Abstract
OBJECTIVE
We tested the ability of a type 1 diabetes (T1D) genetic risk score (GRS) to predict progression of islet autoimmunity and T1D in at-risk individuals.
RESEARCH DESIGN AND METHODS
We studied the 1,244 TrialNet Pathway to Prevention study participants (T1D patients’ relatives without diabetes and with one or more positive autoantibodies) who were genotyped with Illumina ImmunoChip (median [range] age at initial autoantibody determination 11.1 years [1.2–51.8], 48% male, 80.5% non-Hispanic white, median follow-up 5.4 years). Of 291 participants with a single positive autoantibody at screening, 157 converted to multiple autoantibody positivity and 55 developed diabetes. Of 953 participants with multiple positive autoantibodies at screening, 419 developed diabetes. We calculated the T1D GRS from 30 T1D-associated single nucleotide polymorphisms. We used multivariable Cox regression models, time-dependent receiver operating characteristic curves, and area under the curve (AUC) measures to evaluate prognostic utility of T1D GRS, age, sex, Diabetes Prevention Trial–Type 1 (DPT-1) Risk Score, positive autoantibody number or type, HLA DR3/DR4-DQ8 status, and race/ethnicity. We used recursive partitioning analyses to identify cut points in continuous variables.
RESULTS
Higher T1D GRS significantly increased the rate of progression to T1D adjusting for DPT-1 Risk Score, age, number of positive autoantibodies, sex, and ethnicity (hazard ratio [HR] 1.29 for a 0.05 increase, 95% CI 1.06–1.6; P = 0.011). Progression to T1D was best predicted by a combined model with GRS, number of positive autoantibodies, DPT-1 Risk Score, and age (7-year time-integrated AUC = 0.79, 5-year AUC = 0.73). Higher GRS was significantly associated with increased progression rate from single to multiple positive autoantibodies after adjusting for age, autoantibody type, ethnicity, and sex (HR 2.27 for GRS >0.295, 95% CI 1.47–3.51; P = 0.0002).
CONCLUSIONS
The T1D GRS independently predicts progression to T1D and improves prediction along T1D stages in autoantibody-positive relatives.
Introduction
Early identification of individuals at risk for type 1 diabetes (T1D) allows study of the biology of the preclinical stages of T1D and inclusion of those at highest T1D risk in monitoring and prevention trials. Current prediction models for T1D use immunologic and metabolic markers, but these markers change during disease progression and reflect advanced stages in the autoimmune process (1–7), whereas genetic predictors are time-independent and may be assessed only once at study entry. T1D has a significant heritable risk as evidenced by studies of monozygotic twins that demonstrated rates of disease concordance >50%, higher with younger age at diagnosis of the index twin (8,9). Approximately 50% of this heritability is attributable to the HLA region (10), with another >50 loci making smaller contributions to disease risk (reviewed in previous studies [11–13]). Recently, Oram et al. (14) developed and validated a T1D genetic risk score (GRS) that incorporates HLA and non-HLA T1D-associated single nucleotide polymorphisms (SNPs) and was discriminative of T1D from type 2 diabetes, monogenic diabetes, and controls (15). In this study, we tested the prognostic utility of the T1D GRS for differentiating rates of progression of islet autoimmunity and development of clinical T1D in autoantibody-positive relatives of individuals with T1D.
Research Design and Methods
Participants
Type 1 Diabetes TrialNet is a National Institutes of Health–funded international network that aims to prevent T1D (16). TrialNet Pathway to Prevention (PTP) is an observational study that prospectively follows at-risk first- or second-degree relatives of patients with T1D for development of islet autoimmunity and clinical T1D (17). This study included TrialNet PTP participants who had one or more positive, persistently detectable islet autoantibodies and had been genotyped using the Illumina ImmunoChip (n = 1,244). Study participants gave informed consent and the study was approved by ethics committees at each site.
Procedures
Participants were initially screened for autoantibodies to glutamic acid decarboxylase (GAD65), insulin (microinsulin antibody assay [mIAA]), and insulinoma-associated antigen 2 (IA-2A). If any of these was positive, autoantibodies to zinc transporter 8 (ZnT8) and islet cell antibodies (ICA) were tested. Participants were monitored with autoantibody testing, hemoglobin A1c (HbA1c), and oral glucose tolerance test at 6- or 12-month intervals depending on estimated risk (18). T1D was diagnosed in participants with 1) symptomatic hyperglycemia, defined as fasting plasma glucose ≥7.0 mmol/L, 2-h plasma glucose after 75 g oral glucose ≥11.1 mmol/L, a random plasma glucose ≥11.1 mmol/L, or an HbA1c ≥6.5%; or 2) asymptomatic hyperglycemia documented on two separate occasions. Islet-autoantibody (17) and C-peptide (19) assays have been previously described. HLA genotyping was performed as previously described (20). Illumina ImmunoChip genotyping was performed at the Center for Public Health Genomics, University of Virginia. The Diabetes Prevention Trial–Type 1 (DPT-1) Risk Score is a diabetes risk score derived from ICA-positive individuals and validated in TrialNet that combines BMI, age, glucose, and C-peptide (2,21). We stratified our analysis by a metabolic DPT-1 Risk Score of ≤7 or >7 based on previous work (22).
T1D GRS
The T1D GRS was calculated from 30 variants known to be associated with T1D (Supplementary Table 2), ranked and weighted by published odds ratios as previously described (14). We drew odds ratios for each SNP from the largest available meta-analysis study that used T1Dbase (http://t1dbase.org/). Twenty-nine of these variants were directly genotyped, whereas rs11755527 was imputed using IMPUTE2 (r2 = 0.99997). rs2187668 and rs7454108 were used to determine HLA DR haplotype (23). The T1D GRS threshold that was previously shown to optimally discriminate T1D from type 2 diabetes was 0.280 (14). T1D GRS percentiles in a reference T1D population (24) are provided to allow comparisons between different genetic scores. The same methods were used to calculate a 10-SNP score using the top 10 T1D-associated SNPs (Supplementary Table 2), which account for most of the genetic risk. We assessed the predictive power of both the 10-SNP and 30-SNP scores.
Statistical Analyses
We used summary statistics and graphical analyses to assess the distributions and characteristics of the clinical and metabolic measures as well as the T1D GRS, overall and by subgroup. Comparisons between subgroups were made using primarily nonparametric approaches, e.g., Wilcoxon rank sum or Kruskal-Wallis test and the χ2 or Fisher exact test, as appropriate.
Kaplan-Meier methods were used to evaluate the time-to-event distributions for time to progression to T1D and time from single to multiple autoantibody positivity overall and in subgroups (see Supplementary Table 3 for definitions). Cox proportional hazards models were used to test the prognostic influence of these measures on these outcomes in univariate and multivariable settings. Models were adjusted for age, sex, and race/ethnicity. For models of time to conversion from single to multiple autoantibodies, we also adjusted for autoantibody type (i.e., GAD65, insulin, or IA-2A). For time-to-T1D models, we additionally adjusted for DPT-1 Risk Score and the number of positive autoantibodies present at screening. T1D GRS, age, and DPT-1 Risk Score were each evaluated as continuous and dichotomized factors. We assessed whether T1D GRS added predictive power independently over HLA DR3/DR4-DQ8 status by including DR3/DR4-DQ8 in initial multivariate analyses; the HLA DR3/DR4-DQ8 variable was then removed from the final models because the overlap between the two variables (i.e., HLA DR3/DR4-DQ8 is included in the T1D GRS) caused collinearity. Recursive partitioning analyses (risk-stratification method based on classification and regression trees) were used to identify variables and associated cut points that best differentiated outcome-specific risk (rpart package in R) (25). To obtain stable hazard ratio (HR) estimates reflecting meaningful unit changes in the continuous 30-SNP T1D GRS measure, we multiplied this measure by a constant (× 20) when included as a continuous factor in models. All reported HRs for continuous T1D GRS measures reflect this multiplier and reflect HRs associated with an increase of 0.05 in the T1D GRS.
The predictive accuracy of models for time to progression to multiple autoantibodies and to T1D was evaluated for T1D GRS (or HLA), islet autoantibody number, age, and DPT-1 Risk Score using time-dependent area under the curve (AUC) analyses (survAUC in R). Time-integrated AUC measures were calculated for each model in addition to year-specific AUCs on subjects with complete data for the multivariable models, consistent with standard AUC goodness-of-fit measures. In addition, to evaluate if GRS added more to our prognostic models than HLA, we directly compared the GRS versus HLA models as well as comparing them combined with clinical factors (DPT-1 Risk Score, age, autoantibody number). Time-integrated AUC estimates were limited to 7 years given that the third quartile for follow-up in event-free participants in the overall cohort was just over 7 years. Predictive accuracy between models was compared at major time points and reflects comparisons of estimated 5-year AUCs unless stated otherwise (timeROC package in R).
Results
Characteristics of TrialNet participants in this study (n = 1,244) are presented in Supplementary Table 4. The median age at autoantibody determination was 11.1 years (range 1.2–51.8), 48% were male, 81% non-Hispanic white, and 90% first-degree relatives of a patient with T1D. The estimated median follow-up was 5.4 years [95% CI 5.0–5.8]. Of the 291 participants positive for a single antibody, 157 progressed to multiple autoantibody positivity and 55 developed T1D. Of the 953 participants who had multiple antibodies when initially screened, 419 developed T1D.
Overall, the 30-SNP T1D GRS ranged from 0.138 to 0.341 (median = 0.272, corresponding to the 38th–39th percentiles in the reference T1D population [24]). The median T1D GRS for single and multiple autoantibody–positive subjects were 0.266 (30th percentile, range 0.138–0.341) and 0.274 (41st percentile, range 0.169–0.328), respectively.
T1D GRS Is an Independent Predictor of Clinical T1D in Islet Autoantibody–Positive Relatives
The T1D GRS was a significant predictor of risk and rate of progression to T1D in continuous univariate analysis (HR 1.7, 95% CI 1.43–2.0; P < 0.0001) as well as after adjustment for other risk factors (Supplementary Table 5). Of note, with inclusion of the T1D GRS in the multivariable model, HLA DR3/DR4-DQ8 was no longer significant (HR 1.06, 95% CI 0.79–1.41; P = 0.71 [data not shown]). The best predictive model of progression to T1D, with a 7-year time-integrated AUC of 0.794, included GRS, the metabolic DPT-1 Risk Score, age at autoantibody determination, and number of positive autoantibodies (Supplementary Table 5). The GRS remained a significant predictor in this model (HR 1.29, 95% CI 1.06–1.56; P = 0.009). Since we observed a significant interaction between T1D GRS and DPT-1 Risk Score (P = 0.001), as well as between GRS and autoantibody number (P = 0.001), next we also analyzed models of progression to T1D stratified by these features.
Interaction and stratified analyses revealed that GRS is best able to further differentiate T1D risk in those participants with a baseline metabolic DPT-1 Risk Score ≤7.0 (n = 716, which represents 63% of 1,136 participants with DPT-1 Risk Score data available at baseline) (HR 1.66, 95% CI 1.18–2.34; P = 0.003) even after adjusting for age, autoantibody number, sex, ethnicity, and DPT-1 Risk Score (Supplementary Table 5B). Although those with a DPT-1 Risk Score >7 had a higher T1D GRS than those with DPT-1 Risk Score ≤7 (0.274 [SD 0.026] vs. 0.268 [0.028]; P = 0.002), the GRS did not further stratify the risk of T1D in participants who had already developed metabolic abnormalities, as reflected by a DPT-1 Risk Score >7.0 (HR 1.07, 95% CI 0.81–1.41; P = 0.64).
Since ICA and GAD65 autoantibodies may overlap (26), we performed sensitivity analyses with the 167 (out of 1,244) subjects who were only positive for ICA and GAD65 in this cohort and observed that their classification as positive for one versus two autoantibodies yielded similar results and consistent estimates.
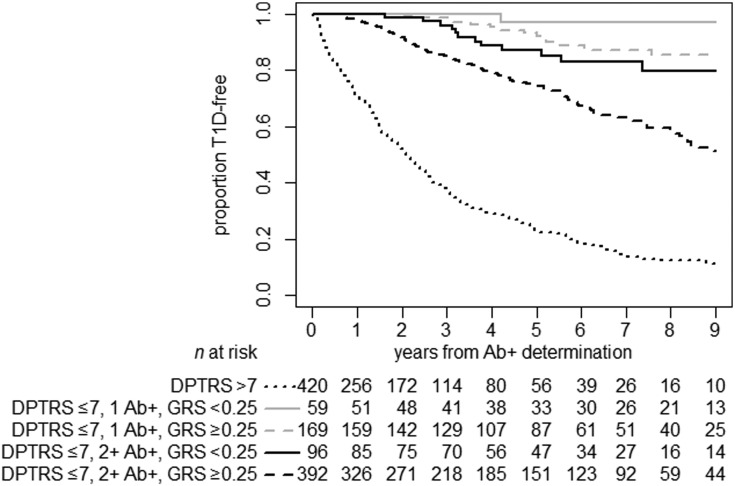
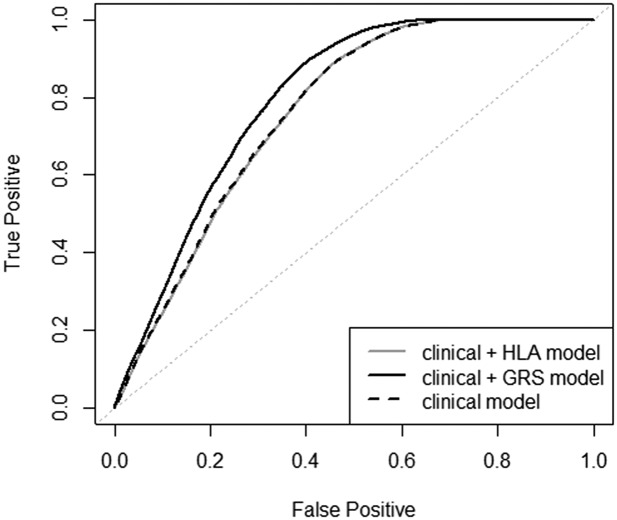
Multivariable recursive partitioning models identified variable cut points and five risk clusters (Fig. 1 and Supplementary Fig. 1). The optimal cut point for GRS in relation to time to progression to T1D was 0.250. DPT-1 Risk Score >7 identified the highest risk group, while in those with DPT-1 Risk Score ≤7 the risk of T1D could be further stratified according to autoantibody number and T1D GRS. To assess the improvement of T1D prediction when T1D GRS was included with established predictors, we calculated time-dependent receiver operating characteristic (ROC) curves integrated across all time points (iAUC) and standard ROC curves for the 2- and 5-year time point. For the overall at-risk cohort with complete data on these factors (n = 1,106, 415 events), the iAUCs were 0.57 for T1D GRS, 0.53 for HLA, 0.59 for autoantibody number, 0.59 for age, and 0.77 for DPT-1 Risk Score. The iAUC for the final composite risk model (i.e., T1D GRS, metabolic DPT-1 Risk Score, age, and autoantibody number) was 0.79. Given that we identified that GRS has the most prognostic utility in participants with DPT-1 Risk Score ≤7, we also evaluated the time-dependent ROC and AUC measures in those with complete data on these factors (n = 696, 132 T1D events). In this subset, we found similar patterns of iAUC for these factors. We observed that the model with GRS combined with the “clinical” variables (i.e., DPT-1 Risk Score, age, and autoantibody number) had significantly better prediction accuracy than the model with HLA combined with the clinical variables, although this was significant at earlier time points (i.e., ROC and AUC estimates for up to 3 years). For example, the 2-year AUC for the clinical + HLA model was 0.78 versus 0.82 for the clinical + GRS model (P < 0.0001) (Fig. 2). Similarly, we observed that, in the participants with lower metabolic risk, the T1D prediction model that combined GRS with the clinical variables DPT-1 Risk Score, age, and autoantibody number performed significantly better than HLA DR3/DR4-DQ8 in addition to the clinical variables (iAUC 0.60 vs. 0.53; P = 0.007).
Figure 1.
Time to T1D in patients’ relatives who were initially without diabetes and islet autoantibody–positive (Ab+), by DPT-1 Risk Score (≤7 vs. >7), number of positive autoantibodies (i.e., single vs. multiple autoantibody positivity), and T1D GRS (<0.250 vs. ≥0.250) (P < 0.0001). While the T1D GRS did not further increase the predictive ability in the group with DPT-1 Risk Score >7, which already had high risk of T1D, it was able to stratify risk in individuals with DPT-1 Risk Score <7, with either a single positive autoantibody or multiple positive autoantibodies. DPTRS, DPT-1 Risk Score.
Figure 2.
Comparison of 2-year AUC for models to predict progression to T1D in participants with DPT-1 Risk Scores ≤7. The clinical model (i.e., DPT-1 Risk Score, age, and islet autoantibody number) in addition to HLA had a 2-year AUC of 0.78, compared with 0.82 for the clinical model in addition to GRS (P < 0.0001).
T1D GRS Is an Independent Predictor of Progression From Multiple Islet Autoantibody Positivity to T1D
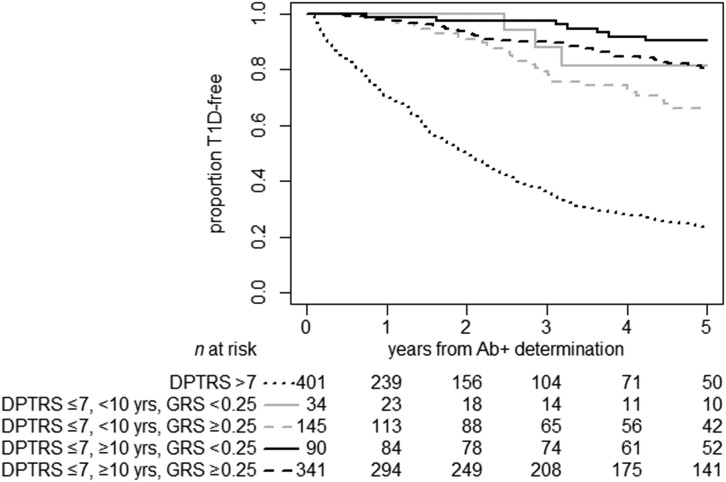
There were 953 participants who were identified as having multiple autoantibody positivity at screening and 157 additional participants who developed multiple positive autoantibodies during follow-up, for a total of 1,110 multiple autoantibody–positive participants in our cohort. After adjusting for age and DPT-1 Risk Score, the T1D GRS was a significant independent prognostic factor for time to progression to T1D as a continuous (P = 0.015) and as a dichotomized variable (cut point = 0.250, P = 0.017) (Supplementary Table 6). Among multiple autoantibody–positive participants with lower metabolic DPT-1 Risk Score, high T1D GRS was a significant factor in multivariable analysis (T1D GRS ≥0.250, HR 2.07, 95% CI 1.21–3.55; P = 0.008) (Supplementary Table 6B). Five-year T1D-free rate estimates were 89% for those with a low T1D GRS (<0.250) versus 77% in participants with high T1D GRS (≥0.250). The risk of progressing from multiple islet autoantibody positivity to T1D could be stratified by the composite grouping of DPT-1 Risk Score, age, and T1D GRS (Fig. 3). Time-to-event ROC and AUC analyses demonstrated that the addition of GRS to the model with age and DPT-1 Risk Score improved the prediction model for T1D in a similar manner to that seen in all autoantibody-positive participants with DPT-1 Risk Score ≤7 (2-year AUC: clinical + HLA 0.68 vs. clinical + GRS 0.73; P < 0.0001). Interestingly, the GRS improved the prediction afforded by HLA DR3/DR4-DQ8 alone (iAUC: 0.65 vs. 0.56, respectively; P = 0.006).
Figure 3.
Time to T1D in multiple islet autoantibody–positive (Ab+) relatives, by DPT-1 Risk Score (≤7 vs. >7), age (<10 vs. ≥10 years), and T1D GRS (<0.250 vs. ≥0.250) (P = 0.0001). While the T1D GRS did not further increase the predictive ability in participants with DPT-1 Risk Score >7, it did stratify risk in individuals with DPT-1 Risk Score <7, aged <10 years or ≥10 years. DPTRS, DPT-1 Risk Score.
T1D GRS Is an Independent Predictor of Progression of Islet Autoimmunity
In our cohort, 157 of the 291 single autoantibody–positive participants progressed to multiple islet autoantibody positivity. Elevated T1D GRS was associated with progression from single to multiple autoantibody positivity, where an increase of 0.050 (e.g., from 0.225 to 0.275) significantly increased risk by 50% (HR 1.49, 95% CI 1.1–2.05; P = 0.015) after adjustment for age, sex, ethnicity, and autoantibody type (Supplementary Table 7).
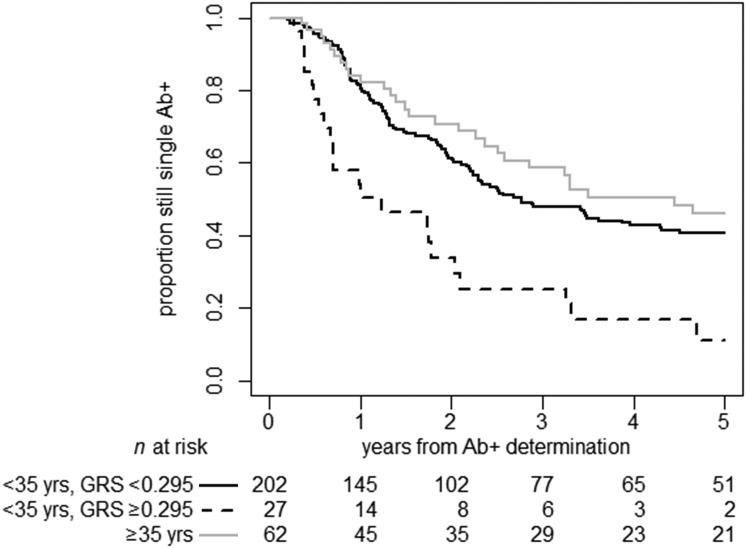
Recursive partitioning identified 0.295 (69th–70th percentiles) as the optimal cut point to discriminate individuals with the highest rate of progression from single to multiple autoantibody positivity. Single autoantibody–positive participants whose T1D GRS exceeded 0.295 had more than two times higher risk of autoantibody progression (HR 2.27, 95% CI 1.47–3.51; P = 0.0002), even adjusting for age, autoantibody type, sex, and ethnicity.
We observed a potential interaction between T1D GRS and age at first autoantibody determination (P = 0.052). In participants <35 years (n = 229), after adjusting for age, sex, ethnicity, and autoantibody type, T1D GRS was a significant predictor of progression to multiple autoantibody positivity, both as a continuous (HR 1.65, 95% CI 1.15–2.37; P = 0.0065) and dichotomous variable, with a cut point of 0.295 (HR 2.57, 95% CI 1.6–4.13; P = 0.0001) but also 0.250 (HR 1.68, 95% CI 1.07–2.64; P = 0.023). On the other hand, in older participants (≥35 years of age when classified as single autoantibody positive), who were at much less risk of T1D overall, the T1D GRS did not significantly inform the risk and prognosis for progression to multiple autoantibody positivity after adjusting for autoantibody type and sex (age was not significant and thus was excluded from the model), although the numbers were relatively smaller (n = 62; HR 0.86, 95% CI 0.25–2.96; P = 0.81) (Fig. 4).
Figure 4.
Time from single to multiple islet autoantibody positivity (Ab+) in relatives of patients, by age (<35 vs. ≥35 years) and T1D GRS group (<0.295 vs. ≥0.295) (P = 0.0001). While the T1D GRS did not further increase the predictive ability in participants aged ≥35 years, it was able to stratify risk in individuals aged <35 years.
In time-dependent ROC analysis, the T1D GRS alone delivered an iAUC of 0.55 compared with 0.53, 0.52, and 0.53 for age, autoantibody type, and HLA DR3/DR4-DQ8 heterozygosity, respectively. The iAUC of a multivariable model that combined age, autoantibody type, and T1D GRS was 0.58.
A Reduced 10-SNP T1D GRS Performed Similarly to the T1D GRS in Predicting Islet Autoimmunity Progression and T1D
We evaluated the performance of a T1D GRS based on the top 10 SNPs (14) (T1D GRS-10), using the same analytic approach as for the 30-SNP measure. In multivariable analysis, the T1D GRS-10 predicted progression to T1D in all subjects (HR 1.16 for each increase of 0.10 in score, 95% CI 1.03–1.31; P = 0.014) and in the subgroup of multiple autoantibody–positive subjects (HR 1.15, 95% CI 1.02–1.30; P = 0.024). Similarly to the 30-SNP score, the 10-SNP GRS was only a significant factor in those with the metabolic DPT-1 Risk Score <7 (P = 0.0026). T1D GRS-10 also predicted progression from single to multiple autoantibody positivity after adjusting for age, sex, ethnicity, and autoantibody type (HR 1.26 for a 0.1 increase in T1D GRS-10, 95% CI 1.03–1.55; P = 0.026). The overall predictive power of T1D GRS-10 was similar to that of the 30-SNP GRS (iAUC = 0.575).
Conclusions
We studied 1,244 relatives of patients with T1D who initially did not have diabetes and were islet autoantibody positive and demonstrated that the T1D GRS is an independent predictor of progression of islet autoimmunity and development of clinical T1D. The T1D GRS improved current prediction models by stratifying risk among individuals who were either single or multiple autoantibody positive. We demonstrated that the combined modeling of the T1D GRS, which includes HLA and non-HLA factors, added to autoantibody and metabolic data offers better prediction of T1D in at-risk relatives. This approach could increase our ability to predict T1D in relatives of patients, as well as to screen and select participants for natural history studies and intervention trials.
This study adds to the recent expanding literature on the applicability of genetic information in the prediction of T1D. The T1D GRS used in the current study was originally developed and validated to distinguish T1D and type 2 diabetes in the Wellcome Trust Case Control Consortium (WTCCC) (n = 3,887) and in a cohort defined by insulin insufficiency (14). The score was also able to discriminate T1D and maturity-onset diabetes of the young and, in neonates, monogenic neonatal diabetes (15). Our present findings extend the use of the T1D GRS to prediction of T1D in relatives at risk. There have been previous attempts to develop genetic scores that integrate genetic information to improve the prediction of T1D (reviewed in a recent study [27]). In particular, it was shown that the combination of HLA and non-HLA genetic factors increases the power of the T1D predictive model (28–31). Winkler et al. (29) developed a genetic score using logistic regression and Bayesian feature selection of the Type 1 Diabetes Genetics Consortium (T1DGC) to define a set of 10 SNPs, including HLA, that identified risk of T1D in first-degree relatives from the BABYDIAB study. Our score, although generated from a log-additive model, contains very similar genetic information, so it is not surprising that the results are consistent. A key additional benefit of our T1D GRS is the inclusion of SNPs tagging other significant HLA risk alleles, e.g., HLA DRB1*15, DRB1*57, and A24. Specifically, DRB1*15:01 (linked to DQB1*06:02) is common in Caucasians and confers strong genetic protection against T1D (20). A score generated by merging the Winkler and colleagues (29,30) and Oram et al. (14) scores has recently been proposed to identify newborns from the general population who will develop islet autoimmunity and T1D. In their study, Bonifacio et al. (32) demonstrated that even in a subset of individuals with high-risk HLA genotypes from The Environmental Determinants of Diabetes in the Young (TEDDY) study, a T1D genetic score predicted development of autoantibodies. Although different characteristics in each cohort (e.g., age, background risk of T1D, proportion of individuals with a relative with T1D) may require adaptations of the T1D GRS, the concept of combining genetic information into a single factor will greatly improve its utility for prediction and trial design. By virtue of being a number, the T1D GRS facilitates incorporation of complex genetic information into prediction models. Importantly, selecting appropriate cut points will optimize the use of the T1D GRS for different goals.
The T1D GRS significantly added predictive power to the current variables used to stratify T1D risk in the TrialNet PTP study. The measurement of autoantibodies and differences in risk associated with autoantibody positivity are well described (33), as are the impact of age and metabolic data (34–36). The fact that the T1D GRS was not a predictor in those with DPT-1 Risk Score >7 demonstrates that, when metabolic abnormalities develop, measures that evaluate these directly become most predictive and, consequently, the role of genetics in risk assessment diminishes. However, the majority of individuals entering TrialNet PTP have a low DPT-1 Risk Score; in this group, the addition of T1D GRS to the currently established predictors (i.e., age, autoantibody number, DPT-1 Risk Score) can best add predictive power and assist in stratification for prevention trials. In the current study, multivariate modeling of autoantibody status, DPT-1 Risk Score, age, and additional demographic factors still leaves the T1D GRS as a significant independent predictor of progression. This observation supports the assessment of all of these features, either in a combined model or a sequential approach, at entry to the TrialNet PTP and other similar studies. Previous studies have shown conflicting results on the ability of genetic factors, age, and autoantibody and metabolic data to predict T1D (36,37). Some of the differences in the role of genetics could be due to the challenges of capturing genetic information; an advantage of the T1D GRS is that it includes SNPs tagging other significant HLA risk alleles, e.g., HLA DRB1*15, DRB1*57, and A24, in addition to non-HLA SNPs. Supporting this notion, in the current study, the T1D GRS was superior to HLA DR3/DR4-DQ8 alone for predicting progression to T1D. Because the T1D GRS further stratified T1D risk beyond that associated with autoantibody number in individuals with low DPT-1 Risk Score, it is plausible that applying the T1D GRS earlier in life would allow discrimination of the individuals who will develop a high DPT-1 Risk Score and T1D.
The unique longitudinal follow-up and monitoring of the TrialNet PTP study also allowed us to further investigate the contribution of the GRS to preclinical stages of T1D. Progression from single to multiple autoantibody positivity was independently predicted by the T1D GRS only in participants <35 years of age, who have higher risk of progression, although the number of individuals ≥35 years old in the analysis was relatively limited and thus we cannot conclusively rule out the influence of the GRS in the subset of individuals ≥35 years old. We had previously observed the protective effect of age on progression to T1D in at-risk adults with a threshold of 35 years of age (38) and the influence of age on the effect of another genetic factor, namely, type 2 diabetes–associated TCF7L2 variants on T1D progression (39). Interestingly, despite having been originally discovered in studies of childhood diabetes, the T1D GRS was able to identify more adult than childhood T1D cases in a recent study of T1D in UK Biobank (40). These results and those from the current study suggest that the genetic factors that regulate the progression of islet autoimmunity may slightly differ by age and further support the emerging notion that age is a key factor in the heterogeneity of T1D pathogenesis. The importance of age in progression through T1D stages is also highlighted by its significant and strong influence in the multivariable models even after adjustment for DPT-1 Risk Score, which includes age as well.
We tested the predictive power of a restricted set of the top 10 SNPs from our score (14), which proved to contain the vast majority of predictive power in the T1D GRS. This is unsurprising owing to the high weights of HLA and the top SNPs in the score. These results may be relevant to large-scale studies where the cost of the T1D GRS per individual may be important.
The study limitations include that it evaluated the performance of T1D GRS only in autoantibody-positive relatives of people with T1D, although recent data (32) suggest that the T1D GRS will be a significant predictor in general population cohorts as well. We tested the T1D GRS and derived score cutoffs within the 1,244 TrialNet PTP participants who had ImmunoChip data; we anticipate that expanding SNP analysis to the whole cohort will validate the current findings. Similarly, TrialNet is a cohort of >80% non-Hispanic whites and, although we were able to control for race/ethnicity, the T1D GRS needs to be specifically tested in other races and possibly modified according to genetic differences. Finally, it is possible that newly discovered variants, better capture of known HLA variants, stage-specific variants (e.g., progression from single to multiple autoantibody positivity), or longer follow-up of the cohort (allowing us to assess whether the rate of progression and its factors change with time) could improve understanding of the long-term predictive power of the T1D GRS.
In summary, the T1D GRS is a strong independent predictor of progression of islet autoimmunity and to clinical T1D in the TrialNet PTP study. Multivariate modeling suggests that the combination of islet autoantibody measurements, DPT-1 Risk Score, age, and T1D GRS into a prediction model may improve assessment of T1D risk. This study, in addition to recent positive analyses in BABYDIAB (29), the Diabetes Autoimmunity Study in the Young (DAISY) (31), and TEDDY (32), suggest that future T1D prediction studies are likely to use a genetic score, such as the T1D GRS, at enrollment. These findings warrant further investigations on the use of the T1D GRS for early assessment of T1D risk, particularly in longitudinal studies.
Supplementary Material
Article Information
Funding. The sponsor of the trial was the T1D TrialNet Study Group, which is a clinical trials network funded by the National Institutes of Health through the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases, and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (cooperative agreements U01 DK061010, U01 DK061034, U01 DK061042, U01 DK061058, U01 DK085465, U01 DK085453, U01 DK085461, U01 DK085466, U01 DK085499, U01 DK085504, U01 DK085509, U01 DK103180, U01 DK103153, U01 DK085476, U01 DK103266, U01 DK103282, U01 DK106984, U01 DK106994, U01 DK107013, U01 DK107014, and UC4 DK106993) and JDRF. J.M.W. was funded by a JDRF Australia Clinical Practitioner Fellowship and National Health and Medical Research Council Fellowship 1078106. R.A.O. was funded by the Diabetes UK Harry Keen Fellowship.
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or JDRF.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. M.J.R. designed the study, interpreted the data, and wrote the manuscript. S.G. contributed to study design, analyzed the data, contributed to data interpretation, and reviewed and edited the manuscript. A.K.S., S.S., J.M.W., M.N.W., P.A., J.S., M.A., and A.P. contributed to data interpretation and manuscript review and editing. R.A.O. contributed to study design, reviewed data, contributed to data interpretation, and reviewed and edited the manuscript. M.J.R., S.G., A.K.S., J.M.W., P.A., J.S., M.A., and A.P. are members of the T1D TrialNet Study Group (see Supplementary Table 1). M.J.R. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented at the 53rd Annual Meeting of the European Association for the Study of Diabetes, Lisbon, Portugal, 11–15 September 2017.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc18-0087/-/DC1.
A complete list of members of the Type 1 Diabetes TrialNet Study Group can be found in the Supplementary Data online.
Contributor Information
Collaborators: Type 1 Diabetes TrialNet Study Group, P. Antinozzi, M. Atkinson, M. Battaglia, D. Becker, P. Bingley, E. Bosi, J. Buckner, P. Colman, P. Gottlieb, K. Herold, R. Insel, T. Kay, M. Knip, J.B. Marks, A. Moran, J. Palmer, M. Peakman, L. Philipson, A. Pugliese, P. Raskin, H. Rodriguez, B. Roep, W. Russell, D.A. Schatz, D. Wherrett, D. Wilson, W. Winter, A. Ziegler, C. Benoist, J. Blum, P. Chase, M. Clare-Salzler, R. Clynes, G. Eisenbarth, C.G. Fathman, G. Grave, B. Hering, F. Kaufman, E. Leschek, J. Mahon, K. Nanto-Salonen, G. Nepom, T. Orban, R. Parkman, M. Pescovitz, J. Peyman, M. Roncarolo, O. Simell, R. Sherwin, M. Siegelman, A. Steck, J. Thomas, M. Trucco, J. Wagner, , Carla J. Greenbaum, Katarzyna Bourcier, Richard Insel, Jeffrey P. Krischer, Ellen Leschek, Lisa Rafkin, Lisa Spain, Catherine Cowie, Mary Foulkes, Heidi Krause-Steinrauf, John M. Lachin, Saul Malozowski, John Peyman, John Ridge, Peter Savage, Jay S. Skyler, Stephanie J. Zafonte, Norma S. Kenyon, Irene Santiago, Jay M. Sosenko, Brian Bundy, Michael Abbondondolo, Timothy Adams, Darlene Amado, Ilma Asif, Matthew Boonstra, Brian Bundy, Cristina Burroughs, David Cuthbertson, Mary Deemer, Christopher Eberhard, Steve Fiske, Julie Ford, Jennifer Garmeson, Heather Guillette, Susan Geyer, Brian Hays, Courtney Henderson, Martha Henry, Kathleen Heyman, Belinda Hsiao, Christina Karges, Nichole Keaton, Amanda Kinderman, Pat Law, Ashely Leinbach, Cristin Linton, Shu Liu, Jennifer Lloyd, Jamie Malloy, Kristin Maddox, Julie Martin, Jessica Miller, Eric Milliot, Margaret Moore, Sarah Muller, Thuy Nguyen, Ryan O’Donnell, Vanessa Oduah, Jennifer Pilger, Amy Roberts, Kelly Sadler, Tina Stavros, Roy Tamura, Keith Wood, Ping Xu, Kenneth Young, Persida Alies, Franz Badias, Aaron Baker, Monica Bassi, Craig Beam, David Boulware, London Bounmananh, Susan Bream, Doug Freeman, Jessica Gough, Jinin Ginem, Moriah Granger, Mary Holloway, Michelle Kieffer, Page Lane, Lavanya Nallamshetty, Yazandra Parrimon, Kate Paulus, Joy Ramiro, AQesha Luvon Ritzie, Archana Sharma, Audrey Shor, Xiaohong Song, Amanda Terry, Jeanne Weinberger, Margaret Wootten, Pamela Harding, Susan McDonough, Paula F. McGee, Kimberly Owens Hess, Donna Phoebus, Scott Quinlan, Erica Raiden, Emily Batts, Chris Buddy, Kristin Kirpatrick, Mary Ramey, Ann Shultz, Chris Webb, Melita Romesco, Judith Fradkin, Emily Blumberg, Gerald Beck, David Brillon, Rose Gubitosi-Klug, Lori Laffel, Robert Veatch, Dennis Wallace, Jonathan Braun, Ake Lernmark, Bernard Lo, Herman Mitchell, Ali Naji, Jorn Nerup, Trevor Orchard, Michael Steffes, Anastasios Tsiatis, Bernard Zinman, Brett Loechelt, Lindsey Baden, Michael Green, Adriana Weinberg, Santica Marcovina, Jerry P. Palmer, Adriana Weinberg, Liping Yu, Sunanda Babu, William Winter, George S. Eisenbarth, Polly Bingley, Raphael Clynes, Linda DiMeglio, George Eisenbarth, Brian Hays, Jennifer Marks, Della Matheson, Henry Rodriguez, Darrell Wilson, Maria J. Redondo, David Gomez, Xiati Zheng, Sandra Pena, Massimo Pietropaolo, Emily Batts, Tyler Brown, Jane Buckner, Angela Dove, Marissa Hammond, Deborah Hefty, Jani Klein, Kristen Kuhns, McKenzie Letlau, Sandra Lord, Marli McCulloch-Olson, Lisa Miller, Gerald Nepom, Jared Odegard, Mary Ramey, Elaine Sachter, Marissa St. Marie, Kimberly Stickney, Dana VanBuecken, Ben Vellek, Christine Webber, Laurie Allen, Jenna Bollyk, Nicole Hilderman, Hebatullah Ismail, Steve Lamola, Srinath Sanda, Heather Vendettuoli, David Tridgell, Roshanak Monzavi, Meredith Bock, Lynda Fisher, Mary Halvorson, Debra Jeandron, Mimi Kim, Jamie Wood, Mitchell Geffner, Francine Kaufman, Robertson Parkman, Christine Salazar, Robin Goland, Raphael Clynes, Steve Cook, Matthew Freeby, Mary Pat Gallagher, Rachelle Gandica, Ellen Greenberg, Amy Kurland, Sarah Pollak, Amy Wolk, Mary Chan, Linda Koplimae, Elizabeth Levine, Kelly Smith, Jeniece Trast, Linda DiMeglio, Janice Blum, Carmella Evans-Molina, Robin Hufferd, Bonnie Jagielo, Christy Kruse, Vanessa Patrick, Mark Rigby, Maria Spall, Kim Swinney, Jennifer Terrell, Lyla Christner, LeeAnn Ford, Sheryl Lynch, Martha Menendez, Patricia Merrill, Mark Pescovitz, Henry Rodriguez, Cielo Alleyn, David Baidal, Steve Fay, Jason Gaglia, Brittany Resnick, Sarah Szubowicz, Gordon Weir, Ronald Benjamin, Debbie Conboy, Andrea deManbey, Richard Jackson, Heyam Jalahej, Tihmar Orban, Alyne Ricker, Joseph Wolfsdorf, Hui H. Zhang, Darrell Wilson, Tandy Aye, Bonita Baker, Karen Barahona, Bruce Buckingham, Kerry Esrey, Trudy Esrey, Garry Fathman, Radhika Snyder, Beenu Aneja, Maya Chatav, Oralia Espinoza, Eliana Frank, Jenny Liu, Jennifer Perry, Rebecca Pyle, Alison Rigby, Kristin Riley, Adriana Soto, Stephen Gitelman, Saleh Adi, Mark Anderson, Ashley Berhel, Kathy Breen, Kathleen Fraser, Andrea Gerard-Gonzalez, Paula Jossan, Robert Lustig, Sara Moassesfar, Amy Mugg, David Ng, Priya Prahalod, Martha Rangel-Lugo, Srinath Sanda, Joshua Tarkoff, Christine Torok, Rebecca Wesch, Ivy Aslan, Jeanne Buchanan, Jennifer Cordier, Celia Hamilton, Louise Hawkins, Thu Ho, Anjali Jain, Karen Ko, Theresa Lee, Shelly Phelps, Stephen Rosenthal, Taninee Sahakitrungruang, Lorraine Stehl, Lisa Taylor, Marcia Wertz, Jenise Wong, Louis Philipson, Rosemary Briars, Nancy Devine, Elizabeth Littlejohn, Tiffany Grant, Peter Gottlieb, Georgeanna Klingensmith, Andrea Steck, Aimon Alkanani, Kimberly Bautista, Ruth Bedoy, Aaron Blau, Betsy Burke, Laraine Cory, MyLinh Dang, Lisa Fitzgerald-Miller, Alex Fouts, Vicky Gage, Satish Garg, Patricia Gesauldo, Raymond Gutin, Cory Hayes, Michelle Hoffman, Kaitlin Ketchum, Nyla Logsden-Sackett, David Maahs, Laurel Messer, Lisa Meyers, Aaron Michels, Stesha Peacock, Marian Rewers, Perla Rodriguez, Flor Sepulbeda, Rachel Sippl, Andrea Steck, Iman Taki, Bao-Khan Tran, Tuan Tran, R. Paul Wadwa, Philip Zeitler, Jennifer Barker, Sandra Barry, Laurie Birks, Leah Bomsburger, Terra Bookert, Leah Briggs, Patricia Burdick, Rosio Cabrera, Peter Chase, Erin Cobry, Amy Conley, Gabrielle Cook, Joseph Daniels, Dominic DiDomenico, Jennifer Eckert, Angelica Ehler, George Eisenbarth, Pamela Fain, Rosanna Fiallo-Scharer, Nicole Frank, Hannah Goettle, Michelle Haarhues, Sherrie Harris, Lauren Horton, John Hutton, Joy Jeffrrey, Rachael Jenison, Kelly Jones, Whitney Kastelic, Maria Amelia King, Debbie Lehr, Jenna Lungaro, Kendra Mason, Heather Maurer, Luy Nguyen, Allison Proto, Jaime Realsen, Kristina Schmitt, Mara Schwartz, San Skovgaard, Jennifer Smith, Brandon Vanderwel, Mary Voelmle, Rebecca Wagner, Amy Wallace, Philip Walravens, Laurie Weiner, Becky Westerhoff, Emily Westfall, Katina Widmer, Hali Wright, Desmond Schatz, Annie Abraham, Mark Atkinson, Miriam Cintron, Michael Clare-Salzler, Jessica Ferguson, Michael Haller, Jennifer Hosford, Diane Mancini, Hank Rohrs, Janet Silverstein, Jamie Thomas, William Winter, Gloria Cole, Roberta Cook, Ryan Coy, Elena Hicks, Nancy Lewis, Jennifer Marks, Alberto Pugliese, Carlos Blaschke, Della Matheson, Natalia Sanders-Branca, Jay Sosenko, Luz Arazo, Ray Arce, Mario Cisneros, Samir Sabbag, Antoinette Moran, Carrie Gibson, Brian Fife, Bernhard Hering, Christine Kwong, Janice Leschyshyn, Brandon Nathan, Beth Pappenfus, Anne Street, Mary Ann Boes, Sarah Peterson Eck, Lois Finney, Theresa Albright Fischer, Andrea Martin, Chenai Jacqueline Muzamhindo, Missy Rhodes, Jennifer Smith, John Wagner, Bryan Wood, Dorothy Becker, Kelli Delallo, Ana Diaz, Barbara Elnyczky, Ingrid Libman, Beata Pasek, Karen Riley, Massimo Trucco, Brian Copemen, Diane Gwynn, Frederico Toledo, Henry Rodriguez, Sureka Bollepalli, Frank Diamond, Emily Eyth, Danielle Henson, Anne Lenz, Dorothy Shulman, Phillip Raskin, Soumya Adhikari, Brian Dickson, Erin Dunnigan, Ildiko Lingvay, Lourdes Pruneda, Maria Ramos-Roman, Philip Raskin, Chanhaeng Rhee, John Richard, Mark Siegelman, Daytheon Sturges, Kathryn Sumpter, Perrin White, Marilyn Alford, Jamie Arthur, M. Larissa Aviles-Santa, Erica Cordova, Renee Davis, Stefani Fernandez, Steve Fordan, Tauri Hardin, Aris Jacobs, Polina Kaloyanova, Ivanna Lukacova-Zib, Sasan Mirfakhraee, Alok Mohan, Hiroshi Noto, Oralenda Smith, Nenita Torres, Diane Wherrett, Diana Balmer, Lesley Eisel, Roze Kovalakovska, Mala Mehan, Farah Sultan, Brenda Ahenkorah, Jose Cevallos, Natasha Razack, Mary Jo Ricci, Angela Rhode, Mithula Srikandarajah, Rachel Steger, William E. Russell, Margo Black, Faith Brendle, Anne Brown, Daniel Moore, Eric Pittel, Alyssa Robertson, April Shannon, James W. Thomas, Kevan Herold, Laurie Feldman, Robert Sherwin, William Tamborlane, Stuart Weinzimer, Jorma Toppari, Tiina Kallio, Maarit Kärkkäinen, Elina Mäntymäki, Tiina Niininen, Birgitta Nurmi, Petro Rajala, Minna Romo, Sointu Suomenrinne, Kirsti Näntö-Salonen, Olli Simell, Tuula Simell, Emanuele Bosi, Manuela Battaglia, Eleonora Bianconi, Riccardo Bonfanti, Pauline Grogan, Andrea Laurenzi, Sabina Martinenghi, Franco Meschi, Matteo Pastore, Luca Falqui, Maria Teresa Muscato, Matteo Viscardi, Harriet Castleden, Nicola Farthing, Sam Loud, Claire Matthews, Jennifer McGhee, Ann Morgan, Joanna Pollitt, Rebecca Elliot-Jones, Carole Wheaton, Mikael Knip, Heli Siljander, Heli Suomalainen, Peter Colman, Felicity Healy, Shelley Mesfin, Leanne Redl, John Wentworth, Jinny Willis, Maree Farley, Leonard Harrison, Christine Perry, Fiona Williams, A. Mayo, J. Paxton, V. Thompson, L. Volin, C. Fenton, L. Carr, E. Lemon, M. Swank, M.K. Luidens, M. Salgam, V. Sharma, D. Schade, C. King, R. Carano, J. Heiden, N.D. Means, L. Holman, I. Thomas, D. Madrigal, T. Muth, C.L. Martin, C. Plunkett, C. Ramm, R.J. Auchus, W. Lane, E. Avots, M. Buford, C. Hale, J. Hoyle, B. Lane, A. Muir, S. Shuler, N. Raviele, E. Ivie, M. Jenkins, K. Lindsley, I. Hansen, D.O. Fadoju, E.I. Felner, B. Bode, R. Hosey, J. Sax, C. Jefferies, S. Mannering, R. Prentis, J.X. She, M. Stachura, D. Hopkins, J. Williams, L. Steed, E. Asatapova, S. Nunez, S. Knight, P. Dixon, J. Ching, T. Donner, S. Longnecker, K Abel, K. Arcara, S. Blackman, L. Clark, D. Cooke, L. Plotnick, P.A. Levin, L. Bromberger, K. Klein, K. Sadurska, C. Allen, D. Michaud, H. Snodgrass, G. Burghen, S. Chatha, C. Clark, J. Silverberg, C. Wittmer, J. Gardner, C. LeBoeuf, P. Bell, O. McGlore, H. Tennet, N. Alba, M. Carroll, L. Baert, H. Beaton, E. Cordell, A. Haynes, C. Reed, K. Lichter, P. McCarthy, S. McCarthy, T. Monchamp, J. Roach, S. Manies, F. Gunville, L. Marosok, T. Nelson, K. Ackerman, J. Rudolph, M. Stewart, K. McCormick, S. May, T. Falls, T. Barrett, K. Dale, L. Makusha, C. McTernana, K. Penny-Thomas, K. Sullivan, P. Narendran, J. Robbie, D. Smith, R. Christensen, B. Koehler, C. Royal, T. Arthur, H. Houser, J. Renaldi, S. Watsen, P. Wu, L. Lyons, B. House, J. Yu, H. Holt, M. Nation, C. Vickers, R. Watling, R. Heptulla, J. Trast, C. Agarwal, D.J. Newell, R. Katikaneni, C. Gardner, A. Del Rio, A. Logan, H. Collier, C. Rishton, G. Whalley, A. Ali, S. Ramtoola, T. Quattrin, L. Mastrandea, A.J. House, M. Ecker, C. Huang, C. Gougeon, J. Ho, D. Pacuad, D. Dunger, J. May, C. O’Brien, C. Acerini, B. Salgin, A. Thankamony, R. Williams, J. Buse, G. Fuller, M. Duclos, J. Tricome, H. Brown, D. Pittard, D. Bowlby, A. Blue, T. Headley, S. Bendre, K. Lewis, K. Sutphin, C. Soloranzo, J. Puskaric, H. Madison, M. Rincon, M. Carlucci, R. Shridharani, B. Rusk, E. Tessman, D.M. Huffman, H. Abrams, B. Biederman, M.D. Jones, V. Leathers, W. Brickman, P. Petrie, D. Zimmerman, J. Howard, L. Miller, R. Alemzadeh, D.V. Mihailescu, R. Melgozza-Walker, N. Abdulla, C. Boucher-Berry, D. Ize-Ludlow, R. Levy, C. Swenson Brousell, R. Scott, H. Heenan, H. Lunt, D. Kendall, J. Willis, B. Darlow, N. Crimmins, D. Edler, T. Weis, C. Schultz, D. Rogers, D. Latham, C. Mawhorter, C. Switzer, W. Spencer, P. Konstantnopoulus, S. Broder, J. Klein, B. Bachrach, M. Gardner, D. Eichelberger, L. Knight, L. Szadek, G. Welnick, B. Thompson, R. Hoffman, A. Revell, J. Cherko, K. Carter, E. Gilson, J. Haines, G. Arthur, B. Bowen, W.B. Zipf, P. Graves, R.A. Lozano, D. Seiple, K. Spicer, A. Chang, J. Fregosi, J. Harbinson, C. Paulson, S. Stalters, P. Wright, D. Zlock, A.E. Freeth, J. Victory, H. Maheshwari, A. Maheshwari, T. Holmstrom, J. Bueno, R. Arguello, J. Ahern, L. Noreika, V. Watson, S. Hourse, P. Breyer, C. Kissel, Y. Nicholson, M. Pfeifer, S. Almazan, J. Bajaj, M. Quinn, K. Funk, J. McCance, E. Moreno, R. Veintimilla, A. Wells, J. Cook, S. Trunnel, D. Transue, J. Surhigh, D. Bezzaire, K. Moltz, E. Zacharski, J. Henske, S. Desai, K. Frizelis, F. Khan, R. Sjoberg, K. Allen, P.P. Manning, G. Hendry, B. Taylor, S. Jones, R. Couch, R. Danchak, D. Lieberman, W. Strader, M.E. Bencomo, T. Bailey, L. Bedolla, C. Roldan, C. Moudiotis, B. Vaidya, C. Anning, S. Bunce, S. Estcourt, E. Folland, E. Gordon, C. Harrill, J. Ireland, J. Piper, L. Scaife, K. Sutton, S. Wilkins, M. Costelloe, J. Palmer, L. Casas, C. Miller, M. Burgard, C. Erickson, J. Hallanger-Johnson, P. Clark, W. Taylor, J. Galgani, S. Banerjee, C. Banda, D. McEowen, R. Kinman, A. Lafferty, S. Gillett, C. Nolan, M. Pathak, L. Sondrol, T. Hjelle, S. Hafner, J. Kotrba, R. Hendrickson, A.P. Cemeroglu, T. Symington, M. Daniel, Y. Appiagyei-Dankah, D.C. Postellon, M.S. Racine, L. Kleis, K. Barnes, S.E. Godwin, H. McCullough, K. Shaheen, G. Buck, L. Noel, M.L. Warren, S. Weber, S.M. Parker, I. Gillespie, B.A. Nelson, C. Frost, J. Amrhein, E.C. Moreland, A. Hayes, J. Peggram, J. Aisenberg, M.E. Riordan, J. Zasa, E. Cummings, K. Scott, T. Pinto, A. Mokashi, K. McAssey, E. Helden, P. Hammond, L. Dinning, S. Rahman, S. Ray, C. Dimicri, S. Guppy, H. Nielsen, C.K. Vogel, C. Ariza, L. Morales, Y.T. Chang, R.A. Gabbay, L. Ambrocio, L. Manley, R. Nemery, W. Charlton, P. Smith, L. Kerr, B. Steindel-Kopp, M. Alamaguer, E. Tabisola-Nuesca, A. Pendersen, N. Larson, H. Cooper-Olviver, D. Chan, D. Fitz-Patrick, T. Carreira, Y. Park, R. Ruhaak, D. Liljenquist, G. Browning, T. Coughenour, M.B. Sulk, E. Tsalikan, M. Tansey, J. Cabbage, N. Dixit, S. Pasha, M. King, K. Adcock, H. Atterberry, L. Fox, K. Englert, N. Mauras, J. Permuy, K. Sikes, T. Berhe, B. Guendling, L. McLennan, L. Paganessi, C. Murphy, M.B. Draznin, M. Kamboj, S. Sheppard, V. Lewis, L. Coates, W. Moore, G. Babar, J. Bedard, D. Brenson-Hughes, J. Cernich, M. Clements, R. Duprau, S. Goodman, L. Hester, L. Huerta-Saenz, A. Karmazin, T. Letjen, S. Raman, D. Morin, W. Bestermann, E.J. Morawski, J.L. White, A. Brockmyer, R. Bays, S. Campbell, A. Stapleton, N. Stone, A. Donoho, H. Everett, H. Hensley, M. Johnson, C. Marshall, N. Skirvin, P. Taylor, R. Williams, L. Ray, C. Wolverton, D.A. Nickels, C. Dothard, P.W. Speiser, M. Pellizzari, L. Bokor, K. Izuora, S. Abdelnour, P. Cummings, S. Paynor, M. Leahy, M. Riedl, S. Shockley, R. Saad, T. Briones, S. Casella, C. Herz, K. Walsh, J. Greening, F. Hay, S. Hunt, N. Sikotra, L. Simons, D.G. Karounos, R. Oremus, L. Dye, L. Myers, D. Ballard, W. Miers, R. Sparks, K.M. Thraikill, K. Edwards, J. Fowlkes, S. Kemp, A. Morales, L. Holland, L. Johnson, P. Paul, A. Ghatak, K. Phelen, H. Leyland, T. Henderson, D. Brenner, E. Oppenheimer, I. Mamkin, C. Moniz, C. Clarson, M. Lovell, A. Peters, V. Ruelas, D. Borut, D. Burt, M. Jordan, S. Castilla, P. Flores, M. Ruiz, L. Hanson, J. Green-Blair, R.J. Sheridan, K.A. Wintergerst, G. Pierce, A. Omoruyi, M. Foster, S. Kingery, A. Lunsford, I. Cervantes, T. Parker, P. Price, J. Urben, I. Doughty, H. Haydock, V. Parker, P. Bergman, S. Duncum, C. Rodda, A.D. Thomas, R. Ferry, D. McCommon, J. Cockroft, A. Perelman, R. Calendo, C. Barrera, E. Arce-Nunez, Y. Martinez, M. De la Portilla, I. Cardenas, L. Garrido, M. Villar, R. Lorini, E. Calandra, G. D’Annuzio, K. Perri, N. Minuto, C. Rebora, R. Callegari, O. Ali, J. Kramer, B. Auble, S. Cabrera, P. Donohoue, R. Fiallo-Scharer, M. Hessner, P. Wolfgram, A. Kansra, N. Bettin, R. McCuller, A. Miller, S. Accacha, J. Corrigan, E. Fiore, R.L. Levine, T.A. Mahoney, C. Polychronakos, V. Gagne, H. Starkman, M. Fox, D. Chin, F. Melchionne, L.A. Silverman, I. Marshall, L. Cerracchio, J. Cruz, A. Viswanathan, J. Wilson, S. Chalew, S. Valley, S. Layburn, A. Lala, P. Clesi, M. Genet, G. Uwaifo, A. Charron, T. Allerton, W. Cefalu, L. Melendez-Ramirez, R. Richards, C. Alleyn, E. Gustafson, M. Lizanna, J. Wahlen, S. Aleiwe, M. Hansen, H. Wahlen, C.J. Levy, A. Bonaccorso, R. Rapaport, Y. Tomer, D. Chia, M. Goldis, L. Iazzetti, M. Klein, C. Levister, L. Waldman, E. Wallach, M.O. Regelmann, Z. Antal, M. Aranda, C. Reynholds, N. Leech, D. Wake, C. Owens, M. Burns, J. Wotherspoon, A. Murray, K. Short, G. Curry, S. Kelsey, J. Lawson, J. Porter, S. Stevens, E. Thomson, S. Winship, L. Wynn, E. Wiltshire, J. Krebs, P. Cresswell, H. Faherty, C. Ross, A. Vinik, P. Barlow, M. Bourcier, M.L. Nevoret, J. Couper, S. Beresford, N. Thalagne, H. Roper, J. Gibbons, J. Hill, S. Balleaut, C. Brennan, J. Ellis-Gage, L. Fear, T. Gray, L. Jones, C. McNerney, L. Pointer, N. Price, K. Few, D. Tomlinson, L. Denvir, J. Drew, T. Randell, P. Mansell, S.A. Bell, S. Butler, Y. Hooton, H. Navarra, A. Roper, G. Babington, L. Crate, H. Cripps, A. Ledlie, C. Moulds, R. Norton, B. Petrova, O. Silkstone, C. Smith, K. Ghai, M. Murray, V. Viswanathan, M. Henegan, O. Kawadry, J.A. Olson, L. Patterson, T. Ahmad, B. Flores, D. Domek, S. Domek, K. Copeland, M. George, J. Less, T. Davis, M. Short, A. Dwarakanathan, P. O’Donnell, B. Boerner, L. Larson, M. Phillips, M. Rendell, K. Larson, C. Smith, K. Zebrowski, L. Kuechenmeister, M. Thevarayapillai, M. Daniels, H. Speer, N. Forghani, R. Quintana, C. Reh, A. Bhangoo, P. Desrosiers, L. Ireland, T. Misla, C. Torres, S. Wells, J. Villar, M. Yu, D. Berry, D. Cook, J. Soder, A. Powell, M. Ng, M. Morrison, Z. Haslam, M. Lawson, B. Bradley, J. Courtney, C. Richardson, C. Watson, E. Keely, D. DeCurtis, M. Vaccarcello-Cruz, Z. Torres, K. Sandberg, H. Hsiang, B. Joy, D. McCormick, A. Powell, H. Jones, J. Bell, S. Hargadon, S. Hudson, M. Kummer, S. Sauder, E. Sutton, K. Gensel, R. Aguirre-Castaneda, V. Benavides Lopez, D. Hemp, S. Allen, J. Stear, E. Davis, T. Jones, A. Roberts, J.A. Dart, N. Paramalingam, L.E. Levitt Katz, N. Chaudhary, K.M. Murphy, S.M. Willi, B. Schwartzman, C. Kapadia, D. Larson, D. McClellan, G. Shaibai, L.A. Kelley, G. Villa, C. Kelley, R. Diamond, M. Kabbani, T. Dajani, F. Hoekstra, M. Magorno, J. Holst, V. Chauhan, N. Wilson, P. Bononi, M. Sperl, A. Millward, M. Eaton, L. Dean, J. Olshan, H. Renna, C. Milliard, D. Snyder, S. Beaman, K. Burch, J. Chester, A. Ahmann, B. Wollam, D. DeFrang, R. Fitch, K. Jahnke, K. Hanavan, B. Klopfenstein, L. Nicol, R.W. Bergstrom, T. Noland, J. Brodksy, L. Bacon, J.B. Quintos, L.S. Topor, S. Bialo, B. Bancroft, A.G. Soto, W. Lagarde, H. Lockemer, T. Vanderploeg, M.A. Ibrahim, M. Huie, V. Sanchez, R. Edelen, R. Marchiando, J. Palmer, T. Repas, M. Wasson, P. Auker, J. Culbertson, T. Kieffer, D. Voorhees, T. Borgwardt, L. DeRaad, K. Eckert, E. Isaacson, H. Kuhn, A. Carroll, M. Schubert, G. Francis, S. Hagan, T. Le, M. Penn, E. Wickham, C. Leyva, K. Rivera, J. Padilla, I. Rodriguez, N. Jospe, J. Czyzyk, B. Johnson, U. Nadgir, N. Marlen, G. Prakasam, C. Rieger, N. Glaser, E.C. Heiser, B. Harris, C. Foster, H. Slater, K. Wheeler, D.L. Donaldson, M. Murray, D.E. Hale, R. Tragus, D.R. Word, J. Lynch, L. Pankratz, W. Rogers, R. Newfield, S. Holland, M. Hashiguchi, M. Gottschalk, A. Philis-Tsimikas, R. Rosal, S. Franklin, S.M. Guardado, N. Bohannon, M. Garcia, T. Aguinaldo, J. Phan, V. Barraza, D. Cohen, J. Pinsker, U. Khan, J. Wiley, L. Jovanovic, P. Misra, M. Wright, D. Cohen, K. Huang, M. Skiles, S. Maxcy, C. Pihoker, K. Cochrane, J. Fosse, S. Kearns, M. Klingsheim, N. Wright, L. Viles, H. Smith, S. Heller, M. Cunningham, A. Daniels, L. Zeiden, J. Field, R. Walker, K.J. Griffin, L. Bartholow, C. Erickson, J. Howard, B. Krabbenhoft, C. Sandman, A. Vanveldhuizen, J. Wurlger, A. Zimmerman, K. Hanisch, L. Davis-Keppen, A. Cotterill, J. Kirby, M. Harris, A. Schmidt, C. Kishiyama, C. Flores, J. Milton, W. Martin, C. Whysham, A. Yerka, T. Freels, J.M. Hassing, J. Webster, R. Green, P. Carter, J. Galloway, D. Hoelzer, S. Roberts, S. Said, P. Sullivan, H.F. Allen, E. Reiter, E. Feinberg, C. Johnson, L.A. Newhook, D. Hagerty, N.H. White, L. Levandoski, J. Kyllo, M. Johnson, C. Benoit, P. Iyer, F. Diamond, H. Hosono, S. Jackman, L. Barette, P. Jones, I. Sills, S. Bzdick, J. Bulger, R. Weinstock, I. Douek, R. Andrews, G. Modgill, G. Gyorffy, L. Robin, N. Vaidya, S. Crouch, K. O’Brien, C. Thompson, N. Thorne, J. Blumer, J. Kalic, L. Klepek, J. Paulett, B. Rosolowski, J. Horner, M. Watkins, J.L. Casey, K. Carpenter, C. Burns, J. Horton, C. Pritchard, D. Soetaert, A.G. Wynne, K. Kaiserman, M. Halvorson, C. Chin, O.Y. Molina, C. Patel, R. Senguttuvan, M. Wheeler, O. Furet, C. Steuhm, D.H. Jelley, S. Goudeau, L. Chalmers, D. Greer, C. Panagiotopoulos, D.L. Metzger, D. Nguyen, M. Horowitz, M.P. Christiansen, E. Glades, C. Morimoto, M. Macarewich, R. Norman, K. Patin, C. Vargas, A. Barbanica, A. Yu, P. Vaidyanathan, W. Osborne, R. Mehra, S. Kaster, S. Neace, J. Horner, G. Reeves, C. Cordrey, L. Marrs, T. Miller, S. Dowshen, D. Doyle, S. Walker, D. Catte, H. Dean, M. Drury-Brown, B. Hackman, M.M.C. Lee, S. Malkani, K. Cullen, K. Johnson, P. Hampton, M. McCarrell, C. Curtis, E. Paul, and Y. Zambrano
References
- 1.Mrena S, Virtanen SM, Laippala P, et al. Models for predicting type 1 diabetes in siblings of affected children. Diabetes Care 2006;29:662–667 [DOI] [PubMed] [Google Scholar]
- 2.Sosenko JM, Krischer JP, Palmer JP, et al.; Diabetes Prevention Trial–Type 1 Study Group . A risk score for type 1 diabetes derived from autoantibody-positive participants in the Diabetes Prevention Trial–Type 1. Diabetes Care 2008;31:528–533 [DOI] [PubMed] [Google Scholar]
- 3.Xu P, Beam CA, Cuthbertson D, Sosenko JM, Skyler JS, Krischer JP; DPT-1 Study Group . Prognostic accuracy of immunologic and metabolic markers for type 1 diabetes in a high-risk population: receiver operating characteristic analysis. Diabetes Care 2012;35:1975–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu P, Wu Y, Zhu Y, et al.; Diabetes Prevention Trial–Type 1 (DPT-1) Study Group . Prognostic performance of metabolic indexes in predicting onset of type 1 diabetes. Diabetes Care 2010;33:2508–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siljander HT, Simell S, Hekkala A, et al. Predictive characteristics of diabetes-associated autoantibodies among children with HLA-conferred disease susceptibility in the general population. Diabetes 2009;58:2835–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steck AK, Johnson K, Barriga KJ, et al. Age of islet autoantibody appearance and mean levels of insulin, but not GAD or IA-2 autoantibodies, predict age of diagnosis of type 1 diabetes: diabetes autoimmunity study in the young. Diabetes Care 2011;34:1397–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steck AK, Vehik K, Bonifacio E, et al.; TEDDY Study Group . Predictors of progression from the appearance of islet autoantibodies to early childhood diabetes: The Environmental Determinants of Diabetes in the Young (TEDDY). Diabetes Care 2015;38:808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redondo MJ, Jeffrey J, Fain PR, Eisenbarth GS, Orban T. Concordance for islet autoimmunity among monozygotic twins. N Engl J Med 2008;359:2849–2850 [DOI] [PubMed] [Google Scholar]
- 9.Redondo MJ, Yu L, Hawa M, et al. Heterogeneity of type I diabetes: analysis of monozygotic twins in Great Britain and the United States. Diabetologia 2001;44:354–362 [DOI] [PubMed] [Google Scholar]
- 10.Cudworth AG, Woodrow JC. Letter: HL-A antigens and diabetes mellitus. Lancet 1974;2:1153. [DOI] [PubMed] [Google Scholar]
- 11.Barrett JC, Clayton DG, Concannon P, et al.; Type 1 Diabetes Genetics Consortium . Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009;41:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groop L, Pociot F. Genetics of diabetes—are we missing the genes or the disease? Mol Cell Endocrinol 2014;382:726–739 [DOI] [PubMed] [Google Scholar]
- 13.Redondo MJ, Steck AK, Pugliese A. Genetics of type 1 diabetes. Pediatr Diabetes 2018;19:346–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oram RA, Patel K, Hill A, et al. A type 1 diabetes genetic risk score can aid discrimination between type 1 and type 2 diabetes in young adults. Diabetes Care 2016;39:337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel KA, Oram RA, Flanagan SE, et al. Type 1 diabetes genetic risk score: a novel tool to discriminate monogenic and type 1 diabetes. Diabetes 2016;65:2094–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skyler JS, Greenbaum CJ, Lachin JM, et al.; Type 1 Diabetes TrialNet Study Group . Type 1 Diabetes TrialNet–an international collaborative clinical trials network. Ann N Y Acad Sci 2008;1150:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahon JL, Sosenko JM, Rafkin-Mervis L, et al.; TrialNet Natural History Committee; Type 1 Diabetes TrialNet Study Group . The TrialNet Natural History Study of the Development of Type 1 Diabetes: objectives, design, and initial results. Pediatr Diabetes 2009;10:97–104 [DOI] [PubMed] [Google Scholar]
- 18.Greenbaum CJ, Mandrup-Poulsen T, McGee PF, et al.; Type 1 Diabetes Trial Net Research Group; European C-Peptide Trial Study Group . Mixed-meal tolerance test versus glucagon stimulation test for the assessment of beta-cell function in therapeutic trials in type 1 diabetes. Diabetes Care 2008;31:1966–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Little RR, Rohlfing CL, Tennill AL, et al. Standardization of C-peptide measurements. Clin Chem 2008;54:1023–1026 [DOI] [PubMed] [Google Scholar]
- 20.Pugliese A, Boulware D, Yu L, et al.; Type 1 Diabetes TrialNet Study Group . HLA-DRB1*15:01-DQA1*01:02-DQB1*06:02 haplotype protects autoantibody-positive relatives from type 1 diabetes throughout the stages of disease progression. Diabetes 2016;65:1109–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sosenko JM, Skyler JS, Mahon J, et al.; Type 1 Diabetes TrialNet and Diabetes Prevention Trial–Type 1 Study Groups . Validation of the Diabetes Prevention Trial–Type 1 Risk Score in the TrialNet Natural History Study [published correction appears in Diabetes Care 2018;41:913]. Diabetes Care 2011;34:1785–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sosenko JM, Skyler JS, Mahon J, et al.; Type 1 Diabetes TrialNet and Diabetes Prevention Trial-Type 1 Study Groups . Use of the Diabetes Prevention Trial-Type 1 Risk Score (DPTRS) for improving the accuracy of the risk classification of type 1 diabetes. Diabetes Care 2014;37:979–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barker JM, Triolo TM, Aly TA, et al. Two single nucleotide polymorphisms identify the highest-risk diabetes HLA genotype: potential for rapid screening. Diabetes 2008;57:3152–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu P, Krischer JP; Type 1 Diabetes TrialNet Study Group . Prognostic classification factors associated with development of multiple autoantibodies, dysglycemia, and type 1 diabetes—a recursive partitioning analysis. Diabetes Care 2016;39:1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atkinson MA, Kaufman DL, Newman D, Tobin AJ, Maclaren NK. Islet cell cytoplasmic autoantibody reactivity to glutamate decarboxylase in insulin-dependent diabetes. J Clin Invest 1993;91:350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Redondo MJ, Oram RA, Steck AK. Genetic risk scores for type 1 diabetes prediction and diagnosis. Curr Diab Rep 2017;17:129. [DOI] [PubMed] [Google Scholar]
- 28.Steck AK, Dong F, Wong R, et al. Improving prediction of type 1 diabetes by testing non-HLA genetic variants in addition to HLA markers. Pediatr Diabetes 2014;15:355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkler C, Krumsiek J, Buettner F, et al. Feature ranking of type 1 diabetes susceptibility genes improves prediction of type 1 diabetes. Diabetologia 2014;57:2521–2529 [DOI] [PubMed] [Google Scholar]
- 30.Winkler C, Krumsiek J, Lempainen J, et al. A strategy for combining minor genetic susceptibility genes to improve prediction of disease in type 1 diabetes. Genes Immun 2012;13:549–555 [DOI] [PubMed] [Google Scholar]
- 31.Frohnert BI, Ide L, Dong F, et al. Late-onset islet autoimmunity in childhood: the Diabetes Autoimmunity Study in the Young (DAISY). Diabetologia 2017;60:998–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonifacio E, Beyerlein A, Hippich M, et al.; TEDDY Study Group . Genetic scores to stratify risk of developing multiple islet autoantibodies and type 1 diabetes: a prospective study in children. PLoS Med 2018;15:e1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Insel RA, Dunne JL, Atkinson MA, et al. Staging presymptomatic type 1 diabetes: a scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care 2015;38:1964–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosi E, Boulware DC, Becker DJ, et al.; Type 1 Diabetes TrialNet Study Group . Impact of age and antibody type on progression from single to multiple autoantibodies in type 1 diabetes relatives. J Clin Endocrinol Metab 2017;102:2881–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bingley PJ, Gale EA; European Nicotinamide Diabetes Intervention Trial (ENDIT) Group . Progression to type 1 diabetes in islet cell antibody-positive relatives in the European Nicotinamide Diabetes Intervention Trial: the role of additional immune, genetic and metabolic markers of risk. Diabetologia 2006;49:881–890 [DOI] [PubMed] [Google Scholar]
- 36.Truyen I, De Pauw P, Jørgensen PN, et al.; Belgian Diabetes Registry . Proinsulin levels and the proinsulin:c-peptide ratio complement autoantibody measurement for predicting type 1 diabetes. Diabetologia 2005;48:2322–2329 [DOI] [PubMed] [Google Scholar]
- 37.Gorus FK, Balti EV, Messaaoui A, et al.; Belgian Diabetes Registry . Twenty-year progression rate to clinical onset according to autoantibody profile, age, and HLA-DQ genotype in a registry-based group of children and adults with a first-degree relative with type 1 diabetes. Diabetes Care 2017;40:1065–1072 [DOI] [PubMed] [Google Scholar]
- 38.Ferrara CT, Geyer SM, Evans-Molina C, et al.; Type 1 Diabetes TrialNet Study Group . The role of age and excess body mass index in progression to type 1 diabetes in at-risk adults. J Clin Endocrinol Metab 2017;102:4596–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Redondo MJ, Geyer S, Steck AK, et al.; Type 1 Diabetes TrialNet Study Group . TCF7L2 genetic variants contribute to phenotypic heterogeneity of type 1 diabetes. Diabetes Care 2018;41:311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas NJ, Jones SE, Weedon MN, Shields BM, Oram RA, Hattersley AT. Frequency and phenotype of type 1 diabetes in the first six decades of life: a cross-sectional, genetically stratified survival analysis from UK Biobank. Lancet Diabetes Endocrinol 2018;6:122–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.