementary Fig. 1) provides further solution state validation of the CmDnm1 tetramer (Fig. 5c and Supplementary Fig. 7). Oval or diamond shaped particles were preferentially observed with symmetry and dimensions similar to the CmDnm1 tetramer crystal structure (front view). Alternatively, figure-of-8 shaped class averages were observed that are consistent with side views of the CmDnm1 tetramer, which incorporates a 90° screw or twist around the particle long axis (Fig. 3a).
Discussion
Here we show the native structure of the mitochondrial fission DLP CmDnm1 from Cyanidioschyzon merolae. The observed hinge 1 closed conformation is reminiscent of the BDLP1 crystal structure in the apo and GDP bound conformation 28. It is also consistent with the conformation of the mitochondrial membrane fusion DLP s-Mgm1 derived from a low resolution EM reconstruction of 2D crystals 11. In this case, an s-Mgm1 homology model based on BDLP1 was fitted into the 2D crystal reconstruction as a back-to-back dimer with now striking similarity to the interface 5 CmDnm1 dimer. Additionally, it has been speculated that Mitofusin 1 may form a hinge 1 closed conformation via an Asp189 HD1-HD2 tether 12. A model for the yeast mitofusin-like protein Fzo1 based on a BDLP1-like closed state has also been validated via site-directed mutagenesis and in vivo functional assays 32. Collectively, this data suggests that the hinge 1 closed conformation is a conserved feature amongst bacterial and eukaryotic membrane fusion DLPs. Close structural and functional homology between CmDnm1 and human DNM1-L suggests that hinge 1 closure may be prevalent amongst other mitochondrial fission DLPs.
The CmDnm1 diamond shaped tetramer is formed via interface 2 and interface 5. Interface 2 facilitates formation of the criss-cross dimer that constitutes the basic assembly unit of the classical dynamin, DNM1-L and MxA/B filament 10,14,15,19,27. Interface 5 has the effect of sequestering a pair of criss-cross dimers oriented back-to-back in the hinge 1 closed conformation. The CmDnm1 tetramer therefore likely represents an off-membrane inactive state. DNM1-L is known to exist in a dimer-tetramer equilibrium in solution 10, whilst classical Dynamin 1 is reported as a monomer, dimer and tetramer 33–36. The architecture and conformation of these dimers and tetramers in solution is unknown. One option is that they form interface 2 dimers that oligomerise as CmDnm1-like tetramers via interface 5. Alternatively, subunits may be arranged similar to the Dynamin 3 tetramer with hinge 1 open and hinge 2 closed 16. Ultimately further studies are required in DNM1-L and the classical dynamins to understand if the CmDnm1 tetramer represents a broadly conserved off-membrane state.
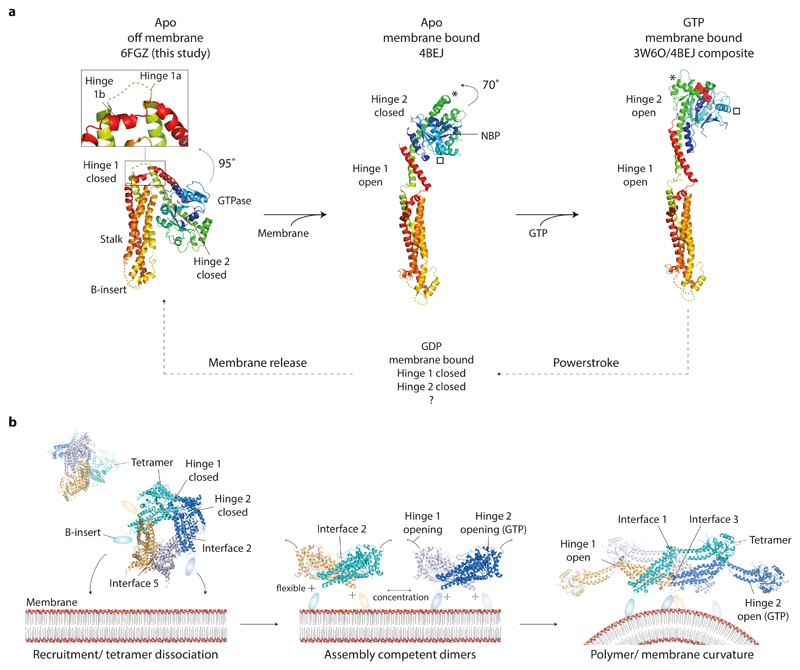
Based on our data, a model for CmDnm1 activation and self-assembly is presented (Fig. 6). The CmDnm1 tetramer structure suggests that when off-membrane and at low concentration, CmDnm1 is in a hinge 1 and hinge 2 closed conformation. Tetramer recruitment to the membrane releases interface 5 and induces transition to interface 2 dimers. Hinge 1 undergoes a 95° rotation to an assembly competent open conformation. In the absence of nucleotide hinge 2 remains closed. This conformation is equivalent to the DNM1-L and Dynamin 1 apo crystal structures, and is consistent with the non-constricted protein-lipid tubes observed for mitochondrial and classical dynamins 17,37. When GTP is bound, hinge 2 shifts to the open conformation by rotating 70° 4,5,18. CmDnm1 would then be competent to G-dimerise and promote membrane fission. This conformation generates the constricted protein-lipid tubes observed for mitochondrial and classical dynamins 17,38.
Figure 6. CmDnm1 nucleotide hydrolysis cycle and activation model.
a, General model of membrane and GTP induced conformational changes in CmDnm1 and DNM1-L. In all panels, the equivalent orientation of the stalk is shown. (Left) Whilst off membrane and in the apo state, CmDnm1 is in a hinge 1 and hinge 2 closed conformation. (Middle) Membrane binding or self-assembly induces hinge 1 opening. In the absence of nucleotide, hinge 2 remains closed as shown by the DNM1-L crystal structure (PDB 4BEJ). (Right) Composite model of DNM1-L G-domain/BSE truncation bound to GMPPCP (PDB 3W6P) with the DNM1-L stalk (PDB 4BEJ). GTP binding induces hinge 2 opening and facilitates G-dimerization. * and show the movement of equivalent structural positions between apo (middle) and GTP bound (right) states. GTP hydrolysis to GDP induces hinge 2 closure and speculatively hinge 1 closure. b, Model for CmDnm1 activation and polymerisation. (Left) CmDnm1 is off membrane and sequestered as an interface 2 and interface 5 tetramer. (Middle) Membrane recruitment yields interface 2 dimers and hinge 1 opening. (Right) Local concentration of interface 2 dimer promotes polymer formation. GTP binding is required to induce hinge 2 transition to the open conformation.
Potential triggers for tetramer dissociation and hinge 1 opening include membrane association, nucleotide binding, or competition for assembly interfaces with neighbouring subunits. In vitro, CmDnm1 polymerisation is closely correlated with concentration suggesting that inter-subunit binding between interfaces 1, 3 or 4 10 may be sufficient to disassemble interface 5 and promote hinge 1 opening. For Dynamin 3, membrane binding is thought to facilitate polymerisation by releasing the PH domain from an autoinhibitory stalk-bound position 16. In human DNM1-L, the B-insert is also known to allosterically modulate oligomer state 39,40. For CmDnm1, the B-insert may therefore play a similar role in facilitating polymerisation through driving conformational change upon membrane binding. Critically, DNM1-L is known to function with multiple regulatory partners such as Fis1 41, mitochondrial fission factor (Mff) 42, and Mitochondrial Dynamics protein of 49 kDa (MiD49) and 51 kDa (MiD51 or MIEF1) 43,44. A candidate Fis1 appears present in the C. merolae genome. Regulatory proteins will therefore likely play a significant role in modulating CmDnm1 oligomerisation dynamics and conformation. For example, the concentration at which the CmDnm1 tetramer dissociates and hinge 1 opens in vivo may be tuneable depending on the specific regulatory partner bound.
The CmDnm1 hinge 1 closed state may also have general ramification for the mechanism of membrane fission. For DNM1-L and classical dynamins, the hydrolysis of GTP to GDP.AlF4- is known to induce hinge 2 transition to the closed state 6. For classical dynamins, this conformational change has been used to explain how filaments may slide past each other in order to induce helix torsion and ultimately membrane fission via a possible ratchet mechanism 20. Filament sliding is one explanation for the observed twisting activity of Dynamin 1 protein-lipid tubes as exemplified by bead rotation 45,46. It also potentially explains why Dynamin 1 filaments form plectonemic supercoils and writhes 46,47. There is currently no structural data for the hinge 1 conformation of DNM1-L or classical dynamin in the GDP.AlF4- or GDP bound state. However, hinge 1 may close, or partially close in these systems, when transitioning to a GDP bound state given the structure of CmDnm1, and the hinge 1 closed state of BDLP1 in the GDP bound state 28. In this case, hinge 1 and hinge 2 closure predicts a substantial radial and axial twist between two G-dimerized subunits. Such a large-scale conformational change may provide the filament torsion and writhing necessary to induce membrane fission. This model also provides an efficient means to induce filament disassembly and membrane release. The hinge 1 closed structure of CmDnm1 suggests that there is still much to be determined regarding the mechanism of DLP-mediated membrane fission.
Online Methods
Cloning, protein expression and purification
The coding sequence for CmDnm1 from C. merolae (UniProt Q84Y91) was cloned into pOPTM vector (pET derivative), which yields an N-terminal MBP fusion with a TEV cleavage site in the linker, and a hexahistidine tag at the C-terminus. Overexpression of CmDnm1 was achieved in E.coli Rosetta (DE3) cells grown in 2xTY media and induced with 1 mM IPTG at OD600= 0.6 with 19°C overnight shaking. Cells were harvested by centrifugation.
For CmDnm1 purification, cells were lysed by sonication in 50 mM Tris-Cl, 300 mM sodium chloride, final pH 7.5. The cell lysate was centrifuged at 31,100 rcf at 4 °C for 45 min. The supernatant was incubated with amylose resin (New England BioLabs) for 3 hrs at 4 °C with gentle stirring, and loaded into a gravity column. The column was washed with 50 mM Tris-Cl, 500 mM sodium chloride, 10% glycerol, final pH 7.5. The MBP-CmDnm1 fusion protein was eluted in 20 mM Tris-Cl, 150 mM sodium chloride, 15 mM maltose, 15% glycerol, 1mM EDTA, 5 mM DTT, final pH 8.5. The MBP was cleaved from CmDnm1 by TEV protease and the products were separated by gel filtration using either HiPrep 16/60 Sephacryl or 26/60 Sephacryl S-300 columns (GE Healthcare) equilibrated in 20mM Tris-Cl, 150 mM sodium chloride, 1 mM EDTA, 5 mM DTT, 12% glycerol, final pH 8.5. To obtain a SEC elution profile of CmDnm1 at 1 µM, fractions were analysed by SDS-PAGE stained with SYPRO Ruby, visualised with UV transillumination at 473 nm, and band intensities quantified using IMAGEJ software. Above 1 µM concentration, CmDnm1 polymerises and elutes in the void. Void fractions were concentrated to ~ 10 mg/ml, flash frozen in liquid nitrogen and stored at -80 °C.
Cloning and mutagenesis was carried out using either restriction enzyme methods or one-step isothermal DNA assembly 48. All mutants were expressed and purified as for native.
Crystallization, structure determination and refinement
10 mg/ml native CmDnm1 was crystallized by sitting drop vapour diffusion incubated at 20°C against a mother liquor of 0.1 M sodium acetate, 1.0 M ammonium phosphate, 0.1 M lithium sulphate, final pH 4.6. After 1-2 weeks crystals were protected with artificial mother liquor mixed with 25 % glycerol and flash frozen in liquid nitrogen. Well-formed crystals grew in the hexagonal space group P6(2)22 with cell dimensions a=188.2 Å, b=188.2 Å, c=163.6 Å. Diffraction data was collected on beamline ID14eh4 at ESRF, Grenoble, at 100K with wavelength 0.9330, and was limited to 7.0 Å. Strategies to improve diffraction yielded either the equivalent crystals or inhibited crystal growth. The strategies included but were not limited to: 1) additive screens including nucleotides, heavy atoms, and detergents 2) seeding 3) construct modification including truncation of the B-insert, targeted loop deletion, His tag removal, surface entropy reduction mutants, incorporation of fusion proteins including maltose binding protein (MBP), thioredoxin, ZapA 49 and T4 lysozyme both at the sequence termini and integrated into loops 4) limited proteolysis using chymotrypsin 5) targeted mutagenesis including the classic self-assembly limiting mutation 15, here GHRS431-434AAAA, which failed to crystallise. The structure was solved by molecular replacement (MR) with PHASER 50. To generate the search models, the human DNM1-L structure (PDB 4BEJ) was separated into two fragments comprising the G-domain and BSE, and the stalk. Side chains were removed to yield poly-alanine main chain. PHASER solved yielding a single solution with well resolved density, TFZ=17.3 and LLG=232. A CmDnm1 homology model based on the DNM1-L MR solution was then generated with SWISS-MODEL 51. Side chains were removed to yield poly-alanine main chain. COOT 52 was used to align the CmDnm1 homology model onto the MR solution. Residues with no supporting build from 4BEJ or with no obvious electron density were removed. The CmDnm1 homology model was then refined with PHENIX 26. Two rounds of rigid body and group ADP refinement was undertaken, with COOT used for minor manual adjustments to the model. The G-domain and BSE, and the stalk each constituted separate rigid bodies. Secondary structure restraints were applied and the poly-alanine CmDnm1 homology model trimmed to match the DNM1-L MR solution (Cα RMSD = 0.5 Å) supplied as a reference model 53. The final model has a Cα RMSD = 1.0 Å from the DNM1-L MR solution. Using PROCHECK 54, 92.9 % of residues are in the most favoured region of the Ramachandran plot with 0.0 % in the disallowed regions. Coordinates have been deposited in the Protein Data Bank with accession code 6FGZ.
Liposome preparation
L-α-phosphatidylserine (Soy PS-870336) was purchased from Avanti Polar Lipids. 5 mg phosphatidylserine was dissolved in 400 μl chloroform, lyophilised by evaporation with nitrogen gas, desiccated for 1 hr and resuspended in 20 mM HEPES, 150 mM sodium chloride, final pH 7.5. After brief sonication with a micro-tip sonicator, liposomes were generated by extrusion through a polycarbonate membrane with pore size 0.2 μm (Mini-Extruder, Avanti Polar Lipids).
GTPase assays
GTPase assays were performed at room temperature in a buffer containing 20 mM HEPES, 150 mM sodium chloride, 2 mM magnesium sulphate, final pH 7.5. CmDnm1 and GTP concentration was at 5 μM and 1 mM, respectively. To test for lipid-accelerated GTP turnover, 1 mg/ml of L-α-phosphatidylserine liposomes was added to the reaction. Free phosphate concentration was determined using a malachite green based kit (Innova Biosciences, UK, #303-0030). Measurements of absorbance were performed at 620 nm using a CLARIOstar (BMG Labtech).
Electron microscopy
Prior to visualization, proteins were diluted to indicated concentrations in a buffer containing 25 mM HEPES, 150 mM sodium chloride, and 2 mM magnesium sulphate, final pH 7.5. For tubulation, 10 μM CmDnm1 was incubated at 22 °C for 2 hrs with 2 mg/ml liposomes. Ring formation was achieved at 8 μM concentration whilst single particles were observed at 0.3 – 1 μM concentration. 1 mM nucleotide was added as desired. The samples were applied to glow-discharged 300-mesh carbon coated copper grids and stained with 2 % uranyl acetate. Images were recorded on either a FEI Tecnai T12 electron microscope with CCD camera, or a FEI Tecnai TF20 FEG electron microscope equipped with a Falcon II direct electron detector (FEI Company). To generate class averages of CmDnm1 rings, datasets were collected between -2 and -3.5 defocus with 2.05 Å pixel size. To generate class averages of CmDnm1 single particles, a dataset was collected between -1.5 and -3.5 defocus with 1.28 Å pixel size. Images were phase flipped using GCTF and initially processed by iterative rounds of 2D classification using RELION to remove low quality particles. This resulted in stacks containing 6806 (apo) and 14,573 (GMPPCP) images for the rings, and 7740 images for the single particles. Multivariate statistical analysis and 2D classification in IMAGIC was then carried out to generate the final class averages.
Protein cross-linking
MTS-based homobifunctional cross-linking reagents were obtained from Toronto Research Chemicals. PD MiniTrap desalting columns with Sephadex G-25 resin (GE Healthcare) were used to remove all DTT from each protein sample before the cross-linking experiment. Cross-linking of CmDnm1 mutants was carried out at 25 °C and 37 °C with protein concentrations of 1 μM and 7.5 μM (validation of the CmDnm1 closed conformation) or 2 μM (validation of the CmDnm1 tetramer) for 20 min. 1 mM Cu(II)-(1,10-phenanthroline)3 (CuP) or 50 μM MTS reagents (MTS-2-MTS or MTS-4-MTS) were added as desired. The theoretical spanning distance of MTS reagents was derived from 55. For analysis of the cross-linked proteins, all unreacted Cys residues were blocked by 10 mM N-ethylmaleimide (NEM) (Sigma-Aldrich) prior to the addition of 4x SDS sample buffer. Samples were resolved by either 7.5 or 10% SDS-PAGE followed by Coomassie or Instant Blue staining. For reversal of cross-linking, samples were incubated for 30 min on ice with 10 mM DTT. To examine for the ability of cross-linked mutants to form rings or tubulate liposomes, CmDnm1 mutant were fully cross-linked at 1 μM and then concentrated to 10 μM. Samples were mixed with liposomes as desired and analysed by negative stain EM.
Protein labelling
The Cys residues at positions 467 in CmDnm14.8 (Q226W/D467C), CmDnm18.5 (Q222W/D467C), CmDnm122.6 (E252W/D467C), CmDnm1CF-D467C (D467C), position 463 in CmDnm1 (Q222W/D463C), and position 410 in CmDnm1 (E252W/S410C), were selectively labelled in the absence of reducing agent using tenfold molar excess of the thiol-reactive iodoacetamide derivative of BODIPY-FL (Life Technologies) or monobromobimane (mBBr) (Thermo Fisher Scientific). After 30 min incubation at room temperature, DTT was added to 5 mM to quench the reaction. The solution was extensively dialysed against buffer containing 20 mM Tris-Cl, 150 mM sodium chloride, 1 mM EDTA and 5 mM DTT, final pH 7.6 to separate unreacted dye molecules. The efficiency of labelling was determined using a molar absorptivity coefficient of 76,000 M-1 cm-1 at 502 nm and 5,000 M-1 cm-1 for BODIPY and mBBr, respectively. Control experiments using the CmDnm1CF protein showed that background labelling is < 2%.
Photoinduced electron transfer (PET)
All PET measurements were carried out with a minimum of three replicates with 1 μM labelled CmDnm1 mutants in buffer containing 20 mM Tris-Cl, 150 mM sodium chloride, 1 mM EDTA and 5 mM DTT, final pH 7.6 using a Fluorolog-3 photon-counting steady-state spectrofluorometer (Horiba Jobin Yvon) equipped with double excitation and emission monochromators and a cooled PMT housing. BODIPY iodoacetamide was excited at 490 nm (2.5 nm bandpass) and emission was registered at 510 nm (2.5 nm bandpass). The mBBr-labelled protein sample was excited at 380 nm (3 nm bandpass) and emission was registered at 490 nm (2.5 nm bandpass). CmDnm1CF-D467C bound to either BODIPY or mBBr was used to establish the F0 value, which corresponds to a lack of PET-induced quenching.
Supplementary Material
Acknowledgements
We thank the beamline staff at the ESRF for data collection support. T. Pape for in-house EM facility support. We acknowledge the gift of Cyanidioschyzon merolae genomic DNA from T. Kuroiwa. We are grateful to A. Chernyatina and J. Liu for manuscript feedback. This work was supported by a Wellcome Trust Fellowship (097328/Z/11/Z) to H.L.
Footnotes
Data availability
The atomic coordinates have been deposited in the Protein Data Bank under accession code 6FGZ. Materials from this study may be requested from H.L.
Author contributions
O.B and H.L designed experiments. H.L initially purified native protein, obtained crystals and determined structure. O.B purified proteins and performed all other experiments including GTPase, cross-linking and PET assays, and generating EM data. H.L and O.B. wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. Journal of Cell Biology. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Molecular Biology of the Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Low HH, Sachse C, Amos LA, Löwe J. Structure of a Bacterial Dynamin-like Protein Lipid Tube Provides a Mechanism For Assembly and Membrane Curving. Cell. 2009;139:1342–1352. doi: 10.1016/j.cell.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kishida H, Sugio S. Crystal structure of GTPase domain fused with minimal stalks from human dynamin-1-like protein (Dlp1) in complex with several nucleotide analogues. Current Topics in Peptide & Protein Research. 2013;14:67–77. [Google Scholar]
- 5.Wenger J, et al. Functional Mapping of Human Dynamin-1-Like GTPase Domain Based on X-ray Structure Analyses. Plos One. 2013;8:e71835. doi: 10.1371/journal.pone.0071835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chappie JS, et al. A Pseudoatomic Model of the Dynamin Polymer Identifies a Hydrolysis-Dependent Powerstroke. Cell. 2011;147:209–222. doi: 10.1016/j.cell.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan LM, et al. Structural basis for mechanochemical role of Arabidopsis thaliana dynamin-related protein in membrane fission. Journal of Molecular Cell Biology. 2011;3:378–381. doi: 10.1093/jmcb/mjr032. [DOI] [PubMed] [Google Scholar]
- 8.Rennie ML, McKelvie SA, Bulloch EMM, Kingston RL. Transient dimerization of human MxA promotes GTP Hydrolysis resulting in a mechanical power stroke. Structure. 2014;22:1433–1445. doi: 10.1016/j.str.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, et al. Conformational dynamics of dynamin-like MxA revealed by single-molecule FRET. Nature Communications. 2017;8:15744. doi: 10.1038/ncomms15744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frohlich C, et al. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. Embo Journal. 2013;32:1280–1292. doi: 10.1038/emboj.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeVay RM, et al. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. Journal of Cell Biology. 2009;186:793–803. doi: 10.1083/jcb.200906098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao YL, et al. MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion. Nature. 2017;542:372–377. doi: 10.1038/nature21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao S, et al. Structure of Myxovirus Resistance Protein A reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity. 2011;35:514–525. doi: 10.1016/j.immuni.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 14.Gao S, et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature. 2010;465:502–507. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- 15.Faelber K, et al. Crystal structure of nucleotide-free dynamin. Nature. 2011;477:556–561. doi: 10.1038/nature10369. [DOI] [PubMed] [Google Scholar]
- 16.Reubold TF, et al. Crystal structure of the dynamin tetramer. Nature. 2015;525:404–409. doi: 10.1038/nature14880. [DOI] [PubMed] [Google Scholar]
- 17.Mears JA, et al. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nature Structural & Molecular Biology. 2011;18:20–25. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin's assembly-stimulated GTPase activity. Nature. 2010;465:435–440. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez FJD, et al. CryoEM structure of MxB reveals a novel oligomerization interface critical for HIV restriction. Science Advances. 2017;3:e1701264. doi: 10.1126/sciadv.1701264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonny B, et al. Membrane fission by dynamin: what we know and what we need to know. Embo Journal. 2016;35:2270–2284. doi: 10.15252/embj.201694613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishida K, et al. Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2146–2151. doi: 10.1073/pnas.0436886100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imotoa Y, et al. Single-membrane-bounded peroxisome division revealed by isolation of dynamin-based machinery. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9583–9588. doi: 10.1073/pnas.1303483110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koch A, et al. Dynamin-like protein 1 is involved in peroxisomal fission. Journal of Biological Chemistry. 2003;278:8597–8605. doi: 10.1074/jbc.M211761200. [DOI] [PubMed] [Google Scholar]
- 24.Narayanan R, Leonard M, Song BD, Schmid SL, Ramaswami M. An internal GAP domain negatively regulates presynaptic dynamin in vivo: a two-step model for dynamin function. Journal of Cell Biology. 2005;169:117–126. doi: 10.1083/jcb.200502042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingerman E, et al. Dnm1 forms spirals that are structurally tailored to fit mitochondria. Journal of Cell Biology. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D-Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ford MGJ, Jenni S, Nunnari J. The crystal structure of dynamin. Nature. 2011;477:561–566. doi: 10.1038/nature10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Low HH, Löwe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- 29.Mattila JP, et al. A hemi-fission intermediate links two mechanistically distinct stages of membrane fission. Nature. 2015;524:109–114. doi: 10.1038/nature14509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doose S, Neuweiler H, Sauer M. Fluorescence quenching by photoinduced electron transfer: a reporter for conformational dynamics of macromolecules. Chemphyschem. 2009;10:1389–1398. doi: 10.1002/cphc.200900238. [DOI] [PubMed] [Google Scholar]
- 31.Mansoor SE, DeWitt MA, Farrens DL. Distance mapping in proteins using fluorescence spectroscopy: the tryptophan-induced quenching (TrIQ) method. Biochemistry. 2010;49:9722–9731. doi: 10.1021/bi100907m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Vecchis D, et al. A membrane-inserted structural model of the yeast mitofusin Fzo1. Scientific Reports. 2017;7:10217. doi: 10.1038/s41598-017-10687-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Binns DD, et al. Correlation between self-association modes and GTPase activation of dynamin. Journal of Protein Chemistry. 1999;18:277–290. doi: 10.1023/a:1021083211267. [DOI] [PubMed] [Google Scholar]
- 34.Muhlberg AB, Warnock DE, Schmid SL. Domain structure and intramolecular regulation of dynamin GTPase. Embo Journal. 1997;16:6676–6683. doi: 10.1093/emboj/16.22.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamoto PM, Tripet B, Litowski J, Hodges RS, Vallee RB. Multiple distinct coiled-coils are involved in dynamin self-assembly. Journal of Biological Chemistry. 1999;274:10277–10286. doi: 10.1074/jbc.274.15.10277. [DOI] [PubMed] [Google Scholar]
- 36.Tuma PL, Collins CA. Activation of Dynamin gtpase is a result of positive cooperativity. Journal of Biological Chemistry. 1994;269:30842–30847. [PubMed] [Google Scholar]
- 37.Chen YJ, Zhang PJ, Egelman EH, Hinshaw JE. The stalk region of dynamin drives the constriction of dynamin tubes. Nature Structural & Molecular Biology. 2004;11:574–575. doi: 10.1038/nsmb762. [DOI] [PubMed] [Google Scholar]
- 38.Zhang PJ, Hinshaw JE. Three-dimensional reconstruction of dynamin in the constricted state. Nature Cell Biology. 2001;3:922–926. doi: 10.1038/ncb1001-922. [DOI] [PubMed] [Google Scholar]
- 39.Strack S, Cribbs JT. Allosteric modulation of Drp1 mechanoenzyme assembly and mitochondrial fission by the variable domain. Journal of Biological Chemistry. 2012;287:10990–11001. doi: 10.1074/jbc.M112.342105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang YF, Gao XL, Garavito RM. Biochemical characterization of human dynamin-like protein 1. Journal of Biochemistry. 2011;150:627–633. doi: 10.1093/jb/mvr102. [DOI] [PubMed] [Google Scholar]
- 41.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. Journal of Cell Biology. 2000;151:367–379. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Molecular Biology of the Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer CS, et al. MiD49 and MiD51, new components of the mitochondrial fission machinery. Embo Reports. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao J, et al. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. Embo Journal. 2011;30:2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morlot S, et al. Membrane shape at the edge of the dynamin helix sets location and duration of the fission reaction. Cell. 2012;151:619–629. doi: 10.1016/j.cell.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roux A, Uyhazi K, Frost A, De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- 47.Danino D, Moon KH, Hinshaw JE. Rapid constriction of lipid bilayers by the mechanochemical enzyme dynamin. Journal of Structural Biology. 2004;147:259–267. doi: 10.1016/j.jsb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 48.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 49.Low HH, Moncrieffe MC, Löwe J. The crystal structure of ZapA and its modulation of FtsZ polymerisation. Journal of Molecular Biology. 2004;341:839–852. doi: 10.1016/j.jmb.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 50.McCoy AJ, et al. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research. 2014;42:252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D-Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Headd JJ, et al. Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution. Acta Crystallographica Section D-Biological Crystallography. 2012;68:381–390. doi: 10.1107/S0907444911047834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 55.Loo TW, Clarke DM. Determining the dimensions of the drug-binding domain of human P-glycoprotein using thiol cross-linking compounds as molecular rulers. Journal of Biological Chemistry. 2001;276:36877–36880. doi: 10.1074/jbc.C100467200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.