Abstract
Messenger RNA encodes cellular function and phenotype. In the context of human cancer it defines the identity of malignant cells and diversity of tumor tissue. We studied 72,501 single cell transcriptomes of human renal tumors and normal tissue from fetal, pediatric and adult kidneys. We matched childhood Wilms’ tumor with specific fetal cell types, thus providing evidence for the hypothesis that Wilms’ tumor cells are aberrant fetal cells. In adult renal cell carcinoma we identified a canonical cancer transcriptome that matched a little known subtype of proximal convoluted tubular cell. Analyses of the tumor composition defined cancer-associated normal cells and delineated a complex VEGF signaling circuit. Our findings reveal the precise cellular identity and composition of human kidney tumors.
Cancer cell identity is defined by morphological appearance, tissue context, and marker gene expression. Single cell transcriptomics refines this cellular identity on the basis of a comprehensive and quantitative read out of messenger RNA. Precise cellular transcriptomes may reveal a tumor’s cell of origin and the transcriptional trajectories underpinning malignant transformation (1).
We sought to define the identity of normal and cancerous human kidney cells from a catalogue of 72,501 single kidney cell transcriptomes, integrated with tumor whole genome DNA sequencing (2). We studied Wilms’ tumor (n=3), clear cell (ccRCC; n=3) and papillary renal cell carcinoma (pRCC; n=1) in relation to healthy fetal (n=2), pediatric (n=3), adolescent (n=2), and adult kidneys (n=5), as well as ureters (n=4; Table S1)
Normal tissue biopsies were taken from macroscopically normal regions of kidneys resected due to cancer (n=10) or for transplantation (n=2). We performed technical replicates of each biopsy and biological replicates, where clinically permissible (Table S1). We processed kidneys immediately following resection, generating single cell solutions enriched for viable cells. We derived counts of mRNA molecules in each cell for further analyses, subject to quality control (2).
We split 72,501 fetal, normal, and tumor cells into immune and non-immune cell compartments (Fig. S1). Using a community detection algorithm (2), transcriptomes were further segregated into distinct clusters of cells (Table S2). We next generated a reference map of normal mature and fetal cells, assigning an identity to each cluster, by cross-referencing cluster-defining transcripts with canonical markers curated from the literature (Table S3). Ambiguous clusters were not included in the reference map and are presented in Fig. S2-S8. Highly specific cluster-defining transcripts (potential cell markers) are appended (Table S4).
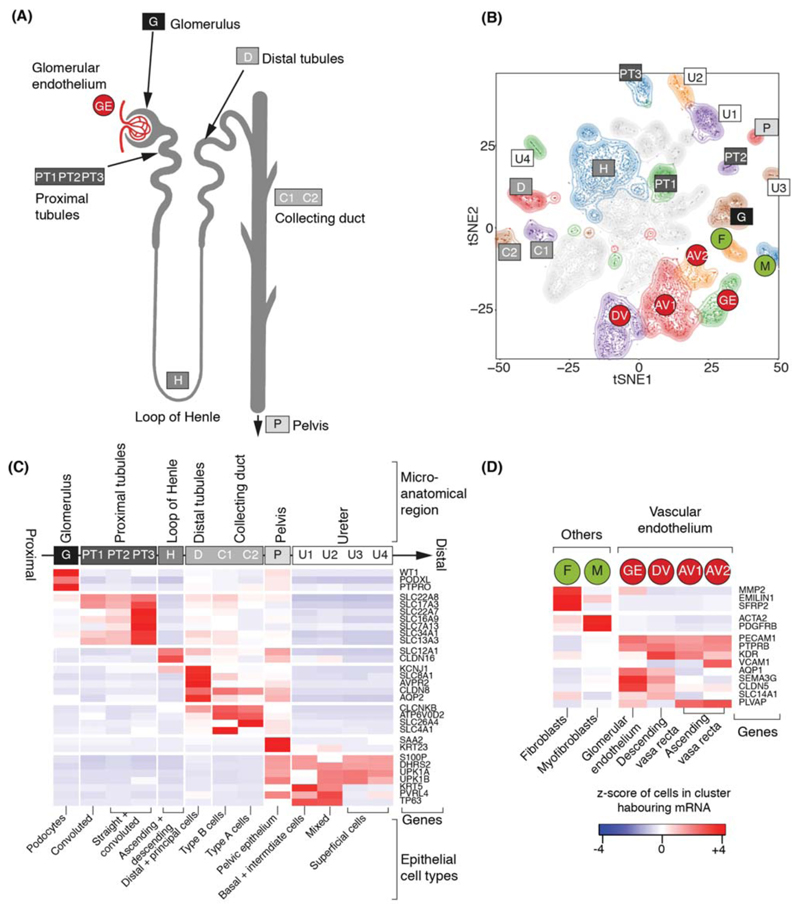
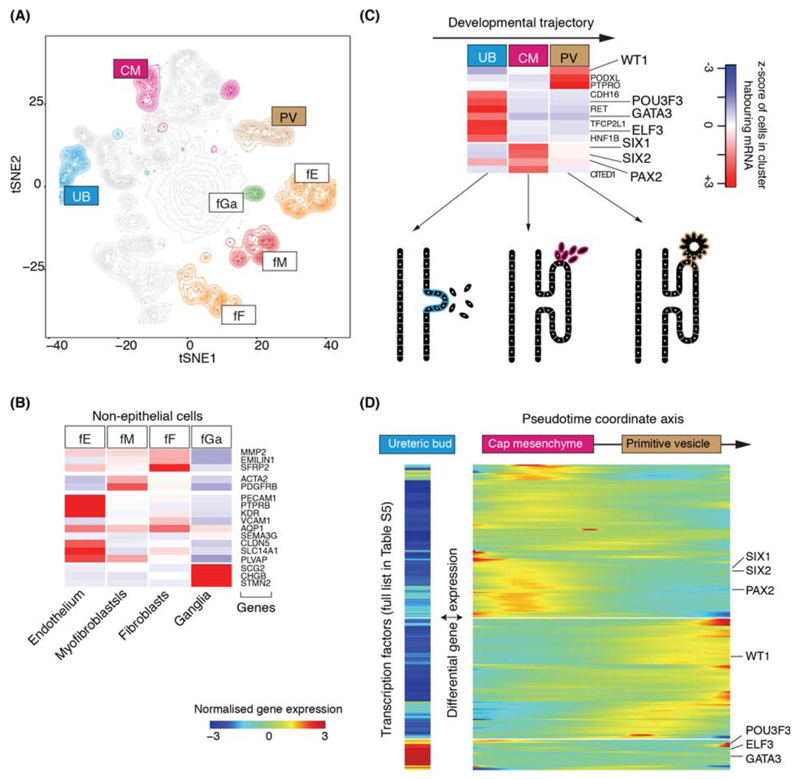
Amongst 42,809 non-malignant cells, 37,951 mature kidney cells represented epithelial cells from distinct micro-anatomical regions of the nephron, with a large proportion of proximal tubular cells (Fig. 1A-C, Fig. S4). Furthermore, there were fibroblasts, myofibroblasts, and vascular endothelial cells, i.e. glomerular endothelium, ascending and descending vasa recta (Fig. 1D, Fig. S2). 4,858 fetal cells grouped into developing nephron cells (ureteric bud, cap mesenchyme, primitive vesicle) and fibroblasts, myofibroblasts, vascular endothelial and ganglion cells (Fig. 2A-C, Fig. S5).
Figure 1. Canonical cell types in normal human kidneys.
(A) Illustration of nephron anatomy with cell clusters marked.
(B) tSNE representation of 8,707 normal epithelial and vascular cells. Clusters are colored, uniquely labelled and emphasized with density contours. Ambiguous clusters are de-emphasized and fully shown in Fig. S2.
(C) Expression of canonical nephron specific genes (Table S3) in clusters from (A). Colors give the fraction of cells expressing each gene in a cluster, scaled to have mean 0 and standard deviation 1 across all clusters.
(D) Expression of clusters in (A) not shown in (C) and their canonical genes.
Figure 2. Fetal cell types and nephrogenesis.
(A) tSNE representation of 4,858 fetal epithelial and vascular cells, colored and labelled as in Fig. 1A.
(B) Expression of markers of clusters in (A), colored as in Fig. 1C.
(C) Expression of nephrogenesis markers from clusters in (A) with illustration of nephron development. Formation of nephrons emanates from the ureteric bud, which induces condensation of the overlying mesenchyme into the cap mesenchyme. The cap mesenchyme then forms the primitive vesicle, the precursor of the glomerulus. The tubular system grows out from both ends of the fetal nephron: ureteric bud and primitive vesicle.
(D) The expression of transcription factor which vary significantly (p<0.01; likelihood ratio test) along the pseudo-time trajectory defined using the CM and PV cells from (C), or differentially expressed between UB versus CM and PV. UB expression is shown in a separated block on the left. Within the right block, pseudo-time increases from left to right and rows are clustered and grouped by hierarchical clustering with canonical transcription factors of nephrogenesis highlighted (see Table S6).
To determine transcriptional programs underlying nephrogenesis, we identified differentially expressed transcription factors in ureteric bud cells against cap mesenchyme and primitive vesicle cells (Fig. 2D). Furthermore, we applied pseudo-timing methods to identify transcription factors that define the transition from cap mesenchyme to primitive vesicle (Fig. 2D). Together, these analyses identified established, and unknown, transcription factors associated with nephron development, included as a reference for subsequent analyses of malignancy (Table S5).
Having established the single cell landscape of healthy kidneys, we characterized the cellular identity of 6,333 non-immune (Fig. S7) and 17,821 immune (Fig. S8) tumor cells from Wilms’ tumor (n=3), ccRCC (n=3) and pRCC (n=1; Table S1). Children had received neoadjuvant cytotoxic treatment prior to nephrectomy, as per British practice. Although this pre-treatment reduced yield (Table S6), recovered cells represent therapeutically relevant surviving cancer cells that determine the degree of adjuvant cytotoxic chemotherapy required (3). We used logistic regression to quantify the similarity between tumor and normal cell clusters, validated through intrinsic control populations (2). That is, the model found myofibroblasts from tumors matched myofibroblasts from mature and fetal kidneys (Fig. 3A) and no match for mast cells, a negative control population inserted into the training data.
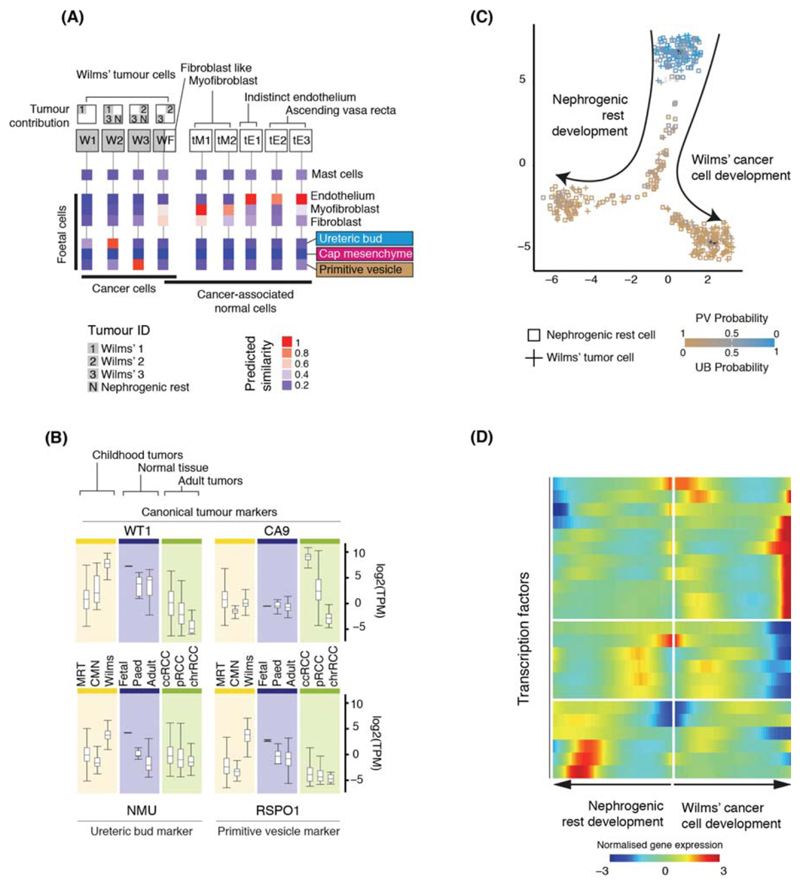
Figure 3. Matching childhood tumors with normal fetal cells.
(A) Similarity of Wilms’ tumor and cancer-associated normal cells to the reference fetal kidney map (Fig. 2A), with mast cells added as a negative control. Square boxes indicate sample contribution. Colors represent the probability that the cluster identified in the column header is “similar” to the fetal cluster identified by the row label (2).
(B) Expression of canonical tumor markers and representative UB and PV specific genes (Table S8) in RNA-seq from childhood cancers (yellow), normal tissue (blue) or adult cancers (green). MRT: malignant rhabdoid tumor; CMN: congenital mesoblastic nephroma. As positive controls, canonical tumor markers are shown: WT1 (Wilms’); CA9 (ccRCC).
(C) Pseudo-time trajectory of all Wilms tumor and nephrogenic rest cell. Color indicates similarity of each cell to the PV or UB fetal population. Jitter has been added to each point’s position with the original position plotted underneath in black (2).
(D) Transcription factors identified as varying significantly along the pseudo-time trajectory in (C). The center of the heatmap corresponds to the cells at the top of (C) and then proceeding left/right along the arrows shown in (C).
This similarity metric may be obfuscated by the phenotypic plasticity of tumor cells. We therefore developed a method to genotype individual cancer cells from mRNA reads using somatic copy number changes (Table S7; Fig. S9) defined by whole genome sequencing (Fig. S10). We validated genotyping calls by phasing single nucleotide polymorphisms across segments with altered copy number, testing for the presence of somatic single nucleotide variants, and comparison to control populations (Fig. S11-S14).
Integrating genotyping and similarity analyses, we found that Wilms’ cells resembled fetal normal cells, evidencing that Wilms’ tumor represents aberrant fetal cells. We found different populations of Wilms’ tumor that matched ureteric bud and primitive vesicle cells (specific developing nephron populations) (Fig. 3A). One cluster (WF), composed of Wilms’ cancer cells and non-cancerous ccRCC fibroblasts, exhibited a fibroblast-myofibroblast transcriptome. In one case, we obtained an anatomically separate perilobar nephrogenic rest, thought to represent a precursor lesion of Wilms’. Like Wilms’ cancer cells, we observed that nephrogenic rest resembled ureteric bud or primitive vesicle. This suggests that the potential to generate the different cell states of the fetal nephron is acquired early, or was not lost, by the developing Wilms’ cancer, although this conclusion is based on only one sample.
To validate the cellular identity of Wilms’ cells, we interrogated bulk transcriptomes of an independent series of 124 Wilms’ tumors for cellular signatures of ureteric bud and primitive vesicle (4, 5). We extracted specific markers expressed within ureteric bud or primitive vesicle cells and unexpressed within non-tumor cells (Table S8, (2)) and probed bulk transcriptomes for these cluster defining transcripts. As comparators to Wilms’ we included fetal, pediatric, and adult normal tissue bulk transcriptomes (n=135) and other childhood kidney tumors: 17 congenital mesoblastic nephroma and 65 malignant rhabdoid tumors. Corroborating the presence of these cells in Wilms’, signatures of primitive vesicle and ureteric bud cells were seen in, and confined to, Wilms’ and normal fetal tissue (Fig. 3B).
Placing Wilms’ tumor cells in pseudo-time revealed two transcriptional programs emanating from the ureteric bud: one branch predominantly describing the development of nephrogenic rest cells and the other of Wilms’ cancer cells (Fig. 3C). There was a significant overlap in the transcription factors underpinning these two programs (Fig 3D; Table S9) and normal nephrogenesis (p<10-4; hypergeometric test). This indicates that developmental relationships exist between Wilms’ tumor cells that have been adopted from normal nephrogenesis. Our analyses reveal the plasticity and fetal identity of Wilms’ cells and transcriptionally defines developmental cell states and trajectories that may harbor targetable vulnerabilities.
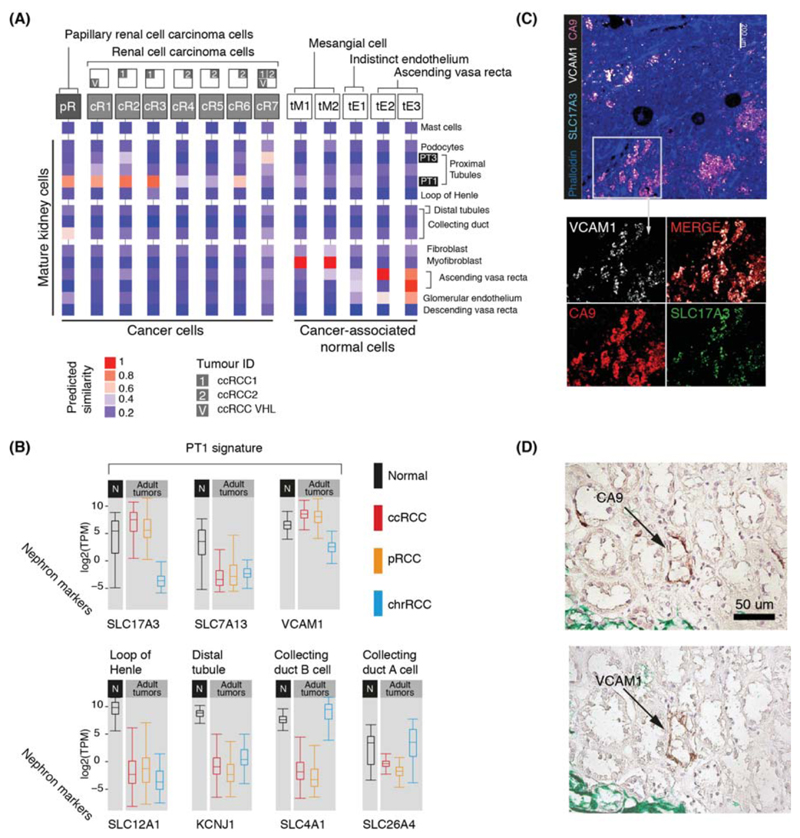
Next we studied ccRCC and pRCC (type 1), including one case of von Hippel Lindau disease-related ccRCC (Table S1). Matching ccRCC and pRCC with normal mature cells, we found that they retained transcriptional features of cluster PT1, a specific subtype of convoluted proximal tubular cell (Fig. 4A). Most (6/7) ccRCC clusters and all pRCC cells matched this particular PT1 cell, indicating that it represents an RCC cell state that transcends the diversity of RCC cells within and across tumors. Little is known about the nearest normal cell correlate of RCC, the PT1 cell, which has been identified to become more abundant in inflamed renal tissue (6).
Figure 4. Matching adult tumors with normal mature kidney cells.
(A) Similarity of adult cancer and cancer-associated normal cells to the mature kidney reference map (Fig. 1B), with mast cells added as a negative control. Square boxes indicate sample contribution. Colors represent the probability that the cluster identified in the column header is “similar” to the normal cluster identified by the row label (2).
(B) Expression of nephron specific genes in bulk RNA-seq as in Fig. 3B. pRCC samples are both type 1 and 2.
(C) Confocal microscopy showing co-localization of PT1 markers (VCAM1, SLC17A3) in ccRCC cells (CA9).
(D) Staining of a proximal tubular ccRCC precursor lesion (CA9) for the PT1 marker, VCAM1.
To validate the identity of the PT1 signature in RCC, we exploited the fact that they were defined by SLC17A3 and VCAM1 with absence of SLC7A13 within our data (Fig. 4B; Fig. S2). We measured these transcripts in an independent series of 1,019 publicly available bulk kidney tumor and normal tissue transcriptomes. High expression of SLC17A3 mRNA distinguished ccRCC and pRCC (type 1 and 2) from other types of RCC (p<10-4; Mann-Whitney test), whereas SLC7A13 mRNA was significantly depleted in ccRCC/pRCC bulk transcriptomes versus normal (p<10-4; Mann-Whitney test), as were mRNAs representing other regions of the nephron (Fig. 4B). VCAM1, specific to PT1 within proximal tubules, was also significantly elevated across RCC bulk transcriptomes (p<10-4; Mann-Whitney test; Fig. 4B), with each individual RCC tumor exhibiting PT1 features (Fig. S15). Confocal microscopy demonstrated co-localization of VCAM1 and SLC17A3 in CA9+ cells, CA9 being a specific marker of ccRCC cells (Fig. 4C). Furthermore, we studied the earliest precursor lesions of ccRCC: CA9+ proximal tubular cells residing in morphologically normal kidney, predisposed to ccRCC through pathogenic germline mutation of VHL. Examining tissue from three individuals, we identified CA9+/VCAM1+ clusters of proximal tubular cells (Fig. 4D). Similarly, tumors arising in these kidneys harbored CA9+/VCAM1+ cells (Fig. S16). As expected, VCAM1 was otherwise sparsely expressed on proximal tubular cells. Together these observations substantiate our proposition that PT1 cells are the nearest normal cell correlate of ccRCC cells. The presence of the PT1 signature in both ccRCC and pRCC may indicate a common origin of these tumors with divergent fates.
Apart from the PT1 signature in pRCC and ccRCC, we found that one ccRCC cell cluster (cR7) matched PT3 cells and that pRCC cells exhibited an additional, weaker match with collecting duct cells (Fig. 4A). Neither signal was enriched in bulk transcriptomes (Fig. 4B). As our study was confined to type 1 pRCC, it is possible that we missed other pRCC cell types.
Finally, we dissected the tumor microenvironment occupied by cancer-associated normal cells, comprised of immune cells, fibroblasts, myofibroblasts and vascular endothelial cells (predominately ascending vasa recta) (Fig. S7, S8, S17). Within these we studied VEGF signaling, an established target in RCC treatment (7, 8). The VEGF signaling circuit in renal tumors involves VEGFA secretion from RCC cells resulting in a response from endothelial cells (7, 8). Measuring expression of the key components of VEGF signaling, we identified tumor infiltrating macrophages as a further source of VEGFA (Fig. S18A), confirmed by confocal microscopy of ccRCC cells and flow cytometry of an independent ccRCC tumor (Fig. S18B,C,D). VEGF-signaling receptors (KDR, FLT1, FLT4) were mainly expressed by one population of ascending vasa recta cells (Fig. S18A, cluster tE1). The other ascending vasa recta cluster, tE2, (Fig. S18A) exhibited lymphangiogenic VEGFC and FLT1. Furthermore, tE2 endothelial cells expressed high levels of ACKR1, a marker of venular endothelium promoting tissue migration of immune cells (9). Overall these findings delineate a complex VEGF signaling circuit within RCC tissue.
By identifying specific normal cell correlates of renal cancer cells, our study moves our understanding of these malignancies beyond a notion of “fetalness” or an approximate micro-anatomical region to a precise cellular, molecularly quantitative resolution. Our findings portray the peak incidence of Wilms’ tumor in early childhood as a corruption of fetal nephrogenesis, in contrast to the life-long development of RCC in mature kidneys. Our study provides a scalable experimental strategy for determining the identity of human cancer cells.
Supplementary Material
One Sentence Summary.
Single cell mRNAs of 72,501 normal and cancerous kidney cells reveals the cellular identity of childhood and adult tumors.
Acknowledgments
We thank Sir Michael Stratton, Peter Campbell, David Rowitch, Manfred Gessler and Manasa Ramakrishna for review of the manuscript; Moritz Gerstung and Valentine Svensson for advice regarding logistic regression. We are indebted to our patients and their families for participating in this research.
Funding: This experiment was principally funded by the St Baldrick’s Foundation (Robert J Arceci International Award to S.B.). Additional funding was received from: Wellcome (S.B., M.H., G.C., C.G.); Cambridge Biomedical Research Campus (biobanking infrastructure; M.R.C.); CRUK Cambridge Centre (biobanking infrastructure); NIHR Blood and Transplant Research Unit (M.R.C.); MRC (M.R.C.); Arthritis Research UK (M.R.C.); The Lister Institute for Preventative Medicine (M.H.); NIHR and Newcastle-Biomedical Research Centre (M.H.); ISAC SRL-EL program; A.F.); joint Wellcome Trust/MRC (S.Lis., S.Lin.); Kidney Cancer UK (M.G.B.T.); Facing up 2 Kidney Cancer (M.G.B.T.); EMBO (R.V.T); Human Frontier Science Program (R.V.T); Children with Cancer UK (S.J.F.).
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Author contributions: S.B. conceived the experiment. M.D.Y., T.J.M. and S.B. analyzed the data, with contributions from F.V.B., B.S., M.D.C.V.H., G.C. and M.C. Samples were curated and / or experiments performed by: F.V.B., J.R.F., M.G.B.T., P.H.M, R.A.B., D.M.O., R.V-T., E.S., K.L., S.Far., C.G., N.R., L.M., T.A., J.N.A., A.C.P.R., I.M., S.F., C.J., D.R., J.N., A.F., J.B., S.Lis., S.Lin. and G.D.S. Pathological expertise was provided by A.Y.W., N.S., and N.C.A.C. created illustrations. T.J.M., M.D.Y. and S.B. wrote the manuscript. M.H., S.A.T., M.C. and S.B. co-directed the study.
Competing interests: None.
Data and materials availability: Raw sequencing data have been deposited in the European Genome-phenome Archive (EGA) under study IDs EGAS00001002171, EGAS00001002486, EGAS00001002325 and EGAS00001002553. Sample specific identifiers can be found in Table S6,S10, a table of mapped UMI counts for each cell and gene combination in Data S1 and metadata about each cell in Table S11. The code necessary to perform the analysis and generate figures can be obtained from https://github.com/constantAmateur/scKidneyTumors.
References
- 1.Ziegenhain C, et al. Comparative Analysis of Single-Cell RNA Sequencing Methods. Molecular cell. 2017;65:631–643.e634. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 2.Supplementary Methods
- 3.Pritchard-Jones K, et al. Omission of doxorubicin from the treatment of stage II-III, intermediate-risk Wilms' tumour (SIOP WT 2001): an open-label, non-inferiority, randomised controlled trial. Lancet (London, England) 2015;386:1156–1164. doi: 10.1016/S0140-6736(14)62395-3. [DOI] [PubMed] [Google Scholar]
- 4.Perlman EJ, et al. MLLT1 YEATS domain mutations in clinically distinctive Favourable Histology Wilms tumours. Nature communications. 2015;6 doi: 10.1038/ncomms10013. 10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gadd S, et al. A Children's Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nature genetics. 2017;49:1487–1494. doi: 10.1038/ng.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seron D, Cameron JS, Haskard DO. Expression of VCAM-1 in the normal and diseased kidney. Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association. 1991;6:917–922. doi: 10.1093/ndt/6.12.917. [DOI] [PubMed] [Google Scholar]
- 7.Ljungberg B, et al. EAU guidelines on renal cell carcinoma: 2014 update. European urology. 2015;67:913–924. doi: 10.1016/j.eururo.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez-Pello S, et al. A Systematic Review and Meta-analysis Comparing the Effectiveness and Adverse Effects of Different Systemic Treatments for Non-clear Cell Renal Cell Carcinoma. European urology. 2017;71:426–436. doi: 10.1016/j.eururo.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 9.Thiriot A, et al. Differential DARC/ACKR1 expression distinguishes venular from non-venular endothelial cells in murine tissues. BMC biology. 2017;15:45. doi: 10.1186/s12915-017-0381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerrelli D, Lisgo S, Copp AJ, Lindsay S. Enabling research with human embryonic and fetal tissue resources. Development (Cambridge, England) 2015;142:3073–3076. doi: 10.1242/dev.122820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozarewa I, et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones D, et al. cgpCaVEManWrapper: Simple Execution of CaVEMan in Order to Detect Somatic Single Nucleotide Variants in NGS Data. Curr Protoc Bioinformatics. 2016;56:15 10 11–15 10 18. doi: 10.1002/cpbi.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menzies A, et al. VAGrENT: Variation Annotation Generator. Curr Protoc Bioinformatics. 2015;52:15 18 11–11. doi: 10.1002/0471250953.bi1508s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proceedings of the National Academy of Sciences. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lun A, Riesenfeld S, Andrews T, Dao TP, Gomes T. On the correct detection of empty droplets in droplet-based single-cell RNA sequencing protocols. 2017 doi: 10.1186/s13059-019-1662-y. ( https://github.com/TimothyTickle/hca-jamboree-cell-identification/blob/master/docs/EmptyDrops_group4_report.pdf) [DOI] [PMC free article] [PubMed]
- 17.Chen YJ, et al. Single-cell RNA sequencing identifies distinct mouse medial ganglionic eminence cell types. Sci Rep. 2017;7 doi: 10.1038/srep45656. 45656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 19.v d Maaten LJP, Hinton GE. Visualizing High-Dimensional Data Using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- 20.v d Maaten LJP. Accelerating t-SNE using Tree-Based Algorithms. Journal of Machine Learning Research. 2014;15:3221–3245. [Google Scholar]
- 21.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajaraman A, Ullman JD. Mining of Massive Datasets. Cambridge University Press; Cambridge: 2011. [Google Scholar]
- 23.Qiu X, et al. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang HM, et al. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 2012;40:D144–149. doi: 10.1093/nar/gkr965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang HM, et al. AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res. 2015;43:D76–81. doi: 10.1093/nar/gku887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman JH, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. 2010. 2010;33:22. [PMC free article] [PubMed] [Google Scholar]
- 28.N Cancer Genome Atlas Research. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.N Cancer Genome Atlas Research et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N Engl J Med. 2016;374:135–145. doi: 10.1056/NEJMoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perlman EJ, et al. MLLT1 YEATS domain mutations in clinically distinctive Favourable Histology Wilms tumours. Nature communications. 2015;6:10013. doi: 10.1038/ncomms10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walz AL, et al. Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer cell. 2015;27:286–297. doi: 10.1016/j.ccell.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colaprico A, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nature communications. 2017;8 doi: 10.1038/ncomms14049. 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hern WM. Correlation of fetal age and measurements between 10 and 26 weeks of gestation. Obstetrics & Gynecology. 1984;63:26–32. [PubMed] [Google Scholar]
- 35.Brunskill EW, et al. Atlas of gene expression in the developing kidney at microanatomic resolution. Developmental cell. 2008;15:781–791. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metsuyanim S, et al. Expression of stem cell markers in the human fetal kidney. PloS one. 2009;4:e6709. doi: 10.1371/journal.pone.0006709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JW, Chou CL, Knepper MA. Deep Sequencing in Microdissected Renal Tubules Identifies Nephron Segment-Specific Transcriptomes. Journal of the American Society of Nephrology : JASN. 2015;26:2669–2677. doi: 10.1681/ASN.2014111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Habuka M, et al. The kidney transcriptome and proteome defined by transcriptomics and antibody-based profiling. PloS one. 2014;9:e116125. doi: 10.1371/journal.pone.0116125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chabardes-Garonne D, et al. A panoramic view of gene expression in the human kidney. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13710–13715. doi: 10.1073/pnas.2234604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han X, Amar S. Secreted Frizzled-related Protein 1 (SFRP1) Protects Fibroblasts from Ceramide-induced Apoptosis. Journal of Biological Chemistry. 2004;279:2832–2840. doi: 10.1074/jbc.M308102200. [DOI] [PubMed] [Google Scholar]
- 41.Matsuyama M, Nomori A, Nakakuni K, Shimono A, Fukushima M. Secreted Frizzled-related Protein 1 (Sfrp1) Regulates the Progression of Renal Fibrosis in a Mouse Model of Obstructive Nephropathy. Journal of Biological Chemistry. 2014;289:31526–31533. doi: 10.1074/jbc.M114.584565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lennon R, et al. Global analysis reveals the complexity of the human glomerular extracellular matrix. Journal of the American Society of Nephrology : JASN. 2014;25:939–951. doi: 10.1681/ASN.2013030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LeBleu VS, et al. Origin and Function of Myofibroblasts in Kidney Fibrosis. Nature medicine. 2013;19:1047–1053. doi: 10.1038/nm.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L, et al. NDUFA4L2 is associated with clear cell renal cell carcinoma malignancy and is regulated by ELK1. PeerJ. 2017;5:e4065. doi: 10.7717/peerj.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Habuka M, et al. The Urinary Bladder Transcriptome and Proteome Defined by Transcriptomics and Antibody-Based Profiling. PloS one. 2015;10:e0145301. doi: 10.1371/journal.pone.0145301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aird WC. Phenotypic Heterogeneity of the Endothelium. Circulation Research. 2007;100:174. doi: 10.1161/01.RES.0000255690.03436.ae. [DOI] [PubMed] [Google Scholar]
- 47.Nawroth R, et al. VE-PTP and VE-cadherin ectodomains interact to facilitate regulation of phosphorylation and cell contacts. The EMBO journal. 2002;21:4885–4895. doi: 10.1093/emboj/cdf497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.