Abstract
Objectives
A carbapenem resistant Providencia rettgeri (PR1) isolate was recovered from a wound infection in Missouri, USA. This isolate possessed an EDTA inhibitable carbapenemase that was unidentified using the Xpert CARBA-R assay. Our objective was to elucidate the molecular determinant of carbapenem resistance in this isolate. We then sought to test the transmissibility of blaIMP-27 loci in clinical P. rettgeri and Proteus mirabilis isolates.
Methods
In October 2016 the novel ambler Class B carbapenemase blaIMP-27, was reported in two different Proteus mirablis (PM185 and PM187) isolates. Broth mating assays for transfer of carbapenemase activity were performed for the three clinical isolates with recipient sodium azide resistant Escherichia coli J53. Antibiotic susceptibility and phenotypic carbapenemase activity testing was performed on the clinical isolates, J53, and transconjugants using the Kirby-Bauer Disk diffusion method according to Clinical & Laboratory Standards Institute guidelines. Plasmid DNA from PM187, PR1, and their transconjugants were used as input for Nextera Illumina sequencing libraries and sequenced on a NextSeq platform
Results
PR1 was resistant to both imipenem and meropenem. PM187 and PR1 could transfer resistance to E. coli via plasmid conjugation (pPM187 and pPR1). pPM187 had a virB/virD4 type IV secretion system (T4SS) whereas pPR1 had traB/traD (T4SS).
Conclusion
2 of 3 blaIMP-27 bearing clinical isolates tested could conjugate resistance into E. coli. The resulting transconjugants became positive for phenotypic carbapenemase production but did not pass clinical resistance breakpoints. blaIMP-27 can be transmitted on different plasmid replicon types that rely on distinct classes of T4SS for horizontal transfer.
Introduction
In January 2016, we isolated a carbapenem resistant Providencia rettgeri (PR1) from a foot wound infection of a patient who visited an outside hospital affiliate of Barnes-Jewish Hospital (Missouri, United States). PR1 was positive for an EDTA-inhibited carbapenemase but no gene was identified by multiplex PCR. Whole genome sequencing (WGS) and antibiotic resistance gene (ARG) identification of the PR1 draft genome identified blaIMP-27. blaIMP-27 was first reported in October 2016 from two Proteus mirabilis strains (PM185 and PM187) from the upper plains region of the United States (1). In December 2016, blaIMP-27 was identified on IncQ plasmids from a variety of swine associated Enterobacteriaceae in the United States (2). Given these recent reports, the greater Midwest region of the United States may be endemic for blaIMP-27, and a potential source for wider geographic dissemination. Accordingly, we acquired PM185 and PM187 to understand, with PR1, the potential for lateral transfer of this resistance gene from P. mirabilis and P. rettgeri into E. coli, and the associated changes in antibiotic resistance (1).
Methods
Bacterial Isolates
The Providencia rettgeri isolate (PR1) was recovered from a chronic foot wound infection clinical culture. The isolate received for evaluation was a de-identified strain. As a result, the study team was not able to obtain patient consent. Proteus mirabilis strains (PM185 and PM187) were provided by Nancy Hanson at Creighton University (1). The sodium azide resistant E. coli J53 strain (ATCC number BAA-2730™) was used as a recipient for transconjugation experiments.
Broth Conjugation
Colonies of PM185, PM187, PR1, and wildtype E. coli J53 were separately suspended in Tryptic Soy Broth (TSB) (Sigma Aldrich, St. Louis, MO) and diluted to 0.05 OD600. 100 μl of PM185, PM187, and PR1 were separately added to 100 μl E. coli J53 (for a 1:1 ratio) and diluted to 5 mL with TSB. Co-cultures were incubated at 37 °C without shaking for 24 hours. 50 μl of co-cultures were suspended onto MacConkey agar plates containing sodium azide (Thermo Fisher Scientific, Waltham, MA) (150 μg/ml) and ceftriaxone (5 μg/ml), spread with glass beads, and incubated for 18 hours at 37 °C. Individual transconjugant colonies were propagated overnight in TSB supplemented with 5 μg/ml ceftriaxone under shaking conditions (220 rpm).
Susceptibility Testing
Each clinical isolate, J53, J53:pPR1, and J53:pPM187 were cultured overnight as described previously. E. coli ATCC 25922 was used as a quality control. Susceptibility testing was performed using Kirby Bauer Disk Diffusion on Mueller Hinton Agar (Hardy Diagnostics) in accordance with CLSI Standards (3).
Plasmid assembly and annotation
We used Illumina sequencing to specifically investigate blaIMP-27 bearing plasmids in PR1 and PM187. Plasmid DNA was obtained using a miniprep kit (Qiagen, Valencia, CA). Plasmid DNA for PR1 and PM187 was processed to remove Illumina adapters (trimmomatic) and contaminating DNA (deconseq). The paired reads were assembled into contigs with SPAdes v3.9.0 (4). Raw reads from the transconjugant minipreps were processed for quality in a similar manner. 100% of the transconjugant reads that aligned to the clinical isolate plasmid assembly using Bowtie2 were assembled into contigs with SPAdes v3.9.0 (5) (4). Gaps were closed by PCR and Sanger-sequencing (Genewiz, South Plainfield, NJ) to yield finished plasmid assemblies (Table S1). Open reading frames were annotated for coding sequence using prokka (6). Antibiotic resistance genes were additionally annotated with Resfams and the ResFinder web server (https://cge.cbs.dtu.dk/services/ResFinder/) (7, 8). pPM187 and pPR1 plasmid maps were made by viewing the gff3 files in DNAPlotter and manually annotated for putative open reading frame function(9). Select T4SS genes were submitted to blastp against the nonredundant protein sequence database on 12/10/17 (10).
Results
PM185 and PM187 were intermediate and susceptible to meropenem and imipenem, respectively (Table 1). Only PR1 was resistant to both carbapenems. PM185 was indeterminate for the carbapenem inactivation method but PM187 and PR1 were both phenotypically positive (Table 1). Southern blot analysis indicates that PR1 has a single copy of blaIMP-27 (Figure S1), similar to PM185 (1). In contrast, PM187 has both chromosomal and plasmid copies of blaIMP-27 (1). Transconjugants were obtained from conjugation assays of PR1 and PM187 with the E. coli J53 recipient but not PM185.
Table 1.
| Zone of Clearance (mm) | Interpretation | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibiotic | PM185 | PM187 | PR1 | J53 | J53:pPM187 | J53:pPR1 | ||||||
| Ampicillin | 24 | S | 6 | R | 14 | R | 21 | S | 17 | S | 17 | S |
| Cefazolin | 9 | R | 9 | R | 6 | R | 25 | S | 8 | R | 9 | R |
| Cefotetan | 11 | R | 14 | I | 6 | R | 33 | S | 11 | R | 10 | R |
| Ceftriaxone | 17 | R | 17 | R | 18 | R | 35 | S | 15 | R | 14 | R |
| Ceftazidime | 23 | S | 20 | I | 22 | S | 30 | S | 17 | R | 15 | R |
| Cefepime | 20 | I | 19 | I | 19 | SDD | 36 | S | 28 | S | 26 | S |
| Meropenem | 20 | I | 22 | S | 6 | R | 32 | S | 25 | S | 27 | S |
| Imipenem | 20 | I | 24 | S | 15 | R | 33 | S | 31 | S | 32 | S |
| Pipercillin-Tazobactam | 33 | S | 26 | S | 31 | S | 30 | S | 30 | S | 31 | S |
| Ampicillin-Sulbactam | 23 | S | 18 | S | 6 | R | 24 | S | 20 | S | 22 | S |
| Ciprofloxacin | 36 | S | 32 | S | 27 | S | 25 | S | 25 | S | 25 | S |
| Levoflocaxin | 35 | S | 30 | S | 26 | S | 25 | S | 25 | S | 25 | S |
| Gentamicin | 23 | S | 15 | S | 16 | S | 25 | S | 25 | S | 26 | S |
| Amikacin | 22 | S | 24 | S | 24 | S | 25 | S | 25 | S | 26 | S |
| Trimethoprim-sulfamethoxazole | 30 | S | 23 | S | 6 | R | 35 | S | 25 | S | 6 | R |
| Colistin | 6 | R | 6 | R | 6 | R | 16 | S | 24 | S | 16 | S |
| Aztreonam | 38 | S | 35 | S | 35 | S | 36 | S | 35 | S | 35 | S |
| Doxycyline | 6 | R | 6 | R | 6 | R | 22 | S | 22 | S | 22 | S |
| Minocycline | 11 | R | 10 | R | 6 | R | 26 | S | 26 | S | 26 | S |
| Tigecycline | 18 | I | 23 | S | 20 | S | 29 | S | 29 | S | 30 | S |
| Carbapenem Inactivation Method | Indeterminate | Positive | Positive | Negative | Positive | Positive | ||||||
Although conjugation did not achieve clinical resistance guidelines, the zone size for meropenem decreased from 32 mm in J53 to 25 mm in J53: pPM187 and 27 mm in J53:pPR1 (Table 1) (27). The zone size for imipenem decreased a lesser amount, from 33 mm in J53 to 31 and 32 mm in J53: pPM187 and J53:pPR1, respectively. Both transconjugants were positive for phenotypic carbapenem production (Table 1).
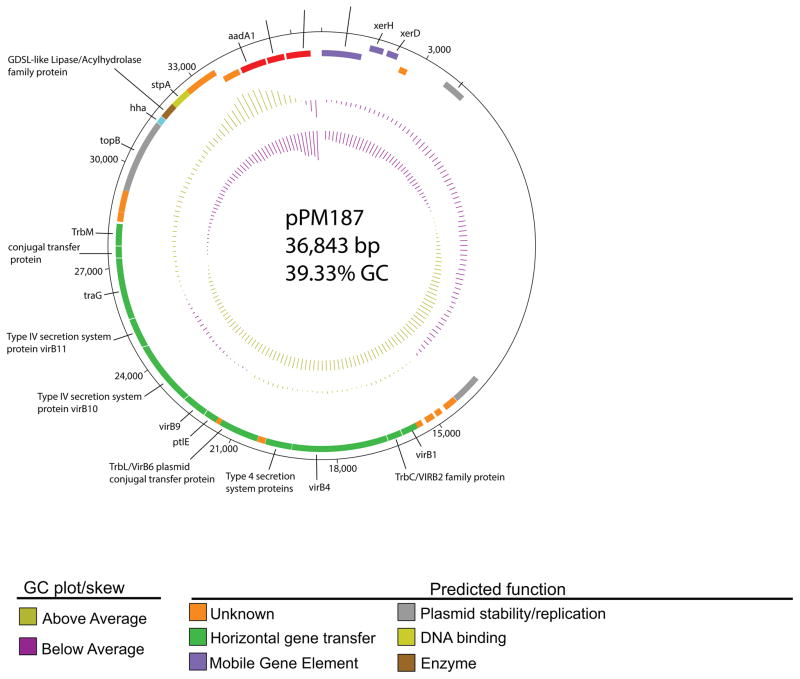
The plasmid from PM187, pPM187 (Genbank NOWA01000087.1), contains a putative virB/D4 IV secretion system operon, providing a potential mechanism for horizontal dissemination (Figure 1A). The virB4 amino acid sequence had 100% identity over its entire length with a conjugal transfer protein (WP_012368868.1) from P. mirabilis HI4320 (11). pPM187 has an IncX8 backbone, a newly discovered IncX family member (12). Unlike pPM187, the assembled blaIMP-27 bearing plasmid, pPR1 (Genbank NOWC01000095.1) did not have a plasmid replicon identified. pPR1 also contained a putative type IV secretion system, though of the tra/trb type (Figure 1B). The traN amino acid sequence had 100% identity across its entire length to the traN (WP_023159916.1) of the blaNDM-1 bearing plasmid pPrY2001 from P. rettgeri 09ACRGNY2001 (13).
Figure 1.
(A) Annotated plasmid diagram from DNAPlotter of pPM187 (36,843 bp) displaying blaIMP-27 co-localized with a Class II integron gene cassette and type IV secretion system. The inner most ring shows GC plot, the second ring shows GC skew, the third ring represents open reading frames in the forward direction, and the fourth ring (adjacent to the nucleotide position counter) indicates open reading frames in the reverse direction. (B) Annotated plasmid diagram from DNAPlotter of pPR1 (107,365 bp) displaying blaIMP-27 co-localized with a Class II integron gene cassette, a tra operon, and additional resistance genes.
Discussion
In this study, we used conjugation experiments to determine that two blaIMP-27 positive clinical isolates, PM187 and PR1, could transfer carbapenemase production to E. coli. We used Illumina sequencing of the transconjugants and clinical isolates to assemble the blaIMP-27 bearing plasmids, pPM187 and pPR1.
E. coli transconjugants with these plasmids (pPR1 and pPM187) gain detectable carbapenemase activity, but this activity does shift the transconjugants past clinical breakpoints for carbapenem resistance. It is possible that regulatory or translational optimization of the conjugated blaIMP-27 bearing plasmid in E. coli is required for clinical resistance (14). In addition to blaIMP-27 expression, it is also possible that porin mutations or efflux activity in the clinical isolates could contribute to phenotypic carbapenem resistance (15).
A previous investigation found that while blaIMP-27 was plasmid-borne in swine-associated Enterobacteriaceae, the IncQ plasmids were not conjugatable. In contrast, the plasmids we have completely sequenced are capable of self-mobilization, likely due to a virB/virD4 T4SS in pPM187 and a traB/traA T4SS in pPR1. The virB4 and traN gene from these T4SS showed similarity to previously described systems from pathogenic P. mirabilis HI4320 and carbapenem resistant P. rettgeri 09ACRGNY2001 (11,13). A limitation of Illumina short-read sequencing is that it generally cannot enable unambiguous assembly of the chromosome and all plasmids. Further work is warranted using long-read sequencing (e.g., from PacBio or Oxford Nanopore) on blaIMP-27 isolates to unequivocally determine chromosomal sequences and compare the non-conjugatable blaIMP-27 IncQ plasmids with pPM187 and pPR1. Although southern blot analysis indicates only a single blaIMP-27 loci exist in PM185 and PR1, this may further enable a comparison between the chromosomal and plasmid (pPM187) platforms of blaIMP-27 in PM187.
blaIMP-27 was unidentified using the FDA-cleared Xpert CARBA-R assay but the first report of blaIMP-27 used the ARM-D™ Multiplex PCR, which indicates some commercially available platforms can assay for blaIMP-27 (11). Therefore, further evaluations of commercial molecular diagnostic tests for blaIMP-27 is warranted.
Supplementary Material
Acknowledgments
The authors would like to thank Center for Genome Sciences & Systems Biology staff Jessica Hoisington-Lopez, Brian Koebbe, & Eric Martin for performing Illumina WGS and operating the High Throughput Computing Facility. The authors would also like to thank Nancy Hanson for generously providing PM185 and PM187. R.F.P presented a portion of this work as a poster at the 2017 American Society for Microbiology Microbe conference in New Orleans, LA. This work was supported in part by a grant to G.D. from the National Institute of General Medical Sciences (NIGMS: http://www.nigms.nih.gov/) of the NIH under award number R01 GM099538. R.F.P was supported by a NIGMS training grant through award T32 GM007067 (PI: James Skeath) and the Monsanto excellence fund graduate fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Disclosures
Dr. Burnham has nothing to disclose. Dr. Dantas has nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Works Cited
- 1.Dixon N, Fowler RC, Yoshizumi A, Horiyama T, Ishii Y, Harrison L, et al. IMP-27, a Unique Metallo191 beta-Lactamase Identified in Geographically Distinct Isolates of Proteus mirabilis. Antimicrob Agents Chemother. 2016;60(10):6418–21. doi: 10.1128/AAC.02945-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mollenkopf DF, Stull JW, Mathys DA, Bowman AS, Feicht SM, Grooters SV, et al. Carbapenemase-Producing Enterobacteriaceae Recovered from the Environment of a Swine Farrow-to- Finish Operation in the United States. Antimicrob Agents Chemother. 2017;61(2) doi: 10.1128/AAC.01298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Institute CLS. Performance standards for antimicrobial susceptibility testing: Twenty-third Informational Supplment M100–S232013 [Google Scholar]
- 4.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 7.Gibson MK, Forsberg KJ, Dantas G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 2015;9(1):207–16. doi: 10.1038/ismej.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–4. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25(1):119–20. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearson MM, Sebaihia M, Churcher C, Quail MA, Seshasayee AS, Luscombe NM, et al. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J Bacteriol. 2008;190(11):4027–37. doi: 10.1128/JB.01981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Q, Su J, McElheny CL, Stoesser N, Doi Y, Wang M. IncX2 and IncX1–X2 Hybrid Plasmids Coexisting in a FosA6-Producing Escherichia coli Strain. Antimicrob Agents Chemother. 2017;61(7) doi: 10.1128/AAC.00536-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mataseje LF, Boyd DA, Lefebvre B, Bryce E, Embree J, Gravel D, et al. Complete sequences of a novel blaNDM-1-harbouring plasmid from Providencia rettgeri and an FII-type plasmid from Klebsiella pneumoniae identified in Canada. J Antimicrob Chemother. 2014;69(3):637–42. doi: 10.1093/jac/dkt445. [DOI] [PubMed] [Google Scholar]
- 14.Zeng X, Lin J. Beta-lactamase induction and cell wall metabolism in Gram-negative bacteria. Front Microbiol. 2013;4:128. doi: 10.3389/fmicb.2013.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potter RF, D’Souza AW, Dantas G. The rapid spread of carbapenem-222 resistant Enterobacteriaceae. Drug Resist Updat. 2016;29:30–46. doi: 10.1016/j.drup.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.