Abstract
DNA methylation is an important heritable landmark conferring epigenetic changes in hybrids and has fascinated biologists and plant-breeders over the years. Although epigenetic changes have been documented in rice and maize hybrids, such investigations have not been reported in pigeonpea. Here, we report genome-wide methylation profiles of pigeonpea sterile and fertile inbred lines and their fertile F1 hybrid at single base resolution. We found that pigeonpea genome is relatively enriched in CG methylation. Identification of differentially methylated regions (DMRs) in the sterile and fertile parent revealed remarkable differences between their methylation patterns. Investigation of methylation status of parental DMRs in hybrid revealed non-additive methylation patterns resulting from trans-chromosomal methylation and trans-chromosomal demethylation events. Furthermore, we discovered several DMRs negatively associated with gene expression in the hybrid and fertile parent. Interestingly, many of those DMRs belonged to transposable elements and genes encoding pentatricopeptide repeats associated proteins, which may mediate a role in modulating the genes impacting pollen fertility. Overall, our findings provide an understanding of two parental epigenomes interacting to give rise to an altered methylome in pigeonpea hybrids, from genome-wide point of view.
Keywords: DNA methylation, TcM, TcDM, pigeonpea, hybrid
1. Introduction
Epigenetic regulation has remained of fundamental interest in understanding gene regulation in eukaryotes. Epigenetic regulation involves differential methylation at cytosine residues and post-translational modifications of histones.1 One of the most comprehensively investigated mechanism of chromatin modification in plants,2 DNA methylation involves covalent transfer of a methyl group to C-5 position of the cytosine ring by DNA methyltransferases and is transmitted across generations.3 DNA methylation acts to regulate transposons and repetitive genome sequences and often results in the suppression of gene activity. 4,5 A major difference between DNA methylation in animals and plants is that in plant genome, DNA methylation occurs at CG, CHH and CHG sequence contexts (where H could be C, T or A).6 It has been shown in Arabidopsis that different enzymes regulate methylation at different sequence contexts. Methyltransferase 1 (MET1) causes and maintains methylation at CG sites6–8 and chromomethylase 3 (CMT3) produces and maintains methylation at CHG sites6,9,10 and this maintenance is further required to preserve methylation patterns over generations. On the other hand, domains rearranged methyltransferase 2 (DRM2) and 24 nt siRNAs, which guide methyltransfrase to specific DNA segments, regulate de novo methylation CHH sites,6,10,11 which is maintained by CMT2.12
De novo methylation of all three different sites involves 24 nt smRNAs (small RNAs), DRM2 and the Dnmt3 homologue involved in RNA-directed DNA methylation (RdDM). In RdDM, RNA is transcribed by the RNA polymerase POLIV, which is then made double stranded by RNA-dependent RNA polymerases (RDRs), cleaved to 24 nt by Dicer-like enzymes and loaded onto argonaute proteins. This effector complex then interacts with other proteins and Polymerase V transcripts to target homologous regions of the genome for methylation by the recruitment of DRM2. The methylation state of CG is maintained during cell replication by MET1, which recognizes hemi methylated CG sites and methylate the newly formed DNA strand. CMT3 maintains CHG methylation, while CHH methylation is recognized by a histone methyltransferase kryptonite (KYP), which can then methylate the H3K9 residues. These two enzymes work in a feedback loop enabling the maintenance of both epigenetic marks, with the absence of either CMT3 or KYP leading to a reduction in CHG methylation. DNA methylation plays an important role during different aspects of plant development and growth. Of these heterosis or hybrid vigour is of vital importance in agriculture. Heterosis is a phenomenon where F1 progeny of a genetic cross exhibits superior characteristics comparedwith its inbred parental lines and has been documented nearly 300 years ago.13 In recent years, several studies have focused on the role of epigenetic regulation in mediating hybrid vigour in model plant Arabidopsis.1,14–17 Previous studies in Arabidopsis have suggested that interaction between similar genomes and different epigenomes of the two different parental ecotypes could result in new transcriptional profiles in hybrids that might contribute to hybrid vigour in the progeny.18,19 Also, it has been proposed that epigenetic regulation via changes in methylation patterns might play a role in mediating differences in gene expression levels that may confer increased biomass in heterotic hybrid offspring of Arabidopsis ecotypes C24 and Landsberg erecta.15 It has also been documented that both DNA methylation patterns and sRNA (small RNA) levels are altered in intraspecific hybrids of Arabidopsis,15,20–22 rice23 and maize.24
Recent advances in DNA methylation profiling techniques have enabled in understanding the correlation between methylation patterns and hybrid vigour in several crop species as well such as rice and Brassica25,26 but knowledge about interaction of methylation patterns and gene expression in legume plants such as pigeonpea has still remained elusive. Pigeonpea (Cajanus cajan (L) Millsp) is a important grain crop with high protein (20–22%) grain legume, mainly cultivated in the tropical and subtropical parts of the world. Pigeonpea contains 11 pairs of chromosomes (2n = 22) and has a genome size of 858 Mbp.27 Globally pigeonpea is cultivated on 5.4 Mha, with an annual production of 4.4 MT and a yield of 8299 hg/ha. Pigeonpea is grown in about 50 countries with India occupying 66% of its area (FAO, 2016, http://www.fao.org/faostat/en/#data/QC). India is also the largest producer and consumer of pigeonpea with an annual production of 2.51 MT.
Pigeonpea is characterized by significant genetic variability. Genetic studies have also revealed the occurrence of non-additive genetic variation in pigeonpea hybrids.28 This particular property has become popular among breeders to produce varieties with improved fertility and been utilized through cytoplasmic male sterility (CMs) system. CMS is a condition in which the ability of a plant to produce fertile pollen is severely impaired. In this condition, rearrangements in mitochondrial genome result in chimeric mitochondrial open reading frames (ORFs), generating non-functionality in anthers. However, the expression of such chimeric mitochondrial ORFs can be suppressed by fertility restorer gene in F1 hybrid, thus restoring fertility. This kind of hybrid breeding technology involves three parents— a male sterile parent, its maintainer and a fertility restorer line (commonly called—A, B and R lines, respectively; reviewed in detail in the literature).29 Few CMS associated mitochondrial orfs have been identified in pigeonpea.30
In this study, we have investigated whether there are any epigenetic changes in F1 progeny of cross between 11 A (male sterile) and 303 R (fertile) inbred lines and whether those changes could potentially mediate an impact on the gene expression patterns including those involved in fertility restoration in the hybrid. We conducted whole genome bisulfite sequencing of 11 A, 303 R parental genotypes and their hybrid to generate whole genome cytosine methylation profiles and found that overall, hybrid methylome was remarkably altered compared with the parental methylome due to the events of transchrosomal methylation and demethylation in hybrid. We next carried out transcriptome profiling of the same plants and compared methylation status of the differentially expressed genes between parental inbreds and the F1 hybrids. We found differences in methylation patterns in the three genotypes, suggesting the involvement of DNA methylation in regulation of gene expression and possibly influencing sterility and fertility.
2. Materials and methods
2.1. Plant material and growth conditions
Pigeonpea lines 11 A and 303 R31 were crossed by hand pollination using 303 R as pollen donor to generate hybrid. Seeds were collected and F1 seed resulting from the cross along with seeds from parental lines were sown again together for further experimental work. Seeds were directly sown in the soil in green house under natural conditions. All the plants were grown at the same time under exactly similar growth conditions to reduce the impact of any environmental variations. Fertility status of all three genotypes was verified by pollen analysis. Pollens were fixed on a glass slide using 2% acetocarmine. Pollens stained with acetocarmine were considered to be fertile. After pollen verification, young leaves of same age from the three lines were harvested, frozen in liquid nitrogen and used to extract DNA and RNA for further work.
2.2. Whole genome bisulfite sequencing library construction and data analysis
High molecular weight genomic DNA was isolated from the leaves using cetyl trimethylammonium bromide method. DNA was fragmented into 100–300 bp using Covaris followed by end repair by adding ‘A’ and True seq methylated adapters were ligated. The ligated DNA fragments were subjected to bisulfite treatment using EZ DNA Methylation-Gold Kit, according to manufacturer’s instructions. The BS libraries were amplified, quality tested using Bio-analyzer (Agilent 2100) and subjected to Ilumina Hi-seq2000 for 90 cycles in paired-end mode. The adaptors and poor quality reads were removed from the raw data using TrimGalore (https://github.com/FelixKrueger/TrimGalore). High quality reads were mapped to the reference genome27,32 using BSMAP33 at default parameters. Methylation information was extracted using Python script. Reads were mapped on chloroplast genome to calculate methylation efficiency and error rate. A bisulfite conversion rate of >99% was obtained for all three samples. In-order to determine true methylated cytosines, P-values were calculated using binomial distribution P= binomial (m, x, error rate), where m= number of methylated reads, n= number of non-methylated reads and x= m + n (coverage on a single C).15 Cytosines with P-value < 0.0025 were considered to be truly methylated. Fraction and mean methylation level was calculated as described previously.34
2.3. Identification of differentially methylated regions
To identify differentially methylated regions (DMRs) at each sequence context (CG, CHG and CHH), genomes were tiled using a sliding window approach with 100 bp window sliding at 100 bp interval for CG and CHG and 150 bp for CHH. Methylation information for each window was calculated using Methyl kit.35
P-value for each window was calculated using Fisher’s exact test. To determine q-values, P-values for each window were corrected using sliding linear model . Following criteria were used for identification of DMRs.
Windows contained at-least three methylated cytosines for CG and CHG; and six for CHH DMRs.
Bases having more than 10X coverage were considered.
Bases having more than 99.9th percentile coverage were discarded to minimize errors arising due to PCR bias.
Windows with methylation difference greater than 20% for CG, CHG and 25% for CHH were selected.
Fisher’s exact test corrected q-values < 0.01.
2.4. Validation of methylation levels using bisulfite sequencing PCR
Two different regions were chosen one for validating methylation levels, one in the intergenic region and the other one in the exonic region. For bisulfite PCR, genomic DNA was extracted from 11 A and F1 hybrid and subjected to bisulfite conversion using EZ DNA Methylation-Gold™ Kit. Primers were designed using METHPRIME36 (Supplementary Table S1). Bisulfite converted DNA was amplified and the fragments were cloned into pGEMT vector. Ten colonies were picked randomly for sequencing. The results were analyzed using QUMA.37
2.5. RNA sequencing library construction and analysis
Total RNA was extracted from the same plants used for BS Seq. Equal amount of total RNA from three biological replicates were pooled. About 1 µg of the total pooled RNA was used to purify poly-A containing mRNA molecules. The purified mRNA was fragmented into smaller pieces and reverse transcribed into cDNA. The cDNA fragments were subjected to end repair by adding single ‘A’ base. True seq adaptors were ligated and cDNA libraries were amplified for 15 cycles. Quality of the library was assessed using Bioanalyzer and the validated library was sequenced by Illumina Hi-Seq 1000 according to manufacturers’ instructions. The quality of the raw reads was determined using FastQc (https://www.illumina.com/products/by-type/informatics-products/basespace-sequence-hub/apps/fastqc.html). Adaptors and poor quality reads were cleaned using Trim Galore. High quality paired end reads were aligned to reference genome (Supplementary Table S2) using Bowtie alignment program,38 then relative abundance of transcripts was calculated using RSEM.39 Transcripts with zero Fragments Per Kilobase of transcript per Million mapped reads (FPKM) value were excluded from analysis. Raw reads count data were imported into R environment and data table containing fragment counts for each transcript were combined and matrix was formed to identify differentially expressed gene using edgeR.40
2.6. GO enrichment analysis
To identify and categorize cluster of differentially expressed genes (DEGs) and genes enriched for methylation in all the three categories (11 A vs. 303 R, 11 A vs. hybrid and 303 R vs. hybrid), GO enrichment analysis was performed using python based program GOATOOLS (https://github.com/tanghaibao/goatools) and Wego.41 Genes with Bonferroni-corrected P-values < 0.05 using GOATOOLS were considered to be enriched.
2.7. Identification of transposable elements
A denovo repeat library was generated using repeatmodeller (http://www.repeatmasker.org/RepeatModeler/; version 1.0.11).This customized library was used to run RepeatMasker (http://www.repeatmasker.org/; version- 4.0.7) to identify and annotate transposable elements (TEs) in pigeonpea at default parameters. TE shorter than 100 bp were excluded from the analysis and the distance between TE, less than 50 bp were conconated (Supplementary Table S3).
2.8. Allele specific methylation analysis using single nucleotide polymorphism
Single nucleotide polymorphisms (SNPs) from parental lines and hybrid were identified using BIS–SNP,42 a variant caller from BS reads. For this, the mapped methylated reads were used as input files along with pigeonpea reference genome.32 SNPs detection was performed using the following parameters: ‘-T BisulfiteGenotyper -stand_call_conf 20 -stand_emit_conf 0 -mmq 30 -mbq 17 -minConv 20’. Spurious SNP with quality score less than 20, more than 120x read coverage, strand bias more than—0.02, quality score by depth less than 1.0, mapping quality zero reads, fraction more than 0.1 and 2 SNPs within the 20 bp bin were further filtered out using VCF post process command.
3. Results
3.1. Genome wide methylation analysis in pigeonpea
To understand the possible roles of epigenetic regulation and the dynamics of DNA methylation during hybrid formation in pigeonpea, we crossed 303 R (Fertile, pollen donor) and 11 A(sterile) lines. Since their F1 progeny restored fertility (Fig. 1A), we hypothesised that it could possibly partially attributed to the new epigenome resulting from the interaction between the two parental epigenomes. Thus, in-order to gain deep insights into the epigenome of the parental lines and their F1 hybrid, we deciphered their respective whole genome methylation pattern at single base resolution. For this high molecular weight genomic DNA was isolated from the leaves of 10 days after flowering (DAF) old plants from three different genotypes, namely 11 A, 303 R and F1 hybrid progeny of 11 A and 303 R. The DNA samples were subjected to bisulfite treatment to generate genomic libraries for high throughput bisulfite sequencing. The reads obtained were processed for removal of adapter and low quality sequences. About 80–100 million cleaned reads (in each of the three genotypes) were mapped on to the reference genome.27,32 We found that more than 80% reads got mapped on to the reference genome. Thus eventually, 46 408 326, 54 030 096 and 43 763 469 reads were obtained for 11 A line, 303 R line and their F1 hybrid progeny, respectively, that mapped uniquely to the reference genome and providing a coverage more than 20X. These were subjected to further analysis of cytosine methylation at single base resolution. Further, to calculate the frequency of cytosine conversion, reads were mapped to un-methylated chloroplast genome. We obtained a conversion rate of more than 99% in all the three genotypes, indicating high efficiency of cytosine conversion in our experiments (Supplementary Table S4).
Figure 1.
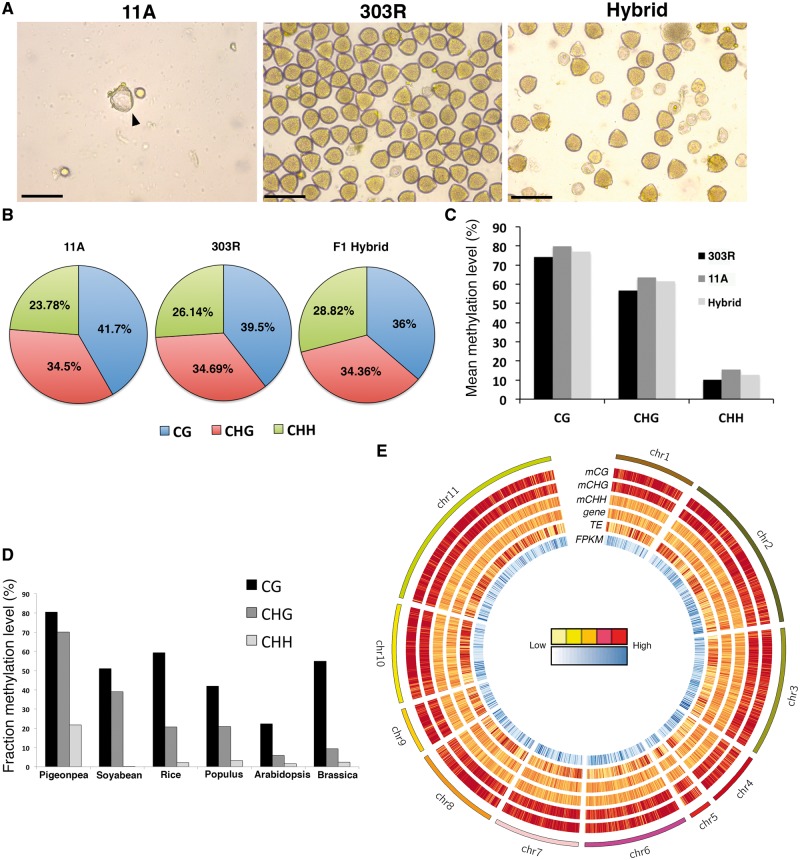
DNA methylation profiles of pigeonpea inbred lines (11 A, 303 R) and their hybrid. (A) Acetocarmine stained pollens of 11A (sterile parent), 303R (fertile parent) and fertile hybrid. Note the density of pollen is less in 11A compared to 303R and hybrid. Also note viable pollens in 303R and hybrid.(B) Genome-wide relative proportions of CG (blue), CHG (red) and CHH (green) methyl cytosine in 11A, 303R and F1 hybrid. (C and D) Fractional methylation levels in the three lines of pigeonpea in CG, CHG and CHH contexts (B) and comparisons with published reports in Soybean, Rice, Populus, Arabidopsis and Brassica (D). (E) A circos plot representing chromosome wise distribution of mCG, mCHG, mCHH, gene, transposons density and FPKM (outer to inner) in F1 hybrid. Yellow to red color relates to lower to higher methylation density in a 100 Kb window (Colored figure is available in the online version of the article). Scale bars 50µm (A).
We then deciphered the fractional proportion of DNA methylation and found that CG methylation was highest in 11 A (41.7%) while CHH methylation was highest in F1 hybrid (28.82%). Methylation in CHG context were similar in all the three genotypes (∼34%) (Fig. 1B). Highest mean methylation was observed in CG context (74%-79%), closely followed by CHG (56%–63%) while mean methylation in CHH context (10–15%) was lowest in both the parents and the hybrid (Fig. 1C). Comparative analysis of fractional methylation levels in pigeonpea, soybean, brassica, oryza and populus revealed that pigeonpea genome was hypermethylated compared with the other crops. Interestingly, CHH methylation levels in pigeonpea genome were significantly higher than those observed in soybean, brassica, oryza and populus25,43 (Fig. 1D). It has been documented that the DNA repeats in the genomes are highly methylated in symmetric contexts.9,44,45 Pigeonpea geneome is highly repetitive (47%) compared with soyabean46 and Arabidopsis.47 Thus, such high levels of DNA methylation in pigeonpea may arise from the occcurance highly repetive genome.
Visualization of chromosome wide distribution of CG, CHG and CHH methylation revealed higher CG and CHG methylation levels compared with CHH while relatively even distribution pattern of CHH methylation (Fig. 1E, Supplementary Fig. S1A and B). High CG and CHG methylation were correlated with transposon rich regions across the genome (Fig. 1E, Supplementary Fig. S1A and B).
3.2. Patterns of DNA methylation in genic and transposable elements
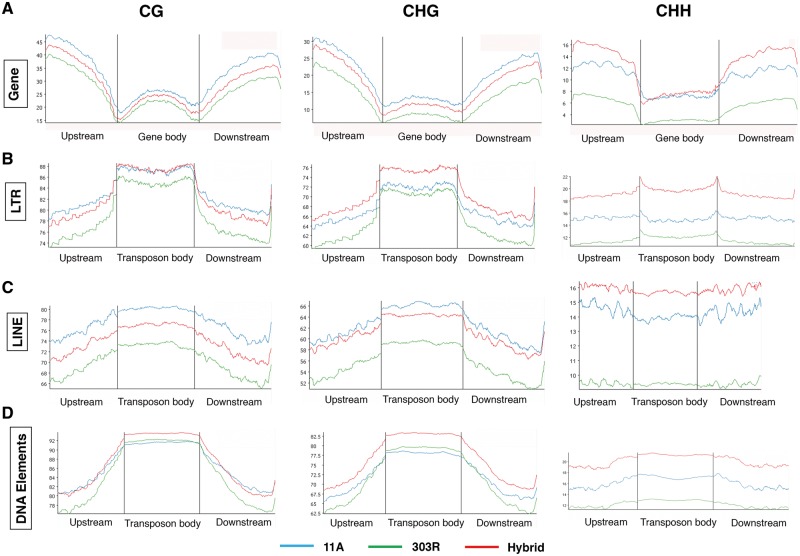
To precisely understand the methylation patterns of the protein coding genes, genes with inserted TE elements were excluded from analysis. We calculated the methylation levels in the genic region and 2 kb upstream and downstream regions of the gene bodies in the two parental lines and their hybrid and found significant methylation differences between the three lines (Wilcoxon rank sum test, P-value < 2.2e-16, n = 26,774). These differences reflect differential methylation patterns in the genic regions of parental lines and their hybrid. Furthermore, upon a detailed investigation of methylation pattern, we observed the occurrence of a single CG methylation peak within the gene body and two troughs near transcriptional start and termination sites in pigeonpea, invariably to earlier reports in other plants.43,48,49 However, contrary to a recent report in rice,48 we found over all low CG methylation levels within the coding regions compared with their flanking regions. A similar trend was observed for CHG methylation, however, the average CHG methylation both within the coding regions and their flanking regions were comparatively lower than CG methylation. CHH methylation however showed a different trend to both CG and CHG methylation in terms of a plateau instead of a single peak within the gene body. Overall CHH methylation both within the genic regions and their flanking regions were significantly attenuated than CG and CHG methylation. However, the hybrid exhibited mid parental range for CG and CHG but a higher CHH methylation compared with parental lines, suggesting altered CHH methylation patterns in the genic regions in pigeonpea hybrids (Fig. 2A).
Figure 2.
Distribution of methyl cytosines in genes and transposable elements in pigeonpea. (A) Line plots showing methylation level in 2Kb flanking (upstream and downstream) and within the gene body in 11A, 303R and F1 hybrid in CG, CHG and CHH contexts. (B–D) Line plots showing methylation level in 2Kb flanking (upstream and downstream) and within the gene body of transposons- long terminal repeats (B); LINE (C) and DNA Elements (D) in 11A, 303R and F1 hybrid in CG, CHG and CHH context. Note higher mCs throughout the transposons compared to genes (Colored figure is available in the online version of the article).
Eukaryote genomes are abundant in TEs, also called mobile elements. TEs are structurally divergent, epigenetically controlled and are mostly involved in genome evolution and gene activity regulation. Thus, we sought to gain a comprehensive insight of the influence of DNA methylation on the transposable elements within the pigeonpea genome. Since no complete dataset on pigeonpea TEs is available, we used repeat masker to identify and classify them. TEs shorter than 100 bp were excluded from analysis. In total, we found 105 733 TEs, which we classified into retro transposons and DNA elements. We next calculated their CG, CHG and CHH methylation levels in all the three genotypes and found that irrespective of their categories, methylation was highly enriched in all the contexts in TEs compared with the gene body. However, interestingly, we found relatively lower CG and CHG methylation levels in the TEs flanking regions than those of the genic regions. By comparing the methylation levels in long terminal repeats within the three genotypes, we found relatively higher methylation levels in F1 hybrid in all the three contexts, compared with the parental lines except for CG methylation in flanking regions where 11 A exhibited higher methylation levels (Fig. 2B). While, in the case of LINE elements, 11 A displayed hyper methylation compared with the F1 hybrid and 303 R in CG and CHG contexts (Fig. 2C). DNA elements were relatively hypermethylated than the class 1 transposons. F1 hybrid displayed uniformly higher methylation irrespective of the mC contexts (Fig. 2D). Overall we observed comparatively lower methylation levels in 303 R, compared with 11 A and F1 hybrid (Fig. 2B–D), suggesting differences in the regulatory mechanisms of TE silencing via methylation in pigeonpea.
3.3. Fertile and sterile pigeonpea inbred lines display locus specific differences in cytosine methylation
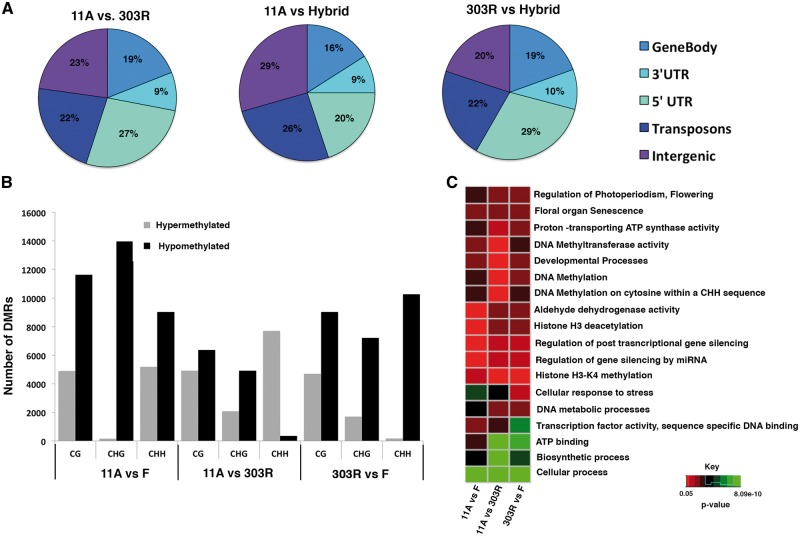
Earlier work in crop plants such as rice and maize have documented differences in methylation levels between a male sterile plant and a fertile plant and revealed hyper-methylation in sterile lines compared with fertile lines.50,51 This suggests differential cytosine methylation could possibly influence fertility and sterility in plants by regulating gene expression. Therefore to investigate whether pigeonpea 11 A and 303 R inbreds exhibit differences in their methylation levels, we compared the methylome of the two lines. Several studies have documented that viewing differential methylation in windows of multiple cytosines seems to reflect more functional consequences rather than viewing at single sites.52,53 Also it is known that CG, CHG and CHH methylation levels are maintained through different regulatory mechanisms.6 Thus, to identify DMRs between 11 A and 303 R lines, we decided to investigate and categorize differential methylation for all the three contexts, CG, CHG and CHH, separately. We averaged the DNA methylation for all the three contexts independently across non-overlapping 100 bp regions (windows), as done previously.54 In-order to categorize a DMR window as CG, CGH or CHH, different conditions were applied. To assign a DMR to CG or CHG category, windows having at-least three cytosines with a minimum of 10X coverage were used. A minimum difference of 20% in CG and in CHG methylation each was chosen. Previously, it has been reported that CHH methylation patterns appeared to be more variable than CG and CHG.54 Since CHH methylation sites are much abundant in genomes and still, the CHH methylation levels were observed to be much lower than the CG and CHG methylation levels (Fig. 1), different criteria were applied for CHH DMR identification. To assign a DMR to CHH category, each window had at-least eight cytosines with minimum of 10X coverage and a minimum of 25% difference in CHH methylation. Eventually, we identified much more CG DMRs (11,300) than CHG (6988) and CHH (8061) DMRs between 303 R and 11 A parental lines (Supplementary Table S5). We also found that the differential methylation between the two parents was more enriched in upstream flanking sequences of the genes than the down stream regions (Fig. 3A). We next assessed relative methylation percentages of the obtained DMRs between the 11 A and 303 R parents (11 A vs. 303 R or Sterile vs. fertile) and found enrichment for hypermethylated CHH DMRs and hypomethylated CG DMRs (Fig. 3B). This suggests that in 11 A line, genes involved in regulation of CG methylation are less active than the RdDM pathway genes and CMT2 that are involved in the regulation CHH methylation (Supplementary Fig. S2).
Figure 3.
Comparative analyses of DMRs between the inbred lines and their hybrid. (A) Pie charts showing the distribution of DMRs in the different genomic regions in 11A vs. 303R, 11A vs. hybrid and 303R vs. hybrid. (B) A bar plot showing number of hypo and hypermethylated CG, CHG and CHH DMRs in 11A vs. 303R, 11A vs. hybrid and 303R vs. hybrid. (C) A heat map showing enrichment of DMR associated genes in 11A vs. 303R, 11A vs. hybrid and 303R vs. hybrid. Color code represents low to high P-values (0.05 to 8.09e-10) (Colored figure is available in the online version of the article).
Interaction of two dissimilar parental epigenomes can result in an altered methylome in their hybrid progeny. Since fertility is restored in the F1 hybrid and earlier studies in Arabidopsis showed higher methylation levels in hybrid compared with parental lines,15,20 we therefore investigated whether there is differential methylation between individual parental lines and the fertile hybrid (Supplementary Table S5). Consistent with previous studies, we found a relative increase in the percentage of methylated cytosines in CG, CHG and CHH DMRs in the hybrid compared with parental lines (Fig. 3B) indicative of occurrence of non-additive methylation in pigeonpea hybrids. We randomly chose hyper and hypo methylated DMRs between sterile parent and F1 hybrid in intergenic region and coding regions. Analysis of their methylation levels using bisulfite sequencing PCR revealed relatively higher methylation levels in intergenic region associated DMR than in the coding region associated DMR (Supplementary Fig. S3) consistent with our in-silico data.
In-order to assess the functional categories of the genes associated with the DMRs between parents and hybrids and to investigate whether DMRs were associated with genes involved in DNA methylation, epigenetic processes or fertility regulation, gene ontology enrichment analysis was performed. We found genes ontology terms for regulation of flowering time, DNA methylation, regulation of gene silencing by miRNA, developmental processes, Histone H4 K4 methylation, DNA methyltransferase, transcription factors, proton transporting ATP synthase activity, regulation of post transcriptional gene silencing were enriched (Fig. 3C). Interestingly, we also observed a strong enrichment of DMRs in gene is associated with aldehyde dehydrogenase activity. A previous study have reported that RF2 encoded aldehyde dehydrogenase activity is required to restore male fertility in maize and also plays a role in anther development.55 Our results thus suggest a possible correlation between differential methylation and fertility or sterility in 11 A, 303 R pigeonpea inbred and their hybrid. Also, genes involved in cellular processes, biosynthetic processes and ATP binding were significantly enriched (Bonferroni corrected P-value < 0.05; Fig. 3C, Supplementary data S1).
Overall, our results indicate that differential methylation could be involved in mediating and regulating several cellular and developmental processes in pigeonpea.
3.4. Trans-chromosomal methylation events alter pigeonpea hybrid methylome
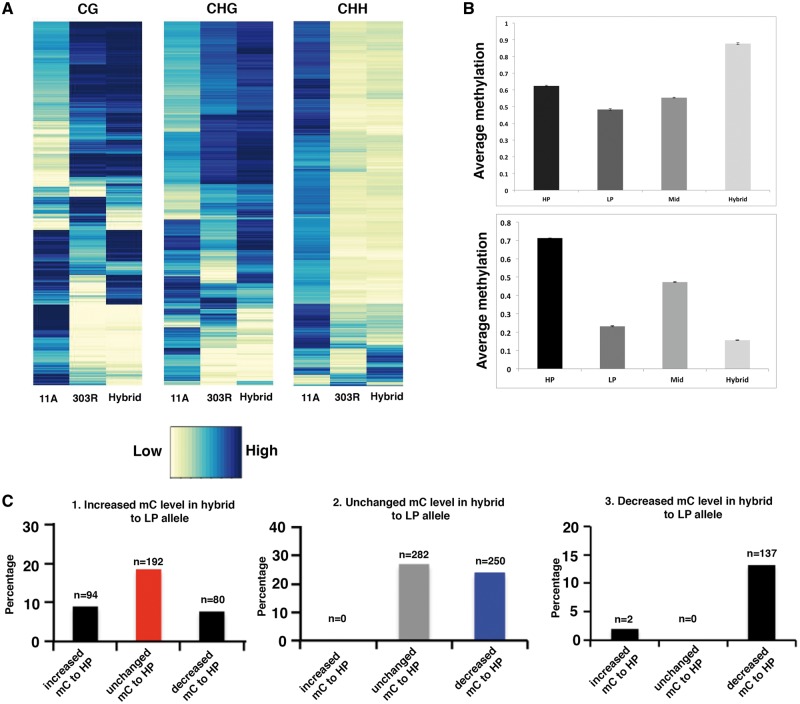
Our results so far show altered methylome in pigeonpea hybrid compared with parents. The changes in hybrid methylome occur at DMRs between the parents and are mediated by either increase or decrease in DNA methylation termed as non-additive methylation.15,20 This increase or decrease occurs via altering methylation status of one parental allele to resemble that of the other parental allele, events known as trans-chromosomal methylation (TCM) and trans-chromosomal demethylation (TCdM).15 We investigated non-additive methylation in F1 hybrid to understand its methylome dynamics. To understand this mechanism, DMRs between the two parental lines were identified and mapped onto the F1 hybrid genome. Average methylation of the mapped loci was calculated in F1 hybrid. Mean parental value (MPV) of the DMRs between parents were obtained and compared with observed methylation in hybrid. Loci having methylation difference of atleast 20% were considered as non-additive methylation sites. Subsequently, we found non-additive methylation in 28% DMRs suggesting methylation interaction at these regions in F1 hybrid. We further analyzed the nature of this non-additive methylation and observed patterns of Trans-chromosomal methylation at 3,911 DMRs and Trans-chromosomal demethylation in 5,377 DMRs (Fig. 4A and B). By investigating the genomic features of the DMRs showing non-additive methylation, we found a higher proportion resided in TE (25%) as compared with the genic regions (15%). These results are in accord with previous findings revealing the roles of TE in genome stability and their regulation by methylation. Furthermore we found that in all the three contexts, the methylome of F1 hybrid resembled more of the 303 R fertile parent than the 11 A sterile. CG and CHG methylation levels of 303 R fertile parent and hybrid were much higher than the 11 A sterile parent. In contrast, CHH methylation levels of 303 R fertile parent and hybrid were lower than that of the 11 A sterile parent (Fig. 4A). Additionally, to decipher the number of direct TcM and TcDM non additive mC clusters, we used an alternative approach of SNPs as described previously.15 For this, we tracked 75,340 mC clusters of parental origin and followed the methylation pattern of parental alleles with differences in methylation level (3,222 out of 75,340) in hybrid using SNPs. We observed 27% mC clusters were stably inherited in hybrids indicative of additive methylation. Among the clusters showing non additive methylation, we observed, TCM events in 18.5% cases where methylation of epi-alleles from low parent increased in the hybrid, whereas methylation of high parent epi-alleles remained unchanged in the hybrid, resulting in an over all increase in methylation in the hybrid. While in 24.1% clusters, TCdM was observed where methylation of the epi-allele from low parent remained unchanged but methylation of the epi-allele from high parent decreased in hybrid, resulting in an overall decrease in methylation in hybrid (Fig. 4C).
Figure 4.
(A) Heat maps showing the methylation status of DMRs of the parents and methylation pattern at those loci in the F1 hybrid. Note in all the mCs context, methylation patterns of F1 hybrid are similar to the fertile parent (303 R) and different to the sterile parent. (B) Bar plot showing non-additive methylation in F1 hybrid. Note increased average methylation in hybrid than the MPV in the upper graph indicating the occurrence of TCM. Similarly, note a decreased average methylation in hybrid than the MPV in the lower upper graph indicating TCdM. (C) Bar plots showing changes in parental allelic methylations in the hybrid. Majority of the non-additive methylation occurred through TCdM (Blue). Red bar represent TcM. Methylation levels of most parental alleles remained unchanged in hybrid (gray) (Colored figure is available in the online version of the article). HP: high parent; LP: low parent.
3.5. Transcriptome profiling in pigeonpea
In order to assess the transcript abundance and differential gene expression between the three genotypes, their whole transcriptome profiling was performed by RNA sequencing. We first compared the transcription between different genotypes (11 A vs. 303 R, 11 A vs. F1 hybrid and 303 R vs. F1 hybrid) to compute genes that were differentially expressed between these lines. The transcripts showing a minimum of 2-fold change and P-value <0.05 were considered differentially expressed. We found 1,621 (460 up, 1,161 down), 1,790 (1,052 up, 738 down) and 1,380 (963 up, 417 down) genes that showed differential expression in 11 A vs. 303 R, 11 A vs. hybrid and 303 R vs. hybrid, respectively. We found that in all the three cases of comparison, the differentially expressed genes were well represented in gene categories like pentatricopeptide repeats (PPRs)- associated proteins involved in fertility restoration,56 methylases, demethylases and histone modifying genes (that play a role in epigenetic regulation) and flowering-associated genes. We performed gene ontology enrichment analysis to identify functional categories of significantly enriched genes (Supplementary Figs S4–S6). Interestingly, we found that gene ontology terms included reproductive processes, developmental processes, protein–DNA complexes and nucleic acid binding proteins that were significantly enriched (Fisher’s exact test P-value <0.05) in all the three comparative scenarios 11 A vs. 303 R, 11 A vs. hybrid and 303 R vs. hybrid.
3.6. Correlation between DMRs and gene expression in pigeonpea inbred and hybrid line
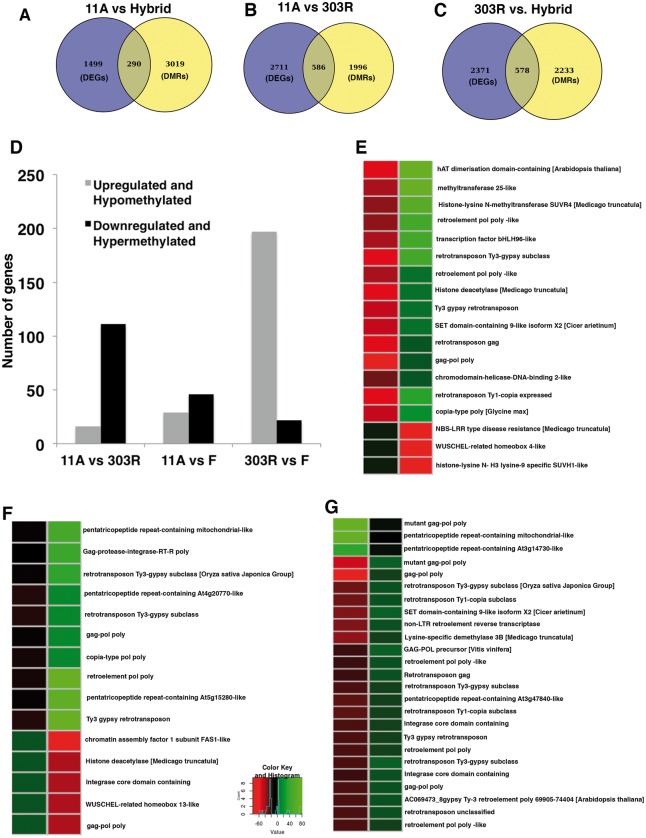
We next asked if the differential methylation in the three different genotypes could potentially influence the gene expression in these plants. For this, first DEGs with DMRs localized in 2 kb flanking (upstream and downstream) and gene body regions were identified as DMR-associated DEGs. Then the correlation between whole genome differential methylation and gene expression was examined (Fig. 5A–C). We next determined the genes that were enriched for DMRs between the three genotypes and found that DMRs were localized in 3,019, 1,996 and 2,233 genes in 11 A vs. hybrid, 11 A vs. 303 R and 303 R vs. hybrid, respectively (Fig. 5A–C). Among these, 290, 586 and 578 genes were found to be both differentially expressed and associated with DMRs in 11 A vs. F, 11 A vs. 303 R and 303 R vs. F, respectively (Fig. 5A–C). Furthermore, we also investigated the correlation between methylation status and expression in the DEGs associated with DMRs and found that 127 (16 hypomethylated and up-regulated; 111 hyper-methylated and downregulated), 75 (29 hypo-methylated and up-regulated; 46 hypermethylated and down-regulated) and 229 (197 hypo-methylated and up-regulated; 22 hyper-methylated and down-regulated) genes showed a negative correlation between their expression and methylation in 11 A vs. R, 11 A vs. Hybrid and 303 R vs. Hybrid, respectively (Fig. 5D and Supplementary data S2). In order to gain insights into the biological significance of the observed negative association between DEGs and DMRs, we investigated the functional categories of those genes. Interestingly we found many DMRs showing negative association with gene expression in fertile hybrid and 303 R lines, compared with 11 A (sterile) line belonged to transposable elements and genes that coded for PPR-associated proteins (Fig. 5E–G). In addition, we also found genes coding for histone deacytylase and methyltransferase (Fig. 5E–G). This suggests that the transcription of those genes might possibly be influenced by methylation.
Figure 5.
Characterization of DMRs associated DEGs in pigeonpea. (A–C) Venn diagrams showing unique and common DEGs (differentially expressed genes) to the genes associated with DMRs in 11A vs. hybrid (A), 11A vs. 303R (B) and 303R vs. hybrid (C). (D) Bar plots showing number of DEGs that were differentially methylated in 11A vs. 303R, 11A vs. hybrid and 303R vs. hybrid. (E–G) Heat maps displaying comparative methylation levels of selective genes that showed negative correlation between their expression and methylation in 11A vs. 303R (E), 11A vs. hybrid (F) and 303R vs. hybrid (G). Color code indicates methylation level (0–1) (Colored figure is available in the online version of the article).
3.7. Association between DNA methylation and transcript abundance
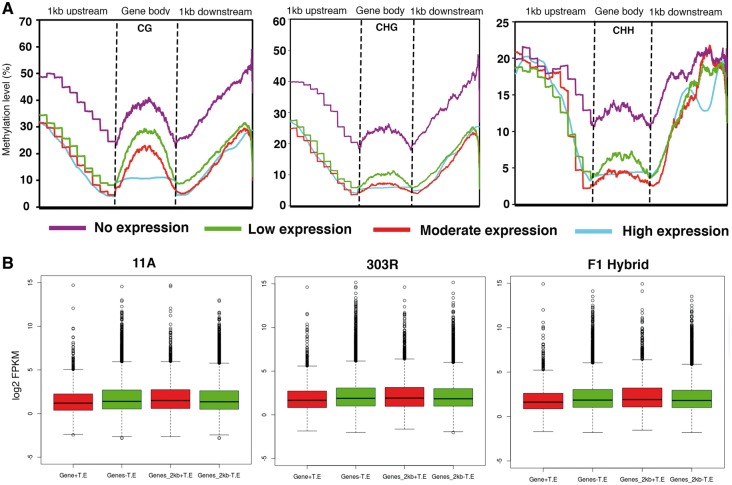
Previous studies have reported a negative association between gene expression and CHG and CHH methylation in plants like cotton.57 Therefore to investigate the occurrence of any relation between changes in methylation levels and gene expression in pigeonpea, we classified genes into four quartiles based on their FPKM values—undetectable expression (FPKM 0-1), low expression (FPKM 1-10), moderate expression (FPKM 10–100) and high expression (FPKM > 100) according to the criteria described in earlier studies58,59 and correlated with changes in their methylation levels. Consistent with previous studies, we found CHG and CHH methylation were negatively correlated with gene expression levels. However, additionally, we also found negative association between CG methylation and gene expression (Fig. 6A and Supplementary Fig. S7). Since transcription is importantly controlled by upstream and downstream regulatory sequences of the genes, we also sought to examine methylation levels in the gene flanking sequences. Similar methylation levels were observed in the sequences upstream and downstream of genes with high, moderate and low expression. However, contrary to this, genes with no expression exhibited high methylation in their flanking regions suggesting that a high methylation in upstream and downstream regulatory sequences might be critical for suppressing transcript levels (Fig. 6A and Supplementary Fig. S7). We also found that the presence of TE elements within the genes or in the vicinity resulted in a comparatively reduced expression relative to the genes without TE elements within or in the neighboring regions (Fig. 6B).
Figure 6.
Correlation between gene expression and DMRs in pigeonpea. (A) Lineplots showing correlation between CG, CHG and CHH DMRs and the gene expression which was divided into four quartiles—no expression (purple); low expression (green); moderate expression (red) and high expression (cyan) in F1 hybrid. (Also see Figure S7 for correlation in 11A and 303R parental lines). Note a negative correlation between gene expression and methylation in all the three contexts. (B) Box plots showing expression levels of genes inserted with and without TE within their gene bodies and 2kb upstream and downstream flanking regions. Gene +TE indicates insertion of transposon. Gene-TE indicates no insertion of transposon (Colored figure is available in the online version of the article).
4. Discussion
Rapid advancements in genome sequencing techniques have enabled genomic comparisons to understand genetic diversities between different plant species or different ecotypes of the same species. However, understanding of epigenetic variation among different ecotypes has become equally important to decipher its role in generating phenotypic variations. A fundamental question of great importance is that how the parental genome combination is regulated in hybrids to give rise to significantly different hybrid genome which results in altered genetic traits. It is widely speculated that many agriculturally important phenomena such as hybrid vigour and fertility restoration may arise due to epigenetic changes between inbred parents and hybrid lines. Thus the field of epigenetic research has expanded its horizon by adopting many crop species as model systems. However, there are still agriculturally important crops such as pigeonpea, in which existence of such mechanisms have remained unexplored. In this study, we investigated and compared epigenetic changes in three different pigeonpea genotypes. We have generated whole genome methylation profiles of sterile (11 A), fertile parents (303 R) and their fertile F1 hybrid, which were unavailable till date. Our analysis of relative fractions of methyl-cytosines in each methylation context in the three pigeonpea lines revealed highest fraction of mCs in CG context, followed by CHG and CHH. Similar pattern of distribution of methylation fractions has been observed previously in rice seedlings60 and Soybean leaves.61 We also found that average methylation levels of the fertile hybrid and the fertile parental line (303 R) were lower than the sterile parent (11 A) suggesting that comparative low DNA methylation of genes may play a role in restoring the fertility in the F1 hybrid.
Mean methylation has been reported to be highest in CG contexts in other plants such as Rice60 and Arabidopsis,15 and followed closely by CHG and CHH. However, our results show that in pigeonpea, mean methylation levels in CHH context are much lower than CG and CHG methylation levels. This suggests a possibility of overall low expression of genes involved in regulation of CHH methylations such as those involved in RdDM pathway that target homologous regions of the genome for methylation62 in pigeonpea compared with Rice and Arabidopsis.
Previously, it has been reported that transcription of genes is affected by methylation in the promoter region but not in the internal regions. However, transposon silencing is regulated by methylation both in the promoter region as well as internal regions.63 Our data demonstrate that fraction of mCs was higher in the transcribed regions of transposons than the transcribed regions of the genes. As shown in Figure 2, the fraction of methyl cytosines in upstream sequences of the transcribed regions of transposon was almost two times higher than fraction of mCs in upstream sequences of genes. Additionally, gene bodies of TEs exhibited higher methylation than their upstream and downstream regions. Contrary to this, genes had lower methylation in the transcribed regions than their flanking regions. As suggested previously for other crops,1,64 this could potentially be a consequence of reduced RdDM pathway activity within the gene body of the transcribed genes compared with TE. Furthermore, we also observed that the fraction of mCs in upstream regions of transposons in fertile 303 R parent was slightly less than sterile 11 A parental line and the fertile hybrid. These observations suggest that one possible epigenetic mechanism of regulating transposon inactivation in pigeonpea could be via methylation and this mechanism could be comparatively weaker in the fertile 303 R line than in the sterile parent and fertile hybrid. Over all, our results comply with previously documented findings that CHH methylation levels are usually lower than CG and CHG, both on a genomic scale and at specific loci.65,66
Further, our findings also reveal the existence of non-additive methylation in F1 hybrid due TCM and TCdM events. TcM mediated increase in DNA methylation were more frequently observed in CG context and TcDM mediated decrease in DNA methylation were more frequently observed in CHH context, consistent with previous findings in Arabidopsis.15 Furthermore, we also dissected the mC changes of differentially methylated parental epi-alleles in the hybrid, using SNPs for tracking the parental epi-alleles in the hybrid. Our analysis revealed that majority of the mC clusters having SNPs showed non- additive methylation in the hybrid when parents had different levels of methylation (HP or LP), consistent with our general mC cluster analysis.
Transposable elements can influence neighboring gene expression, contribute to inheritance of epi-alleles and thus represent the most striking and common example of epigenetic inheritance in plants.67 Consistently, our data also revealed low expression of genes with inserted TEs suggesting similar roles of TEs in pigeonpea.
We also found a strong enrichment of DMRs in a wide spectrum of genes that are associated with chromatin modification and have been reported to regulate flowering, heterosis and reproduction in rice.68 We also found genes involved in DNA methylation that are known to play a role in transposon repression69 and genes involved in histone methylation that help regulate flowering.70
DNA methylation is known to influence gene expression. However, as mentioned above, it has been proposed that DNA methylation and H3K9 methylation in the gene body regions do not affect gene abundance. Despite increased CHG and H3K9 methylation in loss of function mutants of a H3K9 demethylase gene increased in bonsai methylation 1 (ibm1), the expression of their target genes remained unaffected.63 Also, absence of CG gene body methylation in the loss of function MET1 mutants did not alter gene expression.71 Surprisingly, we found a negative correlation between transcript abundance and methylation levels in all the three contexts in the gene body suggesting a more prominent role of DNA methylation in regulating gene expression in pigeonpea. Previous studies in Arabidopsis have reported that methylation in the promoter regions resulted in lower gene expression.71,72 Furthermore, another study in soybean revealed negative correlation between gene expression and DNA methylation in their promoters.73 We also found that high methylation in the 5’ upstream regions of the genes were negatively correlated with gene expression levels, consistent with the idea that DNA methylation in the promoter regions affects gene abundance.61 Our results in pigeonpea together with previous results in soybean73 open new avenues for understanding the relationship between gene expression and gene body DNA methylation and whether they play a direct role in restoring fertility in pigeonpea hybrids requires further investigation.
The raw reads of bisulfite and transcriptomic study were submitted to NCBI Sequence Read Archive under bioproject accession PRJNA435649 and SRP133358 study.
Supplementary Material
Acknowledgements
We thank Council of Scientific and Industrial Research, Government of India for the award of a senior research fellowship, ICAR-IARI, ICAR-NRCPB and NRCPB in house project, New Delhi for supporting A.J. and providing other funds and facilities. We also thank Dr N. Bhatia, The University of Sydney for helpful feedback and ideas on the manuscript.
Authors’ contributions
A.J and K.G conceived and designed the study. A.J. performed all the experimental work and data analysis. H.K. helped in data analysis and interpreting the results. A.J., K.G. wrote the manuscript. A.R. and N.K.S. helped editing the manuscript.
Conflict of interest
None declared.
References
- 1. Henderson I. R., Jacobsen S. E.. 2007, Epigenetic inheritance in plants, Nature, 447, 418–24. [DOI] [PubMed] [Google Scholar]
- 2. Niederhuth C. E., Schmitz R. J.. 2014, Covering your bases: inheritance of DNA methylation in plant genomes, Mol. Plant., 7, 472–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Robertson K. D. 2005, DNA methylation and human disease, Nat. Rev. Genet., 6, 597–610. [DOI] [PubMed] [Google Scholar]
- 4. Suzuki M. M., Bird A.. 2008, DNA methylation landscapes: provocative insights from epigenomics, Nat Rev Genet., 9, 465–76. [DOI] [PubMed] [Google Scholar]
- 5. Bird A. 2007, Perceptions of epigenetics, Nature, 447, 396–8. [DOI] [PubMed] [Google Scholar]
- 6. Law J. A., Jacobsen S. E.. 2010, Establishing, maintaining and modifying DNA methylation patterns in plants and animals, Nat. Rev. Genet., 11, 204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vongs A., Kakutani T., Martienssen R. A., Richards E. J.. 1993, Arabidopsis thaliana DNA methylation mutants, Science, 260, 1926–8., [DOI] [PubMed] [Google Scholar]
- 8. Kankel M. W., Ramsey D. E., Stokes T. L., et al. 2003, Arabidopsis MET1 cytosine methyltransferase mutants, Genetics, 163, 1109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zemach A., McDaniel I. E., Silva P., Zilberman D.. 2010, Genome-wide evolutionary analysis of eukaryotic DNA methylation, Science, 328, 916–9. [DOI] [PubMed] [Google Scholar]
- 10. Cao X., Jacobsen S. E.. 2002, Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing, Curr. Biol., 12, 1138–44. [DOI] [PubMed] [Google Scholar]
- 11. Mosher R. A., Melnyk C. W.. 2010, siRNAs and DNA methylation: seedy epigenetics, Trends Plant Sci., 15, 204–10. [DOI] [PubMed] [Google Scholar]
- 12. Stroud H., Do T., Du J., et al. 2014, Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis, Nat. Struct. Mol. Biol., 21, 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reed H. S. 1942, A short history of the plant sciences. https://www.scribd.com/document/190746815/Reed-Howard-S-1942-a-Short-History-of-the-Plant-Sciences.
- 14. Ng S. Y., Lin L., Soh B. S., Stanton L. W.. 2013, Long noncoding RNAs in development and disease of the central nervous system, Trends Genet., 29, 461–8. [DOI] [PubMed] [Google Scholar]
- 15. Greaves I. K., Groszmann M., Ying H., Taylor J. M., Peacock W. J., Dennis E. S.. 2012, Trans chromosomal methylation in Arabidopsis hybrids, Proc. Natl. Acad. Sci. US. A, 109, 3570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Greaves I. K., Groszmann M., Wang A., Peacock W. J., Dennis E. S.. 2014, Inheritance of trans chromosomal methylation patterns from Arabidopsis F1 hybrids, Proc. Natl. Acad. Sci. USA, 111, 2017–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Greaves I. K., Gonzalez-Bayon R., Wang L., et al. 2015, Epigenetic changes in hybrids, Plant Physiol., 168, 1197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vaughn M. W., Tanurdzic M., Lippman Z., et al. 2007, Epigenetic natural variation in Arabidopsis thaliana, PLoS Biol., 5, e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schmitz R. J., Schultz M. D., Urich M. A., et al. 2013, Patterns of population epigenomic diversity, Nature, 495, 193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen H., He H., Li J., et al. 2012, Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids, Plant Cell, 24, 875–92., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y., Varala K., Moose S. P., Hudson M. E.. 2012, The inheritance pattern of 24 nt siRNA clusters in arabidopsis hybrids is influenced by proximity to transposable elements, PLoS One, 7, e47043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Groszmann M., Greaves I. K., Albertyn Z. I., Scofield G. N., Peacock W. J., Dennis E. S.. 2011, Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor, Proc. Natl. Acad. Sci. U.S.A, 108, 2617–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He G., Zhu X., Elling A. A., et al. 2010, Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids, Plant Cell, 22, 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barber W. T., Zhang W., Win H., et al. 2012, Repeat associated small RNAs vary among parents and following hybridization in maize, Proc. Natl. Acad. Sci. U.S.A, 109, 10444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Parkin I. A., Koh C., Tang H., et al. 2014, Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea, Genome Biol., 15, R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chodavarapu R. K., Feng S., Ding B., et al. 2012, Transcriptome and methylome interactions in rice hybrids, Proc. Natl. Acad. Sci. U.S.A, 109, 12040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh N. K., Gupta D. K., Jayaswal P. K., et al. 2012, The first draft of the pigeonpea genome sequence, J. Plant Biochem. Biotechnol., 21, 98–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saxena K. B., Singh L.. 1990, Low natural outcrossing in ′cleistogamous′ pigeonpea mutant, Int. Pigeonpea Newslett., 11, 9–10. ISSN 0255-786X. [Google Scholar]
- 29. Chen L., Liu Y. G.. 2014, Male sterility and fertility restoration in crops, Annu. Rev. Plant Biol., 65, 579–606. [DOI] [PubMed] [Google Scholar]
- 30. Tuteja R., Saxena R. K., Davila J., et al. 2013, Cytoplasmic male sterility-associated chimeric open reading frames identified by mitochondrial genome sequencing of four Cajanus genotypes, DNA Res., 20, 485–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meshram M. P., Patil A. N.. 2018, Genetics of fertility restoration in A2 cytoplasm based hybrids of pigeonpea. Int. J. Cur. Microbiol. Appl. Sci., ISSN: 2319-7706 565-571. [Google Scholar]
- 32. Varshney R. K., Chen W., Li Y., et al. 2011, Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers, Nat. Biotechnol., 30, 83–9. [DOI] [PubMed] [Google Scholar]
- 33. Xi Y., Li W.. 2009, BSMAP: whole genome bisulfite sequence MAPping program, BMC Bioinformatics, 10, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schultz M. D., Schmitz R. J., Ecker J. R.. 2012, ′Leveling′ the playing field for analyses of single-base resolution DNA methylomes, Trends Genet., 28, 583–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Akalin A., Kormaksson M., Li S., et al. 2012, methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles, Genome Bio.l, 13, R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li L. C., Dahiya R.. 2002, MethPrimer: designing primers for methylation PCRs, Bioinformatics, 18, 1427–31. [DOI] [PubMed] [Google Scholar]
- 37. Kumaki Y., Oda M., Okano M.. 2008, QUMA: quantification tool for methylation analysis, Nucleic Acids Res., 36, W170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Langmead B., Salzberg S. L.. 2012, Fast gapped-read alignment with Bowtie 2, Nat. Methods , 9, 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li B., Dewey C. N.. 2011, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome, BMC Bioinformatics, 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson M. D., McCarthy D. J., Smyth G. K.. 2010, edgeR: a bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics, 26, 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ye J., Fang L., Zheng H., et al. 2006, WEGO: a web tool for plotting GO annotations, Nucleic Acids Res , 34, W293–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu Y., Siegmund K. D., Laird P. W., Berman B. P.. 2012, Bis-SNP: combined DNA methylation and SNP calling for Bisulfite-seq data, Genome Biol., 13, R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Feng S., Cokus S. J., Zhang X., et al. 2010, Conservation and divergence of methylation patterning in plants and animals, Proc. Natl. Acad. Sci. U.S.A., 107, 8689–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lisch D. 2009, Epigenetic regulation of transposable elements in plants, Annu. Rev. Plant Biol., 60, 43–66. [DOI] [PubMed] [Google Scholar]
- 45. Cokus S. J., Feng S., Zhang X., et al. 2008, Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning, Nature, 452, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Qi X., Li M.-W., Xie M., et al. 2014, Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing, Nat. Commun., 5, 4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. The Arabidopsis Genome, I. 2000, Analysis of the genome sequence of the flowering plant Arabidopsis thaliana, Nature, 408, 796. [DOI] [PubMed] [Google Scholar]
- 48. Zhang J., Liu Y., Xia E. H., Yao Q. Y., Liu X. D., Gao L. Z.. 2015, Autotetraploid rice methylome analysis reveals methylation variation of transposable elements and their effects on gene expression, Proc. Natl. Acad. Sci. U.S.A, 112, E7022–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li X., Zhu J., Hu F., et al. 2012, Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression, BMC Genomics, 13, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lu Y., Liu Y., Wang J., Cao M., Rong T.. 2010, Variation and patterns of DNA methylation in maize C-type CMS lines and their maintainers, J. Plant Biochem. Biotechnol., 19, 43–50. [Google Scholar]
- 51. Chen X., Hu J., Zhang H., Ding Y.. 2014, DNA methylation changes in photoperiod-thermo-sensitive male sterile rice PA64S under two different conditions, Gene, 537, 143–8. [DOI] [PubMed] [Google Scholar]
- 52. Schmitz R. J., Schultz M. D., Lewsey M. G., et al. 2011, Transgenerational epigenetic instability is a source of novel methylation variants, Science, 334, 369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Becker C., Hagmann J., Muller J., et al. 2011, Spontaneous epigenetic variation in the Arabidopsis thaliana methylome, Nature, 480, 245–9. [DOI] [PubMed] [Google Scholar]
- 54. Eichten S. R., Briskine R., Song J., et al. 2013, Epigenetic and genetic influences on DNA methylation variation in maize populations, Plant Cell, 25, 2783–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu F., Cui X., Horner H. T., Weiner H., Schnable P. S.. 2001, Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize, Plant Cell, 13, 1063–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang W., Yu C., Hu J., et al. 2015, Pentatricopeptide-repeat family protein RF6 functions with hexokinase 6 to rescue rice cytoplasmic male sterility, Proc. Natl. Acad. Sci. U.S.A, 112, 14984–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Song Q., Guan X., Chen Z. J.. 2015, Dynamic roles for small RNAs and DNA methylation during ovule and fiber development in allotetraploid cotton, PLoS Genet., 11, e1005724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xu W., Yang T., Dong X., Li D. Z., Liu A.. 2016, Genomic DNA methylation analyses reveal the distinct profiles in castor bean seeds with persistent endosperms, Plant Physiol., 171, 1242–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lu X., Wang W., Ren W., et al. 2015, Genome-wide epigenetic regulation of gene transcription in maize seeds, PLoS One, 10, e0139582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Garg R., Narayana Chevala V., Shankar R., Jain M.. 2015, Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response, Sci. Rep., 5, 14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Song Q. X., Lu X., Li Q. T., et al. 2013, Genome-wide analysis of DNA methylation in soybean, Mol. Plant., 6, 1961–74. [DOI] [PubMed] [Google Scholar]
- 62. Matzke M., Kanno T., Daxinger L., Huettel B., Matzke A. J.. 2009, RNA-mediated chromatin-based silencing in plants, Curr. Opin. Cell Biol., 21, 367–76. [DOI] [PubMed] [Google Scholar]
- 63. Inagaki S., Kakutani T.. 2012, What triggers differential DNA methylation of genes and TEs: contribution of body methylation? Cold Spring Harb. Symp. Quant. Biol., 77, 155–60. [DOI] [PubMed] [Google Scholar]
- 64. Zakrzewski F., Schmidt M., Van Lijsebettens M., Schmidt T.. 2017, DNA methylation of retrotransposons, DNA transposons and genes in sugar beet (Beta vulgaris L.), Plant J., 90, 1156–75. [DOI] [PubMed] [Google Scholar]
- 65. West P. T., Li Q., Ji L., et al. 2014, Genomic distribution of H3K9me2 and DNA methylation in a maize genome, PLoS One, 9, e105267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gent J. I., Ellis N. A., Guo L., et al. 2013, CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize, Genome Res., 23, 628–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Martienssen R., Barkan A., Taylor W. C., Freeling M.. 1990, Somatically heritable switches in the DNA modification of Mu transposable elements monitored with a suppressible mutant in maize, Genes Dev., 4, 331–43. [DOI] [PubMed] [Google Scholar]
- 68. Shi J., Dong A., Shen W. H.. 2014, Epigenetic regulation of rice flowering and reproduction, Front. Plant Sci., 5, 803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Higo H., Tahir M., Takashima K., et al. 2012, DDM1 (decrease in DNA methylation) genes in rice (Oryza sativa), Mol. Genet. Genomics, 287, 785–92. [DOI] [PubMed] [Google Scholar]
- 70. Choi S. C., Lee S., Kim S. R., et al. 2014, Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3, Plant Physiol., 164, 1326–37., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang X., Yazaki J., Sundaresan A., et al. 2006, Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis, Cell, 126, 1189–201., [DOI] [PubMed] [Google Scholar]
- 72. Zilberman D., Gehring M., Tran R. K., Ballinger T., Henikoff S.. 2007, Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription, Nat Genet., 39, 61–9. [DOI] [PubMed] [Google Scholar]
- 73. Schmitz R. J., He Y., Valdes-Lopez O., et al. 2013, Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population, Genome Res., 23, 1663–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.