Abstract
The zebrafish genome encodes homologs for most of the proteins involved in inflammatory pathways; however, the molecular components and activation mechanisms of fish inflammasomes are largely unknown. ASC [apoptosis-associated speck-like protein containing a caspase-recruitment domain (CARD)] is the only adaptor involved in the formation of multiple types of inflammasomes. Here, we demonstrate that zASC is also involved in inflammasome activation in zebrafish. When overexpressed in vitro and in vivo in zebrafish, both the zASC and zASC pyrin domain (PYD) proteins form speck and filament structures. Importantly, the crystal structures of the N-terminal PYD and C-terminal CARD of zebrafish ASC were determined independently as two separate entities fused to maltose-binding protein. Structure-guided mutagenesis revealed the functional relevance of the PYD hydrophilic surface found in the crystal lattice. Finally, the fish caspase-1 homolog Caspy, but not the caspase-4/11 homolog Caspy2, interacts with zASC through homotypic PYD–PYD interactions, which differ from those in mammals. These observations establish the conserved and unique structural/functional features of the zASC-dependent inflammasome pathway.
Database
Structural data are available in the PDB under accession numbers 5GPP and 5GPQ.
Keywords: ASC, caspase-1, inflammasome, PYD, zebrafish
Introduction
Innate immunity offers the first line of defense against infection and danger by directly recognizing conserved pathogen- and danger-associated molecular patterns using pattern recognition receptors to initiate immune responses. Cytosolic sensors and adaptors assemble into large macromolecular complexes called inflammasomes [1,2]. Inflammasome malfunction is associated with serious diseases in humans, including type II diabetes, atherosclerosis and gout [3].
Mammalian inflammasome complexes are composed of at least a sensor molecule, an effector molecule (i.e. caspase-1) and the adaptor protein ASC [apoptosis-associated speck-like protein containing a caspase-recruitment domain (CARD)]. Upon stimulation by microbial and damage-associated signals, inflammasomes are assembled and activated, which results in the release of interleukin (IL)-1 and −18. Sensor molecules, such as NLRP3 or AIM2, recruit ASC via homotypic pyrin domain (PYD)–PYD interactions and induce the polymerization of ASC and the assembly of the ASC speck. Then, ASC further interacts with caspase-1 via CARD–CARD interactions. Therefore, the bipartite nature of ASC represents the core structure of the inflammasome. The ASC speck assembles in vivo on the micrometer scale [4]. Extracellular ASC specks released by pyroptotic cells further promote inflammasome activation [5]. The NAIP/NLRC4/caspase-1 inflammasome was previously thought to be an ASC-independent inflammasome; however, a recent study showed that the binding of ASCCARD by VHHASC (a nanobody against the CARD of human ASC) reduced speck formation and IL-1β secretion, but stabilized ASC filaments in the NAIP/NLRC4 inflammasome [6], suggesting that ASC plays important roles in the NLRC4 inflammasome. Thus, ASC plays central roles in many types of inflammasomes.
Detailed information about the structure, assembly and dynamics of inflammasome components at the atomic level is critical for the understanding of inflammation processes. To date, the structures of human ASCPYD and full-length ASC have been determined by NMR spectroscopy [7,8]. In addition, the fiber-like structures of both human and murine ASCPYD were revealed by cryo-EM [9,10]. Molecular dynamics showed that the ASC speck is an organized structure mediated by specific interactions between its PYD and CARD [11]. Mutagenesis studies showed that its CARD is critical for mediating the interaction with the effector caspase-1 [12].
However, there has been no crystal structure of an ASCPYD to date. Previously, we expended significant effort on the crystallization of the full-length ASC protein but achieved little success. The ASC protein is extremely prone to aggregation and is resistant to concentration, especially at protein concentrations higher than 1 mg mL−1, under physiological buffer conditions. However, crystallographers are still attempting to determine the crystal structure of ASC.
To better understand the molecular basis of ASC function, we characterized the structure and function of zebrafish ASC. Zebrafish is the evolutionarily lower organism that encodes an ASC homolog. Most studies of inflammasomes have focused on those in mammals, but little is known about those in teleosts. Structural and mechanistic studies of inflammasomes in lower vertebrates remain of great interest because of the simplicity of the components of these inflammasomes and their high similarity to those of higher organisms; these similarities may provide new insights into the mechanisms and evolution of inflammasome assembly [13]. There are several functional studies on characterization of inflammasomes of gilthead seabream [14–17], goldfish [18] and even zebrafish [19–23]. The zebrafish genome is predicted to have several NACHT, LRR and PYD-containing protein (NLRP) isoforms, including NLP3 (XP_009299711.1) and NLP3X1 (XP_017206752.1). In addition, zebrafish have two PYD-containing caspase homologs, Caspy and Caspy2. Importantly, the zebrafish genome encodes a homolog of ASC. An analysis of genome structure revealed that the zebrafish asc gene contains more introns than do mammalian homologs, suggesting that some introns were lost during evolution. Nonetheless, zASC contains an N-terminal PYD and a C-terminal CARD, predicting functional similarity between mammals and zebrafish. In this study, we determined the crystal structures of the N-terminal PYD and C-terminal CARD of zebrafish ASC independently and elucidated the structural and functional similarities and discrepancies between zebrafish ASC and its mammalian counterparts.
Results
ASC is widely expressed in fish
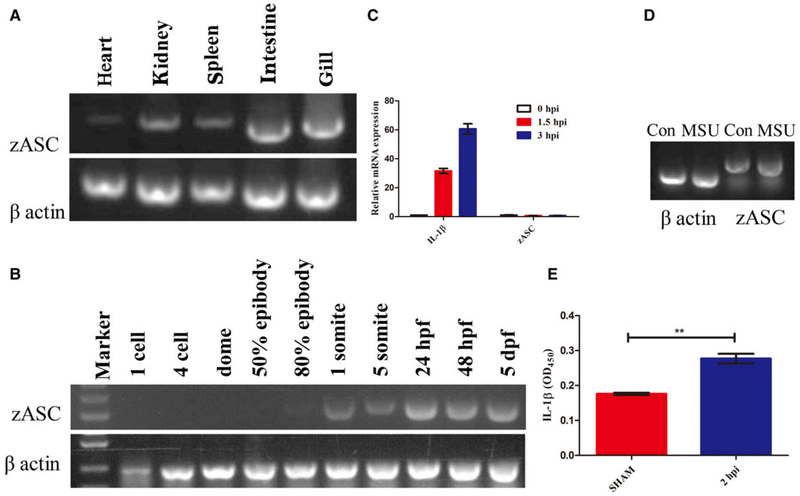
Semi-quantitative PCR was used to determine the relative gene expression levels of zASC in different tissues of adult fish and at different developmental stages of the embryo. As shown in Fig. 1A, zASC mRNA transcripts were ubiquitously expressed in all immune-related tissues, particularly in the kidney, intestine and gill, but there was very little expression in the heart. This pattern is similar to that reported in goldfish [18]. In addition, the zASC gene was expressed beginning at 1 somite (at 10 h post-fertilization) in the early developmental stage (Fig. 1B). A recently study by Kuri et al. [20] showed zebrafish asc expression from morula stage onward, and both results suggest that ASC may play some fundamental role in embryo development in addition to mediating inflammatory responses. Interestingly, the IL-1β mRNA levels were significantly upregulated when zebrafish were treated with the inflammasome activator Staphylococcus aureus (Fig. 1C), and mature, released IL-1β was detected by the ELISA assay (Fig. 1E). However, there were no changes in zASC self-mRNA after exposure to either monosodium urate or S. aureus (Fig. 1C,D).
Fig. 1.
The involvement of zASC in inflammation. Semi-quantitative reverse transcription-PCR analysis of asc gene expression in different tissues in adult zebrafish (A) and at different developmental stages in zebrafish embryos (B). β-actin was used as a control gene. (C) Quantitative expression analysis using Q-RT-PCR of IL-1β and asc in zebrafish embryos at 1.5 and 3 h after infection with Staphylococcus aureus. The expression of il-1β and asc was normalized against an endogenous control gene, elongation factor 1α (EF-1α) (mean ± SEM of at least two experiments, n = 100 zebrafish embryos). (D) Semi-quantitative expression of zebrafish ASC in embryos treated with either PBS or MSU. (E) ELISA of IL-1β levels in zebrafish embryos infected with PBS or S. aureus for 2 h (mean ± SEM of at least three experiments, n = 100 zebrafish embryos). **P = 0.002. hpf, hours post-fertilization; hpi, hours post-infection; MSU, monosodium urate.
zASC forms specks in vitro and in vivo when overexpressed
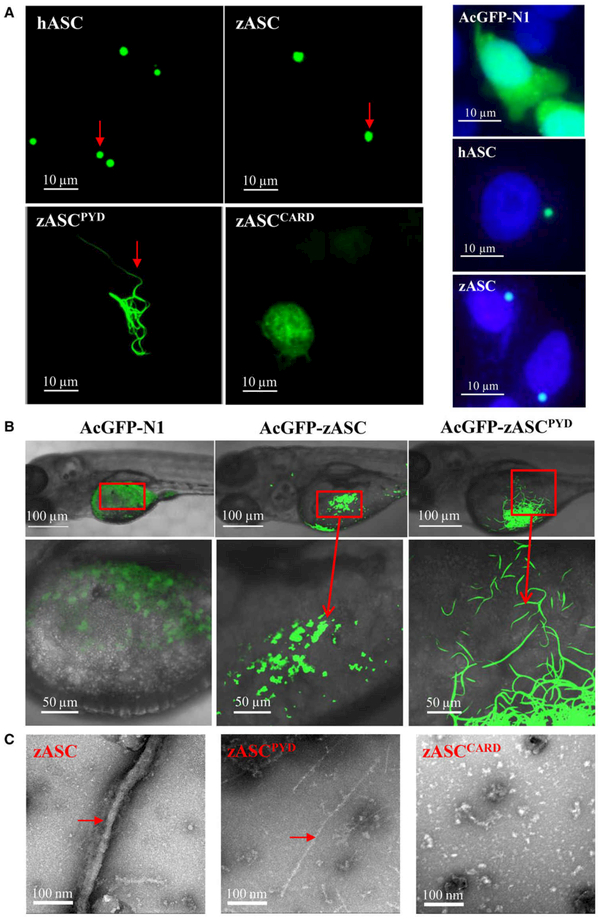
When the ASC speck was first observed over a decade ago, it was described as a hallmark of inflammasome activation [4,24]. The ASC speck can be formed by endogenous or overexpressed full-length ASC protein. In mammals, ASC forms only one speck close to the nucleus per cell [4]. To determine whether zebrafish ASC also forms a speck, we transfected HeLa cells with AcGFP-empty, AcGFP-hASC and AcGFP-zASC plasmids. Here, we used a monomeric AcGFP, which is a superior fusion tag that avoids self-oligomerization. The expression of AcGFP-zASC in HeLa cells resulted in the formation of green specks similar to those in hASC (Fig. 2A, left side, indicated by red arrows) with one speck close to the nucleus per cell (Fig. 2A, right side). In addition, bimolecular fluorescence complementation (BiFC) technology was used in vitro to investigate and visualize zASC interaction in living cells. We found that zASC could interact with itself to bring the fluorescent fragments into close proximity to form a fluorescence signal (see Fig. 6B).
Fig. 2.
Both the PYD and CARD of ASC are required for speck formation. (A) Left side: subcellular localization of the AcGFP-fused hASC, zASC, zASCPYD, zASCCARD and AcGFP-N1 proteins. Bars: 10 μm. Right side: ASC SPECK formation close to nucleus per cell. 4’,6-Diamidino-2-phenylindole (DAPI) staining was used to label nucleus. (B) Representative confocal images of the zebrafish embryo yolk circulation region when zASC and zASCPYD are overexpressed. Bars: 50 and 100 μm. (C) Representative electron micrograph of the zASC, zASCPYD and zASCCARD proteins subjected to negative staining. Bar, 100 nm.
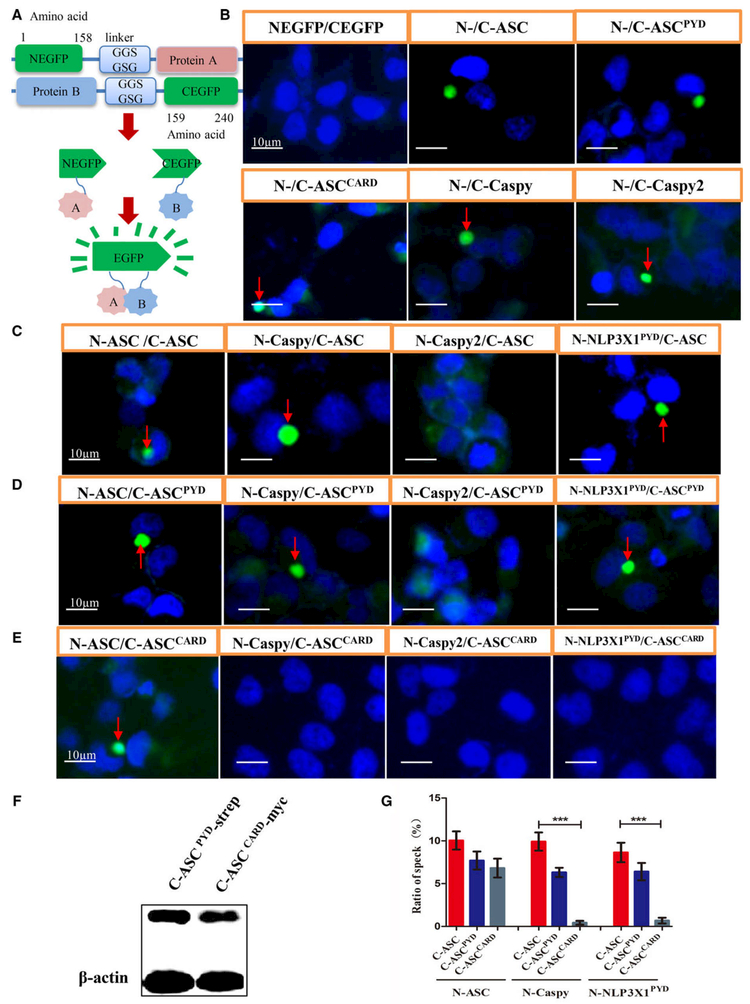
Fig. 6.
Pyrin domain–PYD interactions mediate ASC inflammasome assembly in zebrafish. (A) Cartoon representation of the BiFC assay. If protein A interacts with protein B directly, a fluorescent reporter fragment will exhibit fluorescence signaling. (B) Fluorophore formation was observed after the cotransfection of HeLa cells with NEGFP–zASC, zASCPYD, zASCCARD, zCaspy or zCaspy2 and CEGFP–zASC, zASCPYD, zASCCARD, zCaspy or zCaspy2. (C) Fluorophore formation was observed with ASC–ASC interactions, zASC–zCaspy interactions and zASC–zNLP3X1PYD interactions, but not zASC–zCaspy2 interactions. (D) Fluorophore formation was observed with ASCPYD-mediated interactions with zCaspy or zNLP3X1PYD, but not with zASCCARD-mediated interactions (E). The nucleus was stained with DAPI. Bars: 10 μm. (F) Cell lysates were immunoblotted for CEGFP–ASCPYD–strep and CEGFP–ASCCARD–myc showing similar expression levels of ASCPYD and ASCCARD protein, respectively, when they interact with Caspy. β-Actin was the loading control. (G) Quantification of the prevalence of fluorescence specks. Mean ± SEM of at least three independent experiments. The bar graph shows the results of statistical analysis. n = 10 images from each group. A t test was used for comparison of interaction of Caspy with ASC and ASCPYD, or NLP3X1PYD with ASC and ASCPYD. ***P < 0.001.
The zebrafish model is often used to study innate immunity because of the transparency of its body in early life stages, which allows real-time visualization. Two days after AcGFP-zASC plasmids were microinjected into the one-germ-cell stage of zebrafish, in vivo speck formation was also observed in the yolk circulation area of the embryo (Fig. 2B, indicated by red arrows) while Kuri et al. [20] observed GFP specks in the epidermis with Tg(asc:asc-EGFP) larvae during treatment with copper sulfate. Both the above in vitro and in vivo results demonstrated that zASC oligomerizes as a speck.
Although ASC is the main structural component of the ASC speck, the roles of zASCPYD and zASCCARD in speck formation and downstream signaling are still unclear. To address these questions, we overexpressed zASC, zASCPYD and ASCCARD separately and evaluated their speck formation ability. In contrast to the formation of a large speck, zASCPYD formed less condensed filamentous structures in mammalian cells and zebrafish embryos, even in the absence of activated receptors for the full-length ASC (Fig. 2A,B), while a lack of filaments was observed in zASCCARD-overexpressing cells (Fig. 2A). This finding suggests that both the PYD and CARD of zASC are essential for speck assembly, although the PYD is the major polymerization domain, which is consistent with previous studies of mammalian ASC [25,26].
To further visualize the molecular structure of zASC, we purified monomeric His-NusA-GB1-tagged zASC, zASCPYD and zASCCARD proteins. Upon tobacco etch virus protease treatment to cleave off the His-NusA-GB1 tag in situ, ASC fibers and ASCPYD filaments spontaneously formed in 1 h, while zASCCARD did not (Fig. 2C). Negative-stain electron microscopy images showed that the oligomerization of the full-length zASC led to the formation of fibers that were thicker than zASCPYD filaments (Fig. 2C, indicated by red arrows), suggesting that zASCCARD does have an oligomerization tendency in the context of ASCFL protein.
Structures of the zASCPYD and zASCCARD are highly conserved
After numerous attempts, we decided to use complementary techniques for the crystallization of ASC proteins. The bacterial maltose-binding protein (MBP) has been shown to be the best performing cocrystallization tag [27,28]. In this study, we fused zASCPYD and zASCCARD separately to the C terminus of a MBP tag to facilitate the crystallization of the PYD and CARD. After testing several linker sequences, we successfully crystallized both the MBP–zASCPYD and MBP–zASCCARD fusion proteins, which led to the determination of their structures.
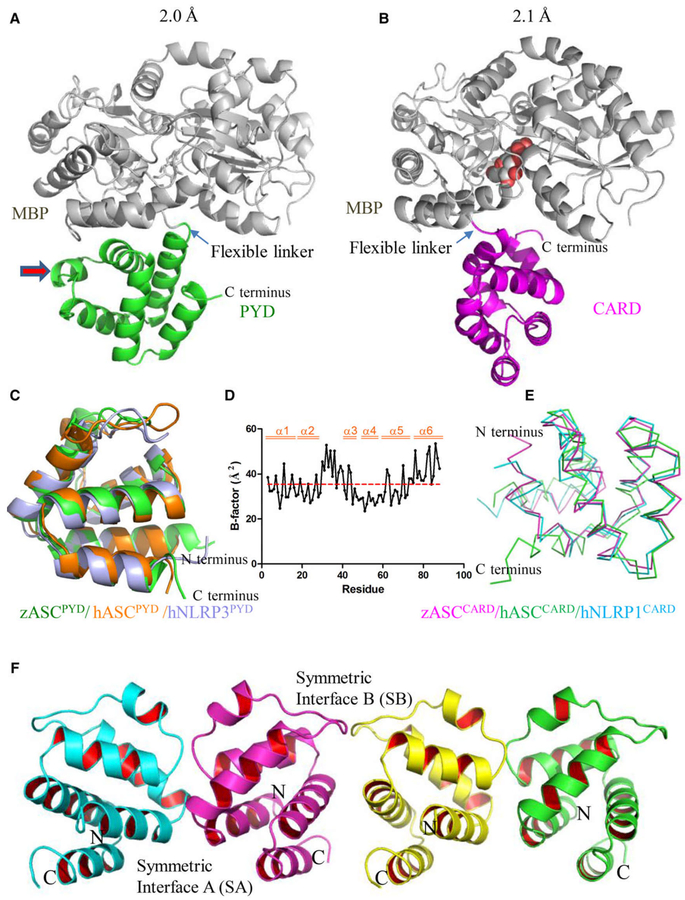
In our structure, the linker region between the N-terminal MBP and C-terminal PYD or CARD adopts a flexible conformation (Fig. 3A,B), which moves the PYD or CARD away from the MBP tag and preserves its native conformation. Both zASCPYD and zASCCARD adopt a six-helix bundle fold similar to other death domain superfamily members (Fig. 3C-E). The zASCPYD has the classical structure of the PYD subfamily, with a long loop (L2/3) between H2 and H3, followed by a short helix (H3) (Fig. 3A, indicated by red arrow). Its L2/3 has higher than average B-factors similar to other PYDs [29], suggesting the high mobility of this region (Fig. 3D).
Fig. 3.
Crystal structure of zebrafish ASCPYD and ASCCARD at atomic resolution. (A) Crystal structure of the PYD of zASC (green) with an MBP tag (gray) at 2.0 Å resolution. The MBP protein was used as a crystallization tag to facilitate the formation of protein crystals. A maltose ligand was built in the ligand-binding pocket of MBP (same for B). (B) Crystal structure of the CARD of zASC (magenta) with an MBP tag (gray) at 2.1 Å resolution. (C) Structural superposition of PYD structures. The crystal structure of zebrafish ASCPYD (green) was superimposed on the NMR structure of human ASCPYD (1UCP, orange) and the crystal structure of human NLRP3PYD (2NAQ, light blue); the overlay yielded RMSD of 1.18 and 1.08 Å, respectively. (D) B-factor analysis of the zASCPYD structure. (E) Structural superposition of the CARD of zebrafish ASC, human ASC (2KN6, green) and human NLRP1 (4IFP, blue). (F) PYD–PYD interaction interface found in the crystal lattice.
Structural comparison
One-dimensional sequence analysis showed that zASC has high sequence homology to mammal orthologs, with 35% sequence identity and 51% similarity to hASC and with 32% sequence identity and 50% similarity to mASC. The three-dimensional structural comparison showed that zASCPYD is similar to hASCPYD and hNLRP3PYD, with RMSD values of 1.18 and 1.08 Å, respectively (Fig. 3C and Table S1). In addition, the structural alignment of zASCCARD with the available hASCCARD and hNLRP1CARD structures yielded RMSD values of 3.07 and 1.01 Å, respectively (Fig. 3E and Table S2).
zASC speck assembly is dependent on PYD–PYD interactions
Interestingly, PYD form continuous lines in the crystal lattice by leaving the MBP tags outside of the PYD string. There are two nearly symmetric PYD–PYD homotypic interaction interfaces in the crystal. We named them symmetric interface A (SA) and symmetric interface B (SB) (Fig. 3F). A continuous PYD string forms by alternating both types of interfaces. Protein interfaces, surfaces and assemblies (PISA) [30] analysis of both interfaces showed that SA has an interface area of 675 Å2 containing 14 hydrogen bonds and eight salt bridges (Table S3), while SB has an interface area of 571 Å2 containing seven hydrogen bonds and six salt bridges. Using PISA, the solvation free energy gain upon the formation of the interface (ΔiG) was calculated for the two interfaces, revealing values of 3.0 and 1.7 kcal mol−1 for SA and SB, respectively.
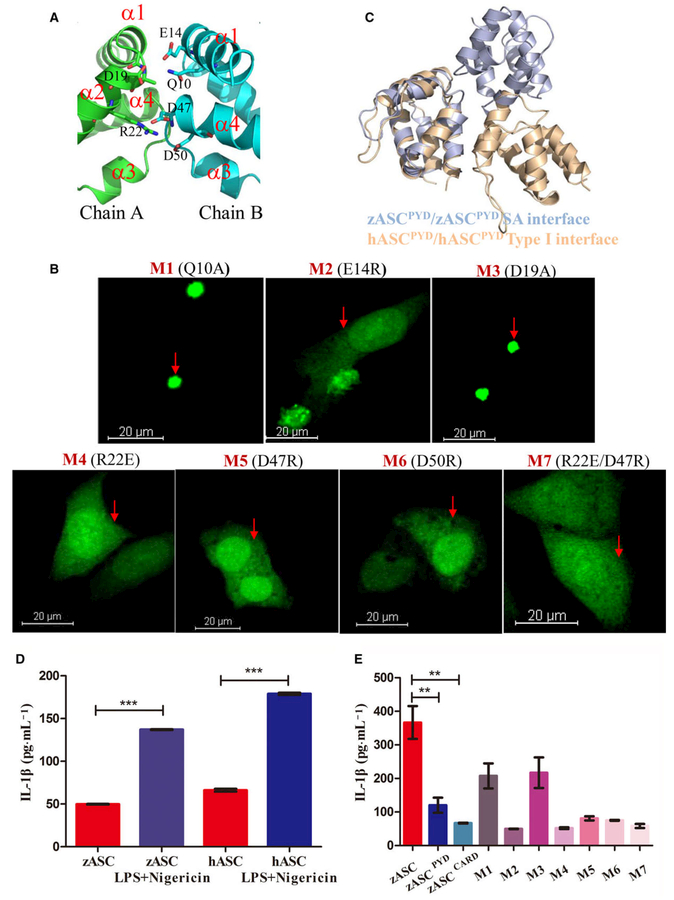
Judging from the interface area and interactions, we conclude that SA is more plausible for homotypic PYD–PYD interactions under physiological conditions. A close inspection of SA reveals that it resides in a region containing H1–H4. In particular, charged residues E14, R22, D47 and D50 are heavily involved in the interface, while the hydrogen bonding between Q10 and D19 is weaker, judging from the distance between them (Fig. 4A and Table S3). In this interface, a motif containing H1–H2 interacts with H3–H4 of a symmetric molecule, and H3–H4 interacts with H1–H2 of a symmetric molecule. While charge–charge interactions between side chains dominate the interactions, some interactions with the main chain are also involved. For example, OE2 of E14 forms a hydrogen bond with N of A18 of the other molecule; NZ of K25 on chain A forms a hydrogen bond with O of K46 on chain B; and NZ of K46 on chain A forms a hydrogen bond with O of E43 on chain B (Table S3).
Fig. 4.
Structure-guided mutagenesis of ASC impairs speck formation and inflammasome activity. (A) Detailed interactions in the SA interface. Critical residues in the PYD–PYD interaction interface are shown as stick models and are labeled. Only one half of the symmetric interface is shown for clarity, in which D19 and R22 of chain A (in green) interact with Q10, E14, D47 and D50 of chain B (cyan). (B) Representative images of HeLa cells after ASC mutant overexpression, including Q10A, E14R, D19A, R22E, D47R and D50R. The nucleus was stained with DAPI, and ASC fused with AcGFP oligomerized as a speck. Scale bar: 20 μm. (C) Comparison of the zASCPYD symmetric interface with the conserved type I interface in mammalian ASCPYD. Zebrafish ASC is colored light blue, while human ASC is colored light brown. (D) Reconstitution of the inflammasome in RAW cells transfected with human ASC and zebrafish ASC, in the absence and in the presence of LPS/Negericin treatment (mean ± SEM of at least two experiments). A t test was used for comparison of LPS/negericin treatment with no-treatment group for statistical analysis. ***P < 0.001. (E) Reconstitution of the inflammasome in RAW cells transfected with zASC, zASCPYD, zASCCARD and zASC mutants. LPS-primed RAW264.7 cells were treated with 10 μM nigericin for additional 2 h (mean ± SEM of at least three experiments). A t test is used for comparison of ASC with ASCPYD group and ASC with ASCCARD group for statistical analysis. **P < 0.01, n = 3. M1, mutant 1 (Q10A); M2, mutant 2 (E14R); M3, mutant 3 (D19A); M4, mutant 4 (R22E); M5, mutant 5 (D47R); M6, mutant 6 (D50R); M7, mutant 7 (R22E/D47R).
To understand the implications of the charged surface in ASC speck formation, we generated structurally guided ASC mutations targeting the interface SA, including Q10A, E14R, D19A, R22E, D47R, D50R and R22E/D47R. Among them, single or double mutations, including R22E, D47R, D50R and R22E/D47R, completely abrogated speck formation, suggesting that these mutants disrupt ASC–ASC interactions in cells (Fig. 4B, indicated by red arrows). Most of these sites overlap with the reported classical type I asymmetric interface, and interface I is the most extensive interface and is required for the propagation of the single layers of the filament [9,10]. In addition, E14 lies in the type III interface, and its mutation (E14R) caused a defect in ASC speck formation (Fig. 4B, indicated by red arrow). In contrast, mutations of some non-conserved residues (Q10A and D19A) displayed ASC speck formation behavior similar to that of the wild-type (WT) protein (Fig. 4B, indicated by red arrows). Taken together, these data indicate that PYD-dependent zASC speck formation in the cell requires the same interface residues identified in our crystal structure.
Comparison with mammalian ASCPYD filaments
Both human and murine ASCPYD assemble into filaments in a conserved mode [9,10]. As a result, we only compared the major interface of human ASC with zebrafish ASC. A PISA analysis of the most dominant type I interface of human ASCPYD filaments between the Ia and Ib surfaces showed that 12 residues are involved in this interface; among them, D51 and D54 are the major residues in Ia, while K21 and R41 are the major residues in Ib. This interface has a surface area of 479.5 Å2 containing five hydrogen bonds and seven salt bridges, and the solvation free energy gain upon formation of the interface (ΔiG) is 0.1 kcal mol−1.
The major interacting residues in the SA interface of zASCPYD, namely, E14, R22, D47 and D50, are equivalent to E13, K21, D48 and D51 of hASCPYD, respectively. Therefore, three of the four major interacting residues of the SA interface of zASCPYD overlap with the hASCPYD type I interface. Despite the conserved residues in the interfaces, the domain orientation is notably different. If one of the subunits is aligned, the second unit interacting with the type I interface would have a different orientation (Fig. 4C).
In addition to the conserved features, there are some notable variations between zASC and mammalian ASC filaments. Interestingly, F59 of human ASC is critical for the human ASC IIb interface, probably stabilizing H4 and forming a hydrophobic interaction with L78 of the neighboring molecule. Its mutation to E abolishes hASC filament formation [10]. The F59 is not conserved in lower vertebrates, with a V in zebrafish and a T in many bony fishes (Fig. 5). Particularly for V/T59 (human ASC numbering), there is predicted to be lack of hydrophobic interactions with the nonconserved C78 in bony fishes. This observation supports that the zASC has distinct surface residues that probably form unique bony-fish-specific filaments.
Fig. 5.
Structure-based sequence analysis of the zebrafish ASC protein. Multiple sequence alignment of ASC proteins by CLUSTAL W program; (*) indicated completely conservative residues, (:) indicated high conservative residues and (.) indicated low conservative residues. Amino acid numbering is indicated on the right. Secondary structure: H, α-helix: the PYD and CARD domains consist of H1–H6 and H7–H12, respectively. Between both domains there is a 30-amino-acid linker, highlighted by underline and purple. GenBank accession numbers for ASC: zebrafish (NP_571570); human (AB023416.2); mouse (NP_075747); Nile tilapia (XP_003451301); Fugu rubripes (XP_011616502); Japanese medaka (XP_011484148); rainbow trout (ACO08655); Atlantic salmon (ACN12209); dog (XP_013969837); panda (XP_002924797); black flying fox (XP_006914227); African elephant (XP_010596841); small-eared galago (XP_003795885); guinea pig (XP_003478302); platypus (XP_001507377).
Mutation of PYD abolishes its inflammasome activity
The murine RAW 264.7 macrophage cell line lacks ASC expression, but it resembles bone-marrow-derived macrophages when the ASC gene is ectopically introduced into it [12]. An ASC-reconstituted RAW 264.7 cell line was used to detect the effects of the mutation of PYD in zASC on inflammasome activity. We used cells with exogenous expression of zASC and mutant cells to analyze the level of mature IL-1β released into the supernatant by ELISA after the cells were treated with lipopolysaccharide (LPS) and nigericin. Similar to human ASC, zebrafish ASC reconstitutes NLRP3 inflammasome activity with LPS and nigericin treatment (Fig. 4D). Furthermore, the analysis of the zASC mutant confirmed that most mutants, except for mutant 1 (Q10A) and mutant 3 (D19A), impaired functional speck formation and abolished nigericin-induced NLRP3 inflammasome activity (Fig. 4E).
zASC interacts with downstream Caspy through PYD–PYD interactions
The zebrafish genome encodes two PYD-containing caspases, Caspy and Caspy2, which are homologs of human caspase-1 and −4/5, respectively [31]. Here, we used the BiFC system (Fig. 6A) to study the in vitro protein interactions. First, the BiFC results confirmed that zASC interacts with itself through both its N-terminal PYD and its C-terminal CARD, which validated our approach (Fig. 6B, indicated by red arrows). We then investigated whether both Caspys can interact with zASC. The data showed that zASC interacts with Caspy but not Caspy2 (Fig. 6C, indicated by red arrow). Additionally, the PYD of ASC (Fig. 6D, indicated by the red arrow), but not the CARD, interacts with Caspy (Fig. 6E,G). C-ASCPYD and C-ASCCARD are expressed at comparable levels when they interact with N-Caspy (Fig. 6F). In combination with previous reports that zASCCARD did not coimmunoprecipitate with Caspy [31], these results showed that zASC recruits the downstream effector Caspy through a homotypic PYD–PYD interaction.
Furthermore, we found that Caspy and Caspy2 could interact with themselves (Fig. 6B, indicated by red arrows), which is similar to the dimerization/oligomerization of mammalian caspase-1 and caspase-11 [32,33].
Finally, although there is no direct NLRP3 ortholog that functions upstream of ASC in fish, a NACHT, LRR and PYD-containing protein 3 isoform X1 (NLP3X1, XP_017206752.1) is predicted to be present in the zebrafish genome. Our data showed that zASC interacts with zNLP3X1 through PYD–PYD interactions as well (Fig. 6C-E, indicated by red arrows, and Fig. 6G).
Discussion
High-resolution structure of ASC
In this work, we extensively studied the structural properties of zASC by determining the crystal structures of the PYD and CARD separately by fusing them to an MBP tag. We also performed an in vitro negative-stain EM study of the ASC signalosome. Based on the crystal structures, we found that the zASCPYD, but not the zASCCARD, tends to form intensive interactions with itself. Furthermore, we demonstrated that the fiber-like structures formed by zASC are primarily mediated by PYD.
Conformation of linker region
The full-length ASC protein has a 30-amino-acid-long linker region between the N-terminal PYD and C-terminal CARD (Fig. 5). This region is less conserved than the PYD and CARD regions among vertebrates. A previous NMR analysis of human ASC showed that the linker region is highly disordered, and the PYD and CARD do not have significant interactions [8]. Indeed, secondary structural prediction by PSIPRED [34] showed that the linker regions of most ASC proteins may potentially have a random coil structure. Taken together, these data led us to conclude that the sequence and length of the linker region are less important for proper ASC function.
PYD–PYD interactions
Charged surface residues have been shown to play critical roles in ASC oligomerization [25]. Moriya et al. found that the ASC molecule is composed of a continuous surface area with positively and negatively charged residues. After examining large-scale surface residue mutations, they further identified Lys21, Lys26, Arg41, Asp48 and Asp51 as critical for human ASC filament formation [25]. In addition, Vajjhala et al. [35] determined the PYD–PYD interface of human ASC using molecular docking calculations. The asymmetric interface contains positively charged residues (Lys21 and Arg41) on one molecule and negatively charged residues (Glu13, Asp48 and Asp51) on the other molecule.
PISA analysis showed that it is not possible to form a stable complex through this SB interface. In the present study, we discovered a symmetric PYD–PYD interface in the crystal primarily formed by E14, D47 and D50 (corresponding to E13, D48 and D51 of human ASC, respectively) on one side and D19 and R22 (corresponding to E18 and K21 of human ASC, respectively) on the other side. These residues are conserved and have been reported to be essential for human and murine ASC filament formation [10,11,26,35]. Our structure-guided mutagenesis studies confirmed that the clustering of zASCPYD filaments and their condensation into a dense speck structure required the intact PYD–PYD interface. Here, charge reversal double mutants (R22E/D47R) did not rescue the defectiveness of the single mutants. We assume another residue (such as D50) may be involved in interface interaction. Additional structural studies,i.e. cryo-EM studies of zASC filaments, are needed to answer whether lower vertebrates use the same process to assemble filaments.
PYD mediate signaling in zebrafish
In mammals, the adaptor ASC bridges the sensor receptor and oligomerizes via PYD–PYD interactions, while the CARD recruits downstream caspase-1 via CARD–CARD interactions [36]. Zebrafish ASC contains an N-terminal PYD and a C-terminal CARD. The largest difference lies in effector molecules. The Caspy protein is a PYD-containing caspase, although it shares the highest homology with CARD-containing caspase-1 in mammals. Our results showed that zASC oligomerizes as a speck via homotypic PYD–PYD interactions, and zASC interacts with the downstream effector molecule Caspy directly via PYD–PYD interactions. Recently, by examining the colocalization of the CaspyPYD with ASCPYD and ASCCARD in vivo, Kuri et al. [20] confirmed PYD interaction between zASC and zCaspy. However, zASC can likely recruit murine pro-caspase-1 to cleave pro-IL-1β in reconstituted RAW macrophages, which may be through CARD.
As an adaptor molecule, zASC is hypothesized to link putative upstream PYD- or CARD-containing proteins to Caspy. The NOD-like family member NLRP3, which is upstream of ASC, is specific to mammals. However, no homolog has been identified in lower vertebrates. Herein, we discovered a gene named NLP3X1 in the zebrafish genome. Interestingly, zASC interacts with NLP3X1 via its PYD. Whether NLP3X1 directly interacts with Caspy and its roles in inflammasome activation warrant further investigation.
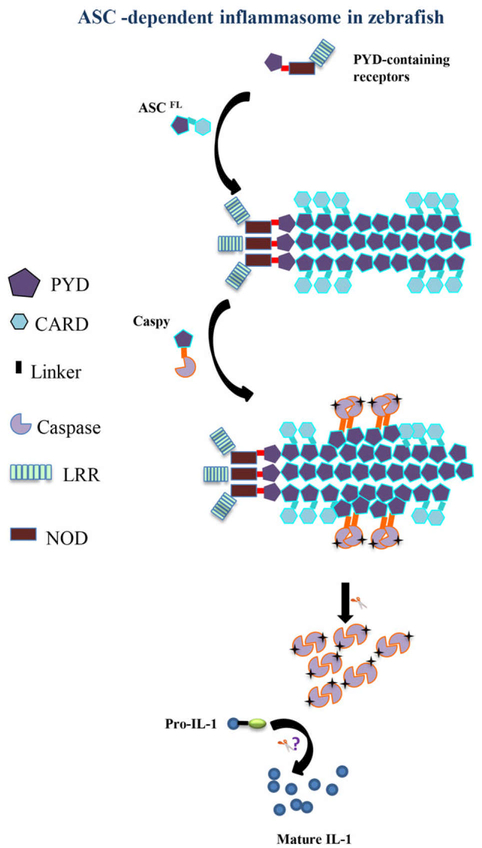
In summary, our in vitro and in vivo studies showed that the monomeric structure and function of ASC are highly conserved in zebrafish. The ASCPYD presents a platform for homo- and hetero-PYD association in inflammasome assembly in lower vertebrates (Fig. 7). However, whether IL-1β cleavage in zebrafish is dependent on Caspy or inflammasome activation needs further study. Future structural studies are urgently needed to clarify similarity and variation of ASC filament assembly in lower vertebrates at the molecular level.
Fig. 7.
Possible model of the ASC-dependent inflammasome in zebrafish. The major difference between the zebrafish and mammalian inflammasomes lies in the interaction modules between ASC and its effector caspases. Zebrafish use PYD–PYD interactions to recruit PYD-containing Caspy.
Materials and methods
Cloning, protein expression and purification
The coding regions of the PYD and CARD regions of the zebrafish ASC gene (NM_131495) were amplified by PCR and separately ligated into an in-house engineered MBP-tagged expression vector in a pET30a backbone [29]. For large-scale expression, the recombinant plasmids were transformed into Escherichia coli Rosetta (DE3) strain (Stratagene, La Jolla, CA, USA) cells. Cultures were grown to a D600 of approximately 1.2 at 37 °C with constant shaking at 180 rpm. After the temperature was dropped to 18 °C, isopropyl β-D-1-thiogalactopyranoside was added to a final concentration of 0.2 mM, and the cells were incubated for an additional 4–5 h. The cells were harvested and resuspended in Ni-binding buffer (500 mM NaCl, 10 mM imidazole and 20 mM Tris/HCl pH 8.0) supplemented with DNase (Biomatik, Cambridge, ON, Canada) and protease inhibitors (Roche, Indianapolis, IN, USA). After sonication, soluble protein was purified from the cell lysate with a HisTrap IMAC column (GE Healthcare, Little Chalfont, Buckinghamshire, UK), followed by a Superdex 200 preparative column (GE Healthcare) using gel filtration buffer (5 mM maltose, 2 mM DTT, 1 mM EDTA, 250 mM NaCl and 20 mM Tris/HCl pH 8.0). The peak corresponding to a monomer fraction was collected, and the purity was revealed by SDS/PAGE.
Crystallization, data collection and structure determination
The purified recombinant MBP-tagged zASCPYD or zASCCARD protein was concentrated to over 50 mg mL−1, and crystallization conditions were screened using both commercially available and in-house-designed crystallization kits. Several hits were readily identified within 2 weeks after the set-up of hanging drops. High-quality zASCCARD crystals were obtained under conditions of 2.0 M ammonium citrate, 0.1 M HEPES pH 7.0, 10% (w/v) maltose and 10% (v/v) glycerol. Additionally, the crystallization condition was used as a cryoprotectant solution to flash-cool the crystals in liquid nitrogen. The zASCPYD crystals were obtained under conditions of 20% poly(ethylene glycol) methyl ether 2000 and 0.1 M NaAcO pH 4.6, with 25% poly(ethylene glycol) 1500, 10% ethylene glycol, and 0.1 M NaAcO pH 4.6 used as the cryoprotectant solution.
X-ray diffraction data were collected at the GM/CA-CAT beamline at the Advance Photon Source and at the BL18U and BL-19U1 beamlines at the Shanghai Synchrotron Radiation Facility. The data were processed using the HKL2000 program suite [37] and XDS [38]. The structure was determined by molecular replacement calculations with PHASER [39] using the same mutant MBP structure (PDB code: 4IFP [40]) as a search template. The structural model was improved by rounds of manual model fitting with COOT and structural optimization and refinement in the PHENIX GUI [41]. The final structural models were validated using the Molprobity server [42] and the RCSB ADIT validation server [43]. Molecular graphics were generated with PYMOL (Delano Scientific LLC, CA, USA). Protein sequence alignments were prepared with MEGA7 [44], CLUSTALX 2.1 [45] and ESPRIPT 3 [46]. The structure superposition and the RMSD values of the structures were calculated with PYMOL.
Construction of zASC and mutagenesis
The entire open reading frames of zebrafish ASC, Caspy and Caspy2 were amplified by PCR from a cDNA library reverse transcribed from total mRNA prepared from the whole body of an adult zebrafish. The inserts were cloned into the pAcGFP-N1 vector (Addgene, MA) to generate pAcGFP-zASC and pAcGFP-zASCPYD. The PYD and CARD of zebrafish ASC, Caspy and Caspy2 were fused to the C-terminus of the EGFP fragment of the pEGFP vector (pNEGFP) and were fused to the N-terminus of the EGFP fragment of the pEGFP vector (pCEGFP), respectively. Point mutations in zASC were introduced using primer-mediated site-directed mutagenesis. The WT and mutant ASC were subcloned into the pAcGFP-N1 vector. All mutants were verified by DNA sequencing.
Transfection and immunofluorescence of tagged proteins
HeLa cells and human embryonic kidney (HEK)-293T cells were grown in DMEM supplemented with glutamine, antibiotics and 10% FBS. To assess the formation of ASC specks, cells were seeded on glass coverslips. The total amount of transfected plasmid DNA was quantified, and 1 × 105 HeLa or 293T cells were transfected with 1 μg of plasmid using Lipofectamine™ 2000 (Thermo Fisher Scientific, Life Technologies, Waltham, MA, USA) according to the manufacturer’s instructions. For the cellular localization assay, HeLa cells were transiently transfected with pAcGFP-zASC. For the BiFC assay, 293T or HeLa cells were cotransfected with pNEGFP and pCEGFP fusion constructs. At 24 h post-transfection, the cells were fixed with 3.7% formaldehyde in PBS at 37 °C for 10 min and were washed three times with PBS for 5 min each time. The samples were mounted with Ultra Cruz Mounting medium containing DAPI (Santa Cruz Biotechnology, Dallas, TX, USA; sc-24941). Images were acquired using a confocal fluorescence microscope (Olympus, Tokyo, Japan).
Reconstruction of inflammasome in RAW 264.7 cells
For the reconstitution of the ASC inflammasome pathway, the murine RAW 264.7 macrophage cell line, which lacks the expression of ASC [47], was transfected with 0.8 μg pAcGFP-ASC or the ASC mutant plasmids in 24-well plate using 0.8 ll EndoFectin™ Max transfection reagent (Gene-Copoeia, Rockville, MD, USA) according to the manufacturer’s instructions. Thus fused ASC or ASC mutant with AcGFP is overexpressed. After 24 h transfection, the NLRP3 inflammasome was activated using 10 μM nigericin treatment for 2 h after priming with 300 ng mL−1 LPS for 6 h. Then, the culture supernatants were collected. The release of mature IL-1β was determined using an IL-1β ELISA kit (R&D Systems, Minneapolis, MN, USA).
Negative-stain electron microscopy
The GB1-NusA-tagged full-length zASC, ASCPYD and zASCCARD fusion proteins were expressed and purified in E. coli using the same protocol as for the MBP-tagged fusion proteins. Two hours after tobacco etch virus pro-tease was added to the purified samples at a concentration of 50 lg mL−1, which is sufficient to cleave off the GB1-NusA tag, the samples were observed using negative-stain EM without purification.
Experimental animal and bacterial strains
Fertilized cells of the AB/WT zebrafish strains were used in this study [48]. All treatments of experimental animals were approved by the Committee on the Ethics of Animal Experiments of the University of Science and Technology of China (USTC) (License Number: USTCACUC1103013) and were in accordance with the guidance and provisions of the Committee on Laboratory Animal Resource Center and the Animal Care and Use Guidelines of the USTC.
GFP-expressing strains of Staphylococcus aureus (NCTC8325) were used and diluted to a uniform concentration of 1000 colony-forming units per nanoliter.
Gene expression analysis
One hundred larvae at the time points indicated and different tissues from adults were pooled and frozen in RNAiso Plus (Takara, Dalian, China). Total RNA was extracted according to standard procedures. RT-PCR was performed using a one-step RT-PCR kit (Takara, China) following the manufacturer’s instructions.
Quantitative real-time PCR was performed with a Bio-Rad iCycler MyIQ2 instrument using the two-step real-time quantitative PCR SYBR Green Supermix kit (Bio-Rad, Hercules, CA, USA). For each mRNA, the gene expression was normalized against the expression of ef1a in each sample. The primers used are shown in Table S4. In all cases, PCR was performed in triplicate and repeated at least twice.
Zebrafish in vivo imaging
For in vivo observations of ASC speck and ASCPYD filament formation, 3 days post-fertilization AB/WT zebrafish were used in imaging after zASC overexpression. Prior to imaging, all larvae were also treated with 0.01% MS222 (3-amino benzoic acid ethyl ester). An Olympus (BX60WI) fluorescence microscope with a × 10 Olympus Plan Fluor objective (NA 0.30) and a × 60 Olympus Plan water-dipping objective (NA 0.9) were used to obtain the fluorescence images. To prevent damage to the living samples, a minimal exposure time was used. IMARIS software (Bitplane, Zurich, Switzerland), ImageJ software (NIH, Bethesda, MD, USA) and Adobe Photoshop CS2 were used for image processing.
IL-1β ELISA assay
The concentration of mature zebrafish IL-1β in the whole body was quantified using a fish ELISA kit according to the manufacturer’s instructions (Cusabio, Wuhan, China). The statistical significance of differences was evaluated using Student’s t test.
Supplementary Material
Acknowledgements
We would like to thank the staff of GM/CA-CAT at Advance Photon Source (APS) and the BL17U, BL18U and BL19U1 beamlines at the Shanghai Synchrotron Radiation Facility (SSRF) for their assistance during data collection [49]. YL is supported by the China Postdoctoral Science Foundation (Grant No.: 2015M582007), the National Natural Science Fund for Young Scholars (Grant No.: 31600598), and the Tenth Batch Special Support of China Postdoctoral Science Foundation (Grant No.: 2015T100454). TJ is supported by the Fundamental Research Funds for the Central Universities, the 100 Talents Program of CAS and the National Natural Science Foundation of China (Grant No.: U1732109).
Accession numbers
Coordinates and structure factors have been deposited in the Protein Data Bank, and the accession codes are 5GPP and 5GPQ respectively (also see Table S5).
Abbreviations
- ASC
apoptosis-associated speck-like protein containing a caspase-recruitment domain
- BiFC
bimolecular fluorescence complementation
- CARD
caspase-recruitment domain
- DAPI
4’,6-diamidino-2-phenylindole
- IL
interleukin
- LPS
lipopolysaccharide
- MBP
maltose-binding protein
- NLP3X1
NACHT, LRR and PYD-containing protein 3-like isoform X1
- PISA
protein interfaces, surfaces and assemblies
- PYD
pyrin domain
- SA
symmetric interface A
- SB
symmetric interface B
- WT
wild-type
Footnotes
Conflict of interest
Authors declare no conflict of interests.
Supporting information
Additional supporting information may be found online in the Supporting Information section at the end of the article:
References
- 1.Schroder K & Tschopp J (2010) The inflammasomes.Cell 140, 821–832. [DOI] [PubMed] [Google Scholar]
- 2.Martinon F, Burns K & Tschopp J (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 10, 417–426. [DOI] [PubMed] [Google Scholar]
- 3.Strowig T, Henao-Mejia J, Elinav E & Flavell R (2012) Inflammasomes in health and disease. Nature 481, 278– 286. [DOI] [PubMed] [Google Scholar]
- 4.Masumoto J, Taniguchi S, Ayukawa K, Sarvotham H, Kishino T, Niikawa N, Hidaka E, Katsuyama T, Higuchi T & Sagara J (1999) ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J Biol Chem 274, 33835–33838. [DOI] [PubMed] [Google Scholar]
- 5.Franklin BS, Bossaller L, De Nardo D, Ratter JM, Stutz A, Engels G, Brenker C, Nordhoff M, Mirandola SR, Al-Amoudi A et al. (2014) The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nat Immunol 15, 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt FI, Lu A, Chen JW, Ruan J, Tang C, Wu H & Ploegh HL (2016) A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly. J Exp Med 213, 771–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liepinsh E, Barbals R, Dahl E, Sharipo A, Staub E & Otting G (2003) The death-domain fold of the ASC PYRIN domain, presenting a basis for PYRIN/PYRIN recognition. J Mol Biol 332, 1155–1163. [DOI] [PubMed] [Google Scholar]
- 8.de Alba E (2009) Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC). J Biol Chem 284, 32932–32941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sborgi L, Ravotti F, Dandey VP, Dick MS, Mazur A, Reckel S, Chami M, Scherer S, Huber M, Bockmann A et al. (2015) Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy. Proc Natl Acad Sci USA 112, 13237–13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H & Egelman EH (2014) Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahillioglu AC, Sumbul F, Ozoren N & Haliloglu T (2014) Structural and dynamics aspects of ASC speck assembly. Structure 22, 1722–1734. [DOI] [PubMed] [Google Scholar]
- 12.Proell M, Gerlic M, Mace PD, Reed JC & Riedl SJ (2013) The CARD plays a critical role in ASC foci formation and inflammasome signalling. Biochem J 449, 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Li Y, Cao X, Jin X & Jin T (2016) Pattern recognition receptors in zebrafish provide functional and evolutionary insight into innate immune signaling pathways. Cell Mol Immunol 14, 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Compan V, Baroja-Mazo A, Lopez-Castejon G, Gomez AI, Martinez CM, Angosto D, Montero MT, Herranz AS, Bazan E, Reimers D et al. (2012) Cell volume regulation modulates NLRP3 inflammasome activation. Immunity 37, 487–500. [DOI] [PubMed] [Google Scholar]
- 15.Angosto D, Lopez-Castejon G, Lopez-Munoz A, Sepulcre MP, Arizcun M, Meseguer J & Mulero V (2012) Evolution of inflammasome functions in vertebrates: Inflammasome and caspase-1 trigger fish macrophage cell death but are dispensable for the processing of IL-1beta. Innate immunity 18, 815–824. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Castejon G, Sepulcre MP, Mulero I, Pelegrin P, Meseguer J & Mulero V (2008) Molecular and functional characterization of gilthead seabream Sparus aurata caspase-1: the first identification of an inflammatory caspase in fish. Mol Immunol 45, 49–57. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Castejon G, Young MT, Meseguer J, Surprenant A & Mulero V (2007) Characterization of ATP-gated P2X7 receptors in fish provides new insights into the mechanism of release of the leaderless cytokine interleukin-1 beta. Mol Immunol 44, 1286–1299. [DOI] [PubMed] [Google Scholar]
- 18.Xie J & Belosevic M (2016) Functional characterization of apoptosis-associated speck-like protein (ASC) of the goldfish (Carassius auratus L.). Dev Comp Immunol 65, 201–210. [DOI] [PubMed] [Google Scholar]
- 19.Tyrkalska SD, Candel S, Perez-Oliva AB, Valera A, Alcaraz-Perez F, Garcia-Moreno D, Cayuela ML & Mulero V (2017) Identification of an evolutionarily conserved ankyrin domain-containing protein, caiap, which regulates inflammasome-dependent resistance to bacterial infection. Front Immunol 8, 1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuri P, Schieber NL, Thumberger T, Wittbrodt J, Schwab Y & Leptin M (2017) Dynamics of in vivo ASC speck formation. J Cell Biol 216, 2891–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vincent WJ, Freisinger CM, Lam PY, Huttenlocher A & Sauer JD (2016) Macrophages mediate flagellin induced inflammasome activation and host defense in zebrafish. Cell Microbiol 18, 591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tyrkalska SD, Candel S, Angosto D, Gomez-Abellan V, Martin-Sanchez F, Garcia-Moreno D, Zapata-Perez R, Sanchez-Ferrer A, Sepulcre MP, Pelegrin P et al. (2016) Neutrophils mediate Salmonella Typhimurium clearance through the GBP4 inflammasome-dependent production of prostaglandins. Nat Commun 7, 12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vojtech LN, Scharping N, Woodson JC & Hansen JD (2012) Roles of inflammatory caspases during processing of zebrafish interleukin-1beta in Francisella noatunensis infection. Infect Immun 80, 2878–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards N, Schaner P, Diaz A, Stuckey J, Shelden E, Wadhwa A & Gumucio DL (2001) Interaction between pyrin and the apoptotic speck protein (ASC) modulates ASC-induced apoptosis. J Biol Chem 276, 39320–39329. [DOI] [PubMed] [Google Scholar]
- 25.Moriya M, Taniguchi S, Wu P, Liepinsh E, Otting G & Sagara J (2005) Role of charged and hydrophobic residues in the oligomerization of the PYRIN domain of ASC. Biochemistry 44, 575–583. [DOI] [PubMed] [Google Scholar]
- 26.Dick MS, Sborgi L, Ruhl S, Hiller S & Broz P (2016) ASC filament formation serves as a signal amplification mechanism for inflammasomes. Nat Commun 7, 11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waugh DS (2016) Crystal structures of MBP fusion proteins. Protein Sci 25, 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin T, Chuenchor W, Jiang J, Cheng J, Li Y, Fang K, Huang M, Smith P & Xiao TS (2017) Design of an expression system to enhance MBP-mediated crystallization. Sci Rep 7, 40991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin T, Perry A, Smith P, Jiang J & Xiao TS (2013) Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J Biol Chem 288, 13225–13235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krissinel E & Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372, 774–797. [DOI] [PubMed] [Google Scholar]
- 31.Masumoto J, Zhou W, Chen FF, Su F, Kuwada JY, Hidaka E, Katsuyama T, Sagara J, Taniguchi S, Ngo-Hazelett P et al. (2003) Caspy, a zebrafish caspase, activated by ASC oligomerization is required for pharyngeal arch development. J Biol Chem 278, 4268–4276. [DOI] [PubMed] [Google Scholar]
- 32.Datta D, McClendon CL, Jacobson MP & Wells JA (2013) Substrate and inhibitor-induced dimerization and cooperativity in caspase-1 but not caspase-3. J Biol Chem 288, 9971–9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S et al. (2011) Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121. [DOI] [PubMed] [Google Scholar]
- 34.Buchan DW, Minneci F, Nugent TC, Bryson K & Jones DT (2013) Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 41, W349–W357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vajjhala PR, Mirams RE & Hill JM (2012) Multiple binding sites on the pyrin domain of ASC protein allow self-association and interaction with NLRP3 protein. J Biol Chem 287, 41732–41743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin T & Xiao TS (2015) Activation and assembly of the inflammasomes through conserved protein domain families. Apoptosis 20, 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zbyszek Otwinowski WM (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W (2010) Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC & Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chuenchor W, Jin T, Ravilious G & Xiao TS (2013) Structures of pattern recognition receptors reveal molecular mechanisms of autoinhibition, ligand recognition and oligomerization. Curr Opin Immunol 26C, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS & Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H, Guranovic V, Dutta S, Feng Z, Berman HM & Westbrook JD (2004) Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr D Biol Crystallogr 60, 1833–1839. [DOI] [PubMed] [Google Scholar]
- 44.Kumar S, Stecher G & Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- 46.Robert X & Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42, W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pelegrin P, Barroso-Gutierrez C & Surprenant A (2008) P2X7 receptor differentially couples to distinct release pathways for IL-1beta in mouse macrophage. J Immunol 180, 7147–7157. [DOI] [PubMed] [Google Scholar]
- 48.Li L, Yan B, Shi YQ, Zhang WQ & Wen ZL (2012) Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration. J Biol Chem 287, 25353–25360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang QS, Yu F, Huang S, Sun B, Zhang KH, Liu K, Wang ZJ, Xu CY, Wang SS, Yang LF et al. (2015) The macromolecular crystallography beamline of SSRF. Nucl Sci Tech 26, 12–17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.