Abstract
Recombination arrest is a necessary step for the evolution of distinct sex chromosomes. Structural changes, such as inversions, may represent the mechanistic basis for recombination suppression and comparisons of the structural organization of chromosomes as given by chromosome-level assemblies offer the possibility to infer inversions across species at some detail. In birds, deduction of the process of sex chromosome evolution has been hampered by the lack of a validated chromosome-level assembly from a representative of one of the two basal clades of modern birds, Paleognathae. We therefore developed a high-density genetic linkage map of the ostrich Z chromosome and used this to correct an existing assembly, including correction of a large chimeric superscaffold and the order and orientation of other superscaffolds. We identified the pseudoautosomal region as a 52 Mb segment (≈60% of the Z chromosome) where recombination occurred in both sexes. By comparing the order and location of genes on the ostrich Z chromosome with that of six bird species from the other major clade of birds (Neognathae), and of reptilian outgroup species, 25 Z-linked inversions were inferred in the avian lineages. We defined Z chromosome organization in an early avian ancestor and identified inversions spanning the candidate sex-determining DMRT1 gene in this ancestor, which could potentially have triggered the onset of avian sex chromosome evolution. We conclude that avian sex chromosome evolution has been characterized by a complex process of probably both Z-linked and W-linked inversions (and/or other processes). This study illustrates the need for validated chromosome-level assemblies for inference of genome evolution.
Keywords: sex chromosomes, inversions, assembly correction, linkage analysis
Introduction
Recombination arrest is a requisite for the formation of distinct sex chromosomes. Recombination initially stops around a sex-determining locus (Nei 1969; Charlesworth and Charlesworth 1978; Charlesworth et al. 2005) and the nonrecombining segment may subsequently expand across most of the chromosome, although this is not always the case (Stock et al. 2011). A common view is that recombination arrest is driven by the emergence of alleles with contrasting fitness effects between sexes, where selection favors establishment of strong linkage disequilibrium between the sex-determining locus and sexually antagonistic alleles (Rice 1987). Mechanistically, cessation of recombination might be prompted by inversion events hindering proper chromosome pairing and/or formation of cross-overs (Becak et al. 1962; Ohno 1967), or by sex-specific genetic modifiers of the recombination rate (Choi and Henderson 2015).
The two independently evolving copies of a gene that once recombined on the proto-sex chromosomes are referred to as gametologs (Garcia-Moreno and Mindell 2000). The degree of neutral sequence divergence between gametologs reflect the evolutionary time since they stopped recombining. Using divergence data from a number of gametologous gene pairs and noting that pairs with similar degree of divergence cluster along the X/Z chromosome, it has been inferred that sex chromosome recombination stopped in discrete events, forming so called “evolutionary strata” (Lahn and Page 1999). Such strata have been documented in sex chromosomes of a variety of species, both in systems with male and female heterogamety, including mammals (Lahn and Page 1999; Sandstedt and Tucker 2004; Van Laere et al. 2008), birds (Handley et al. 2004; Nam and Ellegren 2008; Wright et al. 2012; Yazdi and Ellegren 2014; Zhou et al. 2014), reptiles (Vicoso et al. 2013a), fish (Roesti et al. 2013; White et al. 2015), and plants (Nicolas et al. 2005; Bergero et al. 2007; Wang et al. 2012; Hough et al. 2014), as well as in fungal mating-type chromosomes (Branco et al. 2017). The presence of evolutionary strata could suggest a succession of punctuated events of recombination cessation from large chromosomal rearrangements such as inversions (Bergero and Charlesworth 2009). Y chromosome inversions hypothesized to have been involved in recombination suppression have been reported in, for example, papaya (Wang et al. 2012) and humans (Lemaitre et al. 2009). In contrast, genetic modifiers of recombination would potentially cause a more gradual spread of cessation of recombination (Chibalina and Filatov 2011; Bergero et al. 2013; Natri et al. 2013). However, disentangling these two processes can be challenging given that the resolution of how strata are defined and distributed is often poor. Moreover, structural rearrangements of sex chromosomes that are unrelated to strata formation and sex chromosome evolution would complicate the picture. This appears to be the case in birds where inferred strata are not linearly ordered along the sex chromosomes (Charlesworth et al. 2005; Nam and Ellegren 2008; Wright et al. 2012) as they are on the human X chromosome (Lahn and Page 1999). Generally, structural and other changes are expected to drift to fixation more rapidly on sex chromosomes (than autosomes) due to their reduced effective population size since the cessation of recombination.
In birds, females are the heterogametic sex (females, ZW; males, ZZ). The sex chromosomes of all birds are syntenic and are thus thought to originate from the same ancestral pair of autosomes. Consistent with a single common origin, it has been inferred that avian sex chromosome evolution was initiated prior to the evolution of modern birds (Handley et al. 2004; Nam and Ellegren 2008; Wright et al. 2012; Yazdi and Ellegren 2014); the two basal clades of birds, Paleognathae (ratites and tinamous) and Neognathae (all remaining birds, >99% of all extant species), share a common ancestor that lived about 100 Ma (Jarvis et al. 2014). However, there is an interesting contrast in the structure of sex chromosomes between Paleognathae and Neognathae concerning the level of differentiation between the Z and W chromosomes. In many Neognath species, the W chromosome is highly degenerated (Rutkowska et al. 2012; Zhou et al. 2014; Smeds et al. 2015; Bellott et al. 2017), largely heterochromatic (Stefos and Arrighi 1971), and rich in satellite-like repeats (Saitoh et al. 1991; Itoh and Mizuno 2002) and ampliconic arrays (Backström et al. 2005; Bellott et al. 2017). Recombination between the Z and W chromosomes is restricted to a small pseudoautosomal region (PAR) (Smeds et al. 2014; Zhou et al. 2014; Bellott et al. 2017). In Paleognathae, the Z and W chromosome have maintained recombination along a large segment. (Vicoso et al. 2013b; Yazdi and Ellegren 2014; Zhou et al. 2014). Thus, sex chromosome differentiation started in an early avian ancestor, further advanced in the Neognath lineage, but was for some reason largely halted in the parallel Paleognath lineage.
Using sequence divergence and phylogenetic data of gametologous genes, an inversion scenario for avian sex chromosome evolution has been indicated (Zhou et al. 2014). More formally testing models of sex chromosome evolution requires chromosome-level genome assemblies to allow making inference on large-scale structural rearrangements. Such assemblies are only available for a limited number of bird species, all from Neognathae. In Paleognathae, a short-read assembly is available for the ostrich Struthio camelus (Zhou et al. 2014) and was further improved by optical mapping (Zhang et al. 2015), but uncertainty still remains concerning order and orientation of scaffolds. A chromosome-level assembly of the ostrich Z chromosome (or of any other Paleognath species) is important for studies of avian sex chromosome evolution, for several reasons. First, it would help to reveal the ancestral state of avian sex chromosomes, prior to the divergence of Paleognathae and Neognathae, and prior to the onset of sex chromosome differentiation. Only with this information available will it be possible to identify subsequent inversion events in avian lineages. Second, it would represent a necessary outgroup to infer the ancestral state of Neognathae sex chromosomes. Third, any attempt to address the question of why ratite sex chromosomes have remained largely undifferentiated would need to be based on information of the organization of the Z chromosome in Paleognath species.
With these considerations in mind, we developed a linkage map of the ostrich Z chromosome. A linkage map of ostrich based on microsatellite data is available (Huang et al. 2008), but it does not have sufficient resolution for assembly correction. We used the dense SNP-based genetic map for correcting the order and orientation of scaffolds from the genome assembly. With the access to a corrected assembly, we then aimed at inferring the inversion events leading to the current arrangement of genes on avian sex chromosomes. Finally, we analyzed the relationship between inversions and sex chromosome evolution in avian lineages.
Materials and Methods
Sampling and DNA Preparation
Blood and tissue samples were collected from 258 ostriches representing 22 full-sib families (5–13 offspring per family; in total 214 offspring and 44 parents) from the Oudtshoorn Research Farm of the Western Cape Department of Agriculture, South Africa. Genomic DNA was extracted by a standard proteinase-K digestion and phenol/chloroform purification protocol. The samples were kindly provided by Charlie Cornwallis.
SNP Selection and Genotyping
The optical map-improved ostrich genome assembly (Zhang et al. 2015) (Struthio_camelus.20130116.OM.fa.gz downloaded from http://gigadb.org/dataset/101013; last accessed August 6, 2018) was used as the reference for variant calling. We used two sources of data in order to identify single nucleotide polymorphisms (SNPs): pooled RNA-seq data from male ostrich brain, liver, and gonad (three pools of each organ, each pool containing 2–3 individuals; supplementary table S1, Supplementary Material online) (Adolfsson and Ellegren 2013) and genomic short reads (accession SRR950910) from the individual used for the ostrich genome assembly (which was sequenced at 85× coverage). SNP calling and filtering were done using the GATK (version 3.3.0) best practices for mRNA and DNA sequences (DePristo et al. 2011). RNA-seq data were filtered for genotype quality > 30, and a coverage of ≥ 14 reads in male pools of two individuals and ≥21 reads in pools of three individuals. To increase the chances of obtaining informative markers, we mostly selected markers from an intersect of SNPs found in both the RNA and the genomic data set. All but 12 SNPs were from known protein-coding genes.
We chose a final set of 384 genetic markers, well spread within and among Z chromosome scaffolds (supplementary fig. S1, Supplementary Material online), that were genotyped in the pedigree. Genotyping was performed at the SNP and SEQ Technology Platform, Uppsala University (www.genotyping.se, last accessed August 6, 2018) by targeted resequencing using a custom designed TruSeq Custom Amplicon panel (Illumina Inc.) on a MiSeq sequencing platform (Illumina Inc.). Genotype data were thoroughly filtered according to steps presented in supplementary tables S2 and S3, Supplementary Material online. After filtering, we retained data for 333 SNPs covering 177 genes and 12 intergenic segments across the Z chromosome (supplementary table S4, Supplementary Material online).
Linkage Analysis
Linkage analysis was performed using Lep-MAP2 (Rastas et al. 2015). In order to specify hemizygosity of the nonrecombining region of the Z chromosome in females, ParentCall was run with ZLimit = 2. Lepmap2 filters out markers by comparing the offspring genotype distribution and the expected Mendelian proportions (segregation distortion test). The default value of dataTolerance = 0.01 was used to filter out markers showing strong deviation from Mendelian segregation (chi-square test, P < 0.01). One of the families (Family 56) showed 37 markers with segregation distortion, which could be due to, for example, contamination or sample mix-up, and the family was therefore removed from the final data set.
In order to assign markers into linkage groups, the SeparateChromosomes module was run with Logarithm of odds (LOD) = 10 and sizeLimit = 4, the latter which will remove linkage groups with less than four markers. The JoinSingles module was subsequently run with LOD = 6 and LOD = 3 to assign further markers to linkage groups. Marker order was determined by allowing different recombination probabilities in the two sexes. The OrderMarkers module, which uses the Kosambi mapping function was run 10 times with useKosambi = 1 and minError = 0.01, the latter which is the minimum allowed haplotype error probability. Because genotyping errors can increase map distance, it is recommended to set minError to a value between 0.01 and 0.4 to reduce the error probability (Rastas et al. 2015). Markers with a genotype error rate estimate > 0.01 were removed. Framework and best-order maps were constructed using LOD = 6 and LOD = 3, respectively. Sex-averaged map for the Z chromosome was obtained using the following calculation: Marker order was checked by pairwise LOD plot. Genetic maps were drawn by the program genetic-mapper (Bekaert 2016).
We confirmed marker order and the framework map by CRIMAP (version 2.507) (Green et al. 1990) using the TWOPOINT option in order to calculate pairwise linkage across all pairs of loci. Markers within the same gene with no recombination were grouped together to create haplotypes. Inferring haplotypes for the PAR was done using PedPhase 3.0 (Li and Li 2009). Haplotypes from the non-PAR region were inferred by a custom Python script available at http://github.com/Homap/phasing_nonPAR; last accessed August 6, 2018. In order to build the map, 10 pairs of loci were chosen based on TWOPOINT analysis. For each run, sex-specific maps were generated. To obtain a framework map, we ran BUILD with LOD ≥ 6 followed by FLIPS4. The best-order map was subsequently obtained by iteratively running BUILD with LOD ≥ 5, 4, and 3, in each case followed by FLIPS4. We used CHROMPIC to visualize cross-overs. Markers that were reported in Lepmap2 having a haplotype error estimate > 0.01 were discarded from the CRIMAP analysis. A more detailed description of this analysis is provided in supplementary materials and methods, Supplementary Material online.
Integrating Linkage Map with Ostrich Genome Assembly
We used ALLMAPS (Tang et al. 2015) to integrate the information obtained from the linkage map with the ostrich Z chromosome assembly. Three inputs were used: the male and female linkage maps, and the genomic sequence of ostrich Z chromosome scaffolds. The two linkage maps were merged to generate a weights file and an input bed file. We set map weights to 1. As superscaffold54 was identified to be chimeric in downstream analysis, we used ALLMAPS to split this scaffold with the split option and the chunk parameter set to 4. In order to refine the region where two scaffolds had been incorrectly merged, we intersected the “breakpoint” coordinates with the gaps in the assembly where most misjoins occurred and refined the breakpoints by finding the largest gap in the breakpoint region. This information was then used to split the chimeric scaffold. In the end, all scaffolds on the Z chromosome were concatenated with an arbitrarily chosen gap size of 5,000 bp between scaffolds.
Inferring Inversion Scenarios
In order to infer inversion events on Z chromosome across the avian phylogeny, we obtained orthologous genes with known location on the Z chromosome of ostrich (Struthio camelus), chicken (Gallus gallus) (Gallus_gallus-5), turkey (Meleagris gallopavo) (Turkey_2.01), quail (Coturnix japonica) (Coturnix_japonica_2.0), collared flycatcher (Ficedula albicollis) (FicAlb_1.4.), great tit (Parus major) (Parus_major1.1), zebra finch (Taeniopygia guttata) (TaeGut3.2.4), and on chromosome 2 of green anole lizard (Anolis carolinensis) (AnoCar2.0) as an outgroup. In addition to lizard, we used the annotated genes of turtle (Chrysemys picta) (Chrysemys picta bellii-3.0.3) and alligator (Alligator mississippiensis) (ASM28112v4) to identify inversions across the reptile species. We ran blastp (Altschul et al. 1990) with an e-value ≥1e−10 between ostrich and each of the other species, and filtered the output for reciprocal best hits. For reptiles, blastp was run between lizard and turtle, and between lizard and alligator, to identify reciprocal best hits. We obtained an intersection of genes between lizard, ostrich, and chicken (269 genes), between ostrich and chicken (547 genes) and finally across lizard and the seven avian species (169 genes).
Using the set of 169 genes, we used GRIMM-Synteny 2.02 (Tesler 2002) in order to generate synteny blocks (>100 kb) (Pevzner and Tesler 2003). Synteny blocks are segments that can be converted into regions with conserved gene order without disruption by microrearrangements (Nadeau and Taylor 1984). As an example, an alignment of individual orthologous genes between ostrich and chicken is shown in supplementary fig. S2, Supplementary Material online together with the corresponding synteny blocks. To validate the synteny blocks, we used LASTZ to align chromosome sequences. Investigating the dotplot obtained from the alignment let us visually evaluate the accuracy of identified synteny blocks. We conducted multiple genome rearrangement (MGR 2.01) (Bourque and Pevzner 2002) analysis on the identified synteny blocks to infer inversions in the phylogeny and to reconstruct the ancestral order of genes on the avian sex chromosomes. We used GRIMM 2.02 (Tesler 2002) to infer the minimum number of inversions that can convert one genome to the other (optimal reversal scenario) for each pair of inferred ancestral and current species. GRIMM 2.02 returns one optimal inversion scenario. However, there can be several equally parsimonious scenarios. We used analyzeTraces of baobabLUNA (Braga 2009) to obtain the complete set of inversion events for every pair of inferred ancestral and current species.
Identification of Breakpoints
Unaligned regions between syntenic blocks were considered as breakpoints. We used orthologues genes between ostrich and chicken to refine the breakpoints. To this end, we used the program CASSIS (Baudet et al. 2010), which aims at finding the precise breakpoint location by a local realignment strategy. IntersectBed from BEDTOOLS (Quinlan and Hall 2010) was used for extracting overlaps with known repeats and genes.
Results
Linkage Mapping and the Sex-Specific Recombination Rate in the Pseudoautosomal Segment
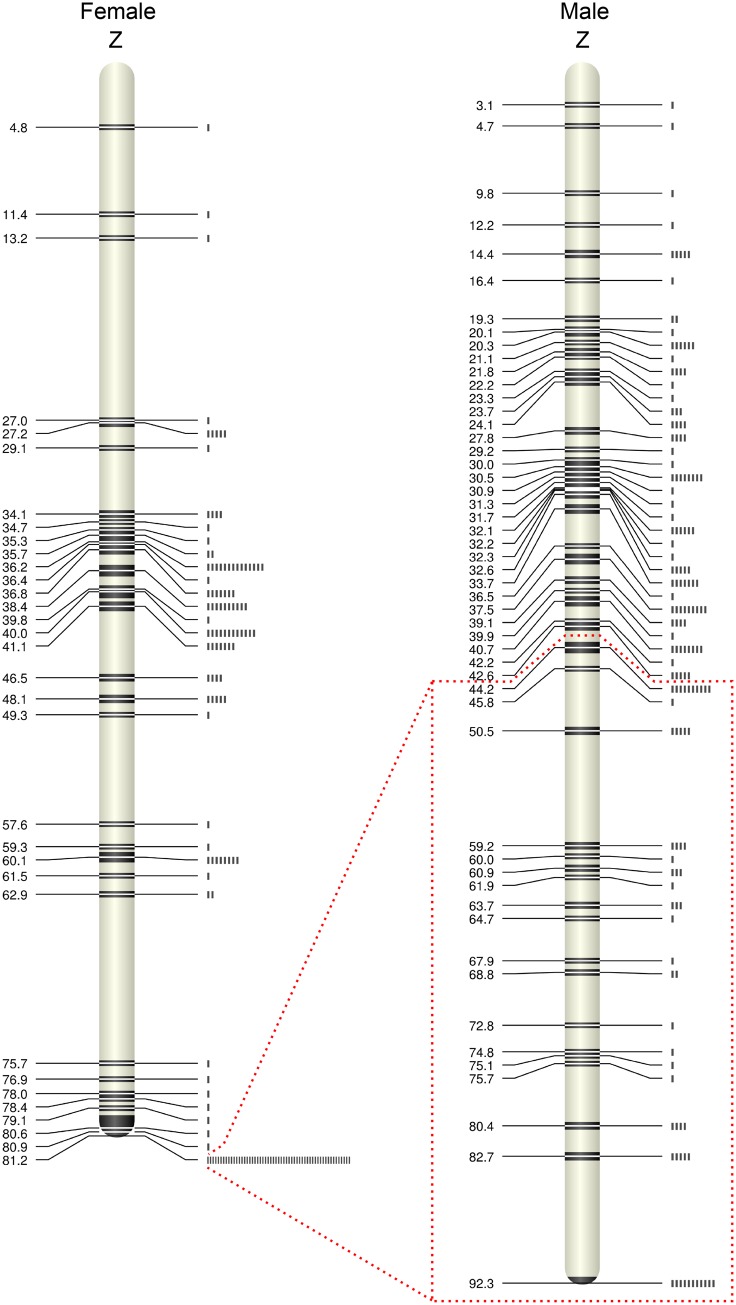
We identified genic SNPs from all putatively Z-linked scaffolds in the ostrich genome assembly and genotyped these in 22 full-sib ostrich families. After filtering based on genotype quality, we obtained a final set of 333 SNPs genotyped in 230 individuals. The markers formed two linkage groups with 262 markers in one linkage group with a male and female genetic map of 92.3 and 81.2 cM, respectively (fig. 1) and a sex-averaged genetic length of 94.2 cM (supplementary fig. S3, Supplementary Material online). Seven markers formed the second linkage group of 6.6 cM (linkage group 2; supplementary fig. S4, Supplementary Material online; remaining markers either had too low polymorphism level to be included in the map, or showed inheritance errors). The finding of two linkage groups from scaffolds thought to originate from the same chromosome was surprising given the number of markers and since the marker density should have been sufficient to place all markers in a single linkage group.
Fig. 1.
—Male and female best-order linkage maps of the ostrich Z chromosome with genetic distances in cM. The box marks the region of the Z chromosome with male but not female recombination. The bars to the right indicate the number of genes at a given genetic position.
The sex chromosomes of most avian lineages have remained remarkably stable in terms of conserved synteny (gene content) since a common sauropod ancestor 275 Ma (Shetty et al. 1999; Griffin et al. 2007; Chapus and Edwards 2009), and are syntenic to human chromosomes 5, 9, and 18 (Fridolfsson et al. 1998; Nanda et al. 2000; Bellott 2010). The latter was confirmed for genes in linkage group 1, while linkage group 2 was found to be syntenic to human chromosomes 3, 6, and 8 (supplementary table S5, Supplementary Material online). By the same token, while linkage group 1 was syntenic to chromosome Z of chicken and other birds, linkage group 2 was syntenic to chicken chromosomes 3 and 12. Linkage group 2 corresponds to the last 12 Mb of superscaffold 54 (scaffolds 816, 79, 289, 179, and 347 in the ostrich assembly prior to optical mapping). Based on linkage and synteny information, we consider this segment to most likely represent an assembly error and therefore removed linkage group 2 from further analysis.
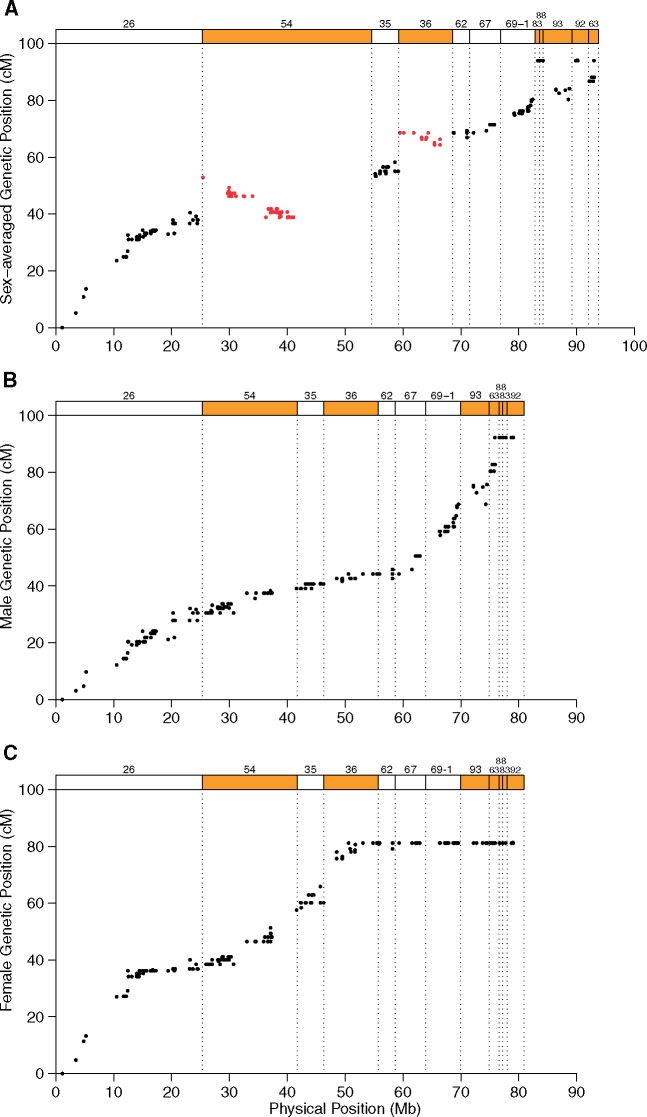
Figure 2A shows the genetic position of markers from linkage group 1 in relation to their location on the ostrich Z chromosome assembly with superscaffolds ordered and oriented as in (Zhang et al. 2015). Note that the segment of superscaffold 54 corresponding to linkage group 2 is evident as a large genomic region without genetic markers. A comparison of the genetic and physical maps revealed three additional assembly errors. The orientation of superscaffolds 36 and 54 (red markers in fig. 2A) are both inverted. Moreover, the order of superscaffolds in the PAR end of the chromosome was reported as 83, 88, 93, 92, and 63 (Zhang et al. 2015; orientation towards chromosome end), but linkage analysis clearly ordered these 93, 63, and 92, with 88 (624 kb) and 83 (782 kb) inseparable from 92.
Fig. 2.
—Genetic position of markers as a function of the physical position on the ostrich Z chromosome. (A) Sex-averaged genetic positions before correction of the physical assembly. Top rectangles represent superscaffolds numbered, ordered and oriented according to (Zhang et al. 2015). Red symbols represent markers from incorrectly assembled scaffolds. (B) Male and (C) female genetic positions with superscaffolds corrected for chimaerism, order, and orientation. Scaffold borders are indicated by dashed lines in all panels.
In figure 2B and C, the relationship between genetic and physical position is shown with the Z chromosome assembly corrected based on the new linkage data. It is clear that linkage group 2 does not reside on the Z chromosome (markers flanking the segment of superscaffold 54 corresponding to linkage group 2 are tightly linked), providing independent validation for the removal of the corresponding segment from the ostrich Z chromosome assembly.
The Z chromosome linkage map spans 92.3 cM in males and 81.2 cM in females (fig. 1). A comparison of the relationship between genetic and physical position in males and females reveals the location of the PAR as the region in which both sexes recombine. It begins in one of the ends of the chromosome (position 0) and extends to ∼52 Mb (fig. 2B and C). In general, the recombination rate in a PAR is expected to be high because of an obligate cross-over in the heterogametic sex, and hence higher in the heterogametic than in the homogametic sex, at least if the sex chromosomes are highly differentiated and the PAR is small. The ostrich PAR is (very) much larger than in many other birds (Smeds et al. 2014; Zhou et al. 2014; Bellott et al. 2017), representing about 62% of the chromosome. Still, there was a much higher PAR recombination rate in females (1.53 cM/Mb) than in males (0.83 cM/Mb). A higher recombination rate in females is particularly observed in a 10 Mb region proximate to the PAR boundary (females: 2.11 cM/Mb vs males: 0.52 cM/Mb, Wilcoxon rank sum test for 200 kb windows: P = 1.32 × 10−6) (fig. 1B and C). When recombination is concentrated to a small PAR, it may affect other genomic features that are related to recombination. We compared gene density (1 Mb windows), repeat density and GC content (50 kb window) between the PAR and the nonrecombining segment (Table 1). Only repeat density differed significantly between the two regions, but the difference was not large (supplementary fig. S5, Supplementary Material online).
Table 1.
Density (mean ± SD) of genomic features in the recombining (PAR) and nonrecombining region of the ostrich Z chromosome
| Element | PAR | Nonrecombining region | Wilcoxon Signed-ranked P value |
|---|---|---|---|
| Gene density | 0.241 ± 0.134 | 0.20 ± 0.135 | 0.16 |
| Repeat density | 0.054 ± 0.031 | 0.071 ± 0.042 | 7.14 × 10−15 |
| GC density | 0.399 ± 0.037 | 0.395 ± 0.03 | 0.62 |
Inference of Inversions on Avian Z Chromosome
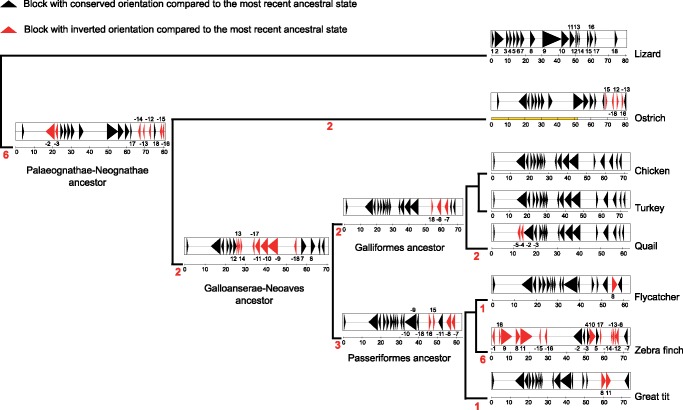
We used the improved ostrich Z chromosome assembly in an attempt to infer sex chromosome rearrangements (inversion events) during avian evolution. Specifically, we compared the order and location of orthologous genes from chromosome 2 of green anole lizard (outgroup to Aves; lizard chromosome 2 and the avian Z chromosome are homologous), and the Z chromosomes of ostrich (Paleognathae), chicken, turkey, quail, collared flycatcher, great tit, and zebra finch (Neognathae). Eighteen synteny blocks were identified, and 25 inversion events were inferred to have occurred across the avian phylogeny (fig. 3). In supplementary figs S6 and S7, Supplementary Material online, we report the inferred order of these events. Six inversions mapped to the long (unrooted) branch between the Paleognathae–Neognathae node and green anole lizard. The short, terminal zebra finch branch clearly had the highest rate of inversions, with six events since its relatively recent split from the flycatcher lineage. GRIMM analysis was further extended using baobabLUNA to obtain all possible inversion scenarios. Two inversion scenarios on the Palaeognathae–Neognathae ancestor and five scenarios on the zebra finch branch were reported. The rest of lineages had only one inversion scenario as reported by GRIMM (supplementary materials and methods, Supplementary Material online). We identified 17 breakpoints with a median size of 9.5 kb between ostrich and chicken. Analyzing the genomic features of breakpoints revealed no significant difference in repeat density or GC content with the rest of the chromosome (data not shown).
Fig. 3.
—Overview of inferred inversion events on the Z chromosome across avian lineages. Each triangle represents a synteny block, which are regions with conserved gene order across the phylogeny. Blocks are numbered according to their order in lizard, thought to represent the organization in a common lizard-bird ancestor. Numbers in red on each branch indicate the number of inferred inversion events in that lineage. A coloring (orientation) and numbering (order) scheme is used to denote a change in synteny blocks. Synteny blocks retaining the orientation of the most recent ancestor in the phylogeny are shown in black, while those that have been inverted compared to the immediate ancestor are shown in red. The numbering scheme represents the position of the block compared to lizard. Minus signs denote that a block has the opposite orientation compared to lizard. The position and length of synteny blocks in ostrich is used to depict the Palaeognathae-Neognathae ancestor, while data from chicken are used for the Galloanserae-Neoaves and Galliform ancestors, and data from flycatcher are used for Passeriform ancestor; in each case a ruler below the chromosome shows the position in Mb. The yellow ruler under ostrich Z indicates the PAR. Note that the gap between blocks is due to lack of homology between lizard chromosome 2 and avian species in those segments.
In the absence of a chromosome-level assembly for other reptiles than the green anole lizard, we cannot directly map the six inversion events separating lizard and birds onto the phylogeny, and hence not infer the organization of the Z chromosome in an early avian ancestor. However, we analyzed scaffold-level assemblies of two other outgroup species, painted turtle and alligator, by aligning orthologous genes from each of these species to lizard chromosome 2 (supplementary fig. S8, Supplementary Material online). Lizard chromosome 2 is colinear with turtle and alligator scaffolds across most of its length—that is colinear within scaffolds, which is at the level we can analyze. There were three minor exceptions, including small inversions at positions 10.7–11.9 Mb and 57.3–58.8 Mb (defined by lizard chromosome 2) between lizard and alligator, and at 61.7–62.7 Mb between lizard and turtle. All these regions are colinear between lizard and ostrich, and the events do therefore not affect the inference of inversions within the avian lineages. These small inversions must have happened in either of the outgroup lineages after the split from an early avian ancestor.
The high degree of conservation in gene order and orientation among lizard, turtle, and alligator in the genomic regions that correspond to the avian Z chromosome indicates that lizard has largely maintained the ancestral organization. In turn, this suggests that the six inversions separating lizard chromosome 2 and avian Z chromosomes occurred in an early avian ancestor prior to the split of Paleaognathae and Neognathae (fig. 3; “Palaeognathae–Neognathae ancestor”). Of these six, two map to an interval between 16.4–22.8 Mb of the ostrich Z chromosome, which is in the still recombining region (PAR). The other four inversions map to the nonrecombining region, between 62.1 and 80.9 Mb, and partly overlap with each other. The remaining two inversions separating lizard and ostrich occurred in the Palaeognath lineage subsequent to the split between Paleaognathae and Neognathae. Both these inversions map to the nonrecombining segment.
In the Neognath lineage subsequent to the split from Palaeognathae, a large inversion occurred in an early ancestor and is thus shared among all Neognath species included in the study (fig. 3; “Galloanserae-Neoaves ancestor”). Since these represent both lineages of the deepest divergence within Neognathae, that is, the split between Neoaves and Galloanserae, the inversion must have happened prior to this divergence. This large inversion maps to 25.5–62.1 Mb of the chicken Z chromosome. The remaining inversions in Neognathae occurred within the boundaries of this large inversion and have further shuffled the gene order and orientation in different avian lineages. For example, two such inversions occurred in a galliform ancestor (fig. 3; “Galliformes ancestor”), followed by two inversions in the quail lineage since its split from chicken and turkey; the latter two species are colinear. Three inversions occurred in the ancestral passeriform lineage, followed by one inversion in the flycatcher lineage and one in the great tit lineage, respectively.
As mentioned above, the Z chromosome has been particularly labile in the zebra finch lineage with six inversions since its split from other passeriform species. One inversion was at position 0.4–1.1 Mb, a region that is otherwise colinear across all other birds and in lizard. The breakpoints of the other five inversions overlapped with one another, with two between 3.9 and 4.7 Mb and three between 65.9 and 67.4 Mb of zebra finch Z chromosome. The rate of intrachromosomal rearrangements in zebra finch autosomes has also been found to be higher than in other bird species (Kawakami et al. 2014).
Discussion
Multiple inversions have occurred on the Z chromosome during avian evolution. This means that a perfect correlation between the degree of divergence of gametologs and their position along the Z chromosome cannot be expected if arrest of recombination has progressed towards the PAR. This makes the interpretation of steps in avian sex chromosome evolution difficult and contrasts to the situation on the human X chromosome where five strata are positioned along the chromosome in order of emergence, with the most recently evolved stratum closest to the PAR (Lahn and Page 1999; Ross et al. 2005). But there are also additional challenges. The resolution in the definition of strata and their boundaries on the avian Z chromosome is low since divergence data is only available for relatively few gametologs (Wright et al. 2012; Zhou et al. 2014; Smeds et al. 2015). Moreover, the degree of divergence estimated as the rate of synonymous substitutions between gametologs does not differ much, or even overlap, among genes located in regions of the Z chromosome postulated to represent different strata (Wright et al. 2012; Zhou et al. 2014). Furthermore, gene trees used to date (relative to the species tree) the cessation of recombination between gametologs have not consistently been presented with strong statistical support (Wright et al. 2014; Zhou et al. 2014; Smeds et al. 2015). With a corrected Z chromosome assembly of ostrich at hand, we revisit the scenario for formation of avian sex chromosomes. Our work adds to the findings recently presented by Zhou et al. (2014) by inferring ancestral states of avian sex chromosome organization, by identification of additional inversion events and by inferring the order and phylogenetic origin of inversion events during avian sex chromosome evolution.
The fact that all birds (but no outgroup lineages) share the same sex chromosomes is a strong indication that all birds also share the same mechanism for sex determination. In turn, this suggests that the avian sex chromosomes started to evolve in an early ancestor of modern birds subsequent to the split from lizards. The Z-linked DMRT1 gene is a likely candidate for being the master regulator of avian sex determination via dosage (Smith et al. 2009; Hirst et al. 2017); birds do apparently not possess a dominant female-determining on the W chromosome, as would have been analogous to the dominant role of Sry on the mammalian Y chromosome. In ostrich, DMRT1 is located at 76.2 Mb, near the chromosome end at 80.9 Mb. One or more of the four Z chromosome inversions that we mapped to the Palaeognathae–Neognathae ancestor lineage in the region corresponding to 62.1–80.9 Mb of the ostrich Z chromosome thus encompassed DMRT1. This event/s may have selected for arrest of recombination between DMRT1 and a sexually antagonistic locus (loci) on the Z chromosome, thereby representing the onset of avian sex chromosome evolution. Whether DMRT1 acted as an autosomal master regulator of sex determination in an early avian ancestor prior to the emergence of Z and W chromosomes is not known and would seem difficult to unveil.
However, we cannot formally demonstrate that a Z-linked inversion/s was responsible for the initial cessation of recombination between avian sex chromosomes. An alternative possibility would be, for example, that an inversion/s occurred on the W chromosome, and these two scenarios are not mutually exclusive. Nor can we say how large genomic region ceased to recombine in the first place and that is irrespective of whether a Z-linked or W-linked inversion, or another mechanism, triggered the process of sex chromosome differentiation. Generally, it is difficult to infer (old) rearrangements on the nonrecombining region of the W chromosome since it is highly degenerated in most avian lineages and since a physically anchored W chromosome assembly is only available for chicken (Bellott et al. 2017).
The 62.1–80.9 Mb segment of the ostrich Z chromosome should correspond to what Zhou et al. (2014) refer to as avian sex chromosome stratum 0. For reasons mentioned above, we do not think it can be taken for certain that the region represents a single stratum, or even that it was subject to discrete rather than gradual recombination arrest. Only two gametologous genes present in the 62.1–80.9 Mb segment have been reported in ostrich, PCGF3Z/W and KCMF1Z/W (Cortez et al. 2014; Yazdi and Ellegren 2014; Zhou et al. 2014). This limited number is consistent with an ancient origin of cessation of recombination in this region since nonrecombining genes tend to gradually degenerate and, in most cases, eventually become lost. In sharp contrast, the density of “surviving” gametologs in the remaining part of the ostrich Z chromosome that do not recombine with the W chromosome (52–62 Mb; Paleognath stratum S1 in Zhou et al. 2014) is much higher, or at least 36 genes in ≈10 Mb. An inverse relationship between the density of gametologs and the age of sex chromosome strata was reported for the human X chromosome (Lahn and Page 1999). No Z chromosome inversions have occurred in 52–62 Mb interval in the ostrich lineage, meaning that cessation of recombination in this region was either prompted by a W chromosome inversion/s or by another mechanism.
A large inversion corresponding to positions 25.5–62.1 Mb of the chicken Z chromosome (i.e. about half of the chromosome) occurred in the Neognath ancestor. Since all investigated Neognath species have a very small PAR, this inversion event constitutes a candidate for significantly extending the nonrecombining region of the W chromosome and forming Neognath stratum S1 (note that this is different from Paleognath stratum S1). If so, this would have left Z-W recombination to only occur in the first 25 Mb of the Z chromosome. Similar to above, however, we cannot exclude that one or more inversions on the W chromosome gave rise to Neognath S1. In any case, the large Z chromosome inversion in a Neognath ancestor means that the ancestral organization of the avian Z chromosome has not been retained and that evolutionary strata may not be linearly ordered according to time of emergence along the chromosome. Several additional Z chromosome inversions within the nonrecombining region that map to internal or terminal lineages of the six Neognath species investigated reinforce this conclusion.
In the above, we have not integrated information about divergence times between gametologs estimated from synonymous substitution rates or phylogenetic analyses of gene trees vs species trees for inferring when recombination between gametologs stopped. Such information has led previous studies to discriminate between additional strata and/or refine the location of strata on the avian Z chromosome (Handley et al. 2004; Wright et al. 2012; Wang et al. 2014; Zhou et al. 2014). While those conclusions may very well be correct, in our view, divergence/phylogenetic data have in some cases not provided unequivocal support for the conclusions made. For example, there is inconsistency among different studies concerning the gametologous gene HNRNPKZ/W (at position 40.07 Mb on the chicken Z chromosome), which has been inferred to be located in galliform-specific stratum III (Wright et al. 2014) as well as in Neognathae S1 (Zhou et al. 2014) and in the oldest stratum emerging in the ancestor of modern birds (Cortez et al. 2014).
The inference of inversions made in this study relied on the use of a genetic map for correction of the physical assembly of the ostrich Z chromosome. Of course, a genetic map is not necessarily error-free. However, we used well-established LOD score thresholds for ordering of markers. Another aspect is that the inference of inversions involved two critical assumptions. The first is the use of simple parsimony principles for building a scenario of series of inversion events leading to the current organization of the Z chromosome in different avian species. However, when several inversions are needed to explain the organization, two or more scenarios are equally likely. Second, we only considered chromosomal rearrangements in the form of inversions. It is possible that a more complex scenario of different types of rearrangements could have taken place. For example, the repositioning of DMRT1 in Neognathae could either be explained by a series of inversions or by a translocation.
Conclusions
Our results highlight the importance of integrating genetic or physical mapping in assembly processes for making reliable inferences of chromosomal evolution. Even though the ostrich Z chromosome assembly prior to our work could be considered to be of good quality given the use of optical mapping, we still found several errors. These included a chimeric superscaffold as well as incorrectly ordered and incorrectly oriented scaffolds. The inverted blocks 12, 13, and 14 in the Galloanserae-Neoaves ancestor (see fig. 3) could only be observed after the correction of disoriented superscaffold 54.
To conclude, our results revealed a complex evolutionary history with multiple inversions during avian sex chromosome evolution. A number of Z chromosome inversions in different avian lineages can explain a ‘scrambled eggs’ pattern of evolutionary strata that are not linearly ordered along the Z chromosome. Some of these inversions may also have been the cause of recombination suppression. Since we also found regions that stopped recombining without the involvement of Z chromosome inversions, avian sex chromosome evolution must also have included W chromosome inversions, or other mechanisms.
Supplementary Material
Acknowledgments
Charlie Cornwallis is kindly acknowledged for access to ostrich DNA samples. We are also grateful to Anel Engelbrecht, Zanell Brand, Maud Bonato, and Schalk Cloete at the Western Cape Department of Agriculture, Elsenburg, South Africa for assistance with providing samples. This study was financed by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. Genotyping was made at the SNP & SEQ Technology Platform, Uppsala University.
Literature Cited
- Adolfsson S, Ellegren H.. 2013. Lack of dosage compensation accompanies the arrested stage of sex chromosome evolution in ostriches. Mol Biol Evol. 30(4):806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Backström N, Ceplitis H, Berlin S, Ellegren H.. 2005. Gene conversion drives the evolution of HINTW, an ampliconic gene on the female-specific avian W chromosome. Mol Biol Evol. 22(10):1992–1999. [DOI] [PubMed] [Google Scholar]
- Baudet C, et al. 2010. Cassis: detection of genomic rearrangement breakpoints. Bioinformatics 26(15):1897–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becak W, Becak ML, Nazareth HR.. 1962. Karyotypic studies of two species of South American snakes (Boa constrictor amarali and Bothrops jararaca). Cytogenetics 1:305–313. [PubMed] [Google Scholar]
- Bekaert M. 2016. Genetic-Mapper: vectorial genetic map drawer. F1000Research. https://github.com/pseudogene/genetic-mapper, last accessed August 6, 2018.
- Bellott DW. 2010. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466(7306):612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, et al. 2017. Avian W and mammalian Y chromosomes convergently retained dosage-sensitive regulators. Nat Genet. 49(3):387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Charlesworth D.. 2009. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 24(2):94–102. [DOI] [PubMed] [Google Scholar]
- Bergero R, Forrest A, Kamau E, Charlesworth D.. 2007. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics 175(4):1945–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Qiu S, Forrest A, Borthwick H, Charlesworth D.. 2013. Expansion of the pseudo-autosomal region and ongoing recombination suppression in the Silene latifolia sex chromosomes. Genetics 194(3):673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Pevzner PA.. 2002. Genome-scale evolution: reconstructing gene orders in the ancestral species. Genome Res. 12(1):26–36. [PMC free article] [PubMed] [Google Scholar]
- Braga MD. 2009. baobabLUNA: the solution space of sorting by reversals. Bioinformatics 25(14):1833–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco S, et al. 2017. Evolutionary strata on young mating-type chromosomes despite the lack of sexual antagonism. Proc Natl Acad Sci USA. 114(27):7067–7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapus C, Edwards SV.. 2009. Genome evolution in Reptilia: in silico chicken mapping of 12, 000 BAC-end sequences from two reptiles and a basal bird. BMC Genomics 10(Suppl 2):S8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D.. 1978. A model for the evolution of dioecy and gynodioecy. Am Nat. 112(988):975–997. [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G.. 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95(2):118–128. [DOI] [PubMed] [Google Scholar]
- Chibalina MV, Filatov DA.. 2011. Plant Y chromosome degeneration is retarded by haploid purifying selection. Curr Biol. 21(17):1475–1479. [DOI] [PubMed] [Google Scholar]
- Choi K, Henderson IR.. 2015. Meiotic recombination hotspots—a comparative view. Plant J. 83(1):52–61. [DOI] [PubMed] [Google Scholar]
- Cortez D, et al. 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508(7497):488–493. [DOI] [PubMed] [Google Scholar]
- DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson AK, et al. 1998. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc Natl Acad Sci USA. 95(14):8147–8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno J, Mindell DP.. 2000. Rooting a phylogeny with homologous genes on opposite sex chromosomes (gametologs): a case study using avian CHD. Mol Biol Evol. 17(12):1826–1832. [DOI] [PubMed] [Google Scholar]
- Green P, Falls K, Crook S.. 1990. Documentation for CRIMAP, Version 2.4. St. Louis, Missouri: Washington Univ. School of Medicine. [Google Scholar]
- Griffin DK, Robertson LB, Tempest HG, Skinner BM.. 2007. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res. 117(1-4):64–77. [DOI] [PubMed] [Google Scholar]
- Handley LL, Ceplitis H, Ellegren H.. 2004. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics 167(1):367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst CE, et al. 2017. Sex reversal and comparative data undermine the W chromosome and support Z-linked DMRT1 as the regulator of gonadal sex differentiation in birds. Endocrinology 158(9):2970–2987. [DOI] [PubMed] [Google Scholar]
- Hough J, Hollister JD, Wang W, Barrett SCH, Wright SI.. 2014. Genetic degeneration of old and young Y chromosomes in the flowering plant Rumex hastatulus. Proc Natl Acad Sci USA. 111(21):7713–7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, et al. 2008. A preliminary microsatellite genetic map of the ostrich (Struthio camelus). Cytogenet Genome Res. 121(2):130–136. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Mizuno S.. 2002. Molecular and cytological characterization of SspI-family repetitive sequence on the chicken W chromosome. Chromosome Res. 10(6):499–511. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346(6215):1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, et al. 2014. A high-density linkage map enables a second-generation collared flycatcher genome assembly and reveals the patterns of avian recombination rate variation and chromosomal evolution. Mol Ecol. 23(16):4035–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC.. 1999. Four evolutionary strata on the human X chromosome. Science 286(5441):964–967. [DOI] [PubMed] [Google Scholar]
- Lemaitre C, et al. 2009. Footprints of inversions at present and past pseudoautosomal boundaries in human sex chromosomes. Genome Biol Evol. 1:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li J. 2009. An almost linear time algorithm for a general haplotype solution on tree pedigrees with no recombination and its extensions. J Bioinform Comput Biol. 7(3):521–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH, Taylor BA.. 1984. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc Natl Acad Sci USA. 81(3):814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K, Ellegren H.. 2008. The chicken (Gallus gallus) Z chromosome contains at least three nonlinear evolutionary strata. Genetics 180(2):1131–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I, et al. 2000. Conserved synteny between the chicken Z sex chromosome and human chromosome 9 includes the male regulatory gene DMRT1: a comparative (re)view on avian sex determination. Cytogenet Cell Genet. 89(1-2):67–78. [DOI] [PubMed] [Google Scholar]
- Natri HM, Shikano T, Merila J.. 2013. Progressive recombination suppression and differentiation in recently evolved neo-sex chromosomes. Mol Biol Evol. 30(5):1131–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. 1969. Linkage modifications and sex difference in recombination. Genetics 63(3):681–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M, et al. 2005. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol. 3(1):e4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1967. Sex chromosomes and sex-linked genes. Berlin (NY: ): Springer. [Google Scholar]
- Pevzner P, Tesler G.. 2003. Genome rearrangements in mammalian evolution: lessons from human and mouse genomes. Genome Res. 13(1):37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastas P, Calboli FCF, Guo B, Shikano T, Merilä J.. 2015. Construction of ultradense linkage maps with Lep-MAP2: stickleback F2 recombinant crosses as an example. Genome Biol Evol. 8(1):78–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. 1987. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex-chromosomes. Evolution 41(4):911–914. [DOI] [PubMed] [Google Scholar]
- Roesti M, Moser D, Berner D.. 2013. Recombination in the threespine stickleback genome–patterns and consequences. Mol. Ecol. 22(11):3014–3027. [DOI] [PubMed] [Google Scholar]
- Ross MT, et al. 2005. The DNA sequence of the human X chromosome. Nature 434(7031):325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowska J, Lagisz M, Nakagawa S.. 2012. The long and the short of avian W chromosomes: no evidence for gradual W shortening. Biol Lett. 8(4):636–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh Y, Saitoh H, Ohtomo K, Mizuno S.. 1991. Occupancy of the majority of DNA in the chicken W chromosome by bent-repetitive sequences. Chromosoma 101(1):32–40. [DOI] [PubMed] [Google Scholar]
- Sandstedt SA, Tucker PK.. 2004. Evolutionary strata on the mouse X chromosome correspond to strata on the human X chromosome. Genome Res. 14(2):267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty S, Griffin DK, Graves JA.. 1999. Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res. 7(4):289–295. [DOI] [PubMed] [Google Scholar]
- Smeds L, et al. 2014. Genomic identification and characterization of the pseudoautosomal region in highly differentiated avian sex chromosomes. Nat Commun. 5(1):5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeds L, et al. 2015. Evolutionary analysis of the female-specific avian W chromosome. Nat Commun. 6(1):7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, et al. 2009. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461(7261):267–271. [DOI] [PubMed] [Google Scholar]
- Stefos K, Arrighi FE.. 1971. Heterochromatic nature of W chromosome in birds. Exp Cell Res. 68(1):228–231. [DOI] [PubMed] [Google Scholar]
- Stock M, et al. 2011. Ever-young sex chromosomes in European tree frogs. PLoS Biol. 9(5):e1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, et al. 2015. ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol. 16(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesler G. 2002. GRIMM: genome rearrangements web server. Bioinformatics 18(3):492–493. [DOI] [PubMed] [Google Scholar]
- Van Laere AS, Coppieters W, Georges M.. 2008. Characterization of the bovine pseudoautosomal boundary: documenting the evolutionary history of mammalian sex chromosomes. Genome Res. 18(12):1884–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D.. 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 11(8):e1001643.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Kaiser VB, Bachtrog D.. 2013. Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution. Proc Natl Acad Sci USA. 110(16):6453–6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JP, et al. 2012. Sequencing papaya X and Y-chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc Natl Acad Sci USA. 109(34):13710–13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. 2014. Temporal genomic evolution of bird sex chromosomes. BMC Evol Biol. 14(1):250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Kitano J, Peichel CL.. 2015. Purifying selection maintains dosage-sensitive genes during degeneration of the threespine stickleback Y chromosome. Mol Biol Evol. 32(8):1981–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AE, Harrison PW, Montgomery SH, Pointer MA, Mank JE.. 2014. Independent stratum formation on the avian sex chromosomes reveals inter-chromosomal gene gonversion and predominance of purifying selection on the W chromosome. Evolution 68(11):3281–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AE, Moghadam HK, Mank JE.. 2012. Trade-off between selection for dosage compensation and masculinization on the avian Z chromosome. Genetics 192(4):1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdi HP, Ellegren H.. 2014. Old but not (so) degenerated–slow evolution of largely homomorphic sex chromosomes in ratites. Mol Biol Evol. 31(6):1444–1453. [DOI] [PubMed] [Google Scholar]
- Zhang JL, Li C, Zhou Q, Zhang GJ.. 2015. Improving the ostrich genome assembly using optical mapping data. Gigascience 4(1):24.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, et al. 2014. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 346(6215):1246338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.