Abstract
The hallmark of nonalcoholic steatohepatitis is hepatocellular inflammation and injury in the setting of hepatic steatosis. Recent work has indicated that dynamic 18F-FDG PET with kinetic modeling has the potential to assess hepatic inflammation noninvasively, while static FDG-PET is less promising. Because the liver has dual blood supplies, kinetic modeling of dynamic liver PET data is challenging in human studies. This paper aims to identify the optimal dual-input kinetic modeling approach for dynamic FDG-PET of human liver inflammation. Fourteen patients with nonalcoholic fatty liver disease were included. Each patient underwent 1-hour dynamic FDG- PET/CT scan and had liver biopsy within six weeks. Three models were tested for kinetic analysis: the traditional two-tissue compartmental model with an image-derived single-blood input function (SBIF), a model with population-based dual-blood input function (DBIF), and a new model with optimization-derived DBIF through a joint estimation framework. The three models were compared using Akaike information criterion (AIC), F test and histopathologic inflammation score. Results showed that the optimization-derived DBIF model improved liver time activity curve fitting and achieved lower AIC values and higher F values than the SBIF and population-based DBIF models in all patients. The optimization-derived model significantly increased FDG K1 estimates by 101% and 27% as compared with traditional SBIF and population-based DBIF. K1 by the optimization-derived model was significantly associated with histopathologic grades of liver inflammation while the other two models did not provide a statistical significance. In conclusion, modeling of DBIF is critical for dynamic liver FDG-PET kinetic analysis in human studies. The optimization-derived DBIF model is more appropriate than SBIF and population- based DBIF for dynamic FDG-PET of liver inflammation.
Keywords: liver inflammation, FDG, dynamic PET, kinetic modeling, dual-blood input function
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) affects approximately 30% of the general population and is emerging as a leading cause of liver-related morbidity and mortality (1,2). Nonalcoholic steatohepatitis (NASH) is a more severe form of NAFLD characterized by hepatocyte inflammation and injury, and can subsequently lead to cirrhosis and associated liver cancer and liver failure (3). NASH develops in 5–10% of NAFLD patients (i.e., 5–10 million people in the United States) and is associated with higher liver-related mortality than is hepatic steatosis alone (4). Differentiation of NASH from simple fatty liver is essential for future patient management in NAFLD, particularly as newer therapies become available (3,5).
The diagnostic hallmark of NASH is hepatocellular inflammation and injury in the setting of hepatic steatosis (3). Imaging methods have been developed to quantify hepatic steatosis (6), e.g. ultrasonography (7), computed tomography (CT) (8) or magnetic resonance imaging (MRI) (9–11). Other methods such as magnetic resonance elastography (MRE) (12) and ultrasound elastography (13) can measure liver stiffness and quantify advanced fibrosis of hepatic tissue (14–16). However, there is currently no effective imaging method for accurate characterization of liver inflammation in clinical practice and clinical trials (17). Liver biopsy followed by clinical histopathology remains the current standard practice (18).
18F-fluorodeoxyglucose (FDG) positron emission tomography (PET) is a widely used and effective method for imaging cell glucose metabolism. Previous studies have reported the use of FDG-PET for imaging infection and inflammation (19,20). Several studies have investigated the use of FDG-PET in the liver (21–27) with focus on evaluation of hepatic steatosis (24,26–32). Contradictory results have been reported (24,26–29) on the relation between FDG standardized uptake value (SUV) and hepatic steatosis. Keramida et al (30–32) have recently pointed out that SUV, if corrected for liver fat content, is increased in hepatic steatosis and may potentially relate to liver inflammation. However, no direct findings have been reported between FDG-PET and hepatic inflammation so far.
Current clinical usage of PET is largely limited to static PET imaging which provides SUV as a semi-quantitative measure of glucose utilization. SUV is related to steatosis (30) and is influenced by many patient factors including body habitus (33). This static way of using PET may not be able to explore the full potential of FDG-PET in imaging of hepatic inflammation. In contrast, dynamic FDG-PET acquires FDG activity images at multiple time points and can exploit quantitative tracer kinetics to characterize and separate the underlying molecular process of glucose metabolism (34). There are already several dynamic FDG-PET studies of liver, e.g., (21,23,32,35–37). However, existing studies mainly examined the overall glucose utilization using either glucose metabolic rate or FDG influx rate Ki. None of these studies have reported relevant or promising results for liver inflammation and NASH assessment. Our recent preliminary work has indicated a new promise (38). While we have not found FDG SUV by static PET or Ki by dynamic PET to be helpful, the FDG K1 parameter, which represents the delivery rate of FDG from blood to hepatic tissue, is promising for characterizing liver inflammation in NASH (38).
The blood input function is essential for kinetic modeling in dynamic FDG-PET (34). Conventionally it is either obtained by invasive arterial blood sampling or noninvasively derived from the left ventricle or aortic regions in dynamic PET images (39–41). In the liver, both the hepatic artery and portal vein provide blood supply to the hepatic tissue (22). The portal vein input is not the same as the hepatic artery input and is difficult to obtain in human subjects (22). Neglecting the influence of portal vein can result in inaccurate kinetic modeling. A common approach to accounting for the effect of dual blood supplies is to use a flow-weighted model of dual-blood input function (DBIF) (21,36,42). The model parameters are often pre-determined by population means that were derived using arterial blood sampling in animal studies (22,37). Due to heterogeneity across patients and potential differences between arterial and image-derived input functions, the population-based DBIF model may become ineffective when arterial blood sampling is not available and in practice, image-derived input function is typically used in human studies.
The objective of this paper is to evaluate and identify an appropriate dual-input kinetic modeling approach for FDG kinetic quantification of liver inflammation using human patient data. Besides the population-based DBIF approach, we also examined a new optimization-derived DBIF model which employs mathematical optimization to jointly estimate the parameters of DBIF and liver FDG kinetics. This model directly utilizes image-derived aortic input function, requires no arterial blood sampling, and is more adaptive to individual patients. We used both statistical information criteria and histopathologic inflammation data to compare these methods.
MATERIALS AND METHODS
Patient Characteristics
Fourteen patients with NAFLD were included in this study. These patients had a liver biopsy as a part of routine clinical care or for enrollment in clinical trials. The University of California Davis Institutional Review Board and the University of California Davis Medical Center Radiation Use Committee approved the study. All patients signed informed consent before participating in the study. Patients with history of alcohol abuse, chronic hepatitis B or C, or other chronic liver disease other than NAFLD were excluded from the study.
Liver Histopathology
Liver biopsies were performed under ultrasound guidance and scored according to the nonalcoholic steatohepatitis clinical research network (NASH-CRN) criteria (43). The NAFLD activity score (NAS, range 0–8) is comprised of severity of steatosis, inflammation, and hepatocellular ballooning. While a NAS score greater than 4 has been reported to correlate with the presence of NASH, the scores of lobular inflammation and ballooning degeneration are noted to represent hepatic inflammation and injury, and are therefore combined to create an overall “hepatic inflammation” score (range 0–5). In this study, an inflammation score higher than 3 was considered indicative of high inflammation, while scores lower than 3 and equal to 3 suggested low and medium inflammation, respectively.
Dynamic 18F-FDG PET/CT
Scan Protocol:
Dynamic 18F-FDG PET studies were performed using the GE Discovery 690 PET/CT scanner at the UC Davis Medical Center. Patients fasted for at least 6 hours. Each patient was injected with 10 mCi 18F- FDG. List-mode time-of-flight data acquisition started right after the intravenous bolus administration and lasted for one hour. At the end of PET scan, a low-dose CT scan was performed for attenuation correction for PET. Dynamic PET data were binned into 49 time frames using the sampling schedule: 30 × 10s, 10 × 60s, and 9 × 300s. Dynamic PET images were then reconstructed using the vendor software with the standard ordered subsets expectation maximization (OSEM) algorithm with 2 iterations and 32 subsets.
Extraction of Blood and Tissue Time Activity Curves (TACs):
Eight spherical regions of interest (ROI), each with 25 mm in diameter, were placed on the eight segments of the liver avoiding any major blood vessels. These ROI placements were tuned and confirmed by an abdominal radiologist. The averaged FDG activity in all 8 ROIs was extracted from the dynamic images to form a global liver TAC of low noise. An additional ROI is placed in the descending aorta region to extract image-derived aortic input function.
Dual-Input Kinetic Modeling
Compartmental Model with Dual-blood Input Function (DBIF):
FDG kinetics commonly follow the two-tissue compartmental model (34) as shown in Figure 1(a). Glucose transporters transport 18F-FDG from blood to hepatic tissue with the rate constant K1 (mL/min/mL) and from hepatic tissue to blood with the rate k2 (1/min). FDG is phosphorylated by hexokinase in cells into FDG 6-phosphate with the rate k3 (1/min) and the dephosphorylation process occurs with the rate k4 (1/min). Cp(t), Cf(t) and Cm(t) represent the FDG concentration in the plasma compartment, free-state FDG in the hepatic tissue compartment, and metabolized FDG 6-phosphate in the tissue, respectively. In traditional kinetic modeling with single-blood input function (SBIF), only blood supply from the aorta is considered and thus the input function Cp(t) is equivalent to the aortic input function CA(t). To account for the effect of dual blood supplies in the liver, a flow-weighted sum of the aortic input CA(t) and portal vein input CPV(t) can be used to model the DBIF (22):
| Eq. 1 |
where fA is the fraction of hepatic artery contributing to the overall liver blood flow. The portal vein input function CPV(t) follows an analytical solution
| Eq. 2 |
which is equivalent to a dispersed version of the aortic input function CA(t) (44). The dispersion is because FDG in the portal vein flows through the gastrointestinal system before entering into the liver. Thus, portal vein can be considered as an additional compartment, as shown in Figure 1(b). As a result, the combined compartmental model is equivalently described by a set of differential equations:
| Eq. 3 |
where
| Eq. 4 |
The total activity that can be measured by PET is the sum of different compartments:
| Eq. 5 |
where vb is the fractional blood volume and θ = [vb,K1,k2,k3,k4,ka,fA]T is a vector collecting all unknown parameters. Note that this DBIF model becomes the traditional SBIF model when ka = 0 and fA = 1.
FIGURE 1:
Kinetic modeling of dynamic liver FDG-PET data with a dual-blood input function from the hepatic artery and portal vein.
Joint Estimation of Kinetic and Input Parameters:
All model parameters are jointly estimated by fitting a measured liver TAC č using the model equation and following nonlinear least square estimation:
| Eq. 6 |
where WRSS(θ) denotes the weighted residual sum of squares of the curve fitting and wm denotes the weighting factor for time frame m. In this study, a uniform weight was used as suggested by Thiele and Buchert (45) and Winterdahl et. al (37). We used the classic Levenberg-Marquardt algorithm with 100 iterations to solve the optimization problem. The fitting process was implemented using C/C++ programing similar to our previous work (46,47). In this paper, we refer this joint estimation as the optimization-derived DBIF approach for clarity. It would become equivalent to the population-based DBIF if the parameters fA and ka are assigned with fixed population means (if known). Based on our initial analysis of the patient data sets, we set the initials of kinetic parameters to vb = 0.01, K1 = 1.0, k2 = 1.0, k3 = 0.01, k4 = 0.01, ka = 1.0, fA = 0.01.
Comparison of Kinetic Models
We compared three input models: the traditional model with SBIF, a model with population-based DBIF and a model with optimization-derived DBIF. In this study, the SBIF was derived from the descending aorta region in dynamic FDG-PET images. To utilize reported population means for the population-based DBIF, we used the following portal vein input model (22,37)
| Eq. 7 |
which is very similar to the exponential-based model in Eq. 2 but not easily integrated into the differential equations for joint estimation. and fA = 0.25 were previously reported for FDG (37).
Comparison Using Statistical Criteria:
The three models were compared using two established statistical criteria for model selection for TACs: the corrected Akaike information criterion (AIC) and the F test (48,49). The AIC is defined by
| Eq. 8 |
where n denotes the total number of unknown parameters. Note that n=5 for the SBIF and population-based DBIF models and n=7 for the optimization-derived DBIF model. Here AIC was corrected for finite sample sizes due to the ratio . A lower AIC value indicates a better selection of model (50).
The F test compares a complex model 2 with a simple model 1 using
| Eq. 9 |
where the degree of freedom (number of unknown parameters) n2 > n1 larger F value indicates better fit. If the p value of F test is 0.05 or less, the model 2 is then considered significantly better than model 1.
Evaluation Using Histopathological Inflammation Data:
Patients in this study were divided into three groups according to their histopathological inflammation scores: low inflammation (<3), medium inflammation (=3), and high inflammation (>3). To examine the capability of FDG kinetic parameters for differentiating different inflammation groups, a two-sample 2-sided t-test at the 0.05 level was used to test the difference of group means and the Mann-Whitney U test was used to test difference of group medians. Other FDG-PET parameters that were reported to be associated with hepatic steatosis were also examined for their potential association with inflammation. These parameters include SUV, fat-adjusted SUV (31), and the metabolic rate of glucose by Keramida’s Patlak analysis (32). The SUV was calculated using the last frame (55–60 minutes) in the dynamic sequence. All statistical analyses were conducted using MATLAB (Natick, MA). P-values less than 0.05 were considered as statistically significant in this study.
RESULTS
Patient Characteristics
A majority of the patients were white (75%) while 25% were Hispanics. Female patients formed 67% of the cohort with 75% of the patients between the ages 40–70 years and 25% between 18–39 years. The mean BMI was 34±6 kg/m2. The mean fasting glucose prior to PET scan was 115±33 mg/dL. The patient population had an equitable spread across NAS score (≥ of 58%). Two thirds of patients had hepatic inflammation score of ≥3.
Demonstration of dual-blood input function
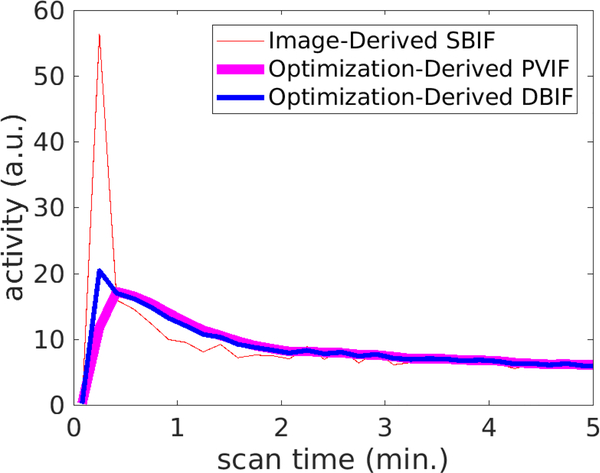
Figure 2 demonstrates an optimization-derived DBIF from a patient data set. Only the first 5 minutes are shown in the figure and the curves after 5 minutes are very similar among others. The DBIF is a weighted combination of the image-derived SBIF and the PVIF derived from the optimization. In this example, the weighting factor fA was 0.19. The DBIF has a much lower peak value than the SBIF because the contribution from hepatic artery is small (19%) while the contribution from portal vein is great (81%).
FIGURE 2:
Example of optimization-derived DBIF and its components from the hepatic artery and the portal vein: image-derived SBIF and portal vein input function (PVIF).
Figure 3 compares the optimization-derived PVIF with two image-derived PVIFs. To measure the PVIF, a spherical ROI was manually placed within the region of the portal vein on the CT image by a radiologist. Both ROI mean and ROI max were used to derive the image-derived PVIF, respectively. The PVIF by ROI mean has lower noise than that of ROI max but has a notable decrease in activity quantitation, which can be explained by the effect of partial volume. The PVIF by ROI max is consistent with the optimization-derived PVIF but the former is compromised by substantial noise, especially in early frames.
FIGURE 3:
Comparison of optimization-derived PVIF minutes; Right: Zoom-in of (A) for the first 5 minutes.
Evaluation of TAC Fit Quality
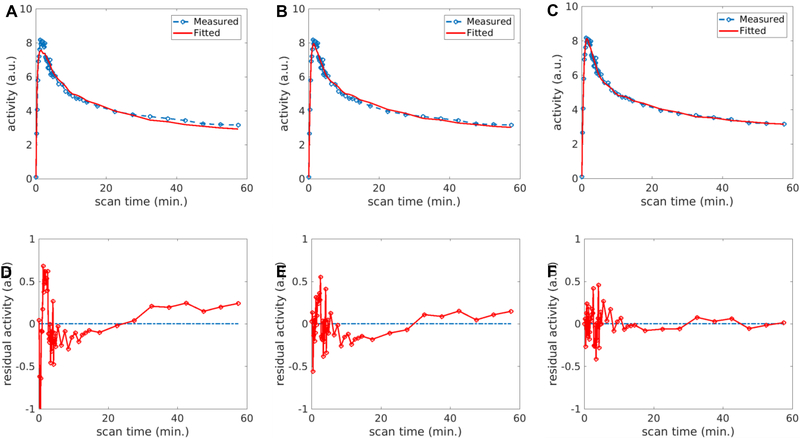
Figure 4A–4C show the fittings of the liver TAC using the image-derived SBIF, population-based DBIF and optimization-derived DBIF, respectively. The residual plots of these fits are shown in Figure 4D–4F. The fitting with SBIF could not fit the early-time peak and late time points due to lack of the portal vein component in the input function. The population-based DBIF provided an improved fit of the peak but still suffered error for late time points due to inaccuracy of the population means for a specific patient data set. A linear trend was observed in the residual plots (Figure 4D and4E) for these two approaches, indicating a systematic bias in the fitting. The optimization-derived DBIF estimated the input parameters from the data and fit both the peak at early time and late time points closely. The linear trend was removed and the residuals became asymptotically normal.
FIGURE 4.
Fit of a liver TAC using different input models: (A) SBIF, (B) population-based DBIF, (C) optimization-derived DBIF; (D-F) corresponding residual plots of the fits in (A-C).
The AIC values of the three fittings were −81, −140 and −159, respectively. AIC values of the three input models are further compared in Figure 5 for all individual patients in the cohort. The AIC difference was −105±23 between SBIF and optimization-derived DBIF, and −59±27 between population-based DBIF and optimization-derived DBIF, respectively. The optimization-derived DBIF model had the lowest AIC amongst the three models in all patients.
FIGURE 5:
AIC values of liver TAC fitting with different input function models.
The results of F test are given in Table 1. The minimum F value among different patients was 80.5 for comparing the optimization-derived DBIF with SBIF and 13.4 for comparing the optimization-derived DBIF with population- based DBIF model, both greater than the F critical value 3.2 calculated with n1 = 5 and n2 = 7 for a p value of 0.05. The p values of F test in individual patients are all small (<0.0001), indicating the optimization-derived DBIF is more appropriate for TAC fitting than the traditional SBIF and population-based DBIF models.
Table 1:
F statistics of model comparison (F critical value is 3.2 at the significance level of 0.05)
| Model Comparison | F values |
p values | ||
|---|---|---|---|---|
| Mean±SD | Min | Max | ||
| Optimization-Derived DBIF vs SBIF |
200.2±102.7 | 80.5 | 389.9 | <0.0001 |
| Optimization-Derived DBIF vs Population-Based DBIF |
67.5±43.7 | 13.4 | 137.7 | <0.0001 |
Change in Kinetic Parameters
The three input models resulted in different estimates in kinetic parameters. The mean and standard deviation of kinetic parameters vb,K1,k2,k3,k4 and Ki = 𝐾1𝑘3/(𝑘2+𝑘3) estimated by the three approaches are listed in Table 2. Results of fA and ka are also included. It is worth noting that a reversible kinetic model (i.e., k4 ≥ 0) was required for accurately modeling FDG TACs in the liver due to possible high level of glucose-6-phosphatase, as discussed in the literature (21,36,51,52). Neglecting k4 in the model reduced TAC fit quality with higher AIC values (results not shown). Compared with the image-derived SBIF, the optimization-derived DBIF significantly increased the mean values of vb (0.0000 vs 0.0229), K1 (0.5112 vs 0.9829), k2 (0.4983 vs 1.1053), k3 (0.0008 vs 0.0141) and Ki (0.0008 vs 0.0119), all with p<0.0001 .The difference in K1 in individual patients was 101% on average with a maximum of 150%, as shown in Figure 6. Compared with the population-based DBIF, the optimization-derived DBIF significantly increased the mean values of k3 (0.0011 vs 0.0141, p<0.0001), k4 (0.0196 vs 0.0534, p=0.0034), Ki (0.0010 vs 0.0119, p<0.0001) and vb (0.0003 vs 0.0229, p<0.0001). Although the mean values of K1 by the optimization-derived DBIF and population-based DBIF models are similar to each other (0.9787 vs. 0.9829), K1 values by the two models were very different in each individual patient (Figure 6). The change was 27% on average with a maximum of 44%.
Table 2:
Mean and standard deviation of FDG kinetic parameters estimated by different input function approaches
| SBIF | Population-Based DBIF |
Optimization-Derived DBIF |
|
|---|---|---|---|
| K1 | 0.5112±0.2064 | 0.9787±0.5509 | 0.9829±0.2730 |
| k2 | 0.4983±0.1760 | 0.9197±0.4222 | 1.1053±0.2488 |
| k3 | 0.0008±0.0010 | 0.0011 ±0.0011 | 0.0141±0.0070 |
| k4 | 0.0512±0.0465 | 0.0196±0.0291 | 0.0534±0.0263 |
| Ki | 0.0008±0.0010 | 0.0010±0.0011 | 0.0119±0.0051 |
| vb | 0.0000±0.0000 | 0.0003±0.0010 | 0.0229±0.0252 |
| fA | - | 0.25 | 0.0440±0.0544 |
| ka (or β) | - | 0.50 | 1.9002±1.2560 |
FIGURE 6:
FDG K1 values estimated by different input function models.
Association with Histopathologic Data
Figure 7 shows the associations of histopathologic grades of liver inflammation with FDG K1 by different input models. The standard boxplots reflect the median, 25% percentile and 75% percentile of K1 in each of low, medium and high inflammation groups. Overall, FDG K1 value decreased as inflammation grade increased. The results of statistical t test for comparing group means and U test for comparing group medians are summarized in Table 3. Neither the SBIF model nor the population-based DBIF model could differentiate low, medium and high inflammation groups (p>0.05). Because of the larger overlap between the low and high inflammation groups in K1 by the SBIF and population-based DBIF models, the p values of the t test were even higher than that of differentiating medium versus high inflammation groups. In comparison, K1 by the optimization-derived DBIF model was better associated with the inflammation grades and differentiated low versus high inflammation groups and medium versus high inflammation groups (p<0.05).
FIGURE 7:
Association of histopathological inflammation score with FDG-K1 estimated by different input models. (A) Image-derived SBIF; (B) Population-based DBIF; (C) Optimization-derived DBIF
Table 3:
P values of t test and U test for comparing different liver inflammation groups using FDG K1 estimated by SBIF, population-based (PB) DBIF, and optimization-derived (OD) DBIF.
| Group Comparison | T test |
U test |
||||
|---|---|---|---|---|---|---|
| SBIF | PB DBIF | OD DBIF | SBIF | PB DBIF | OD DBIF | |
| Low vs. medium inflammation |
0.2733 | 0.3158 | 0.3121 | 0.5556 | 0.5556 | 0.5556 |
| Medium vs. high inflammation |
0.0826 | 0.0941 | 0.0242 | 0.1508 | 0.1508 | 0.0317 |
| Low vs. high inflammation |
0.1206 | 0.1707 | 0.0116 | 0.0635 | 0.1905 | 0.0317 |
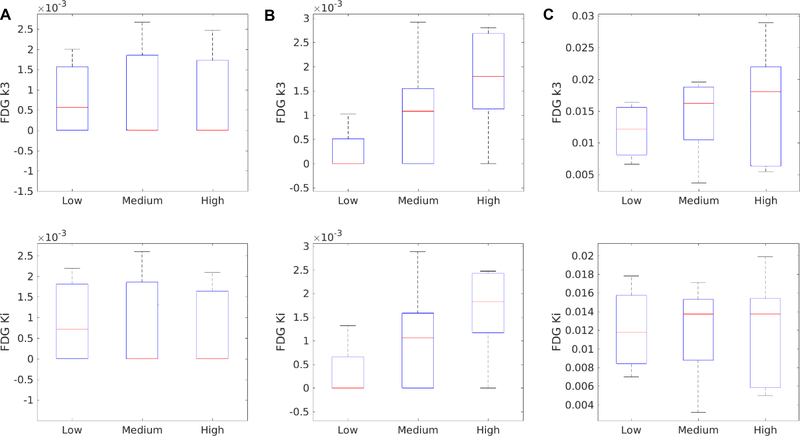
The capability of other kinetic parameters 𝑘2,3,𝑘4 and 𝐾𝑖 for differentiating different inflammation groups are summarized in Table 4. No kinetic parameter was able to differentiate inflammation groups, though k3 and Ki by the population-based DBIF tended to differentiate low versus high inflammation groups. The boxplots of k3 and Ki by different input models are shown in Figure 8. k3 by the population-based DBIF achieved statistical significance (p=0.0444) in the t test but not in the U test (p=0.0794), both p values larger than that of Ki. The optimization-derived DBIF did not improve the p values of k3 and Ki as compared with the population-based DBIF. This may indicate there is no inherent association between these kinetic parameters and liver inflammation, given that the former had better statistical fit quality than the latter as demonstrated by AIC (Figure 5) and F test (Table 2).
Table 4:
P values of T test and U test for comparing different liver inflammation groups using other FDG kinetic parameters estimated by SBIF, population-based (PB) DBIF, and optimization-derived (OD) DBIF.
| Group Comparison |
FDG Parameter |
T test |
U test |
||||
|---|---|---|---|---|---|---|---|
| SBIF | PB DBIF | OD DBIF | SBIF | PB DBIF | OD DBIF | ||
| k2 | 0.5894 | 0.5465 | 0.7848 | 0.7302 | 0.9048 | 1.0000 | |
| Low vs. medium | k3 | 0.9321 | 0.2755 | 0.5661 | 1.0000 | 0.2381 | 0.5556 |
| inflammation | k4 | 0.8121 | 0.2861 | 0.9427 | 1.0000 | 0.4603 | 1.0000 |
| Ki | 0.9367 | 0.3345 | 0.9509 | 1.0000 | 0.5556 | 0.9048 | |
| k2 | 0.1123 | 0.1320 | 0.1021 | 0.1508 | 0.3095 | 0.1508 | |
| Medium vs. high | k3 | 0.9355 | 0.3478 | 0.7661 | 0.8413 | 0.4127 | 0.6905 |
| inflammation | k4 | 0.9975 | 0.7663 | 0.7269 | 1.0000 | 1.0000 | 1.0000 |
| Ki | 0.8636 | 0.3862 | 0.9702 | 0.8413 | 0.4127 | 1.0000 | |
| k2 | 0.3112 | 0.3276 | 0.2042 | 0.7302 | 0.9048 | 0.2857 | |
| Low vs. high | k3 | 0.9955 | 0.0444 | 0.4885 | 1.0000 | 0.0794 | 0.5556 |
| inflammation | k4 | 0.8140 | 0.2223 | 0.7107 | 0.8095 | 0.2540 | 1.0000 |
| Ki | 0.7955 | 0.0580 | 0.9263 | 0.9683 | 0.0794 | 1.0000 | |
FIGURE 8:
Association of liver inflammation score with FDG k3 (top row) and Ki (bottom row) estimated by different input models. (A) Image-derived SBIF; (B) Population-based DBIF; (C) Optimization-derived DBIF.
The associations of liver inflammation grades with SUV, fat-adjusted SUV (31), and the metabolic rate of glucose by Keramida’s Patlak analysis (MRglu-KP) (32) are shown in Figure 9. The p values of t and U tests are reported in Table 5. No association was statistically significant. While SUV had no association with inflammation (p>0.5), the fat-adjusted SUV corrected by CT values (31) tended to increase in high-grade inflammation (p≈0.1). MRglu- KP was calculated using blood glucose level and the Patlak intercept in addition to the Patlak slope (32) and thus had a trend different from Ki in Figure 8. We also applied the correction factor of fat-adjusted SUV to obtain fat-adjusted K1, which however reduced the association between K1 and inflammation (Table 5).
FIGURE 9:
Association of liver inflammation score with different glucose utilization parameters. (A) Standard SUV; (B) Fat-adjusted SUV; (C) Metabolic rate of glucose by Keramida’s Patlak analysis (MRglu-KP).
Table 5:
P values of t test and U test for comparing in different liver inflammation groups using standard SUV, fat-adjusted SUV (fSUV), metabolic rate of glucose by Keramida’s Patlak analysis (MRglu-KP), and fat-adjusted K1 (fK1).
| Group Comparison |
T test |
U test |
||||||
|---|---|---|---|---|---|---|---|---|
| SUV | fSUV | MRglu- KP |
f K1 | SUV | fSUV | MRglu- KP |
fK1 | |
| Low vs. medium inflammation | 0.7678 | 0.7386 | 0.4543 | 0.3110 | 0.9048 | 0.9048 | 0.7302 | 0.4127 |
| Medium vs. high inflammation |
0.9709 | 0.1287 | 0.1139 | 0.1672 | 1.0000 | 0.2222 | 0.2222 | 0.3095 |
| Low vs.high inflammation |
0.7494 | 0.1069 | 0.3690 | 0.0827 | 0.7302 | 0.1111 | 0.5556 | 0.1111 |
DISCUSSION
Determination of liver inflammation in nonalcoholic fatty liver disease patients is crucial in differentiating serious NASH from simple hepatic steatosis. Dynamic FDG-PET with kinetic modeling has the potential to provide a noninvasive imaging biomarker for characterizing hepatic inflammation. Accurate kinetic modeling of dynamic liver FDG-PET data requires consideration of the effect of dual-blood supplies in the liver. Although the input function from the hepatic artery can be derived from the aorta in the dynamic images, it is difficult to derive the portal vein input function from dynamic PET because the size of portal vein (10–15mm) is small compared to the spatial resolution of clinical PET scanner (4–8mm). Partial volume effects, noise and respiratory motion all can contaminate the accuracy of the portal vein input function.
The standard SBIF model simply neglects the portal vein input which theoretically represents more than 80% of the total blood input. Neglecting the PVIF causes overestimation of the peak value of the input function and subsequently results in inaccurate estimation of FDG kinetic parameters. The K1 parameter, which reflects the scale between the blood input function and the tissue TAC (see Eq. 4 and Eq. 5), can hence be underestimated by the fitting process to compensate for the higher peak value of the SBIF. To account for the dual-blood supply effect, we applied the population-based DBIF model (21,36) in this study. It improved TAC fitting with lower AIC and higher F values than SBIF but did not improve association of the FDG biomarker with inflammation grades. This can be explained by the fact that the population model is not adaptive to individual objects and thus might not provide the optimal estimation.
The optimization-derived DBIF model estimated the PVIF adaptively and provided the best performance according to TAC fitting quality and association with histopathological inflammation grades. Because the parameters of the input model were jointly estimated with liver tissue kinetic parameters, the estimation was more adaptive to individual patients and achieved lower AIC and higher F values. In this study, the vb estimates were very small but increased by the new model, though the values were still smaller than expectation in the liver (36). This is possibly explained by the difference between image-derived input function and arterial blood input. Similarly, the fA estimates were far smaller than the population mean 0.25 that was reported based on arterial sampling. This may also be in part due to the capability of being adaptive to individual patient data. The resulting K1 parameter estimates were different from those by population-based DBIF. Both were higher than those by SBIF thanks to the correction on overestimation of the input function peak. Although there was no ground truth of K1 values for validating the estimates, evaluation of statistical information criteria and analysis of association of K1 with liver inflammation grades provided a feasible way to prove the improved performance of the modified DBIF model.
In addition to establishing the kinetic modeling method for dynamic FDG-PET characterization of liver inflammation, this work adds a new contribution to the methodology of dual-input kinetic modeling. We extended and validated the method of DBIF modeling using a joint optimization framework. Previously, Kudomi et al estimated both aortic input function and parameters of input model from multiple regional liver TACs in dynamic FDG-PET (42). Their method, however, is more complex for practical use with multiple steps involved. When the tracer distribution is uniform in the liver, TAC data would also be less helpful for the estimation of the many parameters in their model. Our method is simpler and can be implemented voxel-by-voxel to easily derive parametric images which will allow the future study of liver heterogeneity. The proposed method shares a similar spirit with the work by Chen and Feng (53) designed for modeling a different tracer 11C-acetate. The compartmental model used in (53) was irreversible while a reversible model was required in our study for FDG modeling. Rani et al also used a similar DBIF model for modeling FDG kinetics in rat liver (54). Their model uses a fixed fA and 4 free dispersion parameters (as opposed to 1 parameter in this work), which is more flexible but results in higher variability. Garbarino et al proposed a more complex model for mouse imaging by including the gut as another compartment (35). The model has 9 free parameters if fA is included and requires definition of the gut TAC. Thus, the current work has complemented existing studies to establish the optimization-derived DBIF modeling method. Our method can be easily implemented without additional operations.
Another notable aspect of this work is the validation of dual-input kinetic modeling using histopathological reference. All existing studies of dual-input kinetic modeling were only able to validate the DBIF approaches in animal studies (e.g., pigs (36,37), foxhounds (21), rodents (35,54)) or demonstrated the improvement using statistical TAC fitting quality in human patients (53). No studies had demonstrated an impact on physiological measurements by using a histopathological ground truth. In comparison, the present work provided direct evidence on the impact of DBIF on improving association of the FDG-PET biomarker with histopathological inflammation in human patients.
Using the optimization-derived DBIF model, we found that FDG K1 decreases as liver inflammation grade increases (Figure 7). This result is consistent with previous findings that perfusion reduces in NASH (55,56), given that FDG K1 from a 3-minute early-dynamic FDG-PET scan reflects hepatic blood flow (37). Nevertheless, FDG K1 in this study was derived from one-hour dynamic data. It characterizes the overall delivery rate of FDG from blood to hepatic tissue and may also relate to the expression level of glucose transporters (34), though FDG has a high extraction fraction in the liver as compared to other organs such as the brain. Winterdahl et al. (37) also pointed out that FDG K1 estimated by a scan longer than 3 minutes was less correlated with liver blood flow. It is worth further investigation to determine whether blood flow only or its combination with glucose transporters decides the association of FDG K1 with liver inflammation. Our study also indicated that SUV and other kinetic parameters were not correlated with inflammation grades. FDG k3 and fat-adjusted SUV tended to increase in liver inflammation, but the associations did not achieve statistical significance. Thus, FDG K1 can be the major parameter of interest in future studies.
Earlier studies showed that liver fat content has a modulating effect on hepatic physiology including perfusion, fatty acid metabolism, and glucose metabolism as measured by PET (57) and it is necessary to correct FDG SUV and Ki for the fat content (30). This study showed that fat-adjusted K1 had poorer association with liver inflammation than non-adjusted K1, suggesting a potential effect of liver fat content on hepatic inflammation. This result complements existing observations.
There are limitations with the present study. Histopathological inflammation can vary across different spatial locations in the liver. The statistical analysis of PET data in the study was not specific to the location of biopsy. This is because accurate biopsy location information was difficult to obtain even with ultrasound guidance and was not available in this study. We chose to analyze the PET data based on the whole liver region to achieve robustness to locations and to noise in PET data. It is noted that all the evaluated kinetic models did not achieve a statistical significance (p>0.05) in differentiating low versus medium inflammation levels (Table 3), though the FDG K1 values by the optimization-derived DBIF model tended to differ in the two groups (Fig. 6C). This might be limited by the number of samples (N=14) in this study and is worth further investigation in a larger number of subjects. Note that this study did not include healthy subjects. It is therefore more difficult to reach statistical significance than in other studies comparing patients to healthy subjects. The optimization-derived DBIF model contains two additional parameters as compared with the standard 3-compartment model, which may result in higher variability in parameter estimates. The nonlinear least-square estimation by the Levenberg-Marquardt algorithm may also suffer from local minima. While the better fit quality of the new model has been confirmed by statistical criteria including AIC and F test, our future work will investigate the structural identifiability of the new model using mathematical analysis and investigate practical identifiability using Monte Carlo simulation.
It is possible that k3 is inherently associated with liver inflammation but the association could not be extracted from the data because of noise. k3 may physiologically represent FDG transport instead of phosphorylation, as indicated by Bertoldo et al. in the study of a 4-compartment model in skeletal muscle (58). Also, in contrast to standard Patlak plot which underestimates Ki when k4 is not small, the generalized Patlak plot (59,60) considering the effect of nonzero k4 can be potentially used to improve Keramida’s analysis. In addition, advanced image reconstruction algorithms (61) such as direct reconstruction (62) or the kernel method (63) may provide a hope to reduce the variability of kinetic parameter estimates, given vendors have started to implement sophisticated four-dimensional reconstruction algorithms for whole-body parametric imaging on new PET scanners (64,65). Recent development of high-sensitivity PET scanners such as the EXPLORER (66,67) further provides a new opportunity for obtaining reliable estimation of k3 and even k4 to examine their possible role in characterizing liver inflammation. All these are of interest and value in future studies.
CONCLUSION
In this study, we examined three different kinetic models for analyzing dynamic FDG-PET data for characterizing liver inflammation. Statistical fit quality metrics and analysis of association with histopathology indicated that modeling of DBIF is crucial for accurate kinetic modeling of liver time activity curves. The optimization-derived DBIF model improved the association of FDG K1 with liver inflammation grades and was more appropriate than traditional single-blood input function and population-based DBIF for dynamic FDG-PET kinetic analysis in human NASH studies.
ACKNOWLEDGMENT
The authors thank the anonymous reviewers for their very helpful comments and suggestions which have led to a great improvement to this paper. We also thank Conjing (Ella) Zha and Cole Johnson for their assistances in patient enrollment, and Benjamin Spencer, Heather Hunt, Michael Rusnak, Mike Nguyen and Denise Caudle for their assistances in PET/CT data acquisition. G.W. is supported in part by the UC Davis Comprehensive Cancer Center under NIH grant no. P30 CA093373 and K12 CA138464.
REFERENCES
- 1.Rinella ME. Nonalcoholic Fatty Liver Disease A Systematic Review. JAMA-Journal of the American Medical Association. 2015;313:2263–2273. [DOI] [PubMed] [Google Scholar]
- 2.Loomba R, Sanyal AJ. The global NAFLD epidemic. Nature Reviews Gastroenterology & Hepatology. 2013;10:686–690. [DOI] [PubMed] [Google Scholar]
- 3.Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nature Reviews Gastroenterology & Hepatology. 2013;10:656–665. [DOI] [PubMed] [Google Scholar]
- 4.Musso G, Gambino R, Cassader M, Pagano G. Meta-analysis: Natural history of non-alcoholic fatty liver disease (NAFLD) and diagnostic accuracy of non-invasive tests for liver disease severity. Annals of Medicine. 2011;43:617–649. [DOI] [PubMed] [Google Scholar]
- 5.Wree A, Broderick L, Canbay A, Hoffman HM, Feldstein AE. From NAFLD to NASH to cirrhosis - new insights into disease mechanisms. Nature Reviews Gastroenterology & Hepatology. 2013;10:627–636. [DOI] [PubMed] [Google Scholar]
- 6.Bohte AE, van Werven JR, Bipat S, Stoker J. The diagnostic accuracy of US, CT, MRI and H-1-MRS for the evaluation of hepatic steatosis compared with liver biopsy: a meta-analysis. European Radiology. 2011;21:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mishra P, Younossi Z. Abdominal ultrasound for diagnosis of nonalcoholic fatty liver disease (NAFLD). American Journal of Gastroenterology. 2007;102:2716–2717. [DOI] [PubMed] [Google Scholar]
- 8.Pickhardt PJ, Park SH, Hahn L, Lee SG, Bae KT, Yu ES. Specificity of unenhanced CT for non-invasive diagnosis of hepatic steatosis: implications for the investigation of the natural history of incidental steatosis. European Radiology. 2012;22:1075–1082. [DOI] [PubMed] [Google Scholar]
- 9.Fishbein M, Castro F, Cheruku S, et al. Hepatic MRI for fat quantitation - Its relationship to fat morphology, diagnosis, and ultrasound. Journal of Clinical Gastroenterology. 2005;39:619–625. [DOI] [PubMed] [Google Scholar]
- 10.Reeder SB, Hu HCH, Sirlin CB. Proton density fat-fraction: A standardized mr-based biomarker of tissue fat concentration. Journal of Magnetic Resonance Imaging. 2012;36:1011–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang A, Tan J, Sun M, et al. Nonalcoholic Fatty Liver Disease: MR Imaging of Liver Proton Density Fat Fraction to Assess Hepatic Steatosis. Radiology. 2013;267:422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mariappan YK, Glaser KJ, Ehman RL. Magnetic Resonance Elastography: A Review. Clinical Anatomy. 2010;23:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich-Rust M, Wunder K, Kriener S, et al. Liver Fibrosis in Viral Hepatitis: Noninvasive Assessment with Acoustic Radiation Force Impulse Imaging versus Transient Elastography. Radiology. 2009;252:595–604. [DOI] [PubMed] [Google Scholar]
- 14.Huwart L, Peeters F, Sinkus R, et al. Liver fibrosis: non-invasive assessment with MR elastography. Nmr in Biomedicine. 2006;19:173–179. [DOI] [PubMed] [Google Scholar]
- 15.Yin M, Talwalkar JA, Glaser KJ, et al. Assessment of hepatic fibrosis with magnetic resonance elastography. Clinical Gastroenterology and Hepatology. 2007;5:1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Talwalkar JA, Yin M, Glaser KJ, Sanderson SO, Ehman RL. Early Detection of Nonalcoholic Steatohepatitis in Patients with Nonalcoholic Fatty Liver Disease by Using MR Elastography. Radiology. 2011;259:749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SS, Park SH. Radiologic evaluation of nonalcoholic fatty liver disease. World Journal of Gastroenterology. 2014;20:7392–7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milic S, Stimac D. Nonalcoholic Fatty Liver Disease/Steatohepatitis: Epidemiology, Pathogenesis, Clinical Presentation and Treatment. Digestive Diseases. 2012;30:158–162. [DOI] [PubMed] [Google Scholar]
- 19.Love C, Tomas MB, Tronco GG, Palestro CJ. FDG PET of infection and inflammation. Radiographics. 2005;25:1357–1368. [DOI] [PubMed] [Google Scholar]
- 20.Rudd JHF, Myers KS, Bansilal S, et al. Atherosclerosis inflammation Imaging with F-18-FDG PET: Carotid, iliac, and femoral uptake reproducibility, quantification methods, and recommendations. Journal of Nuclear Medicine. 2008;49:871–878. [DOI] [PubMed] [Google Scholar]
- 21.Brix G, Ziegler SI, Bellemann ME, et al. Quantification of F-18 FDG uptake in the normal liver using dynamic PET: Impact and modeling of the dual hepatic blood supply. Journal of Nuclear Medicine. 2001;42:1265–1273. [PubMed] [Google Scholar]
- 22.Keiding S Bringing Physiology into PET of the Liver. Journal of Nuclear Medicine. 2012;53:425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tragardh M, Moller N, Sorensen M. Methodologic Considerations for Quantitative F-18-FDG PET/CT Studies of Hepatic Glucose Metabolism in Healthy Subjects. Journal of Nuclear Medicine. 2015;56:1366–1371. [DOI] [PubMed] [Google Scholar]
- 24.Borra R, Lautamaki R, Parkkola R, et al. Inverse association between liver fat content and hepatic glucose uptake in patients with type 2 diabetes mellitus. Metabolism-Clinical and Experimental. 2008;57:1445–1451. [DOI] [PubMed] [Google Scholar]
- 25.Iozzo P, Jarvisalo MJ, Kiss J, et al. Quantification of liver glucose metabolism by positron emission tomography: Validation study in pigs. Gastroenterology. 2007;132:531–542. [DOI] [PubMed] [Google Scholar]
- 26.Abikhzer G, Alabed YZ, Azoulay L, Assayag J, Rush C. Altered Hepatic Metabolic Activity in Patients With Hepatic Steatosis on FDG PET/CT. American Journal of Roentgenology. 2011;196:176–180. [DOI] [PubMed] [Google Scholar]
- 27.Lin CY, Lin WY, Lin CC, Shih CM, Jeng LB, Kao CH. The negative impact of fatty liver on maximum standard uptake value of liver on FDG PET. Clinical Imaging. 2011;35:437–441. [DOI] [PubMed] [Google Scholar]
- 28.Abele JT, Fung CI. Effect of Hepatic Steatosis on Liver FDG Uptake Measured in Mean Standard Uptake Values. Radiology. 2010;254:917–924. [DOI] [PubMed] [Google Scholar]
- 29.Bural GG, Torigian DA, Burke A, et al. Quantitative Assessment of the Hepatic Metabolic Volume Product in Patients with Diffuse Hepatic Steatosis and Normal Controls Through Use of FDG-PET and MR Imaging: A Novel Concept. Molecular Imaging and Biology. 2010;12:233–239. [DOI] [PubMed] [Google Scholar]
- 30.Keramida G, Potts J, Bush J, Dizdarevic S, Peters AM. Hepatic steatosis is associated with increased hepatic FDG uptake. European Journal of Radiology. 2014;83:751–755. [DOI] [PubMed] [Google Scholar]
- 31.Keramida G, Potts J, Bush J, Verma S, Dizdarevic S, Peters AM. Accumulation of F-18-FDG in the Liver in Hepatic Steatosis. American Journal of Roentgenology. 2014;203:643–648. [DOI] [PubMed] [Google Scholar]
- 32.Keramida G, Hunter J, Peters AM. Hepatic glucose utilization in hepatic steatosis and obesity. Bioscience Reports. 2016;36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keramida G, Dunford A, Siddique M, Cook GJ, Peters AM. Relationships of body habitus and SUV indices with signal-to-noise ratio of hepatic F-18-FDG PET. European Journal of Radiology. 2016;85:1012–1015. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt KC, Turkheimer FE. Kinetic modeling in positron emission tomography. Quarterly Journal of Nuclear Medicine. 2002;46:70–85. [PubMed] [Google Scholar]
- 35.Garbarino S, Vivaldi V, Delbary F, et al. A new compartmental method for the analysis of liver FDG kinetics in small animal models. EJNMMI Research. 2015;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munk OL, Bass L, Roelsgaard K, Bender D, Hansen SB, Keiding S. Liver kinetics of glucose analogs measured in pigs by PET: Importance of dual-input blood sampling. Journal of Nuclear Medicine. 2001;42:795–801. [PubMed] [Google Scholar]
- 37.Winterdahl M, Munk OL, Sorensen M, Mortensen FV, Keiding S. Hepatic Blood Perfusion Measured by 3-Minute Dynamic F-18-FDG PET in Pigs. Journal of Nuclear Medicine. 2011;52:1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang G, Corwin M, Olson K, et al. Dynamic FDG-PET study of liver inflammation in non-alcoholic fatty liver disease. Journal of Hepatology. 2017;66:S592 (abstract only). [Google Scholar]
- 39.Chen K, Bandy D, Reiman E, et al. Noninvasive quantification of the cerebral metabolic rate for glucose using positron emission tomography, F-18-fluoro-2-deoxyglucose, the Patlak method, and an image-derived input function. Journal of Cerebral Blood Flow and Metabolism. 1998;18:716–723. [DOI] [PubMed] [Google Scholar]
- 40.van der Weerdt AP, Klein LJ, Boellaard R, Visser CA, Visser FC, Lammertsma AA. Image-derived input functions for determination of MRGlu in cardiac F-18-FDG PET scans. Journal of Nuclear Medicine. 2001;42:1622–1629. [PubMed] [Google Scholar]
- 41.Zanotti-Fregonara P, Hines CS, Zoghbi SS, et al. Population-based input function and image-derived input function for C-11 (R)-rolipram PET imaging: Methodology, validation and application to the study of major depressive disorder. Neuroimage. 2012;63:1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kudomi N, Jarvisalo MJ, Kiss J, et al. Non-invasive estimation of hepatic glucose uptake from F-18 FDG PET images using tissue-derived input functions. European Journal of Nuclear Medicine and Molecular Imaging. 2009;36:2014–2026. [DOI] [PubMed] [Google Scholar]
- 43.Kleiner DE, Brunt EM, Van Natta M, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. [DOI] [PubMed] [Google Scholar]
- 44.Iida H, Kanno I, Miura S, Murakami M, Takahashi K, Uemura K. Error analysis of a quantitative cerebral blood flow measurement using H2(15)O autoradiography and positron emission tomography, with respect to the dispersion of the input function. Journal of Cerebral Blood Flow and Metabolism. 1986;6:536–545. [DOI] [PubMed] [Google Scholar]
- 45.Thiele F, Buchert R. Evaluation of non-uniform weighting in non-linear regression for pharmacokinetic neuroreceptor modelling. Nuclear Medicine Communications. 2008;29:179–188. [DOI] [PubMed] [Google Scholar]
- 46.Wang G, Qi J. Generalized Algorithms for Direct Reconstruction of Parametric Images From Dynamic PET Data. IEEE Transactions on Medical Imaging. 2009;28:1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang G, Qi J. An Optimization Transfer Algorithm for Nonlinear Parametric Image Reconstruction From Dynamic PET Data. IEEE Transactions on Medical Imaging. 2012;31:1977–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glatting G, Kletting P, Reske SN, Hohl K, Ring C. Choosing the optimal fit function: Comparison of the Akaike information criterion and the F-test. Medical Physics. 2007;34:4285–4292. [DOI] [PubMed] [Google Scholar]
- 49.Richard MA, Fouquet JP, Lebel R, Lepage M. Determination of an Optimal Pharmacokinetic Model of F-18-FET for Quantitative Applications in Rat Brain Tumors. Journal of Nuclear Medicine. 2017;58:1278–1284. [DOI] [PubMed] [Google Scholar]
- 50.Burnham KP, Anderson DR. Multimodel inference - understanding AIC and BIC in model selection. Sociological Methods & Research. 2004;33:261–304. [Google Scholar]
- 51.Prytz H, Keiding S, Bjornsson E, et al. Dynamic FDG-PET is useful for detection of cholangiocarcinoma in patients with PSC listed for liver transplantation. Hepatology. 2006;44:1572–1580. [DOI] [PubMed] [Google Scholar]
- 52.Choi Y, Hawkins RA, Huang SC, et al. Evaluation of the effect of glucose ingestion and kinetic model configurations of FDG in the normal liver. Journal of Nuclear Medicine. 1994;35:818–823. [PubMed] [Google Scholar]
- 53.Chen S, Feng D. Evaluation of hepatocellular carcinoma with dynamic C-11-acetate PET: A dual-modeling method. IEEE Transactions on Nuclear Science. 2008;55:999–1007. [Google Scholar]
- 54.Rani SD, Nemanich ST, Fettig N, Shoghi KI. Kinetic analysis of FDG in rat liver: Effect of dietary intervention on arterial and portal vein input. Nuclear Medicine and Biology. 2013;40:537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joo I, Lee JM, Yoon JH, Jang JJ, Han JK, Choi BI. Nonalcoholic Fatty Liver Disease: Intravoxel Incoherent Motion Diffusion-weighted MR Imaging-An Experimental Study in a Rabbit Model. Radiology. 2014;270:131–140. [DOI] [PubMed] [Google Scholar]
- 56.Kobayashi M, Suzuki M, Ikeda H, et al. Assessment of hepatic steatosis and hepatic tissue blood flow by xenon computed tomography in nonalcoholic steatohepatitis. Hepatology Research. 2009;39:31–39. [DOI] [PubMed] [Google Scholar]
- 57.Rijzewijk LJ, van der Meer RW, Lubberink M, et al. Liver Fat Content in Type 2 Diabetes: Relationship With Hepatic Perfusion and Substrate Metabolism. Diabetes. 2010;59:2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bertoldo A, Peltoniemi P, Oikonen V, Knuuti J, Nuutila P, Cobelli C. Kinetic modeling of F-18 FDG in skeletal muscle by PET: a four-compartment five-rate-constant model. American Journal of Physiology- Endocrinology and Metabolism. 2001;281:E524–E536. [DOI] [PubMed] [Google Scholar]
- 59.Karakatsanis NA, Zhou Y, Lodge MA, et al. Generalized whole-body Patlak parametric imaging for enhanced quantification in clinical PET. Physics in Medicine and Biology. 2015;60:8643–8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patlak CS, Blasberg RG. Graphical Evaluation of Blood-to-Brain Transfer Constants from Multiple-Time Uptake Data. Generalizations Journal of Cerebral Blood Flow and Metabolism. 1985;5:584–590. [DOI] [PubMed] [Google Scholar]
- 61.Reader AJ, Verhaeghe J. 4D image reconstruction for emission tomography. Physics in Medicine and Biology. 2014;59:R371–R418. [DOI] [PubMed] [Google Scholar]
- 62.Wang G, Qi J. Direct estimation of kinetic parametric images for dynamic PET. Theranostics. 2013;3:802–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang G, Qi J. PET image reconstruction using kernel method. IEEE Transactions on Medical Imaging. 2015;34:61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu J, Panin V, Smith A, et al. Clinical whole body CBM parametric PET with flexible scan modes. Conference Record of IEEE Nuclear Science Symposium and Medical Imaging 2017. [Google Scholar]
- 65.Karakatsanis NA, Casey ME, Lodge MA, Rahmim A, Zaidi H. Whole-body direct 4D parametric PET imaging employing nested generalized Patlak expectation-maximization reconstruction. Physics in Medicine and Biology. 2016;61:5456–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cherry SR, Jones T, Karp JS, Qi JY, Moses WW, Badawi RD. Total-Body PET: Maximizing Sensitivity to Create New Opportunities for Clinical Research and Patient Care. Journal of Nuclear Medicine. 2018;59:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berg E, Zhang X, Bec J, et al. Development and evaluation of mini-EXPLORER: a long axial field-of- view PET scanner for non-human primate imaging. Journal of Nuclear Medicine. 2018;59:doi: 10.2967/jnumed.2117.200519. [DOI] [PMC free article] [PubMed] [Google Scholar]