DMI shows metabolism of acetate and glucose in the brain and liver and reveals the Warburg effect in patients with brain tumors.
Abstract
Currently, the only widely available metabolic imaging technique in the clinic is positron emission tomography (PET) detection of the radioactive glucose analog 2-18F-fluoro-2-deoxy-d-glucose (18FDG). However, 18FDG-PET does not inform on metabolism downstream of glucose uptake and often provides ambiguous results in organs with intrinsic high glucose uptake, such as the brain. Deuterium metabolic imaging (DMI) is a novel, noninvasive approach that combines deuterium magnetic resonance spectroscopic imaging with oral intake or intravenous infusion of nonradioactive 2H-labeled substrates to generate three-dimensional metabolic maps. DMI can reveal glucose metabolism beyond mere uptake and can be used with other 2H-labeled substrates as well. We demonstrate DMI by mapping metabolism in the brain and liver of animal models and human subjects using [6,6′-2H2]glucose or [2H3]acetate. In a rat glioma model, DMI revealed pronounced metabolic differences between normal brain and tumor tissue, with high-contrast metabolic maps depicting the Warburg effect. We observed similar metabolic patterns and image contrast in two patients with a high-grade brain tumor after oral intake of 2H-labeled glucose. Further, DMI used in rat and human livers showed [6,6′-2H2]glucose stored as labeled glycogen. DMI is a versatile, robust, and easy-to-implement technique that requires minimal modifications to existing clinical magnetic resonance imaging scanners. DMI has great potential to become a widespread method for metabolic imaging in both (pre)clinical research and the clinic.
INTRODUCTION
Metabolism plays a key role in the origin or progression of many diseases, including neurodegenerative diseases, diabetes, and cancer (1–4). Clinicians and patients would therefore benefit from a readily available noninvasive imaging modality that can reveal detailed metabolic information. Despite great research progress in recent decades, metabolic imaging options are currently very limited in the clinic. The widely used magnetic resonance imaging (MRI) provides excellent anatomical detail and is nonradioactive but cannot image active metabolism. Proton magnetic resonance spectroscopic imaging (MRSI) can map metabolite levels, but metabolite concentrations are only one aspect of metabolism and do not necessarily reflect the activity of a metabolic pathway (5). Metabolic fluxes in humans have been studied using 13C magnetic resonance spectroscopy (MRS) combined with administration of 13C-labeled substrates to observe downstream 13C-labeled metabolites (6). However, because of its intrinsically low sensitivity, 13C MRS in vivo has been restricted to relatively large single volumes. Both indirectly detecting 13C through 1H-[13C] MRS and using hyperpolarized 13C are successful approaches that largely circumvent the low sensitivity of 13C MRS (7, 8). Yet, both of these methods are technically complex, which may prevent their development into a widespread technique. GlucoCEST is potentially easier to implement in the clinic, yet this technique does not provide direct information on glucose metabolism (9, 10). At the moment, this leaves clinicians with positron emission tomography (PET) as the only available method for metabolic imaging. In the clinic, the most commonly available metabolic substrate for PET is the glucose analog 2-18F-fluoro-2-deoxy-d-glucose (18FDG), which provides high-resolution maps of glucose uptake but does not inform on glucose metabolism. In addition, 18FDG is radioactive, which is a limitation when repetitive scanning is required such as in evaluating disease progression. Clearly, there is a demand for a more comprehensive, nonradioactive, and easily implementable approach to imaging metabolism.
Here, we present a novel approach that meets the current needs for metabolic imaging. Like 13C, deuterium (2H)–labeled substrates can be used to study metabolism with MRSI in vivo. The use of 2H as a tracer for investigating metabolism precedes the discovery of nuclear magnetic resonance (11). More recently, 2H MRS was used to study whole-brain metabolism at ultrahigh field in rats (12), and deuteration of 13C-labeled glucose significantly improves the magnetization lifetime of 13C in hyperpolarized 13C imaging (13, 14). However, to the best of our knowledge, metabolic imaging based on 2H MRS in vivo has not yet been reported. Here, we show how deuterium metabolic imaging (DMI), which is based on 2H MRSI, can noninvasively generate metabolic maps with high spatiotemporal resolution and display the metabolic fate of several 2H-labeled substrates. DMI revealed differences in metabolism of [6,6′-2H2]glucose and [2H3]acetate between normal brain and tumor tissue in a rat glioma model. In the same animal model, DMI also detected therapy-induced changes in glucose metabolism. This method was straightaway translated to humans, and we present metabolic maps acquired after oral administration of [6,6′-2H2]–labeled glucose in healthy human subjects and in patients diagnosed with a glioblastoma multiforme (GBM) brain tumor. To demonstrate the method’s robustness and sensitivity in noncerebral tissues, we administered deuterium-labeled glucose and then used DMI to map 2H-labeled glycogen in both rat and human livers.
RESULTS
In vivo 2H NMR
DMI is based on 2H nuclear magnetic resonance (NMR), which, through the 2H chemical shift, reveals signals of different deuterium-labeled molecules. Figure 1A shows 2H NMR spectra acquired from brain in vivo before administration of a 2H-labeled substrate. Without the 2H-labeled substrate, the only signal detectable in 2H NMR spectra from the brain or liver originates from the 0.0115% natural abundance signal of water, representing a ~10 mM 2H concentration (brain, 10.12 mM; liver, 8.94 mM; see Materials and Methods and Fig. 1A) (15). Sixty minutes after intravenous infusion of [6,6′-2H2]glucose, signals from the 2H-labeled glucose and 2H-lactate and a peak of overlapping 2H-labeled glutamate and glutamine (Glx) can be observed in rat brain in vivo (Fig. 1B). Other 2H NMR spectra in rat brain were acquired postmortem after [6,6′-2H2]glucose infusion (Fig. 1C) and in vivo following [2H3]acetate infusion (Fig. 1D). The labeling patterns in brain metabolites after administration of [6,6′-2H2]glucose or [2H3]acetate accord with their canonical metabolic pathways (fig. S1) (6, 12, 16). Figure 1E shows, from the brain of a healthy human volunteer, a 2H spectrum acquired after oral administration of [6,6′-2H2]glucose. The same 2H-labeled metabolites appear in both human and rat brains (Fig. 1B). The 2H spectrum acquired from the human brain at a lower magnetic field strength (4 T versus 11.7 T) has an inherently lower spectral resolution, resulting in partially overlapping peaks. However, the peaks are sufficiently separated for reliable quantification through spectral fitting. Deuterium NMR spectra acquired in rat and human livers, depicted in Fig. 1F, show the resonance of overlapping [6,6′-2H2]glycogen and [6,6′-2H2]glucose.
Fig. 1. In vivo 2H NMR.
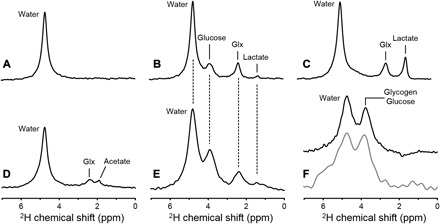
Deuterium NMR spectra acquired without localization from (A) rat brain in vivo at 11.7 T before infusion of any 2H-labeled substrate, (B) rat brain after infusion of [6,6′-2H2]glucose in vivo, (C) rat brain after infusion of [6,6′-2H2]glucose postmortem, and (D) rat brain after infusion of [2H3]acetate in vivo. (E) 2H NMR spectrum acquired from human brain at 4 T, 60 min after oral administration of [6,6′-2H2]glucose. (F) 2H NMR spectra from rat (top, black) and human (bottom, gray) liver after intravenous and oral administration of 2H-labeled glucose, respectively. Each spectrum shown represents 1 min of signal averaging. While the lower magnetic field strength used for human studies leads to a proportionally lower spectral resolution [compare (B) and (E)], the sparsity of the 2H NMR spectra in combination with the absence of large water or lipid signals still allows robust signal identification and quantification (see also Figs. 2 to 5).
The T1 and T2 values of deuterated water and glucose were measured in water and agar phantoms and in rat and human brain in vivo. In addition, the relaxation parameters for Glx were measured in rat and human brain in vivo, whereas deuterated lactate T1, T2, and T2* values were measured in rat brain postmortem (tables S1 and S2). The T1 values of [6,6′-2H2]glucose and its downstream metabolites range from 60 to 350 ms. The water signal showed pronounced biexponential T2 relaxation that likely reflects different tissue compartments, with the longer T2 component representing cerebrospinal fluid and the shorter component originating from intracellular water, as was previously reported in cat brain (17).
DMI of steady-state glucose and acetate metabolism in rat brain
2H NMR, as shown in Fig. 1, can be combined with three-dimensional (3D) spatial phase-encoding gradients and spectral fitting to generate 3D DMI maps of 2H-labeled substrates and metabolites. Figure 2 shows DMI data acquired from a 3D grid of 8-μl voxels from the brain of a glioma-bearing rat after infusion of [6,6′-2H2]glucose. The signals of 2H-labeled glucose, lactate, and Glx were quantified by peak fitting, and the resulting amplitudes represented as color-coded DMI maps were overlaid onto an anatomical MR image (Figs. 2 to 4). Oxidative glucose metabolism is represented by the labeled Glx that appeared to be relatively homogeneous in normal brain, with a marked decrease in the tumor lesion (normal brain, 1.5 ± 0.3 mM; n = 45 voxels; tumor, 0.7 ± 0.2 mM; P < 0.001; n = 8 voxels). The glucose and Glx signals from normal-appearing brain, on the basis of contrast-enhanced MRI (Fig. 2), were in agreement with the DMI data acquired in healthy rats (glucose: normal-appearing brain in tumor-bearing rats, 1.5 ± 0.3 mM (n = 45 voxels); normal brain in healthy rats, 1.7 ± 0.7 mM (n = 81 voxels); P = 0.17; Glx: normal-appearing brain in tumor-bearing rat, 1.5 ± 0.3 mM; normal brain in healthy rat, 1.5 ± 0.5 mM; P = 0.64). DMI showed lower levels of deuterated glucose and higher levels of labeled lactate in tumors compared to normal brain (lactate: tumor, 1.3 ± 0.4 mM; n = 8 voxels; normal brain, 0.6 ± 0.2 mM; P < 0.001; n = 45 voxels; glucose: tumor, 1.2 ± 0.1 mM; normal brain, 1.5 ± 0.3 mM; P = 0.001; see Fig. 2). The observed labeling in the rat brain tumor is consistent with both high glucose uptake and a high rate of glycolysis, which lowers the intracellular glucose level and produces lactate instead of oxidizing the glucose-derived pyruvate. This metabolic phenotype was first described by Otto Warburg and is well known in cancer biology (18, 19). Because the levels of labeled lactate and Glx represent glycolytic and oxidative metabolism, respectively, the ratio of lactate over Glx can be seen as representing the Warburg effect. DMI can consequently be used to spatially map the Warburg effect and showed greatly elevated contrast in tumor lesions (tumor, 1.8 ± 0.8; n = 8 voxels; normal brain, 0.4 ± 0.1; P < 0.001; n = 45 voxels; Fig. 2). During administration of [6,6′-2H2]glucose, noncerebral tissue will also produce small amounts of 2H-labeled lactate, which is released in the blood. The concentration of 2H-labeled lactate in plasma sampled at 60 to 75 min after the start of [6,6′-2H2]glucose infusion was 0.26 ± 0.03 mM (n = 4; fig. S2), which is five times lower than the 2H-labeled lactate level observed with DMI in the brain tumor region. These data confirm that the bulk of the labeled lactate observed in the tumor region was produced locally and not transported from the blood.
Fig. 2. DMI in rat brain after [6,6′-2H2]glucose infusion.
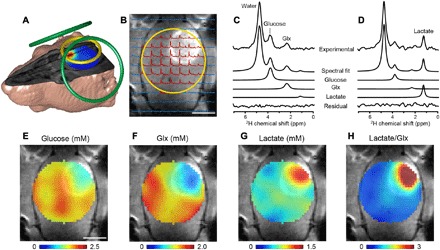
(A) MRI-based 3D rendering of a RG2 glioma rat with radiofrequency (RF) coil positioning for MRI, shimming (green loops), and DMI (yellow loop). The color-coded map overlaid onto the 3D MR image shows the lactate/Glx ratio. (B) Contrast-enhanced T1-weighted MRI in which the tumor appears hyperintense relative to normal brain tissue. The yellow circle shows the DMI surface RF coil position. As RF transmission and reception sensitivity profiles are highest close to the surface coil, the DMI field of view is effectively limited to the RF coil diameter. Localized 2H NMR spectra extracted from an 11 × 11 × 11 MRSI grid with 2 × 2 × 2 mm3 voxels are overlaid onto the MR image. The data shown were acquired between 60 and 90 min after starting the [6,6′-2H2]glucose infusion. Spectra shown in red had sufficient signal for further processing, whereas those in blue were discarded. Experimental 2H NMR spectra from (C) normal-appearing brain and (D) tumor lesion, with the spectral fitting results for the total spectrum, individual peaks, and fitting residue shown below. Note that localized spectra have higher spectral resolution than nonlocalized spectra (Fig. 1), a consequence of the smaller 8-μl volumes’ inherent increased magnetic field homogeneity. (E to H) DMI maps of 2H-labeled glucose and metabolites, based on spectral fitting of individual peaks, expressed in millimolar (E to G). The ratio of lactate/Glx (H) represents the Warburg effect, characterized by high glycolysis and low oxidative metabolism. Scale bars, 5 mm (B and E).
Fig. 3. DMI in human brain after oral [6,6′-2H2]glucose intake.
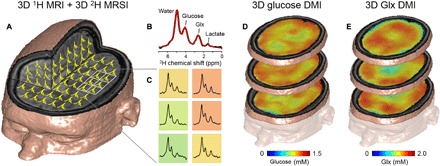
(A) 3D MRI overlaid with 2H MR spectra from a 3D MRSI data set (9 × 13 × 11 matrix) with 20 × 20 × 20 mm3 nominal spatial resolution, acquired between 65 and 90 min after oral [6,6′-2H2]glucose administration. (B) Typical 2H NMR spectrum from a single MRSI voxel overlaid with a spectral fit (red line) indicating the peaks from water, glucose, Glx, and lactate. (C) Grid (2 × 3) extracted from the MRSI with data color-coded by the Glx intensity. (D) 3D maps of 2H-labeled cerebral glucose and (E) Glx levels in millimolar, extrapolated from the 3D MRSI to the 3D MRI grid. Note the seemingly lower level of Glx in areas corresponding to the ventricles.
Fig. 4. DMI visualizes the Warburg effect in a patient with GBM after oral [6,6′-2H2]glucose intake.
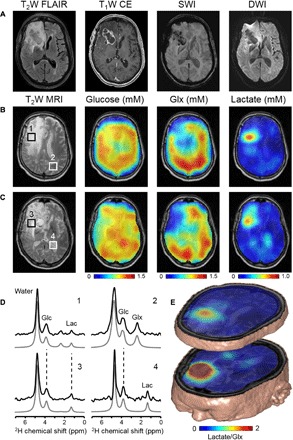
(A) Clinical MR images acquired as standard-of-care in a patient diagnosed with GBM in the right frontal lobe. MR images include (from left to right) T2-weighted fluid-attenuated inversion recovery (T2W FLAIR), T1-weighted contrast-enhanced imaging (T1W CE), susceptibility-weighted imaging (SWI), and diffusion-weighted imaging (DWI). The patient (man, 63 years old) had undergone subtotal resection of the lesion 9 months before the DMI study. He was undergoing treatment on an experimental protocol involving nivolumab or placebo combined with the standard-of-care chemoradiation with temozolomide, followed by adjuvant temozolomide. (B and C) T2W MRI and overlaid DMI maps in two slices that contain the tumor lesion. The MRI and DMI data shown in (C) correspond to the slice position of the clinical MRI scans shown in (A). DMI maps show a homogeneous distribution of 2H-glucose across the slices but lower levels of 2H-labeled Glx and a higher concentration of 2H-labeled lactate in the tumor lesion compared to normal-appearing brain. (D) 2H NMR spectra from selected locations depicted in the T2W MR image, including tissue (1 and 3) within the lesion as seen on T1W CE, (2) from normal-appearing occipital lobe, and (4) containing cerebrospinal fluid from the left lateral ventricle. (E) 3D illustration of combined MRI and DMI of the lactate/Glx ratio representing the spatial distribution of the Warburg effect. The DMI maps show glucose metabolism patterns similar to the RG2 glioma model (Fig. 2)
To explore whether DMI could provide an imaging biomarker for therapy response, we treated glioma-bearing rats with dichloroacetate (DCA). DCA stimulates pyruvate dehydrogenase, which facilitates pyruvate flux into mitochondria for subsequent oxidation, thereby partially reversing the Warburg effect. Treatment with DCA 90 min before [6,6′-2H2]glucose infusion and subsequent DMI appeared to reduce both the area and intensity of the lactate/Glx ratio (fig. S3). Hence, DMI provided a fast, noninvasive way to image a metabolic drug’s effects on glucose metabolism in the brain tumor.
Isotopically labeled acetate has been used as a metabolic substrate to investigate metabolism in animal and human brain and liver (16, 20). Acetate is converted to acetyl–coenzyme A (CoA) and further metabolized in the tricarboxylic acid (TCA) cycle, labeling Glx through fast exchange with α-ketoglutarate. Unlike glucose, acetate does not require pyruvate dehydrogenase activity to enter the TCA cycle. Instead, acetate can be considered a surrogate for fatty acid oxidation. The map of labeled Glx in a glioma-bearing rat shows a relatively homogeneous distribution of acetate oxidation in normal-appearing brain tissue. By contrast, tumor tissue showed higher levels of labeled acetate and lower levels of Glx labeling (acetate, 2.9 ± 0.3 mM; Glx, 0.6 ± 0.2 mM) than normal brain (acetate, 1.0 ± 0.4 mM; Glx, 0.9 ± 0.2 mM; n = 1; fig. S4). These results indicate differences in tumor metabolism of acetate compared to normal brain tissue.
DMI of steady-state glucose metabolism in human brain
We performed DMI on the brains of healthy control subjects who ingested [6,6′-2H2]glucose (0.75 g/kg body weight). To cover a large part of the brain, we collected DMI data using a 3D grid of 2 × 2 × 2 cm3 voxels, and the peak amplitudes of glucose and metabolites are shown as color-coded concentration maps (Fig. 3). The distribution of labeled Glx seemed more prominent in the cortices, rich in gray matter, and lower in regions that, on the basis of anatomical location, contain mostly cerebrospinal fluid. These levels concur with individual 2H NMR spectra from voxels positioned in the cortex and ventricles (fig. S5).
We also performed DMI 60 to 75 min after oral intake of [6,6′-2H2]glucose (0.60 g/kg) (Fig. 4 and fig. S6), on two patients diagnosed with GBM. Figure 4 shows the DMI data collected in patient 1. In normal-appearing brain (for example, a region in the occipital lobe), DMI showed a fairly homogeneous distribution of labeled glucose and Glx, similar to the data from a healthy subject (Fig. 3). The concentrations of 2H-labeled glucose and Glx in normal brain tissue were slightly lower in the patient, most likely because of the smaller oral glucose dose (0.60 g/kg versus 0.75 g/kg in healthy controls). In the lesion area (right frontal lobe), labeled Glx was not observed, whereas 2H-lactate and lactate/Glx maps show striking contrast in both patients (Fig. 4 and fig. S6). These metabolic maps are remarkably similar to the DMI data acquired in the rat model of GBM and demonstrate the distinct differences in glucose metabolism between tumor and normal brain (Fig. 2).
DMI of liver glycogen
The versatility of DMI for imaging metabolism in organs other than the brain is exemplified by imaging labeled glycogen in the liver. As the primary organ for storing glucose in the form of glycogen, the liver plays a major role in whole-body glucose homeostasis. Figure 5D displays a DMI-based map of labeled glycogen 2 hours after intravenous injection of [6,6′-2H2]glucose in a healthy rat. Estimated concentration based on the naturally abundant deuterated water (8.9 mM; see Materials and Methods) indicates levels of labeled glycogen + glucose of 2.7 ± 0.3 mM. No other labeled metabolites are detected in the 2H NMR spectra, which indicates that the liver is predominantly storing glucose instead of using it as an oxidative fuel. However, the concentration of glutamate in rat liver has been reported to be 2 to 3 mM, which is several times lower than in the brain (21). The low level of 2H labeling and the small glutamate pool size could result in deuterium-labeled glutamate levels that are too low to detect with the current 2H volume RF coil setup. We applied a similar approach to healthy human subjects by using a surface RF coil positioned on the liver’s lateral aspect. DMI following oral intake of [6,6′-2H2]glucose also revealed labeled glycogen in human liver, which is homogeneously distributed within the RF coil’s field of view (Fig. 5).
Fig. 5. DMI of liver glycogen.
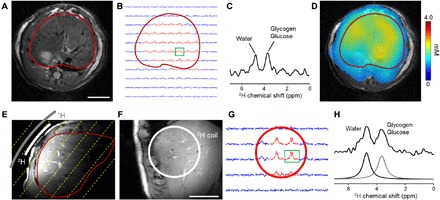
(A) Respiratory-gated, axial MR image of rat abdomen with the liver delineated in red. Scale bar, 10 mm. (B) 2H NMR spectra extracted from a 3D 2H MRSI grid with 4 × 4 × 4 mm3 resolution. Spectra in red have sufficient signal for DMI quantification, whereas those in blue were not considered further. (C) 2H MR spectrum from the voxel indicated in green in (B). (D) Color-coded map of 2H-labeled glucosyl units (in millimolar) after 120 min of [6,6′-2H2]glucose infusion. (E) Axial MR image of the liver acquired with the RF coil placed on the lateral side of the abdomen. RF coils’ position and size indicated in gray (1H) and white (2H). Dashed yellow lines emphasize one dimension of the 3D oblique MRSI grid parallel to the 2H surface coil. The red line delineates the liver. (F) MR image acquired parallel to the RF coils, with the 2H RF coil (white) superimposed. Scale bar, 50 mm. As with the rat brain data, the 2H surface coil’s sensitivity limits DMI data acquisition to positions close to and within the diameter of the coil. (G) 2H NMR spectra extracted from a 3D 2H MRSI grid with 25 × 25 × 25 mm3 resolution, 60 min after oral [6,6′-2H2]glucose intake. Spectra in red have sufficient signal for quantification, whereas those in blue were not considered further. (H) 2H MR spectrum from the voxel indicated in green in (G), with the fitted peaks of 2H-labeled glycogen + glucose and water.
Analysis of 2H label loss
Labeling of substrates with 2H implies the substitution of one or more 1H atoms with 2H. Because the 2H atoms are not part of the molecules’ backbone, there is the possibility that 2H exchanges nonenzymatically with surrounding water or that 2H label is lost in enzymatic reactions. It is clear that 2H label loss in vivo is minimal; otherwise, DMI would simply not be possible because there would be no metabolite labeling to detect. However, absolute quantification of the concentrations of 2H-labeled metabolites benefits from knowing the fractional loss in 2H during metabolism of the substrate of interest. To test the extent of 2H label loss during glycolysis, we incubated mammalian cells (9L, rat gliosarcoma cells) in vitro separately with [6-13C,6,6′-2H2]glucose and [6-13C]glucose and analyzed the labeled lactate produced by these cells using 13C NMR. In a 13C NMR spectrum, 2H-13C J coupling induces a shift and splitting of peaks (fig. S7). The 13C NMR data of a cell culture medium of 9L cells incubated with [6-13C,6,6′-2H2]glucose revealed three species of lactate: [3-13C,3,3′-2H2]lactate (84.7 ± 4.0%), [3-13C,3-2H]lactate (14.4 ± 3.3%), and [3-13C]lactate (0.9 ± 0.8%, corrected for natural abundance; n = 3), indicating 8.1 ± 2.3% 2H label loss occurring during glycolysis. Downstream of glycolysis, and shared by glucose and acetate-derived acetyl-CoA, 2H label loss can be anticipated at the level of citrate synthetase. Citrate is formed through the condensation of acetyl-CoA with oxaloacetate. Here, one of the protons/deuterons from the acetyl-CoA methyl group is cleaved off, representing another mechanism of 2H label loss and resulting in a lower level of 2H labeling in glutamate (and glutamine). The theoretical chance of label loss at citrate synthetase was included in the calculation of the concentration of 2H-labeled Glx (see Materials and Methods).
DISCUSSION
Our results show that DMI, which combines 2H MRSI with administration of deuterium-labeled substrates, can distinguish between normal and aberrant metabolism in vivo. We demonstrated DMI’s potential by mapping the metabolism of either 2H-labeled glucose or acetate in the brain and liver of animal models and human subjects. The DMI-based data allowed 3D visualization of in vivo metabolism with technically simple and robust methods that can be implemented on most clinical MRI scanners. Accordingly, we believe that DMI’s capacity to image metabolism can be of great clinical benefit in a variety of disease settings.
The DMI map of labeled glucose in a patient with GBM showed minimal contrast between the tumor lesion and normal brain due to high glucose uptake in both tissue types, particularly in gray matter. This is a well-described challenge when using 18FDG-PET to evaluate brain tumor glucose metabolism (22). Because DMI can visualize metabolism beyond glucose uptake, maps of 2H-labeled Glx and lactate show striking contrast. The lower levels of labeled Glx and higher levels of labeled lactate in the tumor, observed in both the animal model and the patients with GBM (Figs. 2 and 4), are consistent with the Warburg effect. Using DMI, we can now map the Warburg effect noninvasively with a single ratio-based metric of 2H-labeled lactate/Glx. In this way, DMI overcomes a challenge typically faced by 18FDG-PET in brain tumor applications.
DMI also revealed distinct contrast between glioma and normal brain in the rat brain tumor model after [2H3]acetate infusion. More specifically, acetate levels were higher and acetate oxidation was lower in the tumor compared to normal-appearing brain (fig. S4). Acetate is taken up via the monocarboxylic transporter 1 (MCT1), which is expressed in RG2 glioma and a potential therapeutic target in some cancers (23–25). These data suggest that 2H-labeled acetate levels, as measured with DMI within the glioma, could serve as a noninvasive biomarker of MCT1 expression. Thus, DMI could be used to evaluate the efficacy of a therapy targeting MCT1. However, the RG2 rodent tumor model also has a compromised blood-brain barrier that could facilitate increased acetate uptake independently of MCT1. In addition, the glutamate levels in the RG2 tumor are typically lower than in adjacent normal-appearing brain (25). A smaller Glx pool size could result in a lower absolute level of deuterated Glx and thus affect the image contrast observed with DMI.
We also used DMI to evaluate liver glycogen labeling after [6,6′-2H2]glucose administration, therefore demonstrating this technique’s applicability in organs other than the brain. As with the brain, DMI of the liver is methodologically simple but robust. DMI of liver glycogen storage will be valuable for studying the liver’s role in conditions such as type 2 diabetes and nonalcoholic fatty liver disease. DMI of the liver could also be applied dynamically to measure the rate of glycogen turnover.
Without administration of a 2H-labeled substrate, DMI merely detects the 0.0115% naturally abundant 2H in the large water pool (~10 mM in the brain) and, in some cases, a small signal from lipids (for example, Fig. 1F, rat liver) (15). DMI therefore requires neither additional RF pulses for water suppression nor outer volume suppression to exclude large contaminating signals from adjacent lipid tissue. Further, DMI is minimally affected by magnetic field inhomogeneity because of its low NMR frequency and the intrinsically broad peaks of 2H NMR. In addition, because 2H NMR spectra only contain a few metabolite peaks, some minimal line broadening does not lead to large peak overlap. DMI’s relative invulnerability to magnetic field inhomogeneity is seen in individual 2H NMR spectra of the frontal lobe and tumor areas with iron-containing hemosiderin (Fig. 4 and fig. S5). Together, these aspects make DMI easy to implement and perform, in contrast to technically challenging 1H and 13C MRS(I) (7), thereby broadening its applicability and establishing DMI’s potential.
The feasibility of DMI depends on its sensitivity, which is influenced by the gyromagnetic ratio and magnetic moment of the 2H spin, as well as the relaxation times of the 2H-labeled molecules. The gyromagnetic ratio of 2H is 6.5 times lower than that of 1H, which is the nucleus with the highest NMR sensitivity. This disadvantage is partly offset by the larger magnetic moment generated by 2H, which has a spin quantum number of 1, compared to nuclei, including 1H, with spin ½. Deuterium’s relaxation parameters favor rapid scanning while retaining reasonable spectral resolution. The deuterium T1 relaxation times of 2H-labeled metabolites are approximately 10 times shorter than the corresponding proton T1 values, which, for equivalent scan times, would translate in a √10 increase in signal-to-noise ratio (SNR). Meanwhile, the T2 and T2* relaxation times are relatively long compared to T1, thereby further enhancing the detection of 2H-labeled compounds.
Similar to 17O NMR, the SNR of 2H has an approximate quadratic relation with magnetic field strength, which implies that, at 7 T, the SNR would be three times higher than at 4 T (26). We foresee that DMI can be easily implemented at 7 T, and a spatial resolution approaching 1 × 1 × 1 cm3 can be achieved using an optimized RF coil. Applying NMR in vivo at ultrahigh field can be challenging because the U.S. Food and Drug Administration (FDA) guidelines for the specific absorption rate (SAR). The SAR of DMI at 4 T was estimated at 0.16 W/kg (see Materials and Methods), which is much lower than the FDA guideline of 3 W/kg, and thus, it is straightforward to implement DMI at 7 T. RF inhomogeneity is another complication at 7 T but will not pose a problem because of the low frequency at which DMI operates (45.7 MHz at 7 T).
Both deuterium-labeled substrates and water have been used in clinical research for many years (27, 28). The amounts of 2H administered for DMI (~1 g of 2H for a 70-kg subject) are lower than those used in other research applications (28). Studies in humans focused on cells or molecules with low metabolic turnover rates have used D2O doses that result in 400 times more 2H loading than a typical DMI study (29, 30). The amount of deuterium used for a DMI study is thus orders of magnitude lower than that used in other research applications, for which the dose of 2H has been considered safe (27–30).
In this first implementation of DMI, the focus was limited to data acquisition during isotopic steady state. DMI could, in the future, also be applied dynamically during labeled substrate infusion to assess metabolic turnover. We focused initially on steady-state metabolism because its simplicity favors rapid implementation in the clinic. A limitation of the DMI application in liver was the overlapping peaks of [6,6′-2H2]glucose and labeled glycogen in the 2H NMR spectrum. We therefore cannot accurately quantify the extent of glycogen 2H labeling. Further work is needed to quantify the extent of overlap and/or to explore the use of glucose labeled with 2H at other positions of the molecule (for example, [1-2H]glucose) to avoid the overlap of glucose and glycogen 2H NMR peaks. Future applications of DMI could also focus on in vivo kinetics of specific enzymes, evaluated by exchange of 2H from strategically 2H-labeled substrates with the natural abundant 2H-labeled water pool, although these experiments would only work for reactions that are fast enough to exchange 2H label before magnetization is lost because of relaxation.
Absolute quantification of 2H concentration in the different metabolites requires correction factors to compensate for 2H label loss. The 2H loss from [6,6′-2H2]glucose during glycolysis in vitro is in agreement with a previous report and attributed to the conversion of phosphoenolpyruvate to pyruvate by pyruvate kinase (31). To correct the labeling of Glx for label loss at the citrate synthethase step of the TCA cycle, we used a theoretically derived factor. Future studies could be performed to confirm these factors by well-designed in vivo experiments. However, using the currently known correction factors for label loss, the concentration of 2H-labeled Glx is very close to the concentrations of labeled Glx when using 13C-labeled glucose in vivo in human brain. Therefore, the label loss correction factors determined in vivo are most likely to be close to the theoretical value (32, 33). Another factor in the determination of absolute 2H concentration in vivo is the value of the natural abundance of 2H in tissue water. We used the literature value for natural abundance of 2H in water on Earth (15). Determination of the range of natural abundance 2H levels in tissue water would be beneficial for its use as an internal concentration standard.
Using deuterium to study metabolism was suggested as early as 1935 (11). Global, nonlocalized 2H MRS has been used at ultrahigh magnetic fields to quantify metabolism in a wide range of settings including excised rabbit cornea (34), whole-body mouse lipid synthesis (35), and rat brain in vivo (12). MRI of 2H has also been explored, particularly to measure blood flow and perfusion (36–38). DMI now combines the chemical specificity of 2H NMR with the spatial localization of MRI to produce noninvasively metabolic maps in human tissue. Implementing DMI on existing 3 T clinical MRI scanners only requires minimal technical modifications and the acquisition of an RF coil that operates at the 2H NMR frequency (19.6 MHz). The extensive availability of 3 T MRI scanners should facilitate DMI to become a widespread MRI modality for clinical metabolic imaging.
MATERIALS AND METHODS
MR systems
Animal and high-resolution NMR systems
Animal studies were performed on an 11.7 T Magnex magnet (Magnex Scientific Ltd.) interfaced to a Bruker Avance III HD spectrometer running on ParaVision 6 (Bruker Instruments). The system was equipped with 9.0-cm-diameter Magnex gradients capable of switching 440 mT/m in 180 μs and 0.5-kW RF amplifiers for 1H and 2H transmission.
RF transmission and reception for MRI and shimming during brain studies were performed with two orthogonal 20-mm surface coils driven in quadrature and tuned to the proton NMR frequency (499.8 MHz). Deuterium RF transmission and reception were achieved with a two-turn 14-mm-diameter surface coil tuned to 76.7 MHz and integrated with the proton RF coil according to the design described by Adriany and Gruetter (39).
Liver studies were performed with a 5.9-cm-diameter proton volume coil (6.1 cm in length) for MRI and shimming and a 5.0-cm-diameter deuterium volume coil (6.1 cm in length) for 2H MRS/MRSI. Both coils are constructed according to the design by Bolinger et al. (40), whereby the 2H coil is mounted within the 1H coil. The RF shield is an integral part of the 1H coil to manage capacitance at the high proton frequency (499.8 MHz). High-resolution NMR scans were performed on a 500-MHz MR spectrometer (Bruker Avance, Bruker) using 5-mm probes optimized for 1H and X-nucleus acquisition, respectively.
Human MR system
Human studies were performed on a 4 T Magnex magnet (Magnex Scientific Ltd.) interfaced to a Bruker Avance III HD spectrometer running on ParaVision 6 (Bruker Instruments). The system was equipped with 67-cm-diameter Magnex gradients capable of switching 30 mT/m in 1.1 ms. The gradient system included a full set of third-order shim coils. RF transmission was achieved using 4-kW RF amplifiers for 1H and 2H.
Brain studies were conducted with a 28.5-cm-diameter transverse electromagnetic (TEM) volume coil tuned to the proton frequency (170.5 MHz) for MRI and shimming. 2H RF reception at 26.2 MHz was achieved with a four-coil phased array that was driven as a single RF coil during RF transmission. The four 8 cm × 10 cm rectangular 2H array elements were positioned equidistantly on an 18 cm × 25 cm elliptical former that was positioned within the 1H TEM coil. A custom-built 1H-2H probe consisting of a 9-cm-diameter 2H surface coil with a pair of elliptical 14.5 cm × 11 cm 1H coils arrayed in quadrature for MRI and shimming was used for all human liver studies.
Human subjects
All human studies were approved by the Yale University Institutional Review Board. Healthy control subjects (n = 2) and two patients diagnosed with GBM were recruited to participate in the DMI study. The patients (male; 63 and 67 years old) with GBM had undergone a subtotal resection of the lesion 9 and 1 month(s) before the DMI study. One patient was undergoing treatment on an experimental protocol involving nivolumab or placebo combined with the standard-of-care chemoradiation with temozolomide, followed by adjuvant temozolomide. The other patient had only received surgery before the DMI study.
2H-labeled glucose and acetate administration in vivo
Deuterium-labeled glucose ([6,6′-2H2]glucose) and acetate ([2H3]Na-acetate) were purchased from Cambridge Isotopes Laboratories Inc. Glucose labeled on the sixth carbon position was preferred over [1-2H]glucose because of the higher deuterium content (two 2H atoms) and cost and preferred over [1,2,3,4,5,6,6-2H7]glucose because of cost and to minimize complexity of the 2H NMR spectra.
Human subjects took an oral dose of [6,6′-2H2]glucose dissolved in 200 to 300 ml of water, calculated as 0.75 g/kg body weight, with a maximum of 60 g, regardless of body weight. For infusion studies in rats, labeled glucose was dissolved in water at a concentration of 1 M. Infusions resulted in 1.95 g/kg body weight of [6,6′-2H2]glucose infused during a 120-min study. Infusions were performed via an intravenous line in the femoral vein or an intraperitoneal infusion line. [2H3]acetate was dissolved in water at a concentration of 2 M and was infused as described for glucose, resulting in a total of 2 g/kg body weight of [2H3]acetate infused during a 120-min study. All animal procedures were approved by the Yale University Institutional Animal Care and Use Committee.
Animal experiments
Fischer 344 rats (n = 9) were used for DMI experiments in healthy brain, liver, and glioma after implantation of RG2 cells. The glioma model was established as described previously (25). In short, RG2 glioma cells (American Type Culture Collection) were grown in T75 flasks using standard cell culture conditions (37°C and 5% CO2). RG2 cells were harvested, and ~1250 cells suspended in 5 μl of serum-free, low-glucose Dulbecco’s modified Eagle’s medium (DMEM) were injected in the brain of anesthetized rats.
For DMI experiments, rats were anesthetized with isoflurane using ~60% O2 and ~40% N2O delivered through a nose cone. A heating pad or heated air was used to maintain body temperature at ~37°C. Breathing, heart rate, and blood oxygenation parameters were monitored continuously using a pulse oximeter probe (MouseOx, Starr Life Sciences Corp.) on the foot.
Blood was sampled from an arterial catheter placed in the femoral artery (25). The blood sample was centrifuged (5000 rpm for 5 min), and the plasma was stored at −80°C for later processing. Plasma samples were prepared for high-resolution NMR by adding 160 μl of methanol to 80 μl of plasma to precipitate plasma proteins. This mixture was incubated at −20°C for 20 min, followed by centrifugation at 10,000 rpm for 30 min at 4°C. The supernatant was transferred to a 2-ml vial, and the methanol was evaporated under nitrogen using a TurboVap LV (Biotage) at 45°C for ~60 min. The sample was resuspended in 600 μl of a mixture of water containing phosphate buffer (100 mM), D2O (10%), sodium formate, and imidazole as a chemical shift reference and internal concentration standard, respectively.
In vitro experiments
Rat gliosarcoma cells (9L, American Type Culture Collection) were cultured in T75 flasks using standard cell culture conditions (37°C and 5% CO2). For the 13C/2H labeling experiments, cells were washed with phosphate-buffered saline and incubated with DMEM containing 100% [6-13C]glucose or [6-13C,6,6′-2H2]glucose (Sigma-Aldrich, Isotec). After 6 hours, the incubation medium was collected, [2-13C]glycine was added as an internal concentration standard, and the samples (5 ml) were freeze-dried on a lyophilizer (Labconco). Freeze-dried samples were prepared for NMR analysis by adding 600 μl of a mixture of water containing phosphate buffer (100 mM), D2O (10%), sodium formate, and imidazole as a chemical shift reference and internal concentration standard, respectively.
MR signal acquisition
For human brain studies, subjects were positioned supine within the combined 1H TEM/2H phased-array RF coil. Anatomical MR images were acquired for co-registration with the DMI data. Magnetic field homogeneity was optimized using 3D B0 mapping and second-order spherical harmonic (SH) shims, resulting in an approximately 11-Hz water linewidth in a 70 mm × 90 mm × 40 mm = 252-ml volume placed centrally in the brain. For liver studies, human subjects were positioned supine with the 1H/2H RF coil attached to a rigid support to minimize extraneous movement. The RF coil was placed over the lateral aspect of the liver. Anatomical MR images of the torso were acquired during an end-expiration breath hold to confirm correct positioning of the probe. Magnetic field homogeneity was optimized using respiratory-gated, iterative adjustment of the first-order SH shims across an oblique 30 mm × 40 mm × 40 mm = 48-ml localized volume. The resulting water linewidth was on the order of 30 Hz. DMI liver data were acquired without respiratory triggering.
Animals were positioned prone under the 1H/2H surface coil for brain studies and inside the 1H/2H volume coil for liver studies. Body temperature was maintained at 37°C with a heated water pad (brain studies) or heated air (liver studies). For brain studies, the acquisition of anatomical MR images was followed by 3D B0 mapping (second-order SH shimming), resulting in approximately 25 to 30 Hz across a 7 mm × 5 mm × 10 mm = 750-μl volume. In brain tumor–bearing rats, T1-weighted images were acquired after intravenous or intraperitoneal injection of 200 μl of the T1 contrast agent gadopentetate dimeglumine (Magnevist, Bayer). For liver studies, MR image acquisition, as well as iterative shimming of linear SH shims, was respiration-gated. The magnetic field homogeneity across an oblique 10 mm × 18 mm × 5 mm = 900-μl volume was typically in the 60- to 90-Hz range, as measured from the water linewidth. DMI data were always acquired without respiratory triggering.
2H MR signal acquisition was achieved with a pulse-acquire sequence extended with 3D phase-encoding gradients during the initial 1.1 ms (animal) or 2.7 ms (human) following excitation. For brain and human liver studies, signal excitation was achieved with a 90° rectangular RF pulse of 50-μs (animal) or 500-μs (human) duration. For animal liver studies, signal excitation was achieved with a 2.0-ms Shinnar–Le Roux RF pulses (90°; band width, 3.0 kHz), selecting a 25-mm-thick transverse slab to minimize artifacts due to heart pulsations (41).
Human DMI data from brain were acquired at a nominal 20 mm × 20 mm × 20 mm = 8-ml spatial resolution with spherical k-space encoding. With a repetition time (TR) of 333 ms and eight averages, a typical steady-state DMI acquisition lasted approximately 29 min. DMI from human liver 2H used a 3D 2H MRSI matrix of 11 × 9 × 9 with 25 × 25 × 25 mm3 resolution. Steady-state DMI on the animal brain was acquired at a 2 mm × 2 mm × 2 mm = 8-μl nominal spatial resolution in approximately 35 min (TR = 400 ms; eight averages). The lower sensitivity of the 2H volume coil demanded DMI on the animal liver to be acquired at a 4 mm × 4 mm × 4 mm = 64-μl nominal spatial resolution in approximately 25 min (TR = 400 ms; four averages).
High-resolution 13C NMR spectra of freeze-dried cell culture medium samples were collected using a pulse-acquire sequence with adiabatic 1H-13C decoupling (TR = 20 s; spectral width, 25 kHz; number of averages, up to 3078). High-resolution 1H NMR spectra were acquired from protein-free plasma samples using a pulse-acquire sequence (TR = 20 s; number of averages, 256) with water presaturation and 2H WALTZ decoupling [76.77 MHz; γB1/(2π) = 250 Hz].
T1, T2, and T2* measurements
T1 values of water, glucose, Glx, and lactate were estimated on the basis of a three-parameter fit of the signal amplitude acquired using a nonlocalized inversion-recovery method with 12 logarithmatically spaced inversion times between 10 and 2560 ms and a TR of 2560 ms (42). The inversion-recovery experiments were performed using an adiabatic inversion pulse to assure homogeneous excitation.
Global T2 measurements were performed with a nonlocalized spin-echo sequence with 12 different echo times between 6 and 400 ms. The decaying signal amplitudes were fit with a monoexponential function in case of labeled glucose, Glx, and lactate and a biexponential function for 2H-labeled water. Note that 2H-2H scalar couplings are on the order of 0.1 to 0.2 Hz and therefore do not significantly influence the T2 estimates of the metabolites, which are in the range of 50 to 100 ms (43).
T1 and T2 relaxation measurements were performed after the metabolite 2H label incorporation reached a steady state, typically between 90 and 150 min following the onset of 2H-labeled substrate administration. T2* relaxation times of water were obtained from the 3D DMI data by measuring the frequency width at half maximum (FWHM) amplitude of the phased absorption resonance line. Under the assumption of a Lorentzian line shape, T2* is calculated as 1/(πFWHM).
MR signal processing
All 1H MRI, 2H DMI, and high-resolution NMR data were processed in NMRWizard, a home-written graphical user interface in MATLAB 8.3 (MathWorks). DMI processing included linear prediction of the missing time domain points due to the phase-encoding gradients (6 and 13 points for animal and human studies, respectively) and 5-Hz line broadening, followed by 4D Fourier transformation (three spatial domains and one time domain). The resulting 2H MR spectra were quantified with linear least-squares fitting of up to four Lorentzian lines and a linear baseline. The robustness of spectral fitting was maximized by minimizing the number of unknown parameters in the spectral model. Following linear prediction and phase correction, the phase of all 2H MR signals should be equal and zero. To allow for imperfections in the phase correction, the spectral fitting algorithm allowed a global phase (equal for a signal) constrained to the range [−π/6…+π/6]. Similar to 1H and 13C MRS, the chemical shifts of 2H-labeled metabolites are highly reproducible and insensitive to small changes in the chemical environment. Therefore, the frequencies of all signals were linked to the chemical shift of water, whereby a minor adjustment of ±0.01 ppm was allowed for each resonance. The linewidths of the resonances were linked to the water resonance, whereby differences in intrinsic T2 relaxation (table S1) were taken into account. Signal variations caused by incomplete T1 relaxation and RF transmit B1+ magnetic field inhomogeneity were corrected using T1 relaxation time constants measured in vivo (table S1) and quantitative B1+ maps acquired on a phantom containing water, 1.5% agar, and 1% deuteriumoxide.
Quantification
Corrected amplitudes of fitted water and metabolite peaks were converted to concentration (in millimolar) using the preinfusion, natural abundance water peak as an internal reference equal to 10.12 mM. The latter was determined assuming a 55.5 M water concentration, 80% water content in brain tissue and 70% water content in the liver, and a deuterium natural abundance of 0.0115 % (15, 44). Concentrations of substrates and metabolites were reported as concentration of the labeled molecule by normalizing the 2H NMR signal to the average number of deuterons per molecule. The 2H NMR signal amplitudes of water and [6,6′-2H2]glucose were normalized by dividing the quantified signal by two, whereas the observed 2H-labeled lactate signal was normalized by three and corrected for 2H label loss assumed to be 10% based on our in vitro data. The peak annotated as Glx contains signals from [4,4′-2H2]glutamate, [4-2H]glutamate, [4′-2H]glutamate, [4,4′-2H2]glutamine, [4-2H]glutamine, and [4′-2H]glutamine. The average number of deuterons at the C4 position of Glx is always lower than two because of 2H label loss in the TCA cycle at the conversion of acetyl-CoA to citrate. On the basis of the theoretical chance that both deuterons or only a single 2H atom is transferred from acetyl-CoA to Glx, the average number of deuterons of Glx will be 1.33. In case of [2H3]acetate metabolism, Glx contains a theoretical average of 2.00 deuterons on the C4 position.
The deuterium-labeled metabolite levels calculated on the nominal MRSI grid (for example, 11 × 11 × 11 on rat brain) were interpolated on a per-slice basis to a grid that is fivefold larger per dimension (that is, 55 × 55 × 11 on rat brain). 2D interpolation was achieved by convolving the nominal DMI data with a Gaussian kernel, whereby the convolution also provided an inherent Gaussian smoothing equal to 1.2- to 1.8-pixel widths. Interpolated DMI maps were overlaid with anatomical MR images as amplitude color maps.
Specific absorption rate
Power output for the RF coil used for DMI of human brain was measured at the RF coil connection of each of the four array elements for the nominal amplitude output power level of 550 W used for the DMI acquisition. SAR was estimated using the summed power output, the duty cycle (1.5/1000 ms), and assuming that the RF coil assembly affected 3 kg (60% of a 5-kg human head). This estimation resulted in a SAR of 0.16 W/kg.
Statistics
Differences in concentrations of 2H-labeled metabolites were analyzed using Mann-Whitney U tests in SPSS 24 (IBM). Metabolite concentrations were grouped on the basis of tissue type (normal brain or tumor) by manually drawn regions based on anatomical MRI data and were combined for all animals. The choice for a nonparametric test was based on the differences in data distribution inherent to the much larger number of voxels of normal brain compared to tumor tissue. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank B. Wang and X. Ma for the assistance with animal preparation and I. Prado for the help with the cell culture experiments. Funding: This work was partially supported by NIH funding through grants R01NS087568, R01MH109159, and R01EB14861. As a member of the Yale Cancer Center, H.M.D.F. is indirectly supported by the CCSG/P30 grant (P30 CA016359-34). H.M.D.F. was also supported by a Discovery Award from the American Brain Tumor Association and a Planning Grant from the James S. McDonnell Foundation. Author contributions: H.M.D.F. and R.A.d.G. designed and performed the experiments, analyzed the data, and wrote the manuscript. K.L.B. designed the experiments, analyzed the data, and edited the manuscript. Z.A.C. and R.K.F. recruited the subjects, interpreted the data, and edited the manuscript. P.B.B., S.M., and T.W.N. designed, manufactured, and implemented the components of the 2H RF pipeline. D.L.R. analyzed the data and edited the manuscript. Competing interests: K.L.B. discloses the receipt of consulting fees from Merck and declares no other competing interests. H.M.D.F. and R.A.d.G. are inventors on a U.S. provisional patent application related to this work filed by Yale University (application no. 62/608,861, filed 21 December 2017). All other authors declare that they have no competing interests. Data and materials availability: The in-house developed software NMRWizard (R.A.d.G.) is available for downloading from R.A.d.G.’s Yale University profile webpage. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/8/eaat7314/DC1
Fig. S1. DMI of [6,6′-2H2]glucose and [2H3]acetate metabolism.
Fig. S2. High-resolution 1H NMR of rat plasma.
Fig. S3. DMI detects the effect of DCA on glucose metabolism.
Fig. S4. Difference in 2H-acetate metabolism between normal-appearing rat brain and tumor tissue.
Fig. S5. In vivo 2H MRSI spectra from human brain.
Fig. S6. DMI of the Warburg effect in a patient with GBM.
Fig. S7. High-resolution 13C NMR spectra of 13C- and 2H-labeled lactate in cell culture medium.
Table S1. T1 and T2 relaxation times.
Table S2. T2* of water from the brain in vivo at 4 and 11.7 T.
REFERENCES AND NOTES
- 1.Zilberter Y., Zilberter M., The vicious circle of hypometabolism in neurodegenerative diseases: Ways and mechanisms of metabolic correction. J. Neurosci. Res. 95, 2217–2235 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Morató L., Bertini E., Verrigni D., Ardissone A., Ruiz M., Ferrer I., Uziel G., Pujol A., Mitochondrial dysfunction in central nervous system white matter disorders. Glia 62, 1878–1894 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Pavlova N. N., Thompson C. B., The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samuel V. T., Shulman G. I., Nonalcoholic fatty liver disease as a nexus of metabolic and hepatic diseases. Cell Metab. 27, 22–41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Öz G., Alger J. R., Barker P. B., Bartha R., Bizzi A., Boesch C., Bolan P. J., Brindle K. M., Cudalbu C., Dinçer A., Dydak U., Emir U. E., Frahm J., González R. G., Gruber S., Gruetter R., Gupta R. K., Heerschap A., Henning A., Hetherington H. P., Howe F. A., Hüppi P. S., Hurd R. E., Kantarci K., Klomp D. W. J., Kreis R., Kruiskamp M. J., Leach M. O., Lin A. P., Luijten P. R., Marjańska M., Maudsley A. A., Meyerhoff D. J., Mountford C. E., Nelson S. J., Necmettin Pamir M., Pan J. W., Peet A. C., Poptani H., Posse S., Pouwels P. J. W., Ratai E.-M., Ross B. D., Scheenen T. W. J., Schuster C., Smith I. C. P., Soher B. J., Tkáč I., Vigneron D. B., Kauppinen R. A.; MRS Consensus Group , Clinical proton MR spectroscopy in central nervous system disorders. Radiology 270, 658–679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothman D. L., De Feyter H. M., de Graaf R. A., Mason G. F., Behar K. L., 13C MRS studies of neuroenergetics and neurotransmitter cycling in humans. NMR Biomed. 24, 943–957 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Graaf R. A., Rothman D. L., Behar K. L., State of the art direct 13C and indirect 1H-[13C] NMR spectroscopy in vivo. A practical guide. NMR Biomed. 24, 958–972 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurhanewicz J., Vigneron D. B., Brindle K., Chekmenev E. Y., Comment A., Cunningham C. H., De Berardinis R. J., Green G. G., Leach M. O., Rajan S. S., Rizi R. R., Ross B. D., Warren W. S., Malloy C. R., Analysis of cancer metabolism by imaging hyperpolarized nuclei: Prospects for translation to clinical research. Neoplasia 13, 81–97 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker-Samuel S., Ramasawmy R., Torrealdea F., Rega M., Rajkumar V., Johnson S. P., Richardson S., Gonçalves M., Parkes H. G., Årstad E., Thomas D. L., Pedley R. B., Lythgoe M. F., Golay X., In vivo imaging of glucose uptake and metabolism in tumors. Nat. Med. 19, 1067–1072 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X., Yadav N. N., Knutsson L., Hua J., Kalyani R., Hall E., Laterra J., Blakeley J., Strowd R., Pomper M., Barker P., Chan K., Liu G., McMahon M. T., Stevens R. D., van Zijl P. C. M., Dynamic glucose-enhanced (DGE) MRI: Translation to human scanning and first results in glioma patients. Tomography 1, 105–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoenheimer R., Rittenberg D., Deuterium as an indicator in the study of intermediary metabolism. Science 82, 156–157 (1935). [DOI] [PubMed] [Google Scholar]
- 12.Lu M., Zhu X.-H., Zhang Y., Mateescu G., Chen W., Quantitative assessment of brain glucose metabolic rates using in vivo deuterium magnetic resonance spectroscopy. J. Cereb. Blood Flow Metab. 37, 3518–3530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishkovsky M., Anderson B., Karlsson M., Lerche M. H., Sherry A. D., Gruetter R., Kovacs Z., Comment A., Measuring glucose cerebral metabolism in the healthy mouse using hyperpolarized 13C magnetic resonance. Sci. Rep. 7, 11719 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodrigues T. B., Serrao E. M., Kennedy B. W. C., Hu D.-E., Kettunen M. I., Brindle K. M., Magnetic resonance imaging of tumor glycolysis using hyperpolarized 13C-labeled glucose. Nat. Med. 20, 93–97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris R. K., Becker E. D., Cabral de Menezes S. M., Goodfellow R., Granger P., NMR Nomenclature: Nuclear spin properties and conventions for chemical shifts. IUPAC recommendations 2001. Solid State Nucl. Magn. Reson. 22, 458–483 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Lebon V., Petersen K. F., Cline G. W., Shen J., Mason G. F., Dufour S., Behar K. L., Shulman G. I., Rothman D. L., Astroglial contribution to brain energy metabolism in humans revealed by 13C nuclear magnetic resonance spectroscopy: Elucidation of the dominant pathway for neurotransmitter glutamate repletion and measurement of astrocytic oxidative metabolism. J. Neurosci. 22, 1523–1531 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewy C. S., Ackerman J. J. H., Balaban R. S., Deuterium NMR cerebral imaging in situ. Magn. Reson. Med. 8, 35–44 (1988). [DOI] [PubMed] [Google Scholar]
- 18.Warburg O., Über den Stoffwechsel der Carcinomzellen. Klin. Wschr 4, 534–536 (1925). [Google Scholar]
- 19.Koppenol W. H., Bounds P. L., Dang C. V., Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Befroy D. E., Perry R. J., Jain N., Dufour S., Cline G. W., Trimmer J. K., Brosnan J., Rothman D. L., Petersen K. F., Shulman G. I., Direct assessment of hepatic mitochondrial oxidative and anaplerotic fluxes in humans using dynamic 13C magnetic resonance spectroscopy. Nat. Med. 20, 98–102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brosnan J. T., Man K. C., Hall D. E., Colbourne S. A., Brosnan M. E., Interorgan metabolism of amino acids in streptozotocin-diabetic ketoacidotic rat. Am. J. Physiol. 244, E151–E158 (1983). [DOI] [PubMed] [Google Scholar]
- 22.Kim M. M., Parolia A., Dunphy M. P., Venneti S., Non-invasive metabolic imaging of brain tumours in the era of precision medicine. Nat. Rev. Clin. Oncol. 13, 725–739 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doherty J. R., Cleveland J. L., Targeting lactate metabolism for cancer therapeutics. J. Clin. Invest. 123, 3685–3692 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Floch R. L., Chiche J., Marchiq I., Naiken T., Ilc K., Murray C. M., Critchlow S. E., Roux D., Simon M.-P., Pouysségur J., CD147 subunit of lactate/H+ symporters MCT1 and hypoxia-inducible MCT4 is critical for energetics and growth of glycolytic tumors. Proc. Natl. Acad. Sci. U.S.A. 108, 16663–16668 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Feyter H. M., Behar K. L., Rao J. U., Madden-Hennessey K., Ip K. L., Hyder F., Drewes L. R., Geschwind J.-F., de Graaf R. A., Rothman D. L., A ketogenic diet increases transport and oxidation of ketone bodies in RG2 and 9L gliomas without affecting tumor growth. Neuro Oncol. 18, 1079–1087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu M., Zhang Y., Ugurbil K., Chen W., Zhu X.-H., In vitro and in vivo studies of 17O NMR sensitivity at 9.4 and 16.4 T. Magn. Reson. Med. 69, 1523–1527 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landau B. R., Wahren J., Chandramouli V., Schumann W. C., Ekberg K., Kalhan S. C., Contributions of gluconeogenesis to glucose production in the fasted state. J. Clin. Invest. 98, 378–385 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macallan D. C., Asquith B., Zhang Y., de Lara C., Ghattas H., Defoiche J., Beverley P. C. L., Measurement of proliferation and disappearance of rapid turnover cell populations in human studies using deuterium-labeled glucose. Nat. Protoc. 4, 1313–1327 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Decaris M. L., Li K. W., Emson C. L., Gatmaitan M., Liu S., Wang Y., Nyangau E., Colangelo M., Angel T. E., Beysen C., Cui J., Hernandez C., Lazaro L., Brenner D. A., Turner S. M., Hellerstein M. K., Loomba R., Identifying nonalcoholic fatty liver disease patients with active fibrosis by measuring extracellular matrix remodeling rates in tissue and blood. Hepatology 65, 78–88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Busch R., Neese R. A., Awada M., Hayes G. M., Hellerstein M. K., Measurement of cell proliferation by heavy water labeling. Nat. Protoc. 2, 3045–3057 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Ben-Yoseph O., Kingsley P. B., Ross B. D., Metabolic loss of deuterium from isotopically labeled glucose. Magn. Reson. Med. 32, 405–409 (1994). [DOI] [PubMed] [Google Scholar]
- 32.De Feyter H. M., Herzog R. I., Steensma B. R., Klomp D. W. J., Brown P. B., Mason G. F., Rothman D. L., de Graaf R. A., Selective proton-observed, carbon-edited (selPOCE) MRS method for measurement of glutamate and glutamine 13C-labeling in the human frontal cortex. Magn. Reson. Med. 80, 11–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mason G. F., Petersen K. F., de Graaf R. A., Kanamatsu T., Otsuki T., Rothman D. L., A comparison of 13C NMR measurements of the rates of glutamine synthesis and the tricarboxylic acid cycle during oral and intravenous administration of [1-13C]glucose. Brain Res. Brain Res. Protoc. 10, 181–190 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Aguayo J. B., McLennan I. J., Graham C. Jr., Cheng H.-M., Dynamic monitoring of corneal carbohydrate metabolism using high-resolution deuterium NMR spectroscopy. Exp. Eye Res. 47, 337–343 (1988). [DOI] [PubMed] [Google Scholar]
- 35.Brereton I. M., Doddrell D. M., Oakenfull S. M., Moss D., Irving M. G., The use of in vivo 2H NMR spectroscopy to investigate the effects of obesity and diabetes mellitus upon lipid metabolism in mice. NMR Biomed. 2, 55–60 (1989). [DOI] [PubMed] [Google Scholar]
- 36.Ackerman J. J., Ewy C. S., Becker N. N., Shalwitz R. A., Deuterium nuclear magnetic resonance measurements of blood flow and tissue perfusion employing 2H2O as a freely diffusible tracer. Proc. Natl. Acad. Sci. U.S.A. 84, 4099–4102 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell M. D., Osbakken M., Estimation of myocardial perfusion using deuterium nuclear magnetic resonance. Magn. Reson. Imaging 9, 545–552 (1991). [DOI] [PubMed] [Google Scholar]
- 38.Coleen J. J. H., Ewy C. S., Kim S.-G., Shalwitz R. A., Deuterium magnetic resonance in vivo: The measurement of blood flow and tissue perfusion. Ann. N. Y. Acad. Sci. 508, 89–98 (1987). [DOI] [PubMed] [Google Scholar]
- 39.Adriany G., Gruetter R., A half-volume coil for efficient proton decoupling in humans at 4 tesla. J. Magn. Reson. 125, 178–184 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Bolinger L., Prammer M. G., Leigh J. S. Jr., A multiple-frequency coil with a highly uniform B1 field. J. Magn. Reson. 81, 162–166 (1989). [Google Scholar]
- 41.Pauly J., Le Roux P., Nishimura D., Macovski A., Parameter relations for the Shinnar-Le Roux selective excitation pulse design algorithm [NMR imaging]. IEEE Trans. Med. Imaging 10, 53–65 (1991). [DOI] [PubMed] [Google Scholar]
- 42.R. A. de Graaf, In Vivo NMR Spectroscopy. Principles and Techniques (John Wiley, 2007). [Google Scholar]
- 43.Moskau D., Günther H., 2H,2H-COSY and 2H,2H,13C-RELAY NMR experiments for the analysis of deuteriated compounds. Angew. Chem. Int. Ed. Engl. 26, 156–157 (1987). [Google Scholar]
- 44.Wimmer M., Wilmering B., Sasse D., The relation of rat liver wet weight to dry weight. Histochemistry 83, 571–572 (1985). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/8/eaat7314/DC1
Fig. S1. DMI of [6,6′-2H2]glucose and [2H3]acetate metabolism.
Fig. S2. High-resolution 1H NMR of rat plasma.
Fig. S3. DMI detects the effect of DCA on glucose metabolism.
Fig. S4. Difference in 2H-acetate metabolism between normal-appearing rat brain and tumor tissue.
Fig. S5. In vivo 2H MRSI spectra from human brain.
Fig. S6. DMI of the Warburg effect in a patient with GBM.
Fig. S7. High-resolution 13C NMR spectra of 13C- and 2H-labeled lactate in cell culture medium.
Table S1. T1 and T2 relaxation times.
Table S2. T2* of water from the brain in vivo at 4 and 11.7 T.


