Abstract
Macrophages have well-characterized roles in skeletal muscle repair and regeneration. Relatively little is known regarding the role of resident macrophages in skeletal muscle homeostasis, extracellular matrix remodeling, growth, metabolism and adaptation to various stimuli including exercise and training. Despite speculation into macrophage contributions during these processes, studies characterizing macrophages in non-injured muscle are limited and methods used to identify macrophages vary. A standardized method for the identification of human resident skeletal muscle macrophages will aide in the characterization of these immune cells and allow for the comparison of results across studies. Here, we present an immunohistochemistry (IHC) protocol, validated by flow cytometry, to distinctly identify resident human skeletal muscle macrophage populations. We show that CD11b and CD206 double IHC effectively identifies macrophages in human skeletal muscle. Furthermore, the majority of macrophages in non-injured human skeletal muscle show a ‘mixed’ M1/M2 phenotype, expressing CD11b, CD14, CD68, CD86 and CD206. A relatively small population of CD11b+/CD206- macrophages are present in resting skeletal muscle. Changes in the relative abundance of this population may reflect important changes in the skeletal muscle environment. CD11b and CD206 IHC in muscle also reveals distinct morphological features of macrophages that may be related to the functional status of these cells.
Keywords: Skeletal muscle, Macrophages, Immune cells, Immunohistochemistry, CD68, CD11b, CD206, Flow cytometry
Background
Macrophages are pleotropic immune cells capable of adapting to changes in the local microenvironment. Over the last several years, research has shown that macrophage phenotype is dynamic, existing on a continuum (Mosser and Edwards, 2008, Italiani and Boraschi, 2014, Martinez and Gordon, 2014). However, to date macrophage populations continue to be described using the restrictive M1 and M2 classifications. It is commonly accepted that these designations are an oversimplification of macrophage phenotype and represent opposite extremes of a continuum (Mosser and Edwards, 2008; Gordon et al., 2014 ; Italiani and Boraschi, 2014; Martinez and Gordon, 2014; Murray et al., 2014 ). M1 macrophages are classically activated, have pro-inflammatory functions and are involved in host responses to pathogens and tissue injury. M2 macrophages are alternatively activated, exhibit anti-inflammatory functions and are involved in wound healing and tissue repair. In addition to the functional definition of M1 and M2, cell surface markers have been identified to distinguish between these populations. Surface markers associated with M1 macrophages include CD40, CD64 and the co-stimulatory molecules CD80/CD86 ( Lolmede et al., 2009 ; Ambarus et al., 2012 ), whereas M2 macrophages have been shown to express high levels of CD163, CD206 and galactose receptors ( Lolmede et al., 2009 ; Ambarus et al., 2012 ; Roszer, 2015).
From tissue to tissue, macrophage populations are heterogeneous adopting different functional roles depending on the local environment ( Gordon et al., 2014 ; Italiani and Boraschi, 2014). This, coupled with the macrophage continuum, has led to inconsistencies with regard to identification and nomenclature across fields and across species ( Murray et al., 2014 ). Skeletal muscle macrophages have primarily been studied in rodent models of injury, where the M1 versus M2 macrophage classification has proven useful ( Smith et al., 2008 ; Chazaud et al., 2009 ; Tidball and Villalta, 2010, Kharraz et al., 2013 ; Novak and Koh, 2013; Saclier et al., 2013b ; Rigamonti et al., 2014 ; Tidball et al., 2014 ; Wang et al., 2014 ; Sciorati et al., 2016 ; Varga et al., 2016 ; Mackey and Kjaer, 2017). The skeletal muscle response to injury is characterized by highly orchestrated temporal processes. Initially, M1 macrophages phagocytize damaged skeletal muscle fibers and debris, followed by M2 macrophage-facilitated repair and regeneration ( Chazaud et al., 2009 ; Tidball and Villalta, 2010; Kharraz et al., 2013 ; Saclier et al., 2013a ; Tidball et al., 2014 ; Sciorati et al., 2016 ). Recent work nicely details fiber repair in human skeletal muscle in vivo, showing the presence of macrophages, using a pan-macrophage intracellular marker (CD68+), in regenerating zones along injured fibers (Mackey and Kjaer, 2017). Direct interaction between macrophages and satellite cells (Dumont and Frenette, 2013; Ceafalan et al., 2017 ; Du et al., 2017 ; Wehling- Henricks et al., 2018 ), and defects in skeletal muscle regeneration in the absence of macrophage participation ( Arnold et al., 2007 ; Melton et al., 2016 ), highlight the necessity of these cells for skeletal muscle repair. In vitro, M1 macrophages promote skeletal muscle cell proliferation and M2 macrophages promote differentiation, suggesting that macrophages may play a role in skeletal muscle growth adaptations, as well as repair ( Arnold et al., 2007 ; Saclier et al., 2013b ).
Skeletal muscle is a highly adaptable tissue, able to respond to a wide range of external stimuli, such as exercise, inactivity, hormones and nutritional signals. In contrast to the clearly defined, strongly polarizing responses elicited by acute skeletal muscle injury, the role of tissue resident macrophages during less polarizing processes, such as responses to the aforementioned stimuli, is relatively unknown. Under non-damaging exercise conditions, animal studies report an increase in macrophage populations following aerobic and resistance exercise, linked to both metabolic and growth adaptations ( DiPasquale et al., 2007 ; Ikeda et al., 2013 ). However, the mechanisms by which macrophages in skeletal muscle influence training adaptations remain to be explored. It has also been reported that resident human skeletal muscle macrophage abundance is affected by aging, obesity and diabetes ( Przybyla et al., 2006 ; Hong et al., 2009 ; Varma et al., 2009 ; Tam et al., 2012 ; Fink et al., 2014 ; Reidy et al., 2017 ); however, the inconsistent use of macrophage markers across studies has made the interpretation of these findings difficult. Further, the applicability of the distinctive M1/M2 markers of polarized macrophages to tissue resident macrophages is unclear, as surface markers may not be mutually exclusive on resident macrophages under non-polarizing conditions (Italiani and Boraschi, 2014). Thus, there is a need in the field for a standardized, validated method for identifying and quantifying macrophages in human skeletal muscle. Establishing a simple and reproducible protocol for studying muscle macrophages will aide in the characterization of their role in muscle adaptations to various stimuli, independent of injury.
Although results from studies of skeletal muscle macrophages in animal models are informative, these studies often use macrophage markers that are not directly translatable for use in humans. Even when human homologs do exist, the same surface markers in mouse and rat skeletal muscles often identify different populations in human skeletal muscles, complicating the extrapolation of findings from rodent models to human studies ( Murray et al., 2014 ). For example, CD68 is used as a pan-macrophage marker in humans and an M1 marker in mice. There is a need in the field for a standardized, validated method for identifying and quantifying macrophages in human skeletal muscle. Taking into account the limited mass of frozen muscle tissue available for analyses from human skeletal muscle biopsies (normally in the range of 100 mg), an immunohistochemical method is the most feasible approach to identifying and quantifying human skeletal muscle macrophage populations.
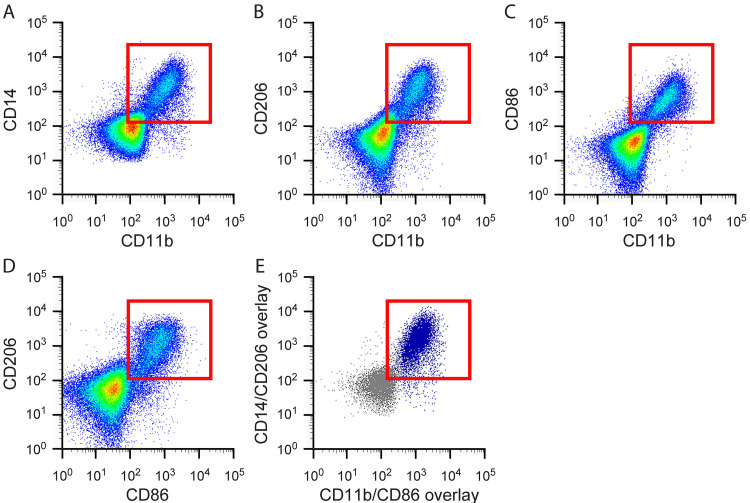
A variety of markers have been used to characterize human macrophages by flow cytometry. The most detailed studies have been performed utilizing peripheral blood mononuclear cells (PBMCs), artificially polarized to an M1 or M2 phenotype ( Martinez et al., 2006 ; Ambarus et al., 2012 ; Iqbal, 2015). These in vitro studies characterize the expression of various marker combinations on M1 and M2 macrophages and provide a good starting point for choosing markers to identify macrophage populations in frozen human skeletal muscle tissue. CD14 has been identified as a monocyte marker, expressed mainly by macrophages but also neutrophils and dendritic cells (Table 1). In blood, CD14 co-staining with CD16 is used to stratify monocytes into three subsets: classical (CD14++/CD16-), intermediate (CD14++/CD16+) and non-classical (CD14+/CD16++) ( Sprangers et al., 2016 ; Boyette et al., 2017 ). It is thought that classical monocytes give rise to tissue macrophages under homeostatic conditions; however, during an inflammatory insult all monocyte populations differentiate into macrophages (Italiani and Boraschi, 2014; Sprangers et al., 2016 ). In tissue, CD16 is predominantly used to identify NK cells, but is also expressed on neutrophils, granulocytes, dendritic cells and some macrophage populations (Table 1). CD11b is a commonly used marker and is expressed on subsets of lymphocytes and monocytes, these include natural killer (NK) cells, granulocytes and macrophages (Table 1). CD68 is expressed by cells in the monocyte lineage, including macrophages, and is the most commonly used macrophage marker in human skeletal muscle tissue (Table 1) ( Stupka et al., 2001 ; Beaton et al., 2002 ; Peterson et al., 2003 ; Crameri et al., 2004 ; Przybyla et al., 2006 ; Crameri et al., 2007 ; Mahoney et al., 2008 ; Mikkelsen et al., 2009 ; Varma et al., 2009 ; Paulsen et al., 2010a ; Paulsen et al., 2010b ; MacNeil et al., 2011 ; Tam et al., 2012 ; Chistiakov et al., 2017 ; Mackey and Kjaer, 2017; Reidy et al., 2017 ). CD68 is a member of the lysosomal/endosomal-associated membrane glycoprotein (LAMP) family of proteins, which are mainly associated with the endosomal/lysosomal compartment. Though largely intracellular, CD68 can traffic to the cell surface. Of note, other cell types have been reported to express CD68, including hematopoietic cells, fibroblasts and endothelial cells (Table 1) ( Kunisch et al., 2004 ; Gottfried et al., 2008 ; Paulsen et al., 2013 ; Chistiakov et al., 2017 ). CD206, the mannose receptor, is a well-accepted macrophage marker in skeletal muscle and is widely used to identify M2 macrophage subsets ( Lolmede et al., 2009 ; Ambarus et al., 2012 ; Italiani and Boraschi, 2014; Roszer, 2015), although CD206 expression by other cell types (including satellite cells) has been reported (Table 1) (Jansen and Pavlath, 2006)[44]. M2 macrophages also express CD163 (Table 1) ( Lolmede et al., 2009 ; Ambarus et al., 2012 ; Roszer, 2015). CD80 and CD86 are co-stimulatory molecules expressed by antigen presenting cells upon activation and have been used to identify M1 macrophage populations (Table 1) (Mosser and Edwards, 2008; Lolmede et al., 2009 ; Ambarus et al., 2012 ). Using three grams of discarded human hamstring muscle from patients undergoing anterior cruciate ligament (ACL) reconstruction surgery, we isolated and labeled mononuclear cells with antibodies against some of the markers described above (CD11b, CD14, CD16, CD86 and CD206) and performed multichannel flow cytometry. Due to the intracellular expression of CD68, we were not able to include CD68 in flow cytometry analyses. Mononuclear cells from skeletal muscle did not express CD16, but co-expressed the other 4 markers tested (Figures 1A-1E). Thus, human skeletal muscle macrophages have a ‘mixed’ phenotype, co-expressing both M1 (CD86) and M2 (CD206) cell surface markers (Figure 1D).
Table 1. Overview of monocyte and macrophage markers.
| Marker | Synonyms | Tissue cell populations | Monocyte/macrophage subset | Notes |
|---|---|---|---|---|
| CD11b | CR3, C3biR, Integrin alphaM, ITGAM, Mac-1, Mo1 | B cells, dendritic cells, granulocytes, macrophages, monocytes, NK cells and T cells | Pan-macrophage | |
| *CD14 | Monocyte differentiation antigen CD14, LPS receptor | B cells, dendritic cells, granulocytes, macrophages and monocytes | Pan-monocyte | *Co-expression with CD16 identifies monocyte subsets |
| *CD16 | FcγRIIIA | B cells, macrophages, monocytes, neutrophils, NK cells T cells |
CD14++/CD16- - classical CD14++/CD16+ - intermediate CD14+/CD16++ - non-classical |
|
| CD68 | gp110, LAMP4, Macrosialin, SCARD1 | B cells, dendritic cells, endothelial cells, fibroblasts, granulocytes, hematopoietic stem cells (LT), macrophages, monocytes, NK cells and T cells | Pan-macrophage | Intracellular expression |
| CD80 | B7, B7-1, B7.1 BB1, CD28LG, CD28LG1, LAB7 | B cells, dendritic cells, macrophages, monocytes and T cells | M1 | Co-stimulatory with CD86 |
| CD86 | B7-2, B7.2, B70, CD28LG2, LAB72 | B cells, dendritic cells, macrophages, monocytes and T cells | M1 | |
| CD163 | D11, D1 1C163A, GHI/61, M130, MM130, RM3/1, SCARI1 | B cells, macrophages and monocytes | M2c | Identifies a subset of CD206+ macrophages in skeletal muscle |
| CD206 | CLEC13D, mannose receptor, MMR, MRC1 | Dendritic cells, certain skin cells, endothelial cells, macrophages and monocytes | M2 | In rare instances, Pax7+ skeletal muscle satellite cells co-express CD206 |
| Markers in non-injured human skeletal muscle | ||||
| CD11b+/CD206- | M1-like macrophages | FC - Also expresses CD14 | ||
| CD11b+/CD206+ | M2-like macrophages | FC - Also express CD14 and CD86 | ||
| CD163+/CD206+ | M2c macrophages | |||
| FC = multichannel flow cytometry analysis | ||||
Figure 1. Flow cytometry from discarded human hamstring muscle showing co-expression of both M1 and M2 macrophage markers.
Mononuclear cells isolated from human skeletal muscle express A. Both pan-monocyte markers CD11b and CD14; B. Both the M2 macrophage marker, CD206, and the pan marker, CD11b; C. Both the M1 marker, CD86, and the pan marker CD11b; D. Both the M2 marker, CD206, and the M1 marker, CD86; E. Overlay of CD206+/CD86+ populations from panel D onto CD14/CD11b flow plot shown in panel A. This overlay shows that CD206+/CD86+ macrophages (denoted in dark blue) also express the pan-monocyte markers CD11b and CD14. Red boxes indicate cells that are double positive for the markers shown.
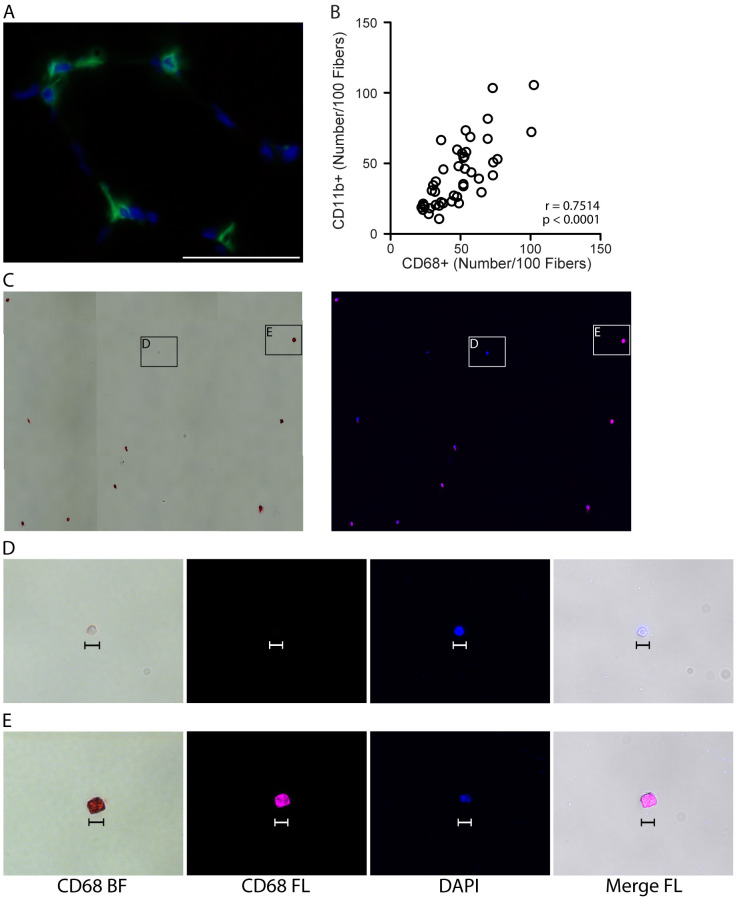
Using these 4 cell surface antibodies against CD11b, CD14, CD86 and CD206 that label skeletal muscle resident macrophages, we sought to develop a simple and reproducible immunohistochemical method for identification of macrophages in fresh frozen human skeletal muscle sections. CD68 is a commonly used pan-macrophage marker in human skeletal muscle. However, CD68 expression is predominantly intracellular, requiring permeabilization steps to perform immunohistochemistry (IHC). These permeabilization steps lead to inconsistent results and compromise staining with additional antibodies for cell surface markers. The cell surface localization of CD11b results in better morphological definition of macrophages and more consistent staining across samples than intracellular CD68 staining. Moreover, CD11b can readily be combined with CD206 and other cell surface markers for IHC. For these reasons, we compared CD11b and CD68 staining, and found CD11b to be comparable to CD68 as a pan-macrophage marker in human skeletal muscle (Figures 2A-2E; CD11b and CD68 antibodies cannot be used to label sections simultaneously for technical reasons, see General Note 12). Furthermore, distinct morphological features of muscle macrophages that may be related to functional status was revealed through CD11b and CD206 double IHC (Figures 8A-8C) ( Durafourt et al., 2012 ; McWhorter et al., 2013 ). We describe here a detailed method for combined IHC using CD11b and CD206 antibodies on frozen human skeletal muscle sections. We also describe in detail our approach to quantifying macrophage subsets in non-injured skeletal muscle. We find the majority of human muscle macrophages are CD11b+/CD206+, whereas a small subset are CD11b+/CD206- (Figures 4A-4C). We were unable to obtain IHC results with antibodies against M1 cell surface markers (CD80 or CD86). Of note, anti-CD163 works well on frozen human skeletal muscle and can be used in place of CD11b and in combination with CD206 with this protocol (see General Note 13). Moreover, combining Pax7 (a satellite cell marker) IHC with CD206 shows very little co-expression, supporting the conclusion that CD206 co-localizes with CD11b in human skeletal muscle and is a valid macrophage marker. This protocol allows reproducible quantification of the relative abundance of CD11b+/CD206+ and CD11b+/CD206- macrophage populations in human skeletal muscle, which can be extended to human skeletal muscle adaptations, aging and disease, enabling comparison of results across studies, across labs and across diverse human populations.
Figure 2. Validating CD11b as a pan-macrophage, cell surface marker in human skeletal muscle.
A. Representative image showing IHC for the pan, intracellular macrophage marker, CD68 (green), with DAPI stained cell nuclei (blue) in human vastus lateralis muscle. Scale bar = 50 µm. B. Correlation of CD11b+ and CD68+ macrophage numbers, identified by IHC on consecutive sections from vastus lateralis muscle, n = 44 muscle samples analyzed. P value determined by Pearson’s correlation, r = Pearson’s correlation coefficient. C. CD14+/CD11b+ mononuclear cells (shown in Figure 1, panel A) were isolated by fluorescence-activated cell sorting (FACS) and stained for CD68. Positive staining is both chromogenic (red when imaged with transmitted light, bright field, BF) and fluorescent (pseudocolored pink, FL). Left image: CD68 BF; positive staining is red. Right: CD68 FL, positive staining is pink. Cell nuclei were labeled with DAPI (blue). D-E. High magnification images showing a CD68- event (D) and CD68+ cell (E) from panel C. CD68- events were small (< 10 µm) and did not appear to be monocytes/macrophages. CD68- structures appeared to stain with DAPI, suggesting that this may be cellular debris (cell nucleus without cytoplasm). Scale bars = 10 µm.
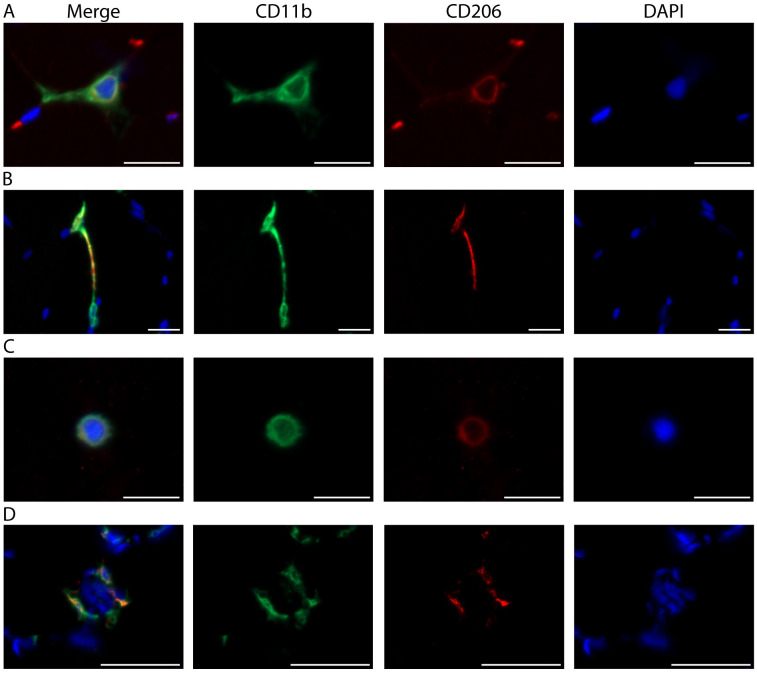
Figure 8. Distinct skeletal muscle macrophage morphologies.
A. Representative image from the vastus lateralis muscle showing a macrophage with classic morphology. Described in other tissues, the outstretched processes of resting macrophages are thought to surveil the local environment ( Olah et al., 2011 ; Durafourt et al., 2012 ). B. A CD11b+/CD206+ macrophage (top) stretching out toward a second CD11b+/CD206- macrophage (bottom). C. Round or amoeboid shaped macrophage. In other tissues, this morphology is thought to be indicative of activation and the production of inflammatory cytokines ( Olah et al., 2011 ; Durafourt et al., 2012 ). A-C) Scale bars = 20 µm. D. Macrophages surround a damaged skeletal muscle fiber (indicated by the presence of central nuclei). Scale bars = 50 µm. A-D) Images were acquired as Z stacks using a 40x objective and cropped to enlarge. CD11b (green), CD206 (red), cell nuclei/DAPI (blue).
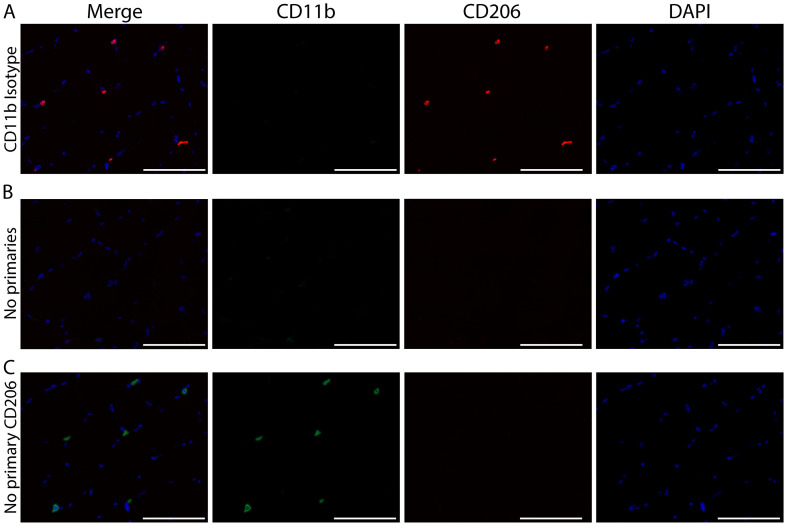
Figure 4. Immunohistochemical controls validating the specificity of CD11b and CD206 staining. Representative images of staining controls.
A) No primary antibody for CD206, B) Isotype control antibody for CD11b and C) No primary antibody for either CD11b or CD206. CD11b (green), CD206 (red) and cell nuclei/DAPI (blue). Scale bars = 100 µm. All images were acquired on serial sections from the same sample with a 20x objective using the same exposure settings. The same display adjustment was applied across all images. A. Images showing a lack of CD206 staining when the primary antibody for CD206 is not applied to the sections. This control shows the specificity of the CD206 antibody; staining is not a product of cross reactivity with reagents used to amplify CD11b or due to non-specific tissue staining. B. Lack of CD11b+ staining when an isotype control antibody is applied. Similar to panel A, this control shows the specificity of the CD11b antibody since the isotype-matched control does not produce non-specific staining. Additionally, this control shows that CD206 staining does not result from cross reactivity to CD11b reagents since no CD11b staining is present but CD206+ cells are clearly identified. C. Images show very low non-specific background staining of skeletal muscle tissue sections from the use of amplification reagents; thus positive macrophage staining can clearly be distinguished from tissue background.
Materials and Reagents
-
Pipette tips:
101-1,000 µl (USA Scientific, TipOne, catalog number: 1122-1832)
0.1-10 µl (USA Scientific, TipOne, catalog number: 1120-3812)
Gloves (VWR, catalog number: 82026-424)
Nalgene Dewar (for liquid nitrogen) (Sigma-Aldrich, catalog number: F9401)
-
Tri-Pour Polypropylene beaker (for cooling isopentane in liquid nitrogen) (VWR, catalog number: 89011-786)
Manufacturer: MEDEGEN MEDICAL PRODUCTS, catalog number: PB5935-400.
MX35 Ultra Low-Profile blades for cryotomy (Thermo Fisher Scientific, catalog number: 3053835)
Superfrost Plus slides (Fisher Scientific, catalog number: 12-550-15)
Shandon Single Cytoslides (Thermo Fisher Scientific, catalog number: 5991056)
Shandon Single Cytofunnel (Thermo Fisher Scientific, catalog number: 5991040)
ImmEdge PAP pen (Vector Laboratories, catalog number: H-4000)
1.5 ml microcentrifuge tubes (USA Scientific, catalog number: 1615-5599)
15 ml conical tube (VWR, catalog number: 89039-666)
24 x 50 mm, No. 1 coverglass (VWR, catalog number: 48393-081)
Kimwipes (KCWW, Kimberly-Clark, catalog number: 34120)
Cork stoppers for making muscle mounts (Fisher Scientific, catalog number: 07-782J)
-
PTFE (Teflon) coated stainless steel spatula (Fisher Scientific, catalog number: 21-401-50A)
Manufacturer: Saint-Gobain Performance Plastics, catalog number: D1069292.
#10 curved blade disposable scalpel (Sklar Surgical Instruments, catalog number: 06-3310)
Dumont #7 curved forceps (Fine Science Tools, catalog number: 11270-20)
Cryo Tongs (Thermo Fisher Scientific, catalog number: 4000388)
Tragacanth gum, powder (Sigma-Aldrich, catalog number: G1128-500G)
Fisher O.C.T compound (Fisher Scientific, catalog number: 23-730-571)
Isopentane (2-methylbutane) (Merck, catalog number: MX0760-1)
Liquid nitrogen (Scott Gross, catalog number: SG #347)
Dry ice (Scott Gross, no catalog number available)
Ice cold acetone, stored at -20 °C (Fisher Scientific, catalog number: A18-4)
Streptavidin/Biotin blocking kit (Vector Laboratories, catalog number: SP-2002)
2.5% normal horse serum (NHS) (Vector Laboratories, catalog number: S-2012)
50% 1x PBS (see 21 and Recipes)/50% glycerol mounting medium (glycerol - VWR, catalog number: BDH1172-1LP) or Vectashield (Vector Laboratories, catalog number: H-1000)
Antibodies for IHC, dilutions made with 2.5% normal horse serum or PBS (Table 2)
ImmPRESS-AP Anti-Mouse IgG (alkaline phosphatase) polymer detection kit (Vector Laboratories, catalog number: MP-5402)
ImmPACT Vector Red Alkaline Phosphatase (AP) Substrate (Vector Laboratories, catalog number: SK-5105)
Antibodies for multichannel flow cytometry (Table 3)
LIVE/DEAD Fixable Blue Dead Cell Stain Kit, for UV excitation (Thermo Fisher Scientific, catalog number: L34961) (Table 3)
UltraComp eBeads Compensation Beads (Thermo Fisher Scientific, catalog number: 01-2222-41)
Polyurethane ice bucket (Fisher Scientific, catalog number: 02-591-45)
-
1x phosphate-buffered saline (PBS) (see Recipes)
Deionized (DI) water
Sodium Chloride (NaCl) (VWR, catalog number: 97061-266)
-
Disodium hydrogen phosphate heptahydrate (Na2HPO4·7H2O) (VWR, catalog number: 200007-704)
Manufacturer: Acros Organic, catalog number: 206515000.
Potassium phosphate monobasic (KH2PO4) (Aldon, catalog number: PP0730-500GR)
Sodium hydroxide 10 N (NaOH) (VWR, catalog number: BDH7247-1)
Hydrochloric acid, 6 N (HCl) (VWR, catalog number: 97064-758)
30% Hydrogen peroxide, ACS, Stabilized (VWR, catalog number: BDH7690-1) (see Recipes)
DAPI for staining cell nuclei (4',6-diamidino-2-phenylindole) (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: D1306) (see Recipes)
Rabbit polyclonal anti-Laminin (Sigma-Aldrich, catalog number: L9393). Use at a dilution of 1:100 in 2.5% NHS
Table 2. Detailed IHC Antibody Information.
| Primary Antibody | Company | Catalog Number | Reactivity | Host /Isotype | Dilution/Diluent |
|---|---|---|---|---|---|
| CD68 | Dako | M0814 | Human/Mouse/Rat/Monkey | Mouse/IgG1 | (1:100)/2.5% NHS |
| CD11b | Cell Sciences | MON1019 | Human | Mouse/IgG1 | (1:100)/2.5% NHS |
| Purified IgG1, κ | BD | 555746 | N/A | Mouse/IgG1 | (1:500)/2.5% NHS |
| C206 | R&D Systems | AF2534 | Human | Goat/IgG (polyclonal) | (1:200)/2.5% NHS |
| CD163 | Hycult Biotech | HM2157 | Human/Monkey | Mouse/IgG1 | (1:50)/2.5% NHS |
| Pax7 | Developmental Studies Hybridoma Bank | Pax7 | Human/Mouse/Rat | Mouse/IgG1 | (1:100)/2.5% NHS |
| Biotinylated goat anti-mouse IgG1 | Jackson ImmunoResearch | 115-065-205 | Mouse IgG1 | Goat/N/A | (1:1,000)/2.5% NHS |
| ImmPRESS –AP | Vector Laboratories | MP-5402 | Mouse IgG | N/A | Neat/no dilution |
| Biotinylated rabbit anti-goat IgG | Vector Laboratories | BA-5000 | Goat | Rabbit/N/A | (1:500)/2.5% NHS |
| Streptavidin HRP (SA-HRP) | Thermo Fisher Scientific | S911 | Biotin | N/A | (1:500)/PBS |
| Streptavidin Alexa Fluor 594 (SA-594) | Thermo Fisher Scientific | S32356 | Biotin | N/A | (1:200)/PBS |
| Superboost TSA Alexa Fluor 488 (TSA 488) | Thermo Fisher Scientific | B40953 | HRP | N/A | (1:500)/PBS |
| ImmPACT Vector Red kit | Vector Laboratories | SK-5105 | Alkaline Phosphatase | N/A | According to manufacturer’s instructions |
Table 3. Detailed antibody information for multichannel flow cytometry.
| Antigen | Fluorophore | Laser | Filter/Bandpass | Ex (nm) | Em (nm) | Concentration (µg/µl) | µl/106 cells (500 µl total volume) | Company | Catalog Number |
| CD11b | Alex Fluor 488 | Blue (488) | 530/30 | 490 | 525 | 0.4* | 25 | BioLegend | 301317 |
| CD14 | Pacific Blue | Violet (407) | 450/50 | 410 | 455 | 0.5 | 20 | BioLegend | 325615 |
| CD16 | VioGreen | Violet (407) | 525/50 | 405 | 520 | NA | 50 | Miltenyi Biotec | 130-113-959 |
| CD86 | Phycoerythrin (PE) | Blue (488) | 575/26 | 496 | 578 | 0.1* | 25 | BioLegend | 305405 |
| CD206 | PerCP/Cy5.5 | Blue (488) | 695/40 | 482 | 695 | 0.1* | 25 | BioLegend | 321121 |
| LIVE/DEAD | Fixable Blue | UV (325) | 450/50 | 350 | 450 | NA | 0.5 | Thermo Fisher Scientific | L34961 |
| Mouse IgG1, κ | Alexa Fluor 488 | Blue (488) | 530/30 | 490 | 525 | 0.2* | 50 | BioLegend | 400132 |
| Mouse IgG1, κ | Pacific blue | Violet (407) | 450/50 | 410 | 455 | 0.5 | 20 | BioLegend | 400131 |
| REA Control (S) | VioGreen | Violet (407) | 525/50 | 405 | 520 | NA | 50 | Miltenyi Biotec | 130-104-608 |
| Mouse IgG2b, κ | Phycoerythrin (PE) | Blue (488) | 575/26 | 496 | 578 | 0.2 | 12.5 | BioLegend | 400311 |
| Mouse IgG1, κ | PerCP/Cy5.5 | Blue (488) | 695/40 | 482 | 695 | 0.2* | 12.5 | BioLegend | 400149 |
| *Concentration is not specific and varies by batch | |||||||||
Equipment
Epifluorescent microscope with automated stage (ZEISS, model: Axio Imager M1)
Cryostat (Thermo Fisher Scientific, model: HM525 NX)
P1000 pipetman (Gilson, catalog number: F123602)
P10 pipetman (Gilson, catalog number: F144802)
P2 pipetman (Gilson, catalog number: F144801)
-
Glass Coplin jars (VWR, catalog number: 470175-194)
Manufacturer: VARIETY GLASS, catalog number: 674.
Humidifying slide chamber (10 slide staining tray with black lid) (Electron Microscopy Sciences, catalog number: 71396-B)
Variable speed 2D rocker, 14 x 12 work surface (USA Scientific, catalog number: 2527-2000)
4 °C refrigerator (Fisher Scientific, model: Isotemp Value Lab, catalog number: 17LREEFSA)
-80 °C freezer (Thermo Fisher Scientific, model: Revco® Elite Plus)
Class II Type A/B3 Biological safety cabinet (NuAire)
LSR II flow cytometer equipped with a 355 nm, 405 nm and 488 nm lasers (BD Biosciences)
Shandon Cytospin 4 Cytocentrifuge (Thermo Fisher Scientific, catalog number: A78300003)
Software
Image capture software (Zeiss, Zen blue)
Image processing software with an event count tool (Zeiss, Zen blue or Zen lite)
Prism7 or equivalent graphing software (GraphPad)
FlowJo v10 (FlowJo)
Procedure
-
Multichannel flow cytometry validation of immunohistochemistry (IHC) macrophage markers
Approximately 3 g of human skeletal muscle tissue is cleaned from a patient’s hamstring tendon that is being utilized for ACL reconstruction surgery (performed in the operating room by trained medical personnel).
Place skeletal muscle tissue in an ice bucket, on ice for transport back to the laboratory, then place inside a UV sterilized biosafety cabinet and perform all possible remaining steps inside the biosafety cabinet.
Split the skeletal muscle specimen into multiple chunks as needed (1-2 g each) to allow for optimal tissue digestion.
Isolate skeletal muscle mononuclear cells according to the established protocol referenced here ( Liu et al., 2015 ).
After obtaining a single cell suspension, centrifuge at 500 × g (rcf) (relative centrifugal force or g-force) for 5 min at 4 °C with gentle braking to pellet cells.
Wash the cell pellet by resuspending with 1 ml sterile (autoclaved) 1x PBS, at this step, cell suspensions from multiple muscle chunks can be recombined back into one sample if desired.
Repeat Step A5 to pellet cells and resuspend in 500 µl of fluorophore-conjugated antibody cocktail (Table 3).
Mix well by pipetting up and down 6-8 times and incubate at 4 °C in the dark for 1 h.
Centrifuge at 500 × g (rcf) for 5 min at 4 °C with gentle braking to pellet cells, then resuspend in sterile 1x PBS to wash.
Repeat Step A9 for a second wash then resuspend cell pellet in 1 ml sterile 1x PBS for FACS.
-
Store in an ice bucket, on ice until fluorescence-activated cell sorting (FACS).
Incubate 1 drop of UltraComp eBeads Compensation Beads with each individual antibody and follow the manufacturer’s protocol to set single channel compensation parameters for each fluorophore-conjugated antibody.
An unstained sample of cell suspension should always be run to determine the location of negative cell populations ( Liu et al., 2015 ).
Determine the dead cells by LIVE/DEAD Fixable blue staining and exclude them from analyses by gating on live cells only.
Set a gate excluding doublets ( Liu et al., 2015 ) using the forward scatter height (y-axis) and forward scatter area (x-axis).
Isotype control antibodies in the appropriate fluorophores are used as a control to ensure antibody staining specificity (Table 3).
FACS sort CD11b+/CD14+ cells for immunocytochemistry (ICC) and collect into 1x PBS.
Following the sort, pellet cells by centrifugation at 500 × g for 5 min at 4 °C and resuspend in 600 µl of 1x PBS.
Load 200 µl of cell suspension into a Shandon Single Cytofunnel and centrifuge with a Shandon Cytospin 4 at 1,000 revolutions per minute (RPM) for 3 min to place cells onto Shandon Single Cytoslides.
Use PAP pen to trace the circle printed on the slide indicating where the sample is located and allow slides to air dry on the bench at room temperature (RT) for 30 min.
Follow Steps C8-C22 of the CD11b/CD206 double IHC protocol below, replacing the primary antibody against CD11b with anti-CD68 (Step C20c) at a 1:100 dilution (Table 2).
Following overnight (ON) incubation with primary anti-CD68 antibody and washes, incubate cells with ImmPRESS-AP, anti-mouse IgG (alkaline phosphatase) polymer detection kit for 1 h at RT, rocking (Table 2).
Discard ImmPRESS-AP reagent and wash cells by pipetting 1x PBS into PAP pen circle, then rock for 5 min at RT. Repeat wash 3 more times for a total of 4 washes.
-
Visualize CD68+ staining by adding alkaline phosphatase substrate using the ImmPACT Vector Red kit according to the manufacturer’s protocol (Table 2).
Staining intensity can be monitored using a standard light microscope to determine the optimal amount of staining time; 10-20 min was found to be sufficient.
Staining will be both chromogenic (appearing red) and fluorescent (visible using a TRITC filter cube).
Repeat Step A18 (above) to wash and coverslip using 50% 1x PBS/50% glycerol.
Image or store for up to a month at 4 °C in the dark (Figure 2).
-
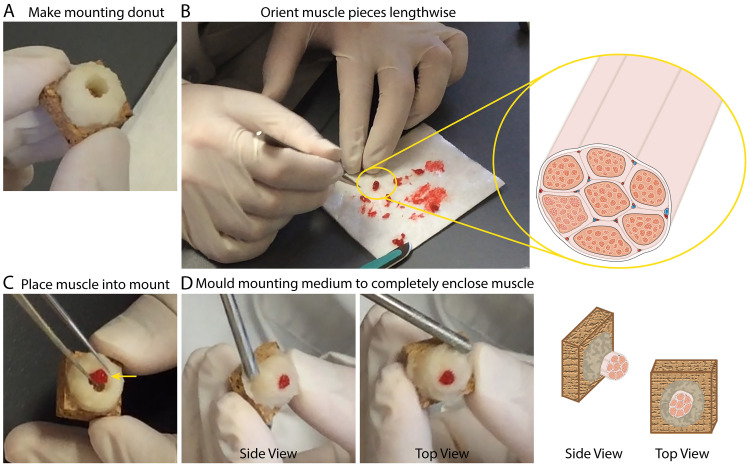
Preparing IHC mounts from a Bergstrom needle human skeletal muscle biopsy ( Tarnopolsky et al., 2011 ; Shanely et al., 2014 )
Note: This method is optional for preparing human skeletal muscle IHC mounts.- Mix tragacanth gum powder and O.C.T compound with a Teflon coated spatula at a ratio of 1 part tragacanth gum powder to 1.5 parts O.C.T to make a thick paste (mounting medium).
- Apply mounting medium to cork and mold into a donut shape, leaving a hole in the center where the muscle specimen will be placed (Figure 3A).
- Following the skeletal muscle biopsy, remove the tissue from the Bergstrom needle and quickly identify skeletal muscle pieces free of fat and connective tissue.
- Gently separate individual pieces, choosing the largest intact pieces for IHC (~0.5-0.75 cm in length and 50-100 mg of total tissue).
- Trim any fat, connective tissue or accessory skeletal muscle fragments with a scalpel to clean the sample.
- Lay each skeletal muscle piece for IHC side by side lengthwise, with the edges on one end aligned (this will become the top of the IHC mount) and trim the other end with a #10 curved blade scalpel to align (bottom of IHC mount) (Figure 3B).
- Using fine-tipped forceps and the Teflon spatula, gently roll the skeletal muscle pieces together into a solid cylindrical shape (muscle cylinder).
- Roll the muscle cylinder onto the tip of the spatula so that the bottom (the edge you trimmed) is aligned with the tip of the spatula.
- Stand the muscle cylinder/spatula up so that skeletal muscle fibers are now running perpendicular to bench and the untrimmed, aligned edge (top) is facing up.
- Gently place the muscle cylinder into the center of the mounting medium donut, against one side of the donut and use forceps to push the muscle cylinder off of the spatula. Remove the spatula. The muscle cylinder is now perpendicular to the surface of the cork with the bottom, trimmed end on the cork and the long axis of the skeletal muscle fibers sticking up (Figure 3C).
- Using the spatula, work the mounting medium in to completely enclose the skeletal muscle specimen, closing gaps and leaving no air pockets between the muscle cylinder and the mounting medium (this protects the muscle specimen from freeze damage).
- Sculpt the top of the mounting medium so that it is flush with the top of the skeletal muscle specimen.
-
Rapidly freeze the IHC mount by placing into liquid nitrogen cooled isopentane for 2 min.
- Isopentane should be a slushy consistency, with solid just beginning to form.
- The cork side of the mount should be facing up and the skeletal muscle specimen facing down, completely submerged in the cooled isopentane.
- Once frozen, use tongs to remove the IHC mount from the cooled isopentane and place on dry ice to rest for 5 min.
- The final IHC mount is now ready to be stored at -80 °C until you are ready to section (Figures 3D-3E).
-
Skeletal Muscle Sectioning and CD11b/CD206 Double IHC protocol
Pull IHC mount from -80 °C freezer and transport to the cryostat in a Styrofoam cooler on dry ice. Set the chamber temperature in the cryostat to -25 °C and place the IHC mount into the cryostat chamber.
Adhere the IHC mount to a chuck by covering the top of the chuck with O.C.T Compound, setting the cork into the O.C.T using forceps and allowing the O.C.T to harden by freezing.
Allow the IHC mount to acclimate to the temperature of the chamber for 1 h prior to sectioning. The chamber temperature may need to be adjusted for each sample in order to achieve flat sections with the greatest number of fibers in cross-section.
-
Cross-section frozen muscle mounts with the cryostat stage angle between 8° and 10°.
The stage angle may also need adjusted for some samples; however, this is less common and stage angles outside of this range are not recommended.
Sample should be adjusted so the top of the mount is perpendicular to the blade when sectioning. In order to obtain cross-sectional fibers, small adjustments in the angle of the chuck/mount may need to be made. Once adjustments are made, quality sections should be cut containing at least 100 muscle fibers and no more than 20% longitudinal fibers.
Using the anti-roll plate to keep sections from curling, cut frozen sections at 7 µm and pick up onto charged Superfrost Plus slides.
Allow sections to dry on the benchtop at RT, then circle samples to be stained using an ImmEdge PAP pen.
Allow PAP pen to dry for an additional 20-30 min.
Fix sections at -20 °C for 3 min by submerging slides in a Coplin jar containing ice cold acetone. Acetone should be stored at -20 °C to maintain temperature.
Quickly dab slide edges onto a paper towel to drain excess acetone and transfer to a Coplin jar filled with 1x PBS at RT.
Rock the slides in 1x PBS for 5 min; repeat wash step two more times for a total of three washes.
Remove slides from Coplin jar, gently flick to remove excess 1x PBS, wipe the back of the slide with a paper towel and place into a humidifying slide chamber containing approximately 1.5 cm of water in the reservoir.
Block endogenous peroxidases by pipetting 3% hydrogen peroxide onto the sections and allow to incubate, rocking at RT for 8 min.
Gently dump the hydrogen peroxide onto a paper towel to discard, replace with 1x PBS and rock for 5 min at RT.
Repeat Step C13 two more times for a total of three washes.
Dump excess 1x PBS onto a paper towel, flick slide, wipe the back with a paper towel and place back in the slide chamber.
-
Perform Streptavidin/Biotin blocking using the kit per the manufacturer’s instructions, carrying out all incubations in the humidifying slide chamber:
Incubate sections with Streptavidin blocking solution for 15 min at RT.
Wash briefly: two times, 2 min with 1x PBS.
Incubate sections with Biotin blocking solution for 15 min at RT.
Remove biotin blocking solution and replace with 1x PBS and incubate for 5 min, at RT, rocking. Repeat this wash with 1x PBS a total of three times.
Dump 1x PBS onto a paper towel, flick slide to remove excess 1x PBS, wipe the back of the slide with a paper towel and return to the slide chamber.
Add enough volume of 2.5% normal horse serum (NHS) to completely cover the sections and rock at 4 °C, overnight.
-
Remove 2.5% NHS, gently dab PAP pen with a kimwipe to re-establish hydrophobic barrier, add the primary antibody for CD11b, diluted 1:100 in 2.5% NHS, and return to the rocker at 4 °C overnight.
To ensure the specificity of the primary antibody against CD11b, an isotype specific control should also be prepared by adding purified mouse IgG1, κ to a section at the same concentration as the primary antibody against CD11b (0.1 mg/ml). The isotype control listed in Table 2 has a concentration of 0.5 mg/ml and should be diluted 1:500 in 2.5% NHS and incubated overnight at 4 °C, rocking (Figure 4A).
To determine signal produced by background staining of the tissue specimen, a no primary antibody control should be included. Prepare the no primary antibody control by covering the section in 2.5% NHS alone (omitting any antibody) and incubating overnight at 4 °C, rocking (Figure 4B).
For identification of macrophages using CD68, substitute anti-CD68 primary antibody, diluted 1:100, for primary antibody against CD11b in this step (Table 2) (Figure 2A).
For identification of specific M2 macrophage populations, substitute anti-CD163 primary antibody, diluted 1:50, for anti-CD11b primary antibody in this step and continue with the staining protocol as written (Table 2).
For identification of satellite cell populations, anti-Pax7 antibody, diluted 1:100, should be substituted for primary antibody against CD11b at this step (Table 2).
Remove primary antibody from sections and wash (as outlined in Step C17 above) four times, 5 min with 1x PBS, rocking at RT.
Dump excess 1x PBS onto a paper towel, flick to remove remaining 1x PBS, wipe the back of the slide with a paper towel, return to the slide chamber.
Add the biotinylated goat anti-mouse IgG1, diluted 1:1,000 in 2.5% NHS, and incubate by rocking for 90 min at RT.
Remove the biotinylated antibody from the sections and repeat the wash in Steps C21 and C22.
Add the SA-HRP, diluted 1:500 in 1x PBS, and incubate for 60 min, rocking at RT.
Remove the SA-HRP from the sections and wash three times, 5 min with 1x PBS, rocking at RT.
After the third wash, repeat Step C22 then add TSA 488, diluted 1:500 in 1x PBS, and incubate for 20 min, rocking at RT.
Remove the TSA 488 and repeat the washes in Step C26.
Repeat Steps C15-C18 to block the streptavidin and biotin used to label CD11b and prevent false co-staining.
Add enough volume of 2.5% NHS to completely cover the sections and rock at RT for at least 60 min (sections can be left in 2.5% NHS longer if desired).
-
Remove 2.5% NHS, gently dab the PAP pen with a Kimwipe to re-establish the hydrophobic barrier, add the primary antibody for CD206, diluted 1:200 in 2.5% NHS, then return the slide chamber to the rocker at 4 °C overnight.
To determine signal produced by background staining of the tissue specimen, a no primary antibody control should be included at this step also. Prepare the no primary antibody control by covering the section in 2.5% NHS alone (omitting any antibody) and incubating overnight at 4 °C, rocking (Figure 4C).
Remove CD206 antibody from sections and repeat Steps C21 and C22 to wash.
Add biotinylated rabbit anti-goat IgG, diluted 1:500 in 2.5% NHS, and incubate for 90 min at RT, rocking.
Remove the biotin from the sections and wash four times, 5 min with 1x PBS, rocking at RT.
Add the SA-594, diluted 1:200 in 1x PBS, and incubate for 60 min, rocking at RT.
Remove the SA-594 from the sections and wash three times, 5 min with 1x PBS, rocking at RT.
Incubate sections with DAPI, diluted 1:10,000 in 1x PBS, for 10 min at RT, rocking.
Remove DAPI from the sections and wash three times, 5 min with 1x PBS.
Dump excess 1x PBS onto a paper towel and add enough volume of PBS/Glycerol or Vectashield mounting medium to cover the sections (20-50 µl/slide).
Gently lower coverslip onto the slide, avoiding the formation of bubbles under the coverslip. Allow the mounting medium to spread by leaving the slides laying coverslip up for 5-10 min.
Drain excess mounting medium from slides by standing vertical on a paper towel for 5 min.
Image muscle sections or store slides at 4 °C protected from light until ready to image. Staining will last for several months if slides are stored properly at 4 °C, in the dark.
Figure 3. Preparing IHC mounts from human skeletal muscle.
. A. Representative image illustrating the donut, made from a mixture of Tragacanth powder and O.C.T. (mounting medium), described under Procedure B Step 2. B. Representative image and illustration depicting the lengthwise orientation of muscle pieces to be mounted for sectioning. C. Representative image showing the placement of a muscle cylinder into the donut made of mounting medium. Yellow arrow serves to illustrate that the muscle is positioned so that it is against one side/wall of the donut. D. Representative images of the finished IHC muscle mount (just prior to freezing); side view (left) and top view (right).
Data analysis
Quantification of skeletal muscle fiber number and macrophage abundance
-
General guidelines for skeletal muscle macrophage analysis
Sections to be analyzed for macrophage numbers should contain at least 50-100 skeletal muscle fibers.
Non-specific staining is common around the edges of skeletal muscle sections; therefore, exclude the edges when counting macrophages.
Do not include longitudinal skeletal muscle fibers in the area to be counted; this skews the number of macrophages/fiber.
Areas containing blood patches, edema or fibrosis will sometimes be observed in damaged tissues (Figure 5B). These areas are usually filled with macrophages and shouldn’t be included in the same analysis as macrophages located in between skeletal muscle fibers (Figure 5A). Whether these macrophages should be quantified depends on the aim of the study.
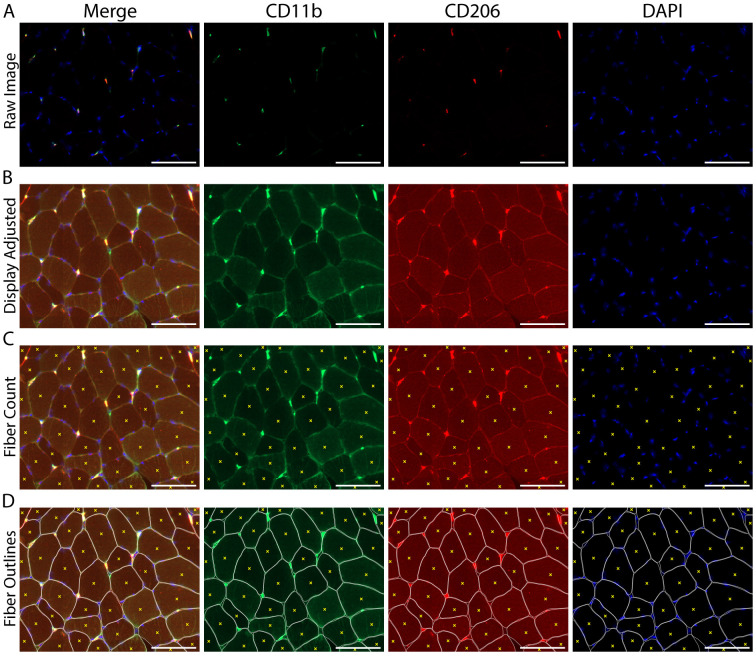
Adjust the image display in any channel so that the background staining is visible and individual skeletal muscle fibers can be distinguished (Figures 6A-6B).
Manually count the number of total skeletal muscle fibers within the region of interest using the “event” tool in Zen image capture software to demarcate individual skeletal muscle fibers (Figures 6C-6D).
-
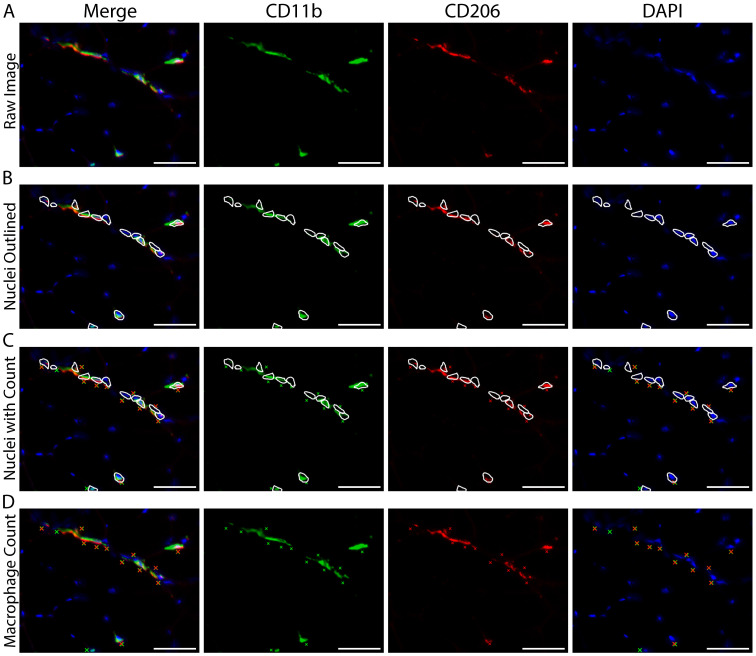
Adjust the image display so that CD11b+ macrophages (green) and DAPI+ cell nuclei (blue) can be clearly observed and cell shape/morphology is distinct (Figures 7A-7B).
Isotype-specific staining controls for antibody specificity should be used to help determine the appropriate display adjustment for identifying true positive staining (Figure 5A).
No primary antibody staining controls for non-specific binding of antibodies/amplification reagents can be used to aid in determining background tissue staining and display adjustment for counting (Figures 4B-4C).
-
Using DAPI as a cell marker, manually identify and count the total number of CD11b+ macrophages located near DAPI (further explanation in the Macrophage Counting Notes section below) (Figure 7C).
Skeletal muscle macrophages are morphologically heterogeneous (Figures 8A-8D). For macrophage counting, we do not discriminate between different morphological groups (i.e., all CD11b+/CD206+ macrophages are counted together regardless of morphology) (see General Note 4).
If DAPI is not present near the staining, we do not count the staining as a positive event, even if the morphology seems apparent.
Staining must be large enough in size to represent a cellular structure (at least the same size as an intact nucleus, > 5 µm) – small dots/patches are usually just non-specific staining of cellular debris by the secondary antibody.
In patches where individual macrophages cannot be distinguished, manually count each DAPI+ nucleus that touches macrophage staining (Figures 9A-9D).
-
After counting total CD11b+ macrophages, go back and manually count the number of these macrophages that also express CD206.
CD11b+/CD206+ macrophages are the mixed M1/M2 skeletal muscle macrophage population.
CD11b+/CD206- macrophages may represent the M1 population.
To determine the number of M1 macrophages, subtract the number of CD11b+/CD206+ macrophages from the total number of macrophages expressing CD11b+. This gives you the number of macrophages only expressing CD11b+ (CD11b+/CD206-).
For final numbers, divide each macrophage count by the total number of fibers in the region counted.
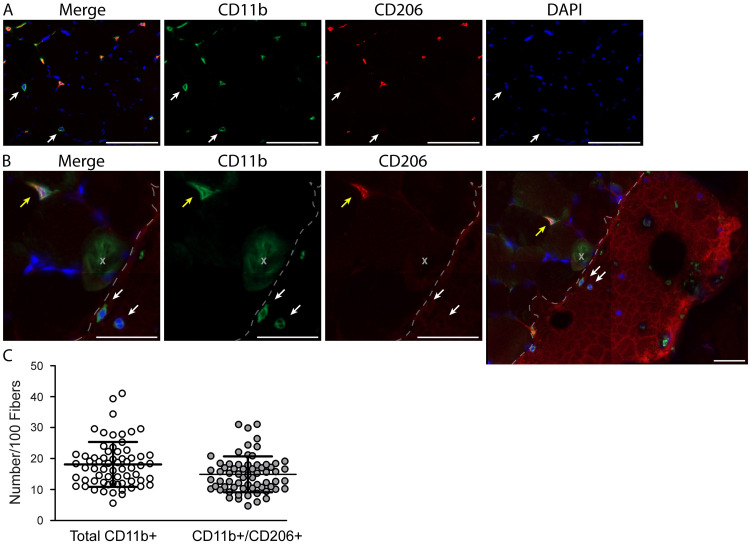
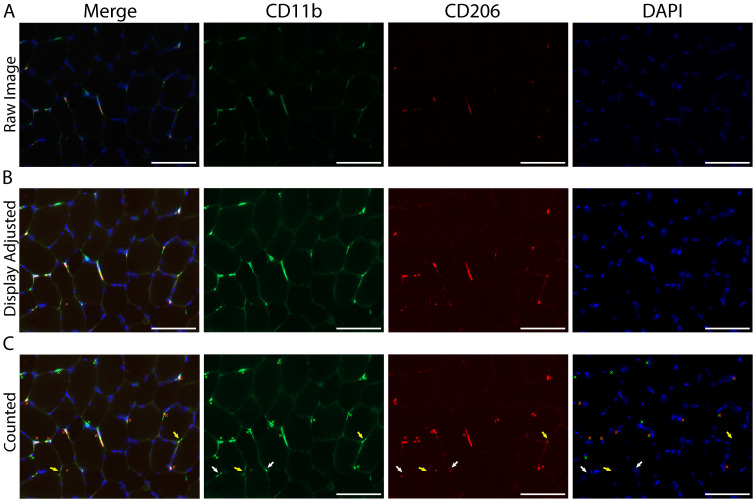
Figure 5. Resident macrophages largely co-express CD11b and CD206 in human vastus lateralis muscle.
A. Representative images showing macrophage staining: CD11b (green), CD206 (red) and DAPI (cell nuclei, blue). The majority of macrophages are positive for both CD11b and CD206; however, CD11b+/CD206- macrophages can be observed (white arrows). Scale bars = 100 µm. B. Muscle section from the vastus lateralis showing a CD11b+/CD206+ (yellow arrow) and two CD11b+/CD206- (white arrows) monocytes. Notice in the larger field of view (at right) the CD11b+/CD206- monocytes are located in a patch of blood at the edge of the muscle section, validating CD206 as a reliable marker of tissue resident macrophages in human skeletal muscle. The patch of blood can be identified by its dried, cracked appearance and high red background signal (border between blood patch and muscle fibers in the section denoted with a dashed line). The gray X is marking an area with green background signal produced by a bubble underneath the section (further outlined in Figure S1). Scale bars = 50 µm. C. Bar graph quantification showing the number of total macrophages (all CD11b+) and CD11b+/CD206+ macrophages per 100 skeletal muscle fibers. Sixty-five muscle biopsies from the vastus lateralis were analyzed. Each dot represents macrophage counts from a single subject/section with overlap between Total CD11b+ and CD11b+/CD206+ groups, illustrating that the majority of macrophages (Total CD11b+) also express CD206 (CD11b+/CD206+). In skeletal muscle samples approximately 82% of the total macrophage population co-express CD11b and CD206.
Figure 6. Image adjustment and counting of skeletal muscle fiber number within a cross-section.
A. Original image, prior to manipulation of the display settings; B. The same image following adjustment of the display to increase the visible background staining, allowing for the identification and counting of skeletal muscle fibers; C. Count of individual muscle fibers, denoted by yellow Xs; D. Fibers were manually outlined to help demarcate individual skeletal muscle fiber borders (white) along with the fiber count from panel C (yellow Xs). A-D) CD11b (green), CD206 (red), cell nuclei/DAPI (blue). Scale bars = 100 µm.
Figure 7. Identifying and quantifying macrophages within a skeletal muscle cross-section.
A. Representative image of macrophage staining in human vastus lateralis: CD11b (green), CD206 (red) and cell nuclei/DAPI (blue). B. Display adjustment of the images in panel A showing enough tissue background staining to clearly identify macrophages from non-specific tissue staining. C. Macrophage count, demarcated with colored Xs: all CD11b+ (green Xs), all CD206+ (red Xs), CD11b+/CD206+ macrophages are marked with both a green and red X. White arrows indicate areas that appear to have positive staining and are near DAPI, but were not counted due to size and/or lack of distinct morphological features determinant of a cellular structure. Other areas were not counted due to a lack of DAPI staining (yellow arrows). Scale bars = 100 µm.
Figure 9. Counting macrophages when individual cells cluster together.
A. Representative image showing an area of macrophage staining with multiple nuclei where individual macrophages cannot be distinguished. B. The image in panel A, with outlines demarcating DAPI+ nuclei touching areas of positive macrophage staining. C. Panel B images including Xs marking each nucleus counted as positive macrophage staining. D. Original image from panel A showing the Xs in panel C marking macrophage counts without the outlines demarcating positive nuclei. A-D) CD11b (green), CD206 (red) and cell nuclei/DAPI (blue). Scale bars = 50 µm.
Notes
-
General Notes
Collection of skeletal muscle biopsies was carried out in accordance with the Declaration of Helsinki. Skeletal muscle biopsies from three separate studies conducted at the University of Kentucky, the University of Alabama at Birmingham and the Geriatric Research Education and Clinical Center, Central Arkansas Veterans Healthcare System were used in this protocol. Subjects at the University of Kentucky provided their written informed consent from protocols approved by the Institutional Review Board and the University of Kentucky. Details regarding subject consent and protocol approval for the other two studies are outlined in the following publications: ( Dennis et al., 2015 ; Long et al., 2017 ).
We have used this protocol to identify macrophage populations in subjects ranging from age 19 to 83 and spanning a wide range of activity levels. We have found the percentage of total CD11b+ macrophages that co-express CD206 (~82%) remains relatively consistent across demographics; however, the abundance (number/fiber) and phenotype may be affected by age, obesity and/or exercise.
We acknowledge that surface marker expression is not sufficient to infer function and further analyses should be employed to determine functional characteristics of macrophage populations.
Skeletal muscle macrophages display heterogeneous morphology; however, we do not currently include macrophage morphology as a variable in our analyses (Figure 8). Though morphology alone is likely not enough to distinguish macrophage populations, we believe macrophage morphology may be an important and telling variable for some studies and may provide further insight into macrophage phenotype beyond surface marker expression alone.
Post sectioning drying time depends on the size of the sample, but 3-4 h are usually sufficient for skeletal muscle biopsies. Following drying, slides can be stored at -20 °C if staining will be done at a later time.
If samples were stored at -20 °C, slides should be allowed to warm to RT for 15-20 min prior to acetone fixation.
Batch controls should be included if multiple sets of staining will be done at different time points within a single study and should include at least one control and one experimental sample.
For all reagents pipetted onto the slide, use enough volume to cover the sections and fill the area you created with the PAP pen, usually between 200 and 500 µl/slide.
During the incubation with 3% hydrogen peroxide, you may see bubbles form on the section indicating that the peroxide is working; a lack of bubbling will not affect the staining outcome but is indicative of poor tissue quality.
Following Step C26, slides can be coverslipped with PBS/Glycerol and staining can be checked prior to moving forward with the protocol. Incubate slides in a Coplin jar with 1x PBS to remove the coverslips (they will fall off), wash 2 or 3 times with 1x PBS and continue with Step C27.
-
Draining slides post coverslip:
Slides should be drained enough that mounting medium is no longer leaking from under the coverslip and the coverslip tightly adheres to the slide.
Take care not to over-drain the slides; this will lead to the formation of air pockets underneath the coverslip.
For long-term storage, slides can be mounted with Vectashield mounting medium.
Slides can be sealed by painting the edges of the coverslip with nail polish to prevent the formation of air pockets underneath the coverslip over time.
Both CD11b and CD68 antibodies are a mouse IgG1 isotype and both require amplification using Superboost Tyramide signal amplification reagents. For these reasons, co-staining of CD11b and CD68 is not possible (false positive double staining occurred).
This protocol can be adapted to identify a subset of CD206+ macrophages expressing CD163 by substituting CD163 primary antibody (see Table 1) in place of CD11b primary antibody and following the remaining protocol as written.
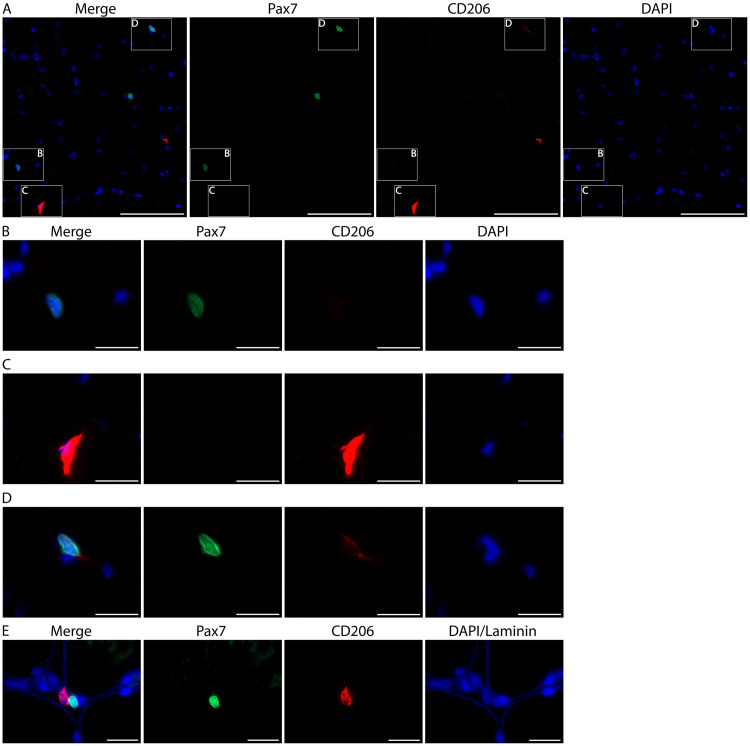
Primary antibody for Pax7 (see Table 1) can be substituted for CD11b primary antibody to label satellite cells; the remaining protocol steps are the same (Figure 10).
-
Macrophage Counting Notes
-
Image capture should be adjusted properly for easy identification of macrophages.
Exposure times should be set so that macrophage and nuclear morphology is clear and discernable and background staining is as minimally visible as possible.
Images should be acquired with a 20x objective or higher magnification.
-
For stitched images:
Section boundaries are easiest to set using DAPI staining to find the edges of the tissue section.
Focus points are best set using the FITC channel (CD11b staining), to ensure that macrophages are in focus with clear morphology.
Staining must be located near DAPI (within 5 µm), but does not have to be directly on top of DAPI staining. If the morphology is a macrophage and it is close but not touching DAPI, we will count the event as a macrophage (Figure 7).
We have found that CD163+ macrophages make-up a subset of total CD206+ macrophages. Thus, if quantifying these populations, CD206+ macrophages should be counted as the ‘parent’ macrophage population (similar to CD11b above).
Establishing a set of counting parameters prior to analyzing data sets is helpful when quantifying macrophages and also helps minimize variance between counters.
-
Choose a handful of images as a ‘guide’ counting set and use these images to set up guidelines for how macrophages will be identified and to train new counters.
It is important to stay consistent between images, groups and studies with regard to the identification of macrophages.
Within one data set it is better to have the same blinded counter to analyze the entire data set due to variation between counters.
We have found that although absolute numbers vary between counters, overall trends with regard to increases or decreases in the abundance of macrophage populations remain consistent if basic guidelines are outlined and followed.
-
Figure 10. Skeletal muscle stem cells (satellite cells) can weakly express CD206 but are easily distinguished from macrophages.
A. Representative image from human vastus lateralis showing a Pax7+ satellite cell (green, B), a CD206+ macrophage (red, C) and a Pax7+/CD206weak satellite cell (D). Cell nuclei are stained with DAPI (blue). Scale bars = 100 µm. B-D. Higher magnification of the boxes from panel A. Note the overexposure of CD206 macrophage staining (C) when the intensity is adjusted so that CD206 expression in satellite cells is visible (D). Images acquired as Z stacks with a 40x objective. Scale bars = 20 µm. E. High magnification images from human vastus lateralis muscle showing a CD206+ muscle macrophage (red) in close proximity to a Pax7+ satellite cell (green). Cell nuclei are stained with DAPI and skeletal muscle fiber borders are stained with Laminin (both in blue). Images were acquired as Z stacks with a 100x oil objective. Scale bars = 20 µm.
Recipes
-
1x PBS (10 mM, pH 7.4)
Mix 69.68 g NaCl, 17.36 g Na2HPO4·7H2O, 2.08 g KH2PO4
Stir to dissolve in DI water
Dilute 10 N NaOH 1:5 with DI water to make a 2 N solution; dilute 6 N HCl 1:3 with DI water to make a 2 N solution
Adjust pH of 1x PBS with 2 N NaOH or HCl
Bring to a final volume of 8 L
1x PBS can be kept at RT for up to 3 months
-
3% hydrogen peroxide
Dilute 30% hydrogen peroxide 1:10 in 1x PBS
-
DAPI for staining cell nuclei
Prepare a 5 mg/ml stock solution by diluting in 1x PBS
Aliquot and store at -20 °C
A working dilution of 1:10,000 in 1x PBS is used for labeling nuclei
Acknowledgments
This work was supported by the National Institute of Aging grant (AG046920) and by the NIH Clinical and Translational Science Award (CTSA) (UL1TR001998) at the University of Kentucky. This work also utilized de-identified samples obtained through Merit Review Award #RX0012030 to Richard A. Dennis from the US Department of Veterans Affairs (VA), Rehabilitation R&D Service. The contents do not represent the views of the VA or US Government. The authors would like to thank Sami Michaels for help with the quantification of macrophages shown in Figure 5. We would also like to thank Doug Long for recruitment of research subjects at the University of Kentucky. Multi-channel flow cytometry was collected with help from the UK Flow Cytometry & Cell Sorting core facility (www.research.uky.edu/core/flow). The UK Flow Cytometry & Cell Sorting core facility is supported in part by the Office of the Vice President for Research, the Markey Cancer Center and an NCI Center Core Support Grant (P30 CA177558) to the University of Kentucky Markey Cancer Center.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Supplementary Data.
References
- 1. Ambarus C. A., Krausz S., M. van Eijk J. Hamann, Radstake T. R., Reedquist K. A., Tak P. P. and Baeten D. L.(2012). Systematic validation of specific phenotypic markers for in vitro polarized human macrophages . J Immunol Methods 375(1-2): 196-206. [DOI] [PubMed] [Google Scholar]
- 2. Arnold L., Henry A., Poron F., Baba-Amer Y., N. van Rooijen A. Plonquet, Gherardi R. K. and Chazaud B.(2007). Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med 204(5): 1057-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beaton L. J., Tarnopolsky M. A. and Phillips S. M.(2002). Contraction-induced muscle damage in humans following calcium channel blocker administration. J Physiol 3): 849-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyette L. B., Macedo C., Hadi K., Elinoff B. D., Walters J. T., Ramaswami B., Chalasani G., Taboas J. M., Lakkis F. G. and Metes D. M.(2017). Phenotype, function, and differentiation potential of human monocyte subsets. PLoS One 12(4): e0176460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ceafalan L. C., Fertig T. E., Popescu A. C., Popescu B. O., Hinescu M. E. and Gherghiceanu M.(2017). Skeletal muscle regeneration involves macrophage-myoblast bonding. Cell Adh Migr 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chazaud B., Brigitte M., Yacoub-Youssef H., Arnold L., Gherardi R., Sonnet C., Lafuste P. and Chretien F.(2009). Dual and beneficial roles of macrophages during skeletal muscle regeneration. Exerc Sport Sci Rev 37(1): 18-22. [DOI] [PubMed] [Google Scholar]
- 7. Chistiakov D. A., Killingsworth M. C., Myasoedova V. A., Orekhov A. N. and Bobryshev Y. V.(2017). CD68/macrosialin: not just a histochemical marker. Lab Invest 97(1): 4-13. [DOI] [PubMed] [Google Scholar]
- 8. Crameri R. M., Aagaard P., Qvortrup K., Langberg H., Olesen J. and Kjaer M.(2007). Myofibre damage in human skeletal muscle: effects of electrical stimulation versus voluntary contraction. J Physiol 1): 365-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crameri R. M., Langberg H., Teisner B., Magnusson P., Schroder H. D., Olesen J. L., Jensen C. H., Koskinen S., Suetta C. and Kjaer M.(2004). Enhanced procollagen processing in skeletal muscle after a single bout of eccentric loading in humans. Matrix Biol 23(4): 259-264. [DOI] [PubMed] [Google Scholar]
- 10. Dennis R. A., Ponnappan U., Kodell R. L., Garner K. K., Parkes C. M., Bopp M. M., Padala K. P., Peterson C. A., Padala P. R. and Sullivan D. H.(2015). Immune function and muscle adaptations to resistance exercise in older adults: study protocol for a randomized controlled trial of a nutritional supplement. Trials 16: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DiPasquale D. M., Cheng M., Billich W., Huang S. A., N. van Rooijen T. A. Hornberger and Koh T. J.(2007). Urokinase-type plasminogen activator and macrophages are required for skeletal muscle hypertrophy in mice. Am J Physiol Cell Physiol 293(4): C1278-1285. [DOI] [PubMed] [Google Scholar]
- 12. Du H., Shih C. H., Wosczyna M. N., Mueller A. A., Cho J., Aggarwal A., Rando T. A. and Feldman B. J.(2017). Macrophage-released ADAMTS1 promotes muscle stem cell activation. Nat Commun 8(1): 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dumont N. A. and Frenette J.(2013). Macrophage colony-stimulating factor-induced macrophage differentiation promotes regrowth in atrophied skeletal muscles and C2C12 myotubes. Am J Pathol 182(2): 505-515. [DOI] [PubMed] [Google Scholar]
- 14. Durafourt B. A., Moore C. S., Zammit D. A., Johnson T. A., Zaguia F., Guiot M. C., Bar-Or A. and Antel J. P.(2012). Comparison of polarization properties of human adult microglia and blood-derived macrophages. Glia 60(5): 717-727. [DOI] [PubMed] [Google Scholar]
- 15. Fink L. N., Costford S. R., Lee Y. S., Jensen T. E., Bilan P. J., Oberbach A., Bluher M., Olefsky J. M., Sams A. and Klip A.(2014). Pro-inflammatory macrophages increase in skeletal muscle of high fat-fed mice and correlate with metabolic risk markers in humans. Obesity(Silver Spring) 22(3): 747-757. [DOI] [PubMed] [Google Scholar]
- 16. Gordon S., Pluddemann A. and Martinez F. Estrada (2014). Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol Rev 262(1): 36-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gottfried E., Kunz-Schughart L. A., Weber A., Rehli M., Peuker A., Muller A., Kastenberger M., Brockhoff G., Andreesen R. and Kreutz M.(2008). Expression of CD68 in non-myeloid cell types. Scand J Immunol 67(5): 453-463. [DOI] [PubMed] [Google Scholar]
- 18. Hong E. G., Ko H. J., Cho Y. R., Kim H. J., Ma Z., Yu T. Y., Friedline R. H., Kurt-Jones E., Finberg R., Fischer M. A., Granger E. L., Norbury C. C., Hauschka S. D., Philbrick W. M., Lee C. G., Elias J. A. and Kim J. K.(2009). Interleukin-10 prevents diet-induced insulin resistance by attenuating macrophage and cytokine response in skeletal muscle. Diabetes 58(11): 2525-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ikeda S., Tamura Y., Kakehi S., Takeno K., Kawaguchi M., Watanabe T., Sato F., Ogihara T., Kanazawa A., Fujitani Y., Kawamori R. and Watada H.(2013). Exercise-induced enhancement of insulin sensitivity is associated with accumulation of M2-polarized macrophages in mouse skeletal muscle. Biochem Biophys Res Commun 441(1): 36-41. [DOI] [PubMed] [Google Scholar]
- 20. Iqbal S. a. K., A (2015). Characterization of in vitro generated human polarized macrophages . J Clin Cell Immunol 6(380). [Google Scholar]
- 21. Italiani P. and Boraschi D.(2014). From Monocytes to M1/M2 Macrophages: Phenotypical vs. Functional Differentiation. Front Immunol 5: 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jansen K. M. and Pavlath G. K.(2006). Mannose receptor regulates myoblast motility and muscle growth. J Cell Biol 174(3): 403-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kharraz Y., Guerra J., Mann C. J., Serrano A. L. and Munoz-Canoves P.(2013). Macrophage plasticity and the role of inflammation in skeletal muscle repair. Mediators Inflamm 2013: 491497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kunisch E., Fuhrmann R., Roth A., Winter R., Lungershausen W. and Kinne R. W.(2004). Macrophage specificity of three anti-CD68 monoclonal antibodies(KP1, EBM11, and PGM1) widely used for immunohistochemistry and flow cytometry. Ann Rheum Dis 63(7): 774-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu L., Cheung T. H., Charville G. W. and Rando T. A.(2015). Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat Protoc 10(10): 1612-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lolmede K., Campana L., Vezzoli M., Bosurgi L., Tonlorenzi R., Clementi E., Bianchi M. E., Cossu G., Manfredi A. A., Brunelli S. and Rovere-Querini P.(2009). Inflammatory and alternatively activated human macrophages attract vessel-associated stem cells, relying on separate HMGB1- and MMP-9-dependent pathways. J Leukoc Biol 85(5): 779-787. [DOI] [PubMed] [Google Scholar]
- 27. Long D. E., Peck B. D., Martz J. L., Tuggle S. C., Bush H. M., McGwin G., Kern P. A., Bamman M. M. and Peterson C. A.(2017). Metformin to Augment Strength Training Effective Response in Seniors(MASTERS): study protocol for a randomized controlled trial. Trials 18(1): 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mackey A. L. and Kjaer M.(2017). The breaking and making of healthy adult human skeletal muscle in vivo . Skelet Muscle 7(1): 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. MacNeil L. G., Baker S. K., Stevic I. and Tarnopolsky M. A.(2011). 17β-estradiol attenuates exercise-induced neutrophil infiltration in men. Am J Physiol Regul Integr Comp Physiol 300(6): R1443-1451. [DOI] [PubMed] [Google Scholar]
- 30. Mahoney D. J., Safdar A., Parise G., Melov S., Fu M., MacNeil L., Kaczor J., Payne E. T. and Tarnopolsky M. A.(2008). Gene expression profiling in human skeletal muscle during recovery from eccentric exercise. Am J Physiol Regul Integr Comp Physiol 294(6): R1901-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martinez F. O. and Gordon S.(2014). The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martinez F. O., Gordon S., Locati M. and Mantovani A.(2006). Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol 177(10): 7303-7311. [DOI] [PubMed] [Google Scholar]
- 33. McWhorter F. Y., Wang T., Nguyen P., Chung T. and Liu W. F.(2013). Modulation of macrophage phenotype by cell shape. Proc Natl Acad Sci U S A 110(43): 17253-17258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Melton D. W., Roberts A. C., Wang H., Sarwar Z., Wetzel M. D., Wells J. T., Porter L., Berton M. T., McManus L. M. and Shireman P. K.(2016). Absence of CCR2 results in an inflammaging environment in young mice with age-independent impairments in muscle regeneration. J Leukoc Biol 100(5): 1011-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mikkelsen U. R., Langberg H., Helmark I. C., Skovgaard D., Andersen L. L., Kjaer M. and Mackey A. L.(2009). Local NSAID infusion inhibits satellite cell proliferation in human skeletal muscle after eccentric exercise. J Appl Physiol(1985) 107(5): 1600-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mosser D. M. and Edwards J. P.(2008). Exploring the full spectrum of macrophage activation. Nat Rev Immunol 8(12): 958-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murray P. J., Allen J. E., Biswas S. K., Fisher E. A., Gilroy D. W., Goerdt S., Gordon S., Hamilton J. A., Ivashkiv L. B., Lawrence T., Locati M., Mantovani A., Martinez F. O., Mege J. L., Mosser D. M., Natoli G., Saeij J. P., Schultze J. L., Shirey K. A., Sica A., Suttles J., Udalova I., J. A. van Ginderachter S. N. Vogel and Wynn T. A.(2014). Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41(1): 14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Novak M. L. and Koh T. J.(2013). Phenotypic transitions of macrophages orchestrate tissue repair. Am J Pathol 183(5): 1352-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Olah M., Biber K., Vinet J. and Boddeke H. W.(2011). Microglia phenotype diversity. CNS Neurol Disord Drug Targets 10(1): 108-118. [DOI] [PubMed] [Google Scholar]
- 40. Paulsen G., Crameri R., Benestad H. B., Fjeld J. G., Morkrid L., Hallen J. and Raastad T.(2010). Time course of leukocyte accumulation in human muscle after eccentric exercise. Med Sci Sports Exerc 42(1): 75-85. [DOI] [PubMed] [Google Scholar]
- 41. Paulsen G., Egner I., Raastad T., Reinholt F., Owe S., Lauritzen F., Brorson S. H. and Koskinen S.(2013). Inflammatory markers CD11b, CD16, CD66b, CD68, myeloperoxidase and neutrophil elastase in eccentric exercised human skeletal muscles. Histochem Cell Biol 139(5): 691-715. [DOI] [PubMed] [Google Scholar]
- 42. Paulsen G., Egner I. M., Drange M., Langberg H., Benestad H. B., Fjeld J. G., Hallen J. and Raastad T.(2010). A COX-2 inhibitor reduces muscle soreness, but does not influence recovery and adaptation after eccentric exercise. Scand J Med Sci Sports 20(1): e195-207. [DOI] [PubMed] [Google Scholar]
- 43. Peterson J. M., Trappe T. A., Mylona E., White F., Lambert C. P., Evans W. J. and Pizza F. X.(2003). Ibuprofen and acetaminophen: effect on muscle inflammation after eccentric exercise. Med Sci Sports Exerc 35(6): 892-896. [DOI] [PubMed] [Google Scholar]
- 44. Przybyla B., Gurley C., Harvey J. F., Bearden E., Kortebein P., Evans W. J., Sullivan D. H., Peterson C. A. and Dennis R. A.(2006). Aging alters macrophage properties in human skeletal muscle both at rest and in response to acute resistance exercise. Exp Gerontol 41(3): 320-327. [DOI] [PubMed] [Google Scholar]
- 45. Reidy P. T., Lindsay C. C., McKenzie A. I., Fry C. S., Supiano M. A., Marcus R. L., LaStayo P. C. and Drummond M. J.(2017). Aging-related effects of bed rest followed by eccentric exercise rehabilitation on skeletal muscle macrophages and insulin sensitivity. Exp Gerontol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rigamonti E., Zordan P., Sciorati C., Rovere-Querini P. and Brunelli S.(2014). Macrophage plasticity in skeletal muscle repair. Biomed Res Int 2014: 560629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roszer T.(2015). Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediators Inflamm 2015: 816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saclier M., Cuvellier S., Magnan M., Mounier R. and Chazaud B.(2013). Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. FEBS J 280(17): 4118-4130. [DOI] [PubMed] [Google Scholar]
- 49. Saclier M., Yacoub-Youssef H., Mackey A. L., Arnold L., Ardjoune H., Magnan M., Sailhan F., Chelly J., Pavlath G. K., Mounier R., Kjaer M. and Chazaud B.(2013). Differentially activated macrophages orchestrate myogenic precursor cell fate during human skeletal muscle regeneration. Stem Cells 31(2): 384-396. [DOI] [PubMed] [Google Scholar]
- 50. Sciorati C., Rigamonti E., Manfredi A. A. and Rovere-Querini P.(2016). Cell death, clearance and immunity in the skeletal muscle. Cell Death Differ 23(6): 927-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shanely R. A., Zwetsloot K. A., Triplett N. T., Meaney M. P., Farris G. E. and Nieman D. C.(2014). Human skeletal muscle biopsy procedures using the modified Bergstrom technique. J Vis Exp(91): 51812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smith C., Kruger M. J., Smith R. M. and Myburgh K. H.(2008). The inflammatory response to skeletal muscle injury: illuminating complexities. Sports Med 38(11): 947-969. [DOI] [PubMed] [Google Scholar]
- 53. Sprangers S., T. J. de Vries and V. Everts (2016). Monocyte heterogeneity: consequences for monocyte-derived immune cells. J Immunol Res 2016: 1475435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stupka N., Tarnopolsky M. A., Yardley N. J. and Phillips S. M.(2001). Cellular adaptation to repeated eccentric exercise-induced muscle damage. J Appl Physiol(1985) 91(4): 1669-1678. [DOI] [PubMed] [Google Scholar]
- 55. Tam C. S., Sparks L. M., Johannsen D. L., Covington J. D., Church T. S. and Ravussin E.(2012). Low macrophage accumulation in skeletal muscle of obese type 2 diabetics and elderly subjects. Obesity(Silver Spring) 20(7): 1530-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tarnopolsky M. A., Pearce E., Smith K. and Lach B.(2011). Suction-modified Bergstrom muscle biopsy technique: experience with 13,500 procedures. Muscle Nerve 43(5): 717-725. [DOI] [PubMed] [Google Scholar]
- 57. Tidball J. G., Dorshkind K. and Wehling-Henricks M.(2014). Shared signaling systems in myeloid cell-mediated muscle regeneration. Development 141(6): 1184-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tidball J. G. and Villalta S. A.(2010). Regulatory interactions between muscle and the immune system during muscle regeneration. Am J Physiol Regul Integr Comp Physiol 298(5): R1173-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Varga T., Mounier R., Horvath A., Cuvellier S., Dumont F., Poliska S., Ardjoune H., Juban G., Nagy L. and Chazaud B.(2016). Highly dynamic transcriptional signature of distinct macrophage subsets during sterile inflammation, resolution, and tissue repair. J Immunol 196(11): 4771-4782. [DOI] [PubMed] [Google Scholar]
- 60. Varma V., Yao-Borengasser A., Rasouli N., Nolen G. T., Phanavanh B., Starks T., Gurley C., Simpson P., McGehee R. E., Jr. P. A. Kern and Peterson C. A.(2009). Muscle inflammatory response and insulin resistance: synergistic interaction between macrophages and fatty acids leads to impaired insulin action. Am J Physiol Endocrinol Metab 296(6): E1300-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang H., Melton D. W., Porter L., Sarwar Z. U., McManus L. M. and Shireman P. K.(2014). Altered macrophage phenotype transition impairs skeletal muscle regeneration. Am J Pathol 184(4): 1167-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wehling-Henricks M., Welc S. S., Samengo G., Rinaldi C., Lindsey C., Wang Y., Lee J., Kuro O. M. and Tidball J. G.(2018). Macrophages escape Klotho gene silencing in the mdx mouse model of Duchenne muscular dystrophy and promote muscle growth and increase satellite cell numbers through a Klotho-mediated pathway. Hum Mol Genet 27(1): 14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.