Abstract
Candida albicans is a normal member of the human microbiota that asymptomatically colonizes healthy individuals, however it is also an opportunistic pathogen that can cause severe infections, especially in immunocompromised individuals. The medical impact of C. albicans depends, in part, on its ability to form biofilms, communities of adhered cells encased in an extracellular matrix. Biofilms can form on both biotic and abiotic surfaces, such as tissues and implanted medical devices. Once formed, biofilms are highly resistant to antifungal agents and the host immune system, and can act as a protected reservoir to seed disseminated infections. Here, we present several in vitro biofilm protocols, including protocols that are optimized for high-throughput screening of mutant libraries and antifungal compounds. We also present protocols to examine specific stages of biofilm development and protocols to evaluate interspecies biofilms that C. albicans forms with interacting microbial partners.
Keywords: Candida albicans, biofilm protocols, biofilm methods, biofilm screens, interspecies biofilms
INTRODUCTION
Candida albicans is a normal member of the human microbiota that asymptomatically colonizes several niches of the body (e.g. skin, ears, nasal cavity, mucosal membranes, gastrointestinal and urogenital tracts) (Douglas, 2003; Gulati et al., 2016; Nobile et al., 2015). C. albicans is also one of the few fungal species that can cause disease in humans, which can range from superficial mucosal and dermal infections to severe disseminated bloodstream and deep-seated tissue infections (Douglas, 2003; Kim et al., 2011; Kullberg et al., 2002; Nobile et al., 2015). These infections are especially serious in immunocompromised individuals (Calderone et al., 2001; Douglas, 2003; Lopez-Ribot, 2005). One important virulence trait of C. albicans is its ability to form biofilms, structured communities of cells that are encased in an extracellular matrix and adhered to a surface (Chandra et al., 2001; Douglas, 2002; Kumamoto, 2002; Lohse et al., 2018; Nobile et al., 2015). C. albicans biofilms can form on both biotic and abiotic surfaces, such as tissues and implanted medical devices, are highly resistant to physical and chemical perturbations, and serve as protected reservoirs that can seed new biofilm infections as well as disseminated (non-biofilm) infections (Douglas, 2002, 2003; Gulati et al., 2016).
C. albicans produces structured biofilms consisting of multiple cell types (spherical yeast-form cells, oval pseudohyphal cells, and cylindrical hyphal cells) (Douglas, 2003; Gulati et al., 2016). C. albicans biofilm formation proceeds through four distinct stages: 1) adherence, where yeast-form cells attach to a surface to seed a biofilm; 2) initiation, where the adhered cells proliferate on the surface to form an anchoring basal layer; 3) maturation, where cells filament and continue to proliferate, leading to a several hundred micron thick biofilm with layers of intercalating hyphae, pseudohyphae and yeast-form cells encased in an extracellular matrix; and 4) dispersion, where yeast-form cells are released from the biofilm to seed new sites (Baillie et al., 1999; Chandra et al., 2001; Gulati et al., 2016; Lohse et al., 2018; Nobile et al., 2015; Uppuluri, Chaturvedi, et al., 2010).
To study biofilm formation in the lab, typical in vitro biofilm assays involve an adherence step where cells first adhere to a solid surface, a wash step to remove non- and weakly-adhered cells, and a maturation step where the adhered cells develop into the biofilm. The final step of the assay entails some sort of measurement of the resulting biofilm (e.g. optical density measurements using a plate reader or microscopic measurements using a confocal scanning laser microscope). For the majority of in vitro C. albicans biofilm assays, the biofilm is exposed to either shaking conditions (using a shaking incubator) or to continuous flow across the biofilm surface (using a microfluidic device) throughout the adherence and maturation steps (Lohse et al., 2017; Tournu et al., 2012). Specific in vitro biofilm assays vary in terms of how the growth of the biofilm is evaluated, such as by dry weight (Hawser et al., 1994; Nobile, Andes, et al., 2006; Nobile et al., 2012; Nobile, Nett, et al., 2006), optical density (Fox et al., 2015; Lohse et al., 2017; Nobile et al., 2014; Uppuluri, Pierce, et al., 2010; Winter et al., 2016), cell viability (Nailis et al., 2010), or direct observations like confocal scanning laser microscopy (Nobile et al., 2012; Nobile et al., 2005). Other variables include the material the biofilm grows on, such as treated or untreated polystyrene plates (Krom et al., 2016; Lohse et al., 2017) or silicone squares (Nobile, Andes, et al., 2006; Nobile et al., 2012; Nobile, Nett, et al., 2006), and the specific stage of biofilm development being observed, such as adherence (Finkel et al., 2012; Lohse et al., 2017; Winter et al., 2016) or dispersion (Lohse et al., 2017; Nobile et al., 2014; Uppuluri, Pierce, et al., 2010). These in vitro biofilm assays can also be used to assess the biofilms formed by different strains, specific mutants of interest (Finkel et al., 2012; Fox et al., 2015; Nobile, Andes, et al., 2006; Nobile et al., 2012; Nobile et al., 2005; Nobile, Nett, et al., 2006; Nobile et al., 2009; Norice et al., 2007; Richard et al., 2005), or upon exposure to antifungal compounds (LaFleur et al., 2011; Lafleur et al., 2013; Pierce et al., 2015; Pierce et al., 2014).
Here we present several validated and commonly-used in vitro biofilm protocols designed to investigate different aspects of C. albicans biofilm formation, each with their individual trade-offs in terms of information generated, throughput, and infrastructure requirements (Figure 1 and Table 1). On the high-throughput end of the spectrum, we present an optical density-based biofilm formation assay using 96- or 384-well microtiter plates that allows for rapid high-throughput screening of large deletion libraries and testing of putative antifungal compounds. We present several variations of this assay, each designed to investigate different aspects of biofilm formation (Figure 1). We also present protocols that allow for the enumeration of live/dead cells within a biofilm, the measurement of biofilm biomass, and the collection of biofilm conditioned media for use in downstream proteomic assays. Additional protocols presented include the formation of sexual biofilms, which are biofilms made by mating-competent cells in response to mating pheromones, and the formation of mixed-species biofilms. Finally, we also present protocols for visualizing C. albicans biofilms using a confocal microscope that allows for assessment of biofilm architecture, and a customizable microfluidic assay that allows for real-time visualization of biofilm formation over time in host-mimicking conditions. The collection of in vitro protocols outlined below are protocols that we have vetted and recommend for the assessment of specific aspects of C. albicans biofilm formation.
Figure 1.
Overview of the steps of a typical in vitro biofilm assay and its relationship to the protocols presented in this chapter. A typical in vitro biofilm assay involves an adherence step, where cells first adhere to a solid surface, a wash step to remove non- and weakly-adhered cells, and a maturation step where the adhered cells develop into the mature biofilm.
Table 1.
Protocols presented in this chapter.
| Assay | Technique Number | Throughput | Specialized Equipment Required* | Result Format |
|---|---|---|---|---|
| Culturing | BP 1 | N/A | N/A | |
| Standard Optical Density | BP 2 | High | Plate Reader | OD Measurement |
| XTT Reduction | AP 1 | High | Plate Reader | OD Measurement |
| Crystal Violet | AP 2 | High | Plate Reader | OD Measurement |
| Inhibition Optical Density | AP 3 | High | Plate Reader | OD Measurement |
| Disruption Optical Density | AP 4 | High | Plate Reader | OD Measurement |
| Dispersal Optical Density | BP 3 | High | Plate Reader | OD Measurement |
| Co-culturing and Analyzing with Standard Optical Density | BP 4 | High | Plate Reader | OD Measurement |
| Colony-Forming Units | AP 5 | Medium | Colonies on Plate | |
| Silicone Square | BP 5 | Medium | Depends on Usage | |
| Cell Adhesion | BP 6 | Medium | Colonies on Plate | |
| Dry Weight | BP 7 | Low | Analytical Balance | Weight of Biofilm |
| Cell Enumeration | BP 8 | Medium | Colonies on Plate | |
| Cell Enumeration Using Florescence Microscopy | AP 6 | Medium | Fluorescent Microscope | Image |
| Pheromone-Stimulated | BP 9 | High | Plate Reader | OD Measurement |
| Microfluidics | BP 10 | Medium | BioFlux EZ1000 | Image / Movie |
| Confocal Imaging | BP 11 | Low | Confocal Microscope | Image |
| Confocal Imaging of Mixed-Species Biofilms | AP 7 | Low | Confocal Microscope | Image |
| Conditioned Media Harvesting | BP 12 | Low | Depends on Usage |
Microplate shaking incubators are required for all C. albicans biofilm assays.
BASIC PROTOCOL 1 - Culturing
In most cases, C. albicans strains are grown in yeast extract peptone dextrose (YEPD; also abbreviated YPD) media at 30°C prior to biofilm assays as this condition has worked for a wide range of C. albicans clinical isolates and deletion strains as well as for other Candida species (e.g. Candida tropicalis and Candida parapsilosis). Alternative media may be substituted for YEPD as needed, although various aspects of the protocol (e.g. duration of growth) may need to be re-optimized as a result. Likewise, strains with severe growth defects may require longer growth periods. This protocol has been optimized for a modest number of strains (i.e. one to dozens at a time), higher throughput options (e.g. overnights grown in deep-well 96-well plates) may be preferable if working with larger numbers of strains at the same time.
Materials
C. albicans strain(s) of interest stored in glycerol
YEPD plates and liquid media (see recipe)
20 ml test tubes and/or 250 ml flasks
Roller drum for test tubes and/or shaker for flasks in a 30°C incubator
-
Cuvettes and spectrophotometer or transparent 96-well plate and OD600-capable plate reader
-
1
Streak C. albicans strains onto YEPD plates from glycerol stocks 2–5 days in advance of assay. Incubate plates at 30°C.
Do not use colonies that are more than 7 days old or store plates at 4°C as C. albicans can acquire aneuploidies under these conditions. Likewise, new plates should be streaked from glycerol stocks rather than by re-streaking cells from an existing plate.
-
1
-
For Small Numbers of Strains/Large Volumes
-
2a
Inoculate a single colony in 25 ml YEPD media in a 250 ml flask from a 2–5 day-old plate. Grow at 30°C with shaking overnight.
-
2a
-
For Large Numbers of Strains/Small Volume
-
2b
Inoculate a single colony in 4 ml YEPD media in a 20 ml test tube from a 2–5 day-old plate. Grow at 30°C with shaking or on a roller drum overnight.
-
3
After 16–18 hr, dilute overnight culture 1:20 or 1:40 and measure OD600 using cuvettes and a spectrophotometer or 96-well plates and a plate reader. Dilute strains as indicated by the specific protocol being used.
If assays will be set up more than 18 hr after overnights were started, remove overnights from the shaker or roller drum in the morning and allow to sit at room temperature until time of use (cultures should not sit for more than 4 hr at room temperature). Vortex the cultures immediately before use. However, if possible, it is better to time the experiment such that the strains will be used prior to 18 hr of overnight growth. Depending on the assay, it may not be necessary to determine the density of the culture(s) prior to use.
-
2b
BASIC PROTOCOL 2 - Standard Optical Density Assay
The Standard Optical Density Assay offers a quick, relatively high-throughput way to screen large numbers of mutant strains for defects in biofilm formation with minimal equipment requirements (Fox et al., 2015; Lohse et al., 2017). The basic version of this assay quantifies biofilm formation based solely on the optical density of the biofilm rather than measurement of cell viability (e.g. metabolic reduction of the tetrazolium salt reagent XTT or the uptake of dyes, such as crystal violet). This assay avoids the extra washing and dye addition steps needed for the XTT Reduction or Crystal Violet assays while avoiding issues with physical disruption of the biofilms, incomplete reagent penetrance, or confounding issues related to the presence of metabolically inactive cells.
Variants of this assay can be used to determine the ability of compounds to prevent the formation of a biofilm or to disrupt an established biofilm. We typically use RPMI-1640 medium for this assay, however other media (e.g. Spider medium, which results in thinner biofilms, and thus can reveal subtler biofilm defects) can also be used. Although this protocol can be modified to grow biofilms on 6-well and 12-well plates, using 4 ml and 2 ml of media per well, respectively, we recommend using 96-well or 384-well formats for high-throughput assays. We recommend the use of six wells per strain or condition in 96-well plate format and eight wells per strain or condition in 384-well plate format, allowing for the evaluation of 16 conditions per 96-well plate and 48 conditions per 384-well plate (including blanks and controls). This protocol has been optimized for use with C. albicans, however it can be adapted for use with other microbial species. When optimizing this protocol for use with other species, changes to the media, temperature, and inoculation amounts are common conditions to vary. Deep well 96-well or 384-well plates may be helpful for pre-aliquoting compound/media mixtures, especially when performing the alternative versions of this assay.
Materials
Transparent, sterile, flat-bottomed, non-tissue culture treated 96-well (BD Falcon 351172) or 384-well (Thermo 242765) microtiter plates
D-PBS (calcium and magnesium salt free), sterile filtered
RPMI-1640 media (with L-Glutamine and MOPS, without sodium bicarbonate, pH 7.0) (see recipe)
Breathe-Easy® sealing membranes (Diversified Biotech BEM-1)
Sterile reagent reservoirs (USA Scientific 2321-2530)
Multi-channel pipettes suitable for 96- or 384-well format
Barrier sterile low retention filter tips 200 μl (GeneMate P-1237-200) for 96-well or for 384-well format (CAPP 5030006C)
Deep Well 96-well (Eppendorf 951031909) or 384-well plates (Axygen P-384-240SQ-C-S)
ELMI DTS-4 shakers (or equivalent shaking incubators capable of holding 96- or 384-well plates, shaking at 200–350 rpm, and holding a temperature of 37°C)
Plate reader with optical density capabilities (600 nm) compatible with 96- or 384-well plate formats (e.g. Tecan Infinite M1000 Pro or BioTek Epoch 2)
-
Vacuum aspirator setup to which 200 μl pipette tips can be attached
-
Following cell density determination for overnight cultures, add cells to wells at a final OD600 = 0.5 (or equivalent to ~1x107 cells/ml) in 200 μl for 96-well plate assays or 1 μl of overnight culture in 90 μl (which gives a final OD600 = 0.15, or equivalent to ~2x106 cells/ml) for 384-well plate assays using RPMI-1640 media.
If starting a large number of wells with the same strain and media, consider diluting cells into media at the desired starting density and pipetting this mixture into wells from a sterile reservoir. -
Seal plates with Breathe-Easy® sealing membranes.
Sealing the plates reduces evaporation and prevents cross-contamination between wells. Shake plate at 37°C for 90 min at 250 rpm (96-well) or 350 rpm (384-well) in an ELMI incubator (or equivalent).
-
Remove membrane and aspirate media.
Change pipette tips on aspirator between wells of different strains and/or conditions. Take care not to scrape the bottom of the well with the tip when aspirating. Wash wells with 200 μl (96-well) or 50 μl (384-well) PBS. Aspirate PBS.
Add 200 μl (96-well) or 90 μl (384-well) of fresh media to each well.
Reseal plate with a new sealing membrane and shake at 37°C for 24 hr at 250 rpm (96-well) or 350 rpm (384-well).
-
Remove membrane and aspirate media from wells (Figure 2).
Use clean pipette tips on the aspirator between wells of different strains and/or conditions. Take care not to scrape the bottom of the well with the tip when aspirating. Note any wells where biofilms detached from the surface during aspiration so that they can be excluded from data analysis. -
Measure OD600 on a 96- or 384-well compatible plate reader (e.g. Tecan Infinite M1000 Pro or BioTek Epoch 2).
The number of reads per well may vary based on the instrument. We recommend obtaining the average density of reads at five independent locations in each well in a 96-well plate or one read from the center of a 384-well plate. Be sure that the wells are still moist for the reading. Dry wells give inaccurate readings. Normalize data by subtracting the OD600 reading of an average blank well (containing media alone) from each experimental and control well. The blank-subtracted OD600 value of each experimental well (normally eight per condition in the 384-well format and six per condition in the 96-well format) is then normalized to the mean blank-subtracted OD600 for the relevant control wells.
Calculate the mean, standard deviation and statistical analyses (normally Student’s unpaired two-tailed t-test assuming unequal variance) for each normalized data set.
-
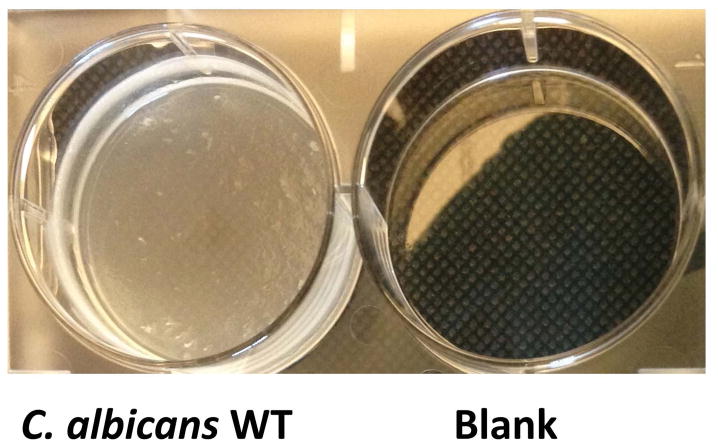
Figure 2.
Typical C. albicans wild-type (WT) biofilm and negative control for the Standard Optical Density Assay in a 6-well plate format (modification of Basic Protocol 2). Shown is a 24 hr biofilm grown in Spider media; the media was aspirated immediately prior to visualization. Typical WT (left) and blank negative control (right) wells are shown. Placing the plate on a textured black background is recommended for the visualization of biofilms.
ALTERNATE PROTOCOL 1 - XTT Reduction Assay
The 2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl)-2H-Tetrazolium-5-Carboxanilide (XTT) assay is a colorimetric assay that detects metabolic activity by measuring the reduction of the tetrazolium salt reagent XTT; alternative versions of this assay use the tetrazolium salt reagent MTT (Krom et al., 2009; Krom et al., 2007; Nett et al., 2011; Ramage et al., 2001). This assay can be performed at the end of the Standard Optical Density Assay (Basic Protocol 2) and be combined with the Inhibition and Disruption Optical Density Assays (Alternate Protocols 3 and 4). One or more variables may need to be optimized depending on the strains or species being used (e.g. exposure times, XTT concentrations). Note that signal may not scale linearly between different strains and/or species, as such care should be taken when making these comparisons (Kuhn et al., 2003). A preliminary time course assay (e.g. reading optical densities every five min for 1 hr after step 5 of this procedure) may be helpful to determine the exposure time best suited to a given strain.
Materials
Materials for Basic Protocol 2
0.5 mg/ml XTT in PBS (XTT Sodium Salt, Sigma X4251)
0.32 mg/ml PMS in water (Phenazine methosulfate (PMS), Sigma P9625)
-
Plate reader with optical density capabilities (492 nm) compatible with 96- or 384-well plate formats (e.g. Tecan Infinite M1000 Pro or BioTek Epoch 2)
Perform steps 1–7 of Basic Protocol 2.
Roughly 15–30 min before the end of the 24 hr growth step, prepare fresh XTT and PMS solutions. Centrifuge before use to remove any insoluble materials and transfer carefully to a new tube. Protect from light.
Mix XTT and PMS at a 9:1 XTT:PMS ratio. Protect solution from light.
-
Remove membrane from plate, aspirate media from wells.
Consider measuring the optical density of the wells, as per step 9 of Basic Protocol 2, as a second form of measurement before proceeding to step 5 of this protocol. -
Quickly add 100 μl (96 well format) or 50 μl (384-well format) of XTT:PMS mixture to wells while taking care to avoid disrupting the biofilm.
Some protocols include one or more PBS washes prior to addition of the XTT:PMS solution. Based on our experiences, omitting this wash step does not significantly affect results and reduces the likelihood of physical disruption of the biofilms. Should wash steps be performed, carefully track wells that have been physically disrupted so that they can be omitted from further analyses. This is a colorimetric assay based on the reduction of XTT, as such it is important to minimize the time between adding the XTT:PMS solution to the first and last wells on a given plate as large differences in reaction times between wells may skew results. -
Incubate plate for 30 min at 37°C in the dark.
The incubation time may need to be increased or decreased depending on the strain(s) and conditions. -
Measure OD492 on a 96- or 384-well compatible plate reader (e.g. Tecan Infinite M1000 Pro or BioTek Epoch 2).
Some protocols suggest transferring the liquid to a new plate prior to optical density measurement. Based on our experiences, this transfer does not appear to be necessary.
ALTERNATE PROTOCOL 2 - Crystal Violet Assay
The Crystal Violet Assay is another colorimetric assay that measures the uptake of a dye (crystal violet) by the biofilm. The amount of dye that ends up in the destaining solution serves as a proxy for the number of viable cells. This assay is less commonly used than the XTT Reduction Assay and requires more time and effort than either the XTT Reduction Assay or the Standard Optical Density Assay. The Crystal Violet Assay can be performed at the end of the Standard Optical Density Assay and can be combined with the Inhibition and Disruption Optical Density Assays (Jin et al., 2003).
Materials
Materials for Basic Protocol 2
0.4% aqueous crystal violet solution (Sigma HT90132-1L)
95% Ethanol
-
Plate reader with optical density capabilities (595 nm) compatible with 96- or 384-well plate formats (e.g. Tecan Infinite M1000 Pro or BioTek Epoch 2)
Perform steps 1–7 of Basic Protocol 2.
After 24 hr growth step, remove membrane, and aspirate media from wells.
-
Wash wells twice with 200 μl (96-well format) or 90 μl (384-well format) PBS.
Change pipette tips on aspirator between wells of different strains and/or conditions. Take care not to scrape the bottom of the well with the tip when aspirating or to physically disrupt the biofilm when adding liquid to the wells. Aspirate PBS, and allow wells to air dry for 45 min.
Stain with 110 μl (96-well format) or 50 μl (384-well format) of 0.4% aqueous crystal violet solution for 45 min.
-
Wash wells four times with 200 μl (96-well format) or 90 μl (384-well format) of water.
Change pipette tips on aspirator between wells of different strains and/or conditions. Take care not to scrape the bottom of the well with the tip when aspirating or to physically disrupt the biofilm when adding liquid to the wells. Destain wells for 45 min in 200 μl (96-well format) or 90 μl (384-well format) 95% ethanol.
Transfer 100 μl (96-well format) or 45 μl (384-well format) of the 95% ethanol destaining solution from each well to a new plate.
Measure OD595 on a 96- or 384-well compatible plate reader (e.g. Tecan Infinite M1000 Pro or BioTek Epoch 2).
ALTERNATE PROTOCOL 3 - Inhibition Optical Density Assays
The Inhibition Optical Density Assays modify the Standard Optical Density Assay to look at the ability of compounds to inhibit biofilm formation during the adherence and/or growth steps of the biofilm assay (Lohse et al., 2017). Solutions containing antifungal agents are included during the 90-min adherence step (Adherence Inhibition and Sustained Inhibition Assays) and/or the 24 hr growth step (Developmental Inhibition and Sustained Inhibition Assays). If it is feasible to perform only one version of the inhibition assays, we recommend that the Sustained Inhibition Assay be chosen as it is the most likely to detect an effect. That said, different compounds may have different effects in the three inhibition assays and as such we recommend that all three versions of the assay be performed when practical. Antifungal compounds are typically solubilized in DMSO. When preparing the antifungal solution in media for the experiment, do not allow total DMSO concentration to exceed 2%. Prepare wells with DMSO at the same concentration as used in the assay, but without the compound of interest, to control for the effect of DMSO on the biofilm (this would serve as the control to use for normalization).
Materials
Materials for Basic Protocol 2
Compound(s) to be tested
Solvent(s) (e.g. DMSO (Sigma D2650))
-
Adherence Inhibition Optical Density Assay
-
1
Include compound(s) being tested in media when adding cells to media during step 1 of Basic Protocol 2.
Compound is present for the 90-min adherence step, normal media is present for the 24 hr growth step. -
2
Perform steps 2–11 of Basic Protocol 2.
-
1
-
Developmental Inhibition Optical Density Assay
-
3
Perform steps 1–5 of Basic Protocol 2.
-
4
Include compound(s) being tested in media when adding to wells during step 6 of Basic Protocol 2.
Normal media is present for the 90 min adherence step, the compound is present for the 24 hr growth step. -
5
Perform steps 7–11 of Basic Protocol 2.
-
3
-
Sustained Inhibition Optical Density Assay
-
6
Include compound(s) being tested in media when adding cells to media during step 1 of Basic Protocol 2.
Compound is present for the 90 min adherence step and the 24 hr growth step. -
7
Perform steps 2–5 of Basic Protocol 2.
-
8
Include compound(s) being tested in media when adding to wells during step 6 of Basic Protocol 2.
-
9
Perform steps 7–11 of Basic Protocol 2.
-
6
ALTERNATE PROTOCOL 4 - Disruption Optical Density Assay
The Disruption Optical Density Assay modifies the Standard Optical Density Assay to look at the ability of compounds of interest to disrupt an established, mature biofilm (Lohse et al., 2017). Although this protocol involves disrupting a 24 hr-old biofilm by exposing it to a compound for 24 hr, the age of the biofilm and the length of exposure to compounds can be modified as needed. This assay can be performed in 96- and 384-well formats, and we recommend the 384-well format whenever possible due to the reduced instances of physical (as opposed to chemical) disruption of biofilms during the disruption step. This protocol involves the removal of the existing media from the biofilms and the addition of fresh media containing the compound(s) being tested. When screening large compound libraries, it may be worth considering adding the compound(s) directly to the existing media rather than removing the existing media and adding fresh media with the compound(s). Antifungal compounds are typically solubilized in DMSO. When preparing the antifungal solution in media for the experiment, do not allow the final DMSO concentration to exceed 2%. Prepare wells with DMSO, at the same concentration as used in the assay but without the compound of interest to control for the effect of DMSO on the biofilm (this would serve as the control to use for normalization).
Materials
Materials for Basic Protocol 2
Compound(s) to be tested
-
Solvent(s) (e.g. DMSO (Sigma D2650))
-
Perform steps 1 through 7 of Basic Protocol 2.
We normally use 8 wells per compound for this assay. Since the same strain is normally used for all of the compounds being tested, the density of the overnight culture is not determined and instead 1 μl of overnight culture (equivalent to ~2x106 cells/ml) is added to the media in each well in the 384-well version of this assay. -
After the 24 hr growth step, remove the membrane and carefully aspirate the media in groups of 6 to 12 wells.
Change pipette tips on aspirator between wells of different strains and/or conditions. Aspirate by slowly lowering tip into well until all liquid has been removed, take care not to scrape the bottom of the well with the tip when aspirating. -
Add 200 μl (96-well) or 90 μl (384-well) of media containing the compound of interest to the wells. Add media slowly to the side of the well opposite the side from which media was aspirated (Figure 3).
Add media slowly to reduce physical disruption of the biofilm, especially in 96-well format. Aspirate and add back media to one group of 6–12 wells at a time in order to avoid exposing biofilms to air and/or desiccation for extended periods of time. Note any wells where the biofilm was disrupted during the aspiration/addition process. Reseal plate and shake at 37°C for an additional 24 hr at 250 rpm (96-well) or 350 rpm (384-well).
After 24 hr, read plate and analyze as per steps 8–11 of Basic Protocol 2.
-
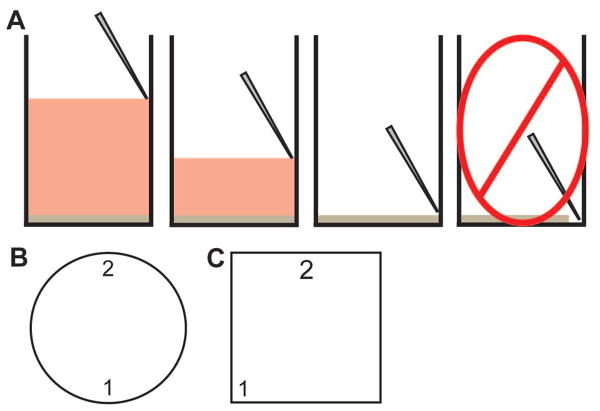
Figure 3.
Standard aspiration techniques and locations in microtiter plates. (A) When aspirating, place the tip against the wall of the well above the surface of the liquid (far left). Slowly lower the tip, taking care to keep it against the well wall (middle left). Stop aspirating when all liquid is gone, which should occur before the tip reaches the bottom of the well and is just above the biofilm (middle right). Do not lower the tip all the way to the bottom of the well as this will physically disrupt any biofilm present (far right). (B–C) Locations to use when aspirating and adding liquid to a well from a 96-well (B) and 384-well (C) microtiter plate. When aspirating at position “1”, add liquid back at position “2”. We normally aspirate from one of the corners of a square well of a 384-well microtiter plate.
BASIC PROTOCOL 3 – Dispersal Optical Density Assays
The Dispersal Optical Density Assays modify the Standard Optical Density Assay to measure the cells that are dispersed during biofilm growth (Lohse et al., 2017; Nobile et al., 2014). These assays utilize optical density to measure the dispersed cells at 24, 48, and 60 hr time points. There are two assays that can be used to assess cell dispersal during biofilm formation, the Sustained Dispersal Assay and the Standard Dispersal Assay. The Sustained Dispersal Assay measurements are obtained while the biofilm grows in the original media with no additional media added, requiring a separate set of samples for each time point. In the Standard Dispersal Assay, in contrast, fresh media is added back to the biofilm after each measurement. These dispersal assays can be customized to study biofilm dispersal differences between different C. albicans strains or to measure the effect of a compound on dispersal during biofilm formation. The assays can be performed in 96- and 384-well plate formats. We recommend the 96-well format whenever possible due to the ease of visually observing dispersed cells floating above the biofilm in the wells. Although slightly more involved than the Standard Optical Density Assay from which it is derived, it is still possible to screen large numbers of mutant strains or compound libraries for defects in biofilm dispersal using these assays. We recommend six wells per strain or condition in 96-well plate format; this allows for evaluation of 16 conditions per 96-well plate (including blanks and controls).
Materials
Materials for Basic Protocol 2
-
Sustained Dispersal Optical Density Assay
-
Perform steps 1 through 7 of Basic Protocol 2.
A separate set of replicates is required for each time point, as once the dispersed cells are measured, media cannot be added back to the well containing the biofilm. -
After the 24 hr growth step, remove the membrane and carefully remove the media with the dispersed cells from the first set of replicates and add this media to a clean 96- or 384-well plate.
Take care not to scrape the bottom of the well or to disturb the biofilm on the walls of the well with the tip during this process. Homogenize the removed media using a multichannel pipette.
Measure OD600 of the removed media (containing dispersed cells) and measure OD600 of the biofilm, from which the media was removed, on a 96- or 384-well compatible plate reader (e.g. Tecan Infinite M1000 Pro or BioTek Epoch 2).
Repeat steps 2–4 for the second set of replicates at the 48 hr time point, followed by the third set of replicates at the 60 hr time point.
Normalize data at each time point by subtracting the OD600 reading of an average blank well (containing media alone) from each experimental and control well from that time point.
Divide the blank-subtracted OD600 value for the media from each experimental well by the blank-subtracted OD600 value of the biofilm of the correlated well to obtain normalized OD600 values. This provides a measurement of dispersed cells per biofilm, and takes into account any variations in biofilms between wells.
Divide the normalized OD600 value for each of the experimental conditions by the normalized OD600 value for the control wells.
Calculate the mean, standard deviation and statistical analyses (typically use a Student’s unpaired two-tailed t-test assuming unequal variance) for each normalized data set.
-
-
Standard Dispersal Optical Density Assay
Perform steps 1 through 7 of Basic Protocol 2.
-
After the 24 hr growth step, remove the membrane and carefully remove the media with the dispersed cells from the wells with biofilms and add this media to a clean 96- or 384-well plate.
Take care not to scrape the bottom of the well or disturb the biofilm on the walls of the well with the tip during this process. -
Add 200 μl (96-well) or 90 μl (384-well) of fresh media to the original wells. Add media slowly to the side of the well opposite the side from which media was removed (Figure 3).
Add media slowly to reduce physical disruption of the biofilm, especially in 96-well format. Reseal plate and shake at 37°C at 250 rpm (96-well) or 350 rpm (384-well).
Homogenize the removed media using a multichannel pipette.
Measure OD600 of the removed media (containing dispersed cells) on a 96- or 384-well compatible plate reader (e.g. Tecan Infinite M1000 Pro or BioTek Epoch 2).
After an additional 24 hr, repeat steps 2–6.
After an additional 12 hr, remove the membrane and carefully remove the media with the dispersed cells from the wells with biofilms and add the media to a clean 96- or 384-well plate.
Homogenize the removed media using a multichannel pipette.
Measure OD600 of the removed media (containing dispersed cells) and measure the OD600 of the biofilm well from which the media was removed using a 96- or 384-well compatible plate reader (e.g. Tecan Infinite M1000 Pro or BioTek Epoch 2).
Normalize data at each time point by subtracting the OD600 reading of an average blank well (containing media alone) from each experimental and control well from that time point.
Normalize the blank-subtracted OD600 value for the media from each experimental well to the blank-subtracted OD600 value of the biofilm of the correlated well by dividing the former by the latter. This provides a measurement of dispersed cells per biofilm at a given time point, and takes into account any variations in biofilms between wells.
Calculate the standard deviation and statistical analyses (typically use a Student’s unpaired two-tailed t-test assuming unequal variance) for each normalized data set.
BASIC PROTOCOL 4 - Co-culturing and Analyzing Candida Mixed-Species Biofilms Using the Standard Optical Density Assay
This protocol is optimized for culturing C. albicans mixed-species biofilms with Escherichia coli, however, the culture conditions indicated can be applied or adapted for growing other microbes together with C. albicans. This protocol is a refined version of the protocol reported by Fox et al. (Fox et al., 2014), for co-culturing C. albicans with different anaerobic and aerobic bacteria. In the following protocol, E. coli was chosen for co-culture with C. albicans as both species are already known to interact in the context of a biofilm (Fox et al., 2014), are prevalent in the human gut as well as in other parts of the body, and can be easily cultured in the laboratory. The 6- and 12-well polystyrene plate biofilm assay described in this protocol can be modified to use 96- and 384-well plate formats for higher throughput screening. Likewise, the protocol can be modified to accommodate the requirements of many of the other protocols derived from the Standard Optical Density Assay. We note that this protocol does not assess the relative abundance of each of the two species (in terms of cell numbers) within the mature biofilm or the viability of either species within that biofilm. If these determinations are of interest, we recommend combining this assay with one or both versions of the Cell Enumeration Assay.
Materials
Materials for Basic Protocol 2
E. coli or other microbial strain(s) of interest
Luria-Bertani (LB) medium and plates for E. coli growth or appropriate media for other microbes under study
Bacto heart infusion (BHI) medium (BD B237500), supplemented with 5% Fetal bovine serum (FBS) (BHI-FBS)
6-well or 12-well microtiter plates, non-tissue culture-treated (Falcon 351146 and 351143)
-
5 ml serological pipets (VWR 89130-896)
-
1
Streak E. coli strains onto LB plates from glycerol stocks 2–5 days in advance of assay. Incubate plates at 37°C.
Plates containing E. coli can be stored at 4°C for up to one week.
-
1
-
For Small Numbers of Strains/Large Volumes
-
2a
Inoculate a single E. coli colony in 25 ml LB media in a 250 ml flask from a 2–5 day old plate. Grow at 37°C with shaking overnight.
-
2a
-
For Large Numbers of Strains/Small Volumes
-
2b
Inoculate a single E. coli colony in 4 ml LB media in a 20-ml test tube from a 2–5 day old plate. Grow at 37°C with shaking or using a roller drum overnight.
-
3
After 12–16 hr, dilute overnight culture 1:20 or 1:40 and measure OD600 using cuvettes and a spectrophotometer or 96-well plates and a plate reader.
If assays will be set up more than 16 hr after overnights were started, remove overnights from the shaker or roller drum in the morning and allow to sit at room temperature until time of use (cultures should not sit for more than 2 hr at room temperature). Vortex the cultures immediately before use. However, if possible, it is better to time the experiment such that the strains will be used prior to 16 hr of overnight growth. -
4
For culturing C. albicans strains, follow Basic Protocol 1.
-
5
Following cell density determination for overnight cultures, add cells to wells at a final OD600 = 0.5 (C. albicans) or OD600 = 0.0125 (E. coli) (equivalent to ~1x107 cells/ml for each species) in 4 ml (for 6-well plate assays) or 2 ml (for 12-well plate assays) in BHI-FBS media.
This protocol seeds the same number of bacterial and fungal cells together at the start of the experiment in a 1:1 ratio, however this ratio can be altered as needed. When working with a species for the first time, we recommend the use of a serial dilution and colony forming unit (CFU) count to correlate OD600 values with an approximate number of cells/ml so that the 1:1 ratio can be achieved (see Alternate Protocol 5). It is also recommended that each species be grown as biofilms individually in BHI-FBS as a single-species biofilm control to compare to the mixed-species biofilms. -
6
Shake plate at 37°C for 90 min at 200 rpm (6-well plate) or 250 rpm (12-well plate) in an ELMI incubator (or equivalent).
-
7
Aspirate media and wash wells with 2 ml (12-well) or 4 ml (6-well) PBS.
Change pipette tips on aspirator between wells of different strains and/or conditions. Take care not to scrape the bottom of the well with the tip when aspirating. -
8
Aspirate PBS and add 2 ml (12-well) or 4 ml (6-well) of fresh media to each well.
-
9
Shake the plate at 37°C for 24 hr at 200 rpm (6-well) or 250 rpm (12-well).
-
10
Aspirate media from wells.
Change pipette tips on aspirator between wells of different strains and/or conditions. Take care not to scrape the bottom of the well with the tip when aspirating. Note any wells where biofilms detached from the surface during aspiration so that they can be excluded from data analysis. -
11
Measure OD600 on a 6- or 12-well compatible plate reader (e.g. Tecan Infinite M1000 Pro or BioTek Epoch 2).
The number of reads per well may vary based on the instrument. We recommend taking the average density of five reads in each section of a five by five grid (i.e. twenty-five independent locations) in each well in a 6-well plate or 12-well plate. Be sure that the wells are still moist for the reading as dry wells give inaccurate readings. -
12
Normalize data by subtracting the OD600 reading of an average blank well (containing media alone) from each experimental and control well. The blank-subtracted OD600 value of each experimental well is then normalized to the mean blank-subtracted OD600 for the relevant control wells.
-
13
Calculate the standard deviation and statistical analyses (typically use a Student’s unpaired two-tailed t-test assuming unequal variance) for each normalized data set.
-
2b
ALTERNATE PROTOCOL 5 - Colony-Forming Units Assay
The starting cell concentration for a biofilm assay as well as the ratio of fungal or bacterial species used to seed mixed-species biofilms significantly affects the nature of the resulting biofilms. As such, accurate quantitation of microbial samples is an essential precursor to growing different species in a biofilm. The Colony-Forming Units (CFU) Assay presented here is a basic method to correlate an unknown OD600 concentration with a specific number of viable cells.
Materials
C. albicans, E. coli, or other microbial strains of interest
YEPD plates (see recipe)
Luria-Bertani (LB) medium and plates for E. coli growth or appropriate media for other microbes under study
D-PBS (calcium and magnesium salt free), sterile filtered
-
1.5 ml microcentrifuge tubes
Culture strains following steps 1 through 3 of Basic Protocol 1 (for fungi) or Basic Protocol 4 (for bacteria) with modifications as appropriate for the species in question.
-
Following cell density determination for overnight cultures, dilute cells to a final OD600 = 0.5 (for fungi) or OD600 = 0.0125 (for bacteria) (roughly equivalent to ~1x107 cells/ml) in 1 ml PBS.
These OD600 values are an approximate estimate for a cell concentration of 1x107 cells/ml to allow for a serial dilution series that will more accurately correlate OD600 with viable cells per ml. -
Perform a series of six sequential 10x dilutions by adding 100 μl of cell- containing solution to 900 μl of PBS. Vortex thoroughly and change pipette tips after each dilution.
The resulting dilution series should range from a high of 1x107 to a low of 1x101 cells/ml. -
Vortex tubes thoroughly and plate 100 μl of tubes 3 through 7 on YEPD plates.
We recommend performing at least two separate dilution series, each based on separate initial OD600 measurements, for each strain. Incubate the plates at 30°C for 24–72 hr, or other conditions as appropriate for the strain(s).
-
Count the colonies on the plate and use the serial dilution factor to estimate the number of cells in the original culture for each replicate in a set.
When counting colonies, the plates should be examined within 24 hr to ensure that colonies can be distinguished before they overgrow if too many colonies were plated. Plates should also be examined after 48–72 hr to allow for the scoring of slower growing strains or species. This set of time points allows for adequate growth while ensuring that colonies are easily distinguished from one another.
BASIC PROTOCOL 5 – Silicone Square Assay
For some protocols, it may be of interest to form C. albicans biofilms on silicone squares rather than (or in addition to) polystyrene plates to test the effects of an alternate surface on biofilm formation (Nobile, Andes, et al., 2006; Nobile et al., 2012; Nobile et al., 2005; Nobile, Nett, et al., 2006). Silicone is a common material used for implanted medical devices, such as catheters, heart pumps, and surgical reconstructive components. Biofilms formed using the Silicone Square Assay can be imaged using confocal microscopy or weighed to determine the dry weight (biomass) of the biofilm formed. For the latter, it is essential to pre-weigh the silicone squares prior to use.
Materials
RPMI-1640 media (with L-glutamine and MOPS, without sodium bicarbonate, pH 7.0) (see recipe) or other medium of interest
Autoclaved silicone squares (1.5 mm x 1.5 mm) cut from medical-grade silicone sheets (Cardiovascular Instrument PR72034-06N, Bentec Medical Inc.)
12-well microtiter plates, non-tissue culture-treated (Falcon 351143)
D-PBS (calcium and magnesium salt free), sterile filtered
Dissecting forceps with a fine tip (VWR 82027-404)
200 proof ethanol (CAS 64-17-5)
-
ELMI DTS-4 shakers (or equivalent shaking incubators capable of holding 12-well plates, shaking at 200 rpm, and holding a temperature of 37°C)
-
Place one autoclaved silicone square per well into a 12-well plate using sterile forceps.
Weigh each silicone square before placing them in the wells if you plan to use them for dry weight assays. Track which square went into each well. Include positive (e.g. wild-type strain) and negative (e.g. known biofilm-defective mutant strain) control wells in addition to a blank (uninoculated) control well. -
Following cell density determination for overnight cultures, add cells to silicone squares at a final OD600 = 0.5 (equivalent to ~1x107 cells/ml) in 2 ml RPMI-1640 media.
Care must be taken to note the direction (facing up) of the silicone square in the well, as this is the surface where the cells will start to adhere. In all the future steps, this side should always face upwards. Shake plate at 37°C for 90 min at 200 rpm in an ELMI incubator (or equivalent).
-
Add 2 ml of PBS to wells in a new 12-well plate. Gently lift the silicone square from the edges using forceps and place in well containing PBS.
Carefully lift the silicone squares by holding just the edges, so as to not disturb the adhered cells. Wash the silicone square in PBS by holding it from the edges using the forceps and by gently agitating the square by moving it up and down within the PBS 2–3 times.
Place the silicone square into the well of a new 12-well plate containing 2 ml of fresh RPMI-1640 media.
-
Shake plate at 37°C for 24 hr at 200 rpm in an ELMI incubator (or equivalent) (Figure 4).
The resulting biofilm (Figure 4) can be visualized using confocal microscopy or used to measure dry weight (biomass) of the biofilm. To measure biofilm dry weights after growth on silicone squares, simply weigh the silicone square after allowing the squares containing the biofilm to dry in an ELMI at 37 °C for 4 hr; no shaking is necessary for this step. Subtract out the weight of the silicone square without the biofilm from the weight of the silicone square containing the biofilm to obtain the biofilm dry weight.
-
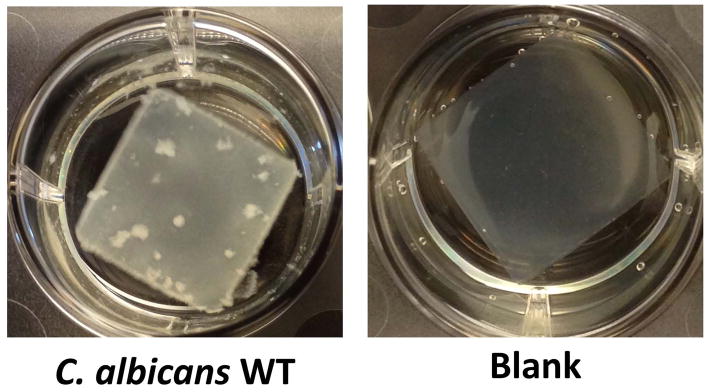
Figure 4.
Typical C. albicans wild-type (WT) biofilm and negative control for the Silicone Square Assay (Basic Protocol 5). The biofilm was allowed to develop on a silicone square for 24 hr in Spider media in a 6-well plate. The biofilm is clearly visible on the surface of the silicone square with WT (left). A blank silicone square is shown as a negative control (right).
BASIC PROTOCOL 6 – Cell Adhesion Assay
Initial cell adhesion to a surface is a crucial step in normal biofilm development and is the first step of biofilm formation. The Cell Adhesion Assay measures the number of cells that adhere to an abiotic surface, such as a microtiter plate, to seed a biofilm. This assay mimics the adherence step of the Standard Optical Density Assay and can be used to measure the effect of a compound(s) on cell adhesion or compare the ability of strains to adhere to a surface (Lohse et al., 2017; Winter et al., 2016). Note that this assay has reduced throughput compared to the Standard Optical Density Assay as it involves serial dilutions and plating to count colony forming units (CFUs) to determine the number of adhered cells. We recommend that this assay be performed in a 96-well plate format, as this format provides an ideal surface area for cell adherence. We recommend the use of four wells per strain or condition in the 96-well plate format, allowing for the evaluation of 24 conditions per 96-well plate (including blanks and controls). The assay can be customized for 6-well and 12-well plate formats, however the serial dilutions should be optimized to correlate with the well surface area of the 6- and 12-well plates.
Materials
Materials for Basic Protocol 2
Compound(s) to be tested
1.5 ml microcentrifuge tubes
-
YEPD plates (see recipe)
Perform steps 1 through 4 of Basic Protocol 2 for the 96-well plate.
-
Wash the wells twice with 200 μl PBS. Aspirate PBS.
This step removes non- and weakly-adhered cells. -
Add 200 μl PBS and vigorously resuspend the adhered cells.
Scrape the bottoms and edges of each well with a pipette tip to dislodge the cells that remain adhered to the surface. Use the pipette to homogenize the suspension. Perform serial dilutions in PBS by setting up two tubes for 10x dilutions for each well to be tested. Add 200 μl of the PBS with resuspended cells to 1800 μl of PBS to obtain a 10x dilution. Vortex the solution thoroughly.
-
Perform a second 10x dilution by adding 100 μl of the above solution to 900 μl of PBS. Vortex the solution thoroughly and plate 100 μl of the solution onto a YEPD plate.
A 1000-fold serial dilution normally provides an optimal number of cells for counting for wild-type conditions. The serial dilution may need to be optimized for different compound(s) and/or strains, depending on their effect on cell adherence. If cell adherence is too high, such that single colonies cannot be effectively counted, perform another 10x dilution. If cell adherence is too low, such that not enough colonies are obtained, plate 100 μl from the first dilution tube. Incubate the YEPD plates at 30°C for 24–72 hr.
-
Count the colonies on each plate and use the serial dilution factor to determine the number of adherent cells for each replicate in a set.
When counting colonies, the plates should be examined within 24 hr to ensure that colonies can be distinguished before they overgrow if too many cells were plated. Plates should also be examined after 48–72 hr to allow for the scoring of slow growing mutant strains. This set of time points allows colonies adequate time to grow while ensuring that they are easily distinguished from one another. Calculate the mean, standard deviation and statistical analyses (typically use a Student’s unpaired two-tailed t-test assuming unequal variance) for each data set.
BASIC PROTOCOL 7 - Dry Weight Assay
This assay measures the biomass or dry weight of C. albicans biofilms grown on the bottom of polystyrene plates in 6-well and 12-well formats (Hawser et al., 1994; Nobile, Andes, et al., 2006; Nobile et al., 2012; Nobile, Nett, et al., 2006). We typically use three replicates per strain or condition. We do not recommend the 96-well or 384-well formats for this assay, as the total mass of biofilm harvested from wells from these plates is much less than for 6- or 12-well plates, and the reduced starting biomass makes it difficult to assess the effects of treatment with compound(s) or to assess differences between distinct strains (e.g. mutant strains of interest). Although this assay is effective in detecting severe biofilm defects, the Dry Weight Assay is less sensitive relative to other more recently developed biofilm assays, such as the Standard Optical Density Assay, and thus is not ideal for detecting minor differences in biofilm formation. The Dry Weight Assay is also notoriously ineffective at detecting enhanced biofilm formation, and should not be used to assess such strains. This assay also has lower throughput than many of the other biofilm assays presented. For these reasons, we recommend the Dry Weight Assay as a secondary assay to further assess candidates identified in other in vitro biofilm assays.
Materials
RPMI-1640 media (with L-Glutamine and MOPS, without sodium bicarbonate, pH 7.0) (see recipe)
6-well or 12-well microtiter plates, non-tissue culture-treated (Falcon 351146 and 351143)
D-PBS (calcium and magnesium salt free), sterile filtered
Mixed Cellulose Esters Membrane (Millipore AAWG02500)
Millipore Filter Device for 25mm discs (Millipore XX10025400) or equivalent
70% ethanol
Vacuum aspirator setup to which 1000 μl tips can be attached
Dissecting forceps with a fine tip (VWR 82027-404)
Analytical scale
-
ELMI DTS-4 shakers (or equivalent shaking incubators capable of holding 6- and 12-well plates, shaking at 200–250 rpm, and holding a temperature of 37°C)
-
Following cell density determination for overnight cultures, add cells to wells at a final OD600= 0.5 (or equivalent to ~1x107 cells/ml) in 2 ml (for 12-well plate assays) or 4 ml (for 6-well plate assays).
Although the protocol listed here uses RPMI-1640 media, other biofilm inducing media can alternatively be used. The authors recommend leaving one well blank (containing media alone with no cells) as a contamination control and having at least three replicates per condition. Shake plate at 37°C for 90 min at 200 rpm (6-well plate) or 250 rpm (12-well plate) in an ELMI incubator (or equivalent).
-
Aspirate media and wash wells with 2 ml (12-well) or 4 ml (6-well) PBS.
Change pipette tips on aspirator between wells of different strains and/or conditions. Take care not to scrape the bottom of the well with the tip when aspirating. Aspirate PBS and add 2 ml (12-well) or 4 ml (6-well) of fresh media to each well.
Shake the plate at 37°C for 24 hr at 200 rpm (6-well) or 250 rpm (12-well).
Remove plate and aspirate media from the wells.
-
Vigorously disrupt the biofilm and resuspend all cells in 2 ml (12-well) or 4 ml (6-well) PBS.
Scrape the bottoms and edges of each well with a pipette tip to dislodge biofilm and cells that remain adhered to the surface. Use the pipette to homogenize the biofilm suspended in PBS. Clean the Millipore filtration device (or equivalent) parts with 70% ethanol, and rinse thoroughly with sterile water.
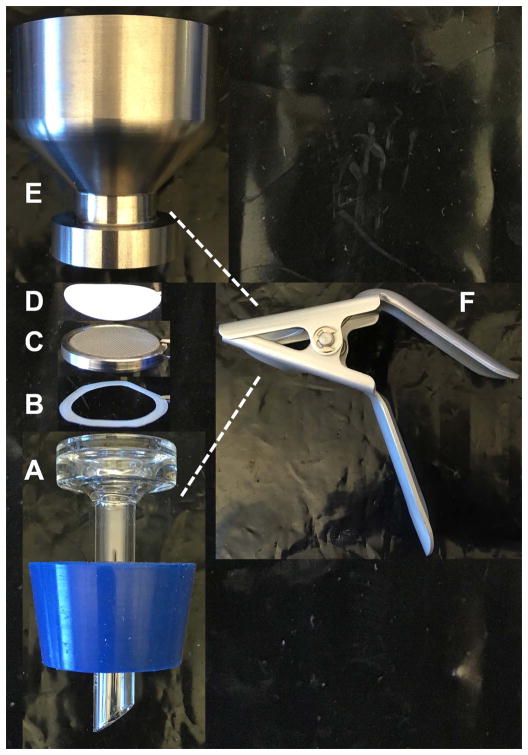
Assemble the filtration device by first placing the plastic ring on the waste flask to maintain a tight seal. Then add the wire filter sieve, a mixed cellulose ester membrane, followed by the funnel (in this order). Lastly, attach the clamp to keep all components together (Figure 5).
Attach a vacuum aspirator setup to the filtration device.
-
Turn the vacuum on and transfer the disrupted biofilm of one well (from step 7) to the funnel to adhere to the membrane. Apply the biofilm solution towards the center of the membrane.
If additional biofilm is left behind in the well, an additional 1 ml of PBS can be added to the well to suspend the remaining biofilm. Then transfer this additional disrupted biofilm to the membrane. Turn off the vacuum, disassemble the filtration device, and carefully remove the wet membrane using forceps. Only use the edges of the membrane, to avoid disturbing the accumulated cells.
-
Place the membrane in a clean well of a 6-well plate.
Use a 6-well plate for this step as the membrane is too large to sit flat in a 12-well plate. -
Repeat steps 8 through 14 for all wells, including the blank control wells.
The filter paper from the blank wells will be used to determine the average weight of the mixed cellulose ester membranes containing any media components. Note that the weights of the individual ester membranes are nearly identical, and thus it is not necessary to pre-weigh each ester membrane prior to use. -
Place the 6-well plates with the cellulose ester membranes in an ELMI at 37 °C for 4 hr; no shaking is necessary for this step.
It is important to allow the membranes to dry thoroughly and uniformly as uneven wetness can lead to inaccurate weight measurements. Weigh the cellulose ester membranes containing biofilm cells individually. Subtract the average of the blank wells from all replicates.
Calculate the mean, standard deviation and statistical analyses (typically use a Student’s unpaired two-tailed t-test assuming unequal variance) for each data set.
-
Figure 5.
Assembly of the vacuum filtration apparatus used for the Dry Weight Assay (Basic Protocol 7). The filtration device is assembled by placing the rubber cork on a flask (A), followed by the plastic O-ring (B), the wire filter sieve (C), and the mixed cellulose ester membrane (D) (in this order from bottom to top). The funnel (E) is then added on top and the clamp (F) is attached to keep all components together. The clamp should cover the top of the glass piece (A) as well as the base of the funnel (E).
BASIC PROTOCOL 8 – Cell Enumeration Assay
This assay determines the viability of cells within a biofilm after exposure to a compound of interest, such as an antifungal agent, to assess if the compound has microbicidal properties against the cells within the biofilm. This protocol involves growing a wild-type biofilm and exposing the biofilm to the compound being tested for 24 hr, although the assay can be modified to use different exposure times based on the needs of the researcher. The addition of Letheen Broth, which enhances biofilm homogenization, avoids the difficulties of obtaining inaccurate colony counts when biofilms are not effectively broken apart, which can occur when biofilms are grown in certain media (e.g. RPMI-1640). We recommend that this assay be performed in 96-well plate formats as this allows for a larger number of live cells present compared to biofilms formed in 384-well plate formats. We recommend four wells per strain or condition in 96-well plate format, allowing for evaluation of 24 conditions per 96-well plate (including blanks and controls). This assay can also be used to look at the relative abundance and viability of the individual species present in a mixed-species biofilm (Fox et al., 2014). When working with mixed-species biofilms, it is sometimes possible to get efficient homogenization without the use of Letheen Broth depending on the media and species used (this should be microscopically verified for each specific case). Should Letheen Broth prove necessary, we note that it is effective at breaking apart both fungal and bacterial biofilms.
Materials
Materials for Basic Protocol 2 and Alternate Protocol 4
Compound(s) to be tested
Solvent(s) (e.g. DMSO (Sigma D2650))
Letheen Broth (Difco 268110)
Sodium thiosulfate (Hach 2267301)
-
1.5 ml Microcentrifuge tubes
Perform steps 1 through 7 of Basic Protocol 2 followed by steps 2–4 of Alternate Protocol 4.
After 24 hr, carefully remove the plate from the shaker and remove the membrane.
-
Carefully aspirate the media in groups of 6–12 wells and gently add 200 μl PBS to wash the biofilms.
At this point, the biofilms are very brittle. To wash the biofilms, add PBS to the wells in a drop-wise manner from one corner of the well to avoid mechanical disruption (Figure 3). Single channel pipettes are recommended over multi-channel pipettes for better control at this step. -
Carefully aspirate the PBS and add 200 μl of fresh PBS to the wells. Vigorously disrupt the biofilm to resuspend the cells.
Scrape the bottoms and edges of each well with a pipette tip to dislodge the biofilm and cells that remain adhered to the surface. Use the pipette to homogenize the biofilm suspended in PBS. Dilute the resuspended biofilm into 2 ml of Letheen Broth supplemented with 0.1% sodium thiosulfate and vigorously vortex to further homogenize the biofilm.
Perform serial dilutions by setting up a series of four tubes for 10x dilutions in PBS for each well. Add 100 μl of the cells resuspended in Letheen Broth and sodium thiosulfate to 900 μl of PBS to obtain a 10x dilution (tube 1). Vortex thoroughly.
Perform a second 10x dilution, by adding 100 μl of tube 1 to 900 μl of PBS (tube 2). Vortex thoroughly.
-
Repeat step 7 twice to obtain tubes 3 and 4 (diluting tube 2 into tube 3 and tube 3 into tube 4). Vortex tube 4 thoroughly and plate 100 μl of the solution on a YEPD plate. There should be one plate per replicate, thus a total of four plates for each strain or condition.
These dilutions have been optimized for a wild-type C. albicans biofilm with no treatment (control). When testing a new compound or strain, it is recommended that 100 μl from tube 3 should also be plated on a separate YEPD plate. This ensures that colonies will be obtained even if there is a higher instance of cell death relative to the wild-type strain. Incubate the plates at 30°C for 24–72 hr.
-
Count the colonies on the plate within 24 hr and between 48–72 hr and use the serial dilution factor to estimate the number of cells in the biofilm for each replicate in a set.
When counting colonies, the plates should be examined within 24 hr to ensure that colonies can be distinguished before they overgrow if too many colonies were plated. Plates should also be examined after 48–72 hr to allow for the scoring of slow growing strains. This set of time points allows for adequate growth while ensuring that colonies are easily distinguished from one another. Calculate the mean, standard deviation and statistical analyses (typically use a Student’s unpaired two-tailed t-test assuming unequal variance) for each data set.
ALTERNATE PROTOCOL 6 – Cell Enumeration Using Florescence Microscopy
The survival of cells in C. albicans biofilms treated with antifungal agents can also be quantified using fluorescent live/dead stains rather than colony counts of serial dilutions. This protocol has been adapted from LaFleur et al. (LaFleur et al., 2006) to work with the Disruption variant of the Standard Optical Density Assay, and requires less time and resources compared to the serial dilution method. Additionally, the visualization step better accounts for any cells that are still adhered to one another, which would be overlooked in the serial dilution version of this protocol. This protocol require access to a microscope with fluorescent capabilities. We recommend that this assay be performed in 96-well plate format and that three wells per strain or condition be tested; this allows for evaluation of 32 conditions per 96-well plate (including blanks and controls).
Materials
Materials for Basic Protocol 2, Basic Protocol 8, and Alternate Protocol 4
Fluorescein diacetate (Sigma-Aldrich, F7378-5G)
Microscope slides (Fisher 12-550-123) or equivalent
Coverslips (Fisher 12-542-B) or equivalent
Fluorescent microscope with ability to detect GFP
Microcentrifuge (Eppendorf miniSpin plus or equivalent)
-
ImageJ Software (NIH)
Perform steps 1 through 7 of Basic Protocol 2.
Prepare RPMI-1640 with the antifungal agent of interest and 100 μg/ml fluorescein diacetate (live/dead stain).
-
After the 24 hr growth step, remove the membrane and carefully aspirate the media in groups of 6–12 wells.
Change pipette tips on aspirator between wells of different strains and/or conditions. Aspirate by slowly lowering tip into well until all liquid has been removed. Take care not to scrape the bottom of the well with the tip when aspirating. -
Add 200 μl of media containing the compound of interest and fluorescein diacetate to the wells. Add media slowly to the side of the well opposite the side from which media was aspirated (Figure 3).
Add media slowly to reduce physical disruption of the biofilm, especially in 96-well plate format. Aspirate and add back media to one group of 6–12 wells at a time in order to avoid exposing biofilms to air and/or desiccation for extended periods of time. Note any wells where the biofilm was disrupted during the aspiration/addition process. Reseal plate and shake at 37°C for an additional 24 hr at 250 rpm.
After 24 hr, carefully remove the plate from the shaker and remove the membrane.
-
Carefully aspirate the media in groups of 6 to 12 wells and gently add 200 μl PBS to wash the biofilms (Figure 3).
At this point, the biofilms are very brittle. To wash the biofilms, add PBS to these wells in a drop-wise manner from one corner of the well to avoid mechanical disruption. Single channel pipettes are recommended over multi-channel pipettes for better control at this step. -
Carefully aspirate the PBS and add 200 μl of fresh PBS to the wells. Vigorously disrupt the biofilm to resuspend the cells.
Scrape the bottoms and edges of each well with a pipette tip to dislodge the biofilm and cells that remain adhered to the surface. Use the pipette to homogenize the biofilm suspended in PBS. If Letheen Broth is needed to break apart the biofilm, dilute the resuspended biofilm into 2 ml of Letheen Broth supplemented with 0.1% sodium thiosulfate and vigorously vortex to further homogenize the biofilm. Otherwise, proceed to step 10.
Vortex vigorously and transfer the disrupted biofilm from each well to a 1.5 ml microcentrifuge tube.
Centrifuge the biofilm-containing tubes in a microcentrifuge for 5 min at 9000xg to separate cells from excess dye in the solution.
Discard the supernatant and resuspend the pellet in 200 μl PBS.
-
Take 1 μl of the resuspended pellet, and place on a clean microscope slide. Place a glass coverslip on top of the sample.
Make one slide per replicate well, for a total of three slides for each strain or condition being tested. Visualize slide using a fluorescence microscope, utilizing the GFP settings.
-
Use the 20X objective to focus on the cells in the biofilm and take an image in both the bright-field and GFP settings (Figure 6).
C. albicans treated with a fluorescent dye can auto-fluoresce, leading to a bright background (e.g. due to cell debris or the biofilm matrix) and bright cells dispersed over the viewing field. To take clear images, adjust the fluorescence excitation wavelength exposure time to reduce background. Dead cells will fluoresce green, while those that do not fluoresce are live cells (Figure 6). -
Overlay the bright-field and GFP images to observe the contrast between the live and dead cells. Take three representative images of each slide.
Capture images with approximately 50–100 cells in the viewing field. Avoid including large clumps of cells in these images as the cells within them are often indistinguishable as separate entities and cannot be counted accurately. Using a cell counting software (ImageJ or equivalent), add the total number of live/dead cells in each captured image. There will be a total of nine images per data set.
Calculate the mean, standard deviation and statistical analyses (typically use a Student’s unpaired two-tailed t-test assuming unequal variance) for each data set.
Figure 6.
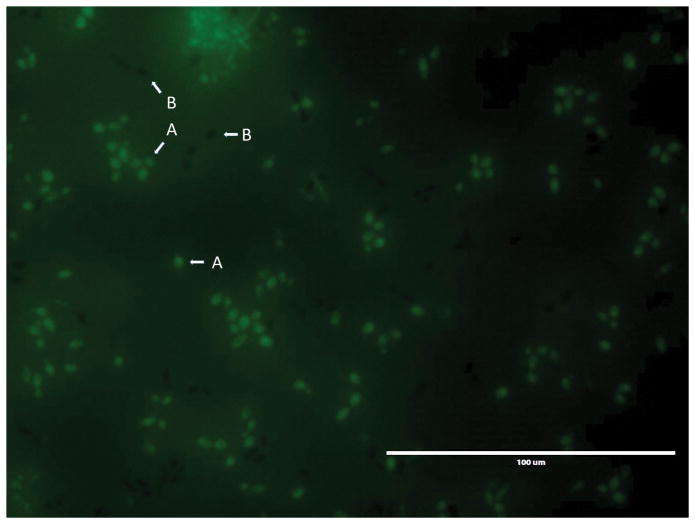
A typical image generated by Cell Enumeration Using Florescence Microscopy (Alternative Protocol 6) using fluorescein staining. The figure depicts both dead (A, green fluorescent) and live (B, dark non-fluorescent) C. albicans cells. Note also the faint background fluorescence which provides contrast to easily visualize the non-fluoresecent live cells. Cells were imaged using standard GFP imaging settings on an EVOS-FL microscope. Scale bar = 100 μm.
BASIC PROTOCOL 9 – Pheromone-Stimulated Assay
Recent work has shown that both mating type and pheromone signaling impact biofilm formation in C. albicans. C. albicans can exist as a, α, or a/α mating types, depending on the mating-type like (MTL) loci present. With the exception of this protocol, the biofilm assays described in this chapter pertain to biofilm formation of exclusively MTL-heterozygote a/α C. albicans cells. Biofilm formation by a/α cells has been reported to be more efficient than that by a or α cells, indicating that genes present at the MTL loci affect biofilm development (Srikantha et al., 2012; Yi et al., 2011). In recent years, alternative C. albicans biofilms that are formed by MTL-homozygous white cells in response to pheromone released by cells of the opposite mating type have been characterized, and these biofilms are often referred to as ‘sexual biofilms’ since they require the presence of mating pheromone (Daniels et al., 2006; Lin et al., 2013; Soll, 2014; Yi et al., 2011). The Pheromone-Stimulated Assay presented below has been modified from the protocol initially reported by Lin et al (Lin et al., 2013). This assay is optimized for use with flat-bottomed, non-tissue culture-treated 12-well plates. It has been validated for use on clinically isolated white a and α C. albicans strains, and can be compared to a/α control strains used in the same assay. The Pheromone-Stimulated Assay will allow researchers to test the biofilm forming abilities of MTL-homozygous strains in the presence of natural or synthetic pheromones of the opposite mating type, or the biofilm abilities of co-cultures of both a and α cells. In the 12-well format, three replicate wells are typically used per condition (blanks and controls included). Include blank wells containing media alone, in addition to wells with cells not exposed to pheromone, as controls. We recommend that this assay be performed using α-factor mating pheromone. Results using a-factor are much more variable due to its increased hydrophobicity.
Materials
C. albicans strain(s) of interest
YEPD plates and liquid media (see recipe)
Spider Media, pH 7.2 (see recipe)
Lee’s Media (Bedell et al., 1979; Lin et al., 2013), pH 6.8 (see recipe)
D-PBS (calcium and magnesium salt free), sterile filtered
Synthetic α pheromone, sequence GFRLTNFGYFEPG (Bennett et al., 2003), 13aa, 90% purity (LifeTein)
12-well microtiter plates, non-tissue culture-treated (Falcon 351143)
10 ml test tubes, 250 ml, and/or 500 ml flasks
Plate reader with optical density capabilities (600nm) compatible with 12-well plate format (e.g. Tecan Infinite M1000 Pro or BioTek Epoch 2)
-
Roller drum for test tubes and/or shaker for flasks in a 25°C incubator
-
1
Streak C. albicans strains onto YEPD plates from glycerol stocks 2–5 days in advance of assay. Incubate plates at 30°C.
Do not use colonies that are more than 7 days old or store plates at 4°C as C. albicans can acquire aneuploidies under these conditions. Likewise, new plates should be streaked from glycerol stocks rather than by re-streaking cells from existing plates.
-
1
-
For Small Numbers of Strains/Large Volumes
-
2a
Inoculate a single colony in 25 ml Spider media in a 250 ml flask (or 50 ml media in a 500 ml flask) from a 2–5 day-old plate. Grow at 25°C with shaking overnight.
-
2a
-
For Large Numbers of Strains/Small Volumes
-
2b
Inoculate a single colony in 4 ml Spider media in a 20 ml test tube from a 2–5 day-old plate. Grow at 25°C with shaking or on a roller drum overnight.
-
3
After 16–18 hr, dilute overnight culture 1:20 or 1:40 and measure OD600 using cuvettes and a spectrophotometer or 96-well plates and a plate reader.
If assays will be set up more than 18 hr after overnights were started, remove overnights from the shaker or roller drum in the morning and allow to sit at room temperature until time of use (cultures should not sit for more than 4 hr at room temperature). Vortex the cultures immediately before use. However, if possible, it is better to time the experiment such that the strains will be used prior to 18 hr of overnight growth. -
4
Following cell density determination for overnight cultures, add cells to a final OD600= 2.5 (or equivalent to ~5x107 cells/ml) in 1 ml in Lee’s media.
-
5
Add synthetic C. albicans α pheromone to each well at a final concentration of 10 μg/ml (6.6 μM). Distribute the pheromone throughout the well by gently agitating the plate.
-
6
Incubate the plate for 24 hr at 25°C under static conditions.
MTL-homozygous cells adhere very weakly before pheromone stimulation. As a result, this assay does not utilize the initial 90-minute shaking adherence step and PBS wash employed by conventional biofilm assays. Pheromone-stimulated biofilms are generally less robust that conventional biofilms, and thus are grown statically to minimize biofilm disruption. -
7
Gently aspirate the media from each well.
Both the pheromone-stimulated and control biofilms will be very fragile. Try not to disturb the biofilm and leave a small layer of supernatant covering the biofilm. -
8
Gently wash the biofilms with 1 ml of PBS. To do this, press the pipette tip against the side of the well. Slowly add buffer while rotating the plate, ensuring even flow of PBS from all sides of the well. Once finished, immediately aspirate the PBS to prevent non-adhered cells from settling back down.
Biofilms formed by MTL-homozygous C. albicans strains are generally very fragile. Be particularly careful during the wash step as entire biofilms can be washed away accidentally. Process one well at a time for this step. -
9
Measure OD600 on a 12-well compatible plate reader (e.g. Tecan Infinite M1000 Pro or BioTek Epoch 2).
The number of reads per well may vary based on the instrument. We recommend taking the average density of five reads in each section of a five by five grid (i.e. twenty-five independent locations) in each well. Be sure that the wells are still moist for the reading as dry wells give inaccurate readings. -
10
Normalize data by subtracting the OD600 reading of an average blank well (containing media alone) from each experimental and control well. The blank-subtracted OD600 value of each experimental well is then normalized to the mean blank-subtracted OD600 for the relevant control wells.
-
11
Calculate the standard deviation and statistical analyses (typically use a Student’s unpaired two-tailed t-test assuming unequal variance) for each normalized data set.
-
2b
BASIC PROTOCOL 10 - Temporal Assessment of C. albicans Biofilm Formation Using a Microfluidics Device
This assay allows for the observation of biofilm formation as a biofilm develops over time using customizable conditions that mimic those of the host, such as those encountered in vascular catheters (Gulati et al., 2017; Lohse et al., 2017; Winter et al., 2016). This assay is highly customizable, and alterations in temperature, media, and flow rates can be made to suit the needs of the researchers. The assay can be used to test the effects of antifungal compounds or to compare biofilm development between different strains. In our experiences, there is good correlation between biofilm formation of mutant strains observed using this assay with that of the in vivo central venous rat catheter model (Andes et al., 2004; Gulati et al., 2017). Although this assay provides highly predictive data pertaining to biofilm development in vivo, it requires costly specialized equipment (the BioFlux 1000z) and has reduced throughput compared to many of the other methods described. We typically use two channels per strain or condition in a specialized 48-well plate (containing 24 channels), allowing for the evaluation of 12 conditions per plate (including blanks and controls).
Materials
RPMI-1640 media (with L-glutamine and MOPS, without sodium bicarbonate, pH 7.0) (see recipe)
Compound(s) to be tested
70% isopropanol
Lens paper (VWR 5284-001)
Cuvettes and spectrophotometer or transparent 96-well plate and OD600-capable plate reader
BioFlux 1000z (Fluxion Biosciences)
48-well plate 0–20 dyne (Fluxion Biosciences 910-0047)
Montage Software (Fluxion, Version 7.8.4.0)
-
ImageJ Software (NIH)
-
Initialize the microfluidic device and set the temperature to 37°C. Heat the plate holder and the plate to 37°C. Preheat RPMI-1640 media at 37°C until the start of the experiment.
Media used for the experiment should be preheated to reduce the chances of air bubbles developing in the microfluidic device. Although the protocol listed here uses RPMI-1640 media, other biofilm-inducing media can also be used. -
Clean the interface plate and the 0–20 dyne 48-well plate using 70% isopropanol and lens paper.
The interface plate connects to the microfluidic system and the 48-well plate. Make sure to remove all liquids when cleaning to avoid blurry or dirty images and videos during the course of the experiment. -
Place the plate on the microfluidic device and add 600 μl of pre-heated RPMI-1640 to the desired inlet wells. We recommend at least two replicate wells per condition(s) to be tested.
For a 12 hr experiment 600 μl is sufficient. For longer experiments, add an additional 50 μl/hr, but do not exceed the maximum well volume of 1500 μl. Mount the clean interface onto the microfluidics plate, align it, and lock it in position using the levers to ensure that the system is airtight.
-
Using the computer interface for the program, flow the media from the inlet to the outlet wells at 1 dyn/cm2 for 5–10 min to prime the microfluidic channels.
The microfluidic channels connect the inlet and outlet wells. Priming the channels with media is essential to remove all air and avoid air bubbles that can interfere with the experiment. -
Following cell density determination for overnight cultures, dilute cells in a microcentrifuge tube to a final OD600 = 0.5 (or equivalent to ~1x107 cells/ml) in 200 μl of pre-warmed RPMI-1640 media and preheat the tube at 37°C until the cells are added to the plate.
C. albicans cells will begin forming hyphae as soon as they are resuspended in RPMI-1640 media at 37°C. In order to capture images before filamentation begins, it is important to make the cell dilutions immediately prior to adding the cells to the microfluidic channels. -
Remove the interface plate and add 50 μl of the cell cultures from step 6 to each outlet well.
Take care to add the cells to the bottom of the well and not to introduce any air bubbles as these will disrupt the flow of media and biofilm formation. Place the interface plate back on the microfluidic plate and lock it in place.
-
Flow the media from the outlet to the inlet wells at 2 dyne/cm2 for 3 seconds to seed the cells in the microfluidic channels.
This step will flush C. albicans yeast cells into the viewing chamber, where the images will be acquired. It is important that the flow is stopped quickly to prevent contamination of the inlet wells supplying fresh media. If these wells are contaminated, the channels supplying media will be blocked with biofilms, confounding the results. Incubate cells without flow for 20 min to allow for cell adherence to the inner surface of the microfluidic channels.
-
During the 20 min adherence step, set up the stage positions on the computer software to allow for image acquisition during the experiment. Calibrate the plate position and focus on each microfluidic channel to obtain the best image and save the settings.
Three stage positions per channel are recommended, distributed evenly through the channel. -
Set the acquisition for 145 total images at an interval of 5 min. We recommend capturing images at 50% Brightfield and 50% camera, with an exposure of 12–20 ms, 0.6 gain and 20MHz digitizer.
It is recommended that the experiment should be set up in dark conditions, to avoid light interference during image capture. Begin image acquisition to capture the initial stages of biofilm development.
After the first round of images is captured, flush away weakly adhered cells by flowing media from inlet to outlet wells at 1 dyne/cm2 for 5 min.
Change the flow rate to 0.5 dyne/cm2 from inlet to outlet wells and leave the experiment running for 12 hr undisturbed and under dark conditions.
-
After 12 hr, use the computer software to view images and create a time-lapse video of biofilm formation (Figure 7 and videos S1–S2).
See Gulati et al., 2017 for additional details on software acquisition settings (Gulati et al., 2017).
-
Figure 7.
Typical images for C. albicans wild-type (WT) and a known biofilm-defective mutant strain from the Microfluidics Assay (Basic Protocol 10). Using the BioFlux 1000Z, biofilms were grown for 12 hr post-adherence in Spider media under dynamic flow (0.5 dyne/cm2) at 37°C. Representative 0 (immediately post-adherence and initial wash), 4, 8, and 12 hr images (top to bottom) are shown for the WT strain (left column) and a known biofilm-defective mutant strain (right column). Scale bars = 50 μm. The corresponding time-lapse videos of biofilm formation for these two strains were acquired at 15 frames/sec and are provided in Videos S1–S2.
BASIC PROTOCOL 11 – Confocal Imaging of C. albicans Biofilms
Confocal Scanning Laser Microscopy (CSLM) is a valuable tool in assaying biofilm formation as it allows for visualization of the biofilm architecture as well as measurement of the thickness of the biofilm (Nobile et al., 2012; Nobile et al., 2005). An array of different labeling methods are available to fluorescently stain biofilms. Polysaccharide-based staining methods, such as concanavalin A and Calcofluor-white which bind to components of the fungal cell wall, are the most commonly used stains for C. albicans biofilms. Other stains that bind to DNA, proteins, and lipids are also available. The protocol described utilizes concanavalin A-Alexa Fluor 594 to stain C. albicans biofilms. Biofilms formed on microtiter plates or silicone squares can be visualized using this assay. For microtiter plates, we recommend that biofilms be formed on 6-well plates and that a blank well be included to monitor contamination during experimental setup. For CSLM imaging of both silicone squares and microtiter plates, two replicates per strain or condition are recommended. In this protocol, we describe imaging using an LSM 700 confocal microscope (Carl Zeiss). Other confocal microscopes may also be used, with the settings appropriate for each instrument. Many of the details in steps 9–13 will differ depending on the system used and as such these steps should be taken as general guidelines. This protocol is derived from Nobile and Mitchell, 2005 and Nobile et al., 2012 (Nobile et al., 2012; Nobile et al., 2005).
Materials
C. albicans strain(s) of interest
RPMI-1640 media (with L-Glutamine and MOPS, without sodium bicarbonate, pH 7.0) (see recipe)
D-PBS (calcium and magnesium salt free), sterile filtered
Concavalin A-Alexa Fluor 594 (Thermo Fisher C11253)
Transparent, sterile, flat-bottomed, non-tissue culture treated 6-well microtiter plates (BD Falcon 351146)
Serological pipettes (VWR 89130-896)
ELMI DTS-4 shakers (or equivalent shaking incubators capable of holding 6-well plates, shaking at 200 rpm, and holding a temperature of 37°C)
Vacuum aspirator setup to which 1000 μl pipette tips can be attached
-
Confocal microscope
Following cell density determination for overnight cultures, add cells to wells at a final OD600 = 0.5 (equivalent to ~1x107 cells/ml) in 4 ml (for 6-well plate assay).
Shake plate at 37°C for 90 min at 200 rpm in an ELMI incubator (or equivalent).
-
Aspirate media and wash wells with 4 ml PBS.
Change pipette tips on aspirator between wells of different strains and/or conditions. Take care not to scrape the bottom of the well with the tip when aspirating. Aspirate PBS and add 4 ml of fresh media to each well.
Shake the plate at 37°C for 24 hr at 200 rpm.
-
Add 20 μl of 10 mg/ml (50 μg/ml final concentration) of Concavalin A directly to each well containing a C. albicans biofilm.
Take care not to disturb or disrupt the biofilm with the tip. After the addition of the dye, the plate should be kept in the dark throughout the experiment. Shake the plate at 37°C for 60 min at 200 rpm in an ELMI incubator (or equivalent).
Place the plate on the stage of the microscope inside a dark room. Take care not to disrupt the biofilm while moving the plate.
To visualize the Concavalin A-Alexa Fluor 594 (red fluorophore), use a 555 nm diode laser with a main beam splitter 405(T80/R20)/488/555/639 on the first channel and a pass band of 550 nm.
-
Visualize biofilms using a water-dipping 40x objective.
A water dipping 63x objective may also be used. It is essential that water-dipping objectives be used with this assay, as the oil-dipping objectives are not compatible for use with the biofilm media. -
Obtain Z-Stacks at 652 x 652 pixels (160 μm x 160 μm) with a pinhole of 1 Airy unit, imaging every 0.5 μm. The stack thickness of a wild-type C. albicans biofilm should be between 240–290 μm.
We recommend obtaining 4–6 Z-stacks at different locations from each well. Make sure that Z-stacks begin from the bottom of the biofilm and not just at the limit of dye penetration. The raw data, obtained as .czi files, are analyzed using ZEN software (Carl Zeiss). The ZEN software is used to measure the depth/thickness of biofilms.
In addition, the .czi files are analyzed using the project stacks function of ImageJ to generate top-views and side-views of each stack (Figure 8).
Figure 8.
Typical Confocal Scanning Laser Microscopy (CSLM) images of a C. albicans wild-type (WT) biofilm grown and imaged following Basic Protocol 11. Both the top-view (left) and side-view (right) of the WT biofilm are shown. Biofilms were grown in Spider media in 6-well plates prior to staining with concanavalin A-Alexa Fluor 594 conjugate. A mixture of yeast and hyphal cells are visible in the top-view (left) but predominantly hyphae are visible in the side-view (right). The extracellular matrix is not visible in either image as it does not efficiently take up the stain. The stain does not penetrate to the base of the biofilm, hence the decrease in color deeper into the biofilm and the inability to visualize the yeast-form cells attached to the solid surface at the base of the substrate. Scale bars = 10 μm.
ALTERNATE PROTOCOL 7 – Culturing and Confocal Imaging of C. albicans Mixed-Species Biofilms
Microbes in nature and in the human host are predominantly found as mixed-species biofilms, composed of many different microbes. This protocol allows for the assessment of the interactions of multiple species in the context of biofilms. Mixed-species biofilms formed with a Standard Optical Density Assay or Silicone Square Assay can then be visualized using confocal microscopy. This allows for visualization of the architecture of the mixed species biofilm, determination of localization of each species within the biofilm, and measurement of the thickness or depth of the biofilm. Below we describe a protocol for assaying C. albicans and E. coli dual-species biofilms derived from Nobile and Mitchell, 2005, Nobile et al., 2012 and Fox et al., 2014 (Fox et al., 2014; Nobile et al., 2012; Nobile et al., 2005).
Materials
Materials for Basic Protocols 4 and 11
-
SYTO 9 (5mM in DMSO) (Thermo Fisher S34854)
Set up single and dual-species biofilms using a 6-well plate according to Basic Protocol 4, steps 1 through 9.
Add 20 μl of 10 mg/ml (50 μg/ml final concentration) of Concavalin A-Alexa Fluor 594 directly to each well of C. albicans cultured alone or C. albicans cultured together with E. coli (or other bacterial species of interest).
-
Add 4 μl of Syto 9 nucleic acid stain (5 μM final concentration) to wells of E. coli cultured alone or C. albicans cultured together with E. coli.
Take care not to disturb or disrupt the biofilm with the tip. After the addition of the dye, the plate should be kept in dark conditions throughout the experiment. Shake the plate at 37°C for 60 min at 200 rpm in an ELMI incubator (or equivalent).
Analyze biofilms using a confocal microscope as described in Basic Protocol 11, using a 488 nm diode laser on a second channel to visualize the bacterial cells.
BASIC PROTOCOL 12 - Harvesting Conditioned Media from C. albicans Biofilms for Proteomic Analyses
The biofilm conditioned media assay offers a quick, relatively high-throughput way to harvest conditioned media from biofilms for use with various proteomic analyses (Winter et al., 2016). This media can then be processed and used in a variety of assays (e.g. multiplex substrate profiling by mass spectrometry (MSP-MS), and trypsin digests followed by shotgun proteomics) (O’Donoghue et al., 2012).
We recommend the use of RPMI-1640 media for this assay, however other media can also be used. We recommend avoiding media that contains serum and/or peptides (e.g. Spider media, YEPD media) as they prevent accurate quantification and normalization of samples and can interfere with subsequent analyses. We typically use 11 wells (approximately 44 ml, plus a 4 ml uninoculated control well) per strain, however a larger number of wells may be needed for samples that give low protein yields and/or for analyses that require more material. Depending on the comparisons to be made, it may be necessary to collect conditioned media from the same strain(s) grown under planktonic conditions. This protocol has been optimized for C. albicans, however it is possible to adapt it to work with other Candida species. When optimizing for other species, changes to the media have the greatest effects on the assay.
Materials
Transparent, sterile, flat-bottomed, Non-tissue culture treated 6-well microtiter plates (BD Falcon 351146)
60 ml Syringes (BD Falcon 309654)
0.45 μm PES membrane sterile syringe filters (Thermo 725-2545)
Sterile 3 ml transfer pipets (BD Falcon 357575)
50 ml screw cap tubes (Axygen Scientific SCT-50ML-25-S or equivalent)
D-PBS (calcium and magnesium salt free), sterile filtered
RPMI-1640 media (with L-Glutamine and MOPS, without sodium bicarbonate, pH 7.0) (see recipe)
Liquid nitrogen
ELMI DTS-4 shakers (or equivalent shaking incubators capable of holding 6-well plates, shaking at 200 rpm, and holding a temperature of 37°C)
Vacuum aspirator setup to which 200 μl pipette tips can be attached
Electric pipettor (e.g. Drummond Pipet-Aid) and sterile 25 ml Pipettes
Refrigerated centrifuge capable of spinning 50 ml screw cap tubes
-
Cold room
-
Add 4 ml of room temperature RPMI-1640 media to each well of a 6-well plate.
We typically use 12 wells (2 plates) for the initial experiment with each strain for this assay. -
Following cell density determination for overnight cultures, add cells to wells at a final OD600 = 0.5 (equivalent to ~1x107 cells/ml) in 4 ml RPMI-1640 media. Do not inoculate the first well on the last plate.
We typically do not inoculate the first well on the last plate of a batch in order to control for contamination. When planktonic controls are needed, we inoculate to a final OD600 = 0.05 in 25 ml in a 125 ml flask. Shake plate at 37°C for 90 min at 200 rpm in an ELMI incubator (or equivalent).
-
Aspirate media. Try to remove any floating clumps of cells that are visible.
Change pipette tips on aspirator between wells of different strains and/or plates. Take care not to scrape the bottom of the well with the tip when aspirating. -
Wash wells with 4 ml PBS. Aspirate PBS, trying to remove any floating clumps of cells that are visible.
Add PBS slowly to the side of the well, taking care to minimize disruption of cells on the surface of the well. -
Add 4 ml of fresh media to each well.
Add media slowly to the side of the well, taking care to minimize disruption of cells on the surface of the well. Shake plate at 37°C for 24 hr at 200 rpm in an ELMI incubator (or equivalent).
-
Pre-label 50 ml screw cap tubes. Chill screw cap tubes, 3 ml transfer pipets, syringes, and syringe filters on ice.
We typically label and chill items used in the harvesting step 20 min before starting that step. -
Remove plates from ELMI shakers. Use sterile 3 ml plastic pipettors to scrape any cells off of the sides of the wells. Disrupt the biofilm at the bottom of the well, collect both the biofilm and the liquid using the pipettor, and transfer both to the chilled, prelabeled 50 ml tube. Keep the 50 ml tube(s) on ice during this step.
Change pipettors between plates. Materials from two plates should fit in a single 50 ml screw cap tube. We typically harvest from all wells of one plate before moving onto the next plate. Remove cells from the sides of all wells on a plate before collecting the liquid and biofilms from the wells. Depending on the strength of the biofilm, it may prove necessary to use the pipette to rigorously scrape the bottom of the well. There should not be visible cloudiness on the bottom of the well after the biofilms and conditioned media are removed. -
Spin tubes for 10 min at 3750 rpm in a chilled centrifuge. After the spin, put tubes on ice and transfer supernatant to new, pre-chilled tubes, taking care to minimize disruption of the pellets. Keep new tubes with supernatants on ice.
The robustness of the pellets can vary greatly between different strains, species, or conditions. Pour gently and do not worry about collecting all of the supernatant if this would risk transferring the cell pellet. Move to cold room if not already there.
-
Pull plunger from 60 ml syringe and place it back in syringe packaging to keep sterile. Attach 0.45 μm PES filter to the syringe. Pour conditioned media into syringe, reinsert plunger, and filter into fresh, pre-chilled 50 ml screw cap tube. Flash freeze tube in liquid nitrogen.
Filter and flash freeze one tube before starting on the next tube. Take care not to accidently dislodge the filter from the syringe when filtering the media. -
Store frozen samples at −80°C pending further processing.
Ideally processing and quantification of samples should be performed within a few days of harvesting. Details of further processing steps will vary depending on the requirements of subsequent proteomic analyses. When preparing conditioned media for MSP-MS or trypsin digests followed by shotgun proteomics, we typically partially thaw the sample in lukewarm tap water (until about ~50% ice left) and then thaw the remainder on ice. We then use 10000 MWCO/15 ml spin units to concentrate samples down (1 per tube) to approximately 0.5–1 ml in a refrigerated centrifuge. Samples are then diluted back to 15 ml with PBS and concentrated back to approximately 1 ml. Samples are then aliquoted, flash frozen with liquid nitrogen, and stored at −80°C. Protein concentrations are determined using the Bradford assay in order to allow for normalization of input into later steps. If possible, use fresh samples for assays rather than ones that have been previously thawed and refrozen.
-
REAGENTS AND SOLUTIONS
YEPD media (2% BactoTM peptone, 2% dextrose, 1% yeast extract). Store at room temperature.
YEPD agar plates (2% BactoTM peptone, 2% dextrose, 1% yeast extract, 2% Agar). Store at room temperature.
Spider media (10 g/l nutrient broth (Difco), 10 g/l mannitol, 4 g/l K2HPO4, pH 7.2). Store at room temperature.
RPMI-1640 (RPMI-1640 with L-Glutamine, without sodium bicarbonate MP Biomedicals, 0910601) with 34.5 g/l MOPS (Sigma, M3183), pH 7.0 (Sodium Hydroxide); 0.22 μm filter sterilized. Store at 4°C and protect from light.
0.5mg/ml XTT in PBS (XTT Sodium Salt Sigma X4251), make fresh prior to use and protect from light.
0.32mg/ml PMS in water (Phenazine methosulfate (PMS), Sigma P9625), make fresh prior to use and protect from light.
0.4% aqueous crystal violet solution. Store at room temperature and protect from light.
Lee’s media – 28.96% (NH4)2SO4, 1.15% MgSO4, 14.48% K2HPO4, 28.96% NaCl, 2.89% L-alanine, 7.53% L-leucine, 5.79% L-lysine, .57% L-methionine, .43% L-ornithine, 2.8% L-phenylalanine, 2.8% L-Proline, 2.8% L-Threonine, .49% L-arginine, (NH4)2SO4 50 g/l, MgSO4 2 g/l, K2HPO4 25 g/l, NaCl 50 g/l, 200 nM ZnSO4, 250 nM CuSO4, 1 μM FeCl3, 1 mM MgCl2, Biotin 10 mg/l and Dextrose 1.25 g/l. pH 6.8. Store at room temperature.
0.1% sodium thiosulfate, make fresh prior to use (from a 10% stock that is stored at room temperature)
Letheen broth (Meat peptone 10 g/l, beef extract 5g/l, Polysorbate 80 5g/l, sodium chloride 5g/l, Lecithin 0.7 g/l). Store at room temperature.
Fluorescein diacetate (5 mg/ml in DMSO). Store at −20°C and protect from light.
COMMENTARY
Background Information
Culturing
We have presented common growth conditions that have worked across a number of strains and isolates in a large number of laboratory groups. YEPD media is easy and inexpensive to make and can be stored for a long time. The growth conditions described consistently result in similar biofilms for a given strain.
Optical Density Assays
These assays have been developed and optimized for medium to high throughput screening of deletion strain and potential antifungal compound libraries as a precursor to more time-consuming and/or cost-intensive assays (Lohse et al., 2017). The small volume, at least compared to many other biofilm assays, reduces the amount of test compound needed, which is particularly useful when testing expensive or limited compounds of interest. The dynamic ranges of these assays are also capable of detecting both increases and decreases in biofilm formation. They are also designed to minimize equipment requirements to one or more shaking incubators and a plate reader with optical density capabilities. Although we have presented several specific protocol variations (e.g. a 24 hr biofilm growth assay, a biofilm disruption assay, and several biofilm inhibition assays), the core protocol can be modified to study other stages of biofilm development (e.g. 4, 8, or 48 hr) or other metrics, such as synergistic drug reactions. Likewise, the related Dispersal Optical Density Assay can be modified to look at dispersal at other time points (Nobile et al., 2014).
The metric of biofilm formation varies between biofilm thickness (Standard Optical Density Assay), metabolic activity (XTT Reduction Assay), and cell viability (Crystal Violet Assay) in the different assays, as such it may be informative to perform more than one variant. The Standard Optical Density Assay has the advantage of the minimal number of processing steps, both reducing the time involved and lessening the potential for inadvertent disruptions of the biofilm. It is important to remember, however, that there may be limitations to each of these approaches (e.g. not distinguishing between live and dead cells, missing metabolically inactive persister cells, and artifacts due to dyes failing to penetrate the full depth of the biofilm).
Mixed Species Assays
C. albicans interacts with a wide range of bacterial species that occupy the same niches in humans (e.g. the oral cavity, skin, and the gastrointestinal and urinogenital tracts) (Gulati et al., 2016; Nobile et al., 2015). C. albicans has been shown to form mixed species biofilms with several bacterial species both in vitro on abiotic surfaces and in vivo on biotic surfaces within the host (Harriott et al., 2011; Morales et al., 2010). These interactions have significant clinical implications; for example, it has been observed that C. albicans biofilms can provide a protective anaerobic niche for pathogenic anaerobic bacteria, allowing these bacteria to grow in unexpected aerobic locations of the host when residing within biofilms with C. albicans (Fox et al., 2014). Likewise, C. albicans biofilms can protect bacteria from exposure to antibiotics (Harriott et al., 2009, 2010). The modification of the Standard Optical Density Assay to study C. albicans mixed-species biofilm formation with interacting bacterial partners offers opportunities to gain insights into cross-kingdom interactions occurring in the context of a biofilm. Furthermore, the base assay can be modified to study additional bacterial species as well as other aspects of biofilm development. Finally, these mixed species biofilm assays can be combined with the Cell Enumeration Assays in order to evaluate the relative abundance and viability of each species within the biofilm.
Silicone Square Assay
In vitro C. albicans biofilm assays are most commonly performed on polystyrene microtiter plates. However, silicone is a common material used for implanted medical devices, such as catheters, heart pumps, and surgical reconstructive components. Silicone squares can be easily cut and customized for different experimental needs, and are thus quite versatile for non-high-throughput experimentation. The use of silicone squares is, therefore, a useful and potentially informative alternative substrate that can be worth testing in certain biofilm assays (Nobile et al., 2012; Nobile et al., 2005).
Dry Weight Assay
In vitro optical density assays offer a proxy for the number of cells and/or the amount of biomass present in a biofilm. The Dry Weight Assay directly measures the amount of biomass that forms during the course of biofilm development and accounts for the presence of extracellular matrix in a way many assays cannot. Since this assay is low throughput, largely due to the vacuum filtration steps, it is used more often for validation of candidates initially identified by other methods.
Cell Enumeration Assays
Optical density assays offer, at best, crude proxies for the viability of cells in a biofilm, a limitation that can be of concern when screening potential antifungal compounds for microbicidal properties. An alternative assay, the Cell Enumeration Assay, utilizes serial dilution and colony forming units to estimate the number of live cells present during biofilm formation. This approach has been utilized effectively in the Cell Adhesion Assay to estimate the number of cells that adhere to seed a biofilm (Lohse et al., 2017; Winter et al., 2016). However, traditionally, it has been a challenge to obtain CFUs from a mature biofilm as biofilms are resilient to physical stresses, and as such do not homogenize sufficiently, resulting in a substantial underestimation in the number of viable cells present. The addition of Letheen Broth, which contains emulsifying components, allows the biofilm to break up and homogenize more effectively (without lysing the cells within the biofilm), resulting in a more accurate estimation of cell numbers. The alternate Cell Enumeration Assay, which utilizes a fluorescent dye (e.g. fluorescein diacetate for C. albicans) to distinguish between live and dead cells, avoids the need to fully homogenize the biofilm by relying on the visual distinction between dead cells which emit a bright green fluorescence and live cells which are generally very dim (Figure 6). This version of the Cell Enumeration Assay avoids the need for serial dilution, significantly reducing the workload and enabling the screening of more strains and/or conditions at one time. We note that this pair of assays is also helpful for looking at the relative abundance and viability of individual species within a mixed-species biofilm.
Pheromone-Stimulated Assay
This assay was originally established in Lin et al. (Lin et al., 2011) and optimized in Lin et al. (Lin et al., 2013). It examines the ability of mating pheromone to stimulate biofilm formation of C. albicans MTL-homozygous strains, which would generally not result in robust biofilms in the absence of pheromone. The assay has been used to test MTL-homozygous strains using natural or synthetic pheromones of the opposite mating type, or co-cultures of both a cells and α cells. In general, this assay works better using α-Factor rather than a-Factor due to α-Factor’s greater solubility in water. We note that even minor alterations to the steps of this assay can result in significant changes to the biofilms formed.
Microfluidics Assay
Most of the C. albicans biofilm assays included in this chapter (e.g. the Optical Density, Silicone Square, and Dry Weight Assays) assess biofilm formation at a single time point. To better assess C. albicans biofilms throughout their development, we recommend the real-time microfluidic assay using the BioFlux Microfluidic device (Gulati et al., 2017). This assay is an effective tool to assess biofilm defects between different strains of C. albicans and to assess the effect of antifungal compounds on biofilm formation over time (Lohse et al., 2017; Winter et al., 2016). This assay is particularly useful to validate results from high-throughput screens. In our experiences, the results from this assay have correlated well with in vivo experiments using the rat central venous catheter model (Andes et al., 2004; Winter et al., 2016).
Confocal Imaging
C. albicans forms highly structured biofilms that can be visualized by Confocal Scanning Laser Microscopy (CSLM). This technique allows for examination of the architecture of the biofilm as well as measurement of the biofilm thickness. CSLM has been utilized to visualize C. albicans biofilms in several studies (Nobile et al., 2012; Nobile et al., 2005), mostly utilizing concanavalin A conjugated with different Alexa Fluor ™ dyes (most commonly Alexa Fluor 594). Recently, CSLM has also been used to study dual-species biofilm formation by staining fungal cells with concanavalin A dyes and bacterial strains with SYTO™ dyes (most commonly, SYTO™ 9) (Fox et al., 2014). CSLM can be used to visualize biofilms formed on both microtiter plates and silicone squares.
Conditioned Media Harvesting for Proteomics
The conditioned media harvesting protocol presented here was developed to be as similar as possible to standard biofilm assays, while also satisfying the sample requirements of subsequent proteomic analyses. This protocol has worked effectively with a number of different strains and species (Winter et al., 2016).
Critical Parameters and Troubleshooting
Culturing
Standard microbiology sterile technique is critical for all protocols in this chapter using microbial cells and we commonly include one or more uninoculated negative controls to verify that media has not been contaminated. Especially when working with a small number of strains, we recommend preparing multiple overnight cultures of each strain so that there are backup cultures in the event of individual culture contamination. When culturing C. albicans, it is critical to be aware that aneuploidies can be acquired if C. albicans is treated in ways commonly used for S. cerevisiae and many bacterial species. In particular, it is important not to use colonies that are more than 7 days old, not to store plates at 4°C, and not to re-streak cells from existing plates. Likewise, strains should be used prior to 18 hr of overnight growth. When using a spectrophotometer for the first time, it is important to first obtain CFU counts to correlate to optical density values. As overnight cultures will be very dense, it is important to dilute samples at least 20-fold to accurately determine the optical density. When working with mutant strains, slow growth phenotypes may result in reduced biofilm formation for reasons unrelated to the ability to form biofilms. As such, it is important to note any mutant(s) whose overnight cultures have significantly lower densities. In certain situations, these mutant strains could be allowed additional time to form a biofilm to fully assess biofilm development in a particular assay.
Optical Density Assays
A wide range of experimental and even equipment-related factors can affect the output of the various optical density based biofilm assays. To help address this, we strongly recommend the inclusion of both positive and negative controls on every plate as well as controls for any solvents (e.g. DMSO, methanol) used to dissolve compounds. In general, we recommend validating the phenotypes of all mutants of interest using independently derived isolates and/or addback strains. Likewise, mutants should also be evaluated for other relevant phenotypes (e.g. adherence defects, filamentation defects) to classify possible reasons for defects in biofilm formation as well as to eliminate false positives (e.g. slow growth in general or in the media being used). Attention should also be paid to the dynamic range of biofilm phenotypes observed when using a given media; if the wild-type strain forms only a thin biofilm when using a certain media, then it may be difficult to detect a reduction in biofilm formation with this media. Alternatively, media that supports robust biofilm formation may mask subtle defects of mutant strains. In these cases, altering the media or specific nutrients (e.g. carbon sources) may be helpful.
There are a number of specific issues to be aware of when performing these assays. Variation in media, the volume of media, incubation times, and the way in which media is added or removed from wells can affect biofilm formation. As such, care should be taken to be as consistent as possible with each step. A related problem stems from the inadvertent physical disruption of biofilms when removing or adding liquid (e.g. the biofilm detaching during aspiration steps, during the addition of liquid in the Disruption Optical Density Assay, or during replacement of the media in the Standard Dispersal Assay), which can range in scale from small disruptions at the edge of a well to the complete loss of a biofilm in a well. Any wells that are physically disrupted should be noted and excluded from subsequent analyses; the ability to exclude the occasional disrupted well is part of the reasoning behind having 6–8 replicate wells of each condition. To avoid inadvertent disruption, it is important to remove or add media slowly and to avoid touching the bottom of the well with pipette tips. Incomplete sealing of plates or peculiarities of specific incubators can result in position specific effects on biofilm formation; in general, we recommend repeating assays where this is noticeable. In order to reduce effects from prolonged exposure to air and the biofilm drying out, we recommend processing only one plate at a time, performing the Disruption Assay on only 6–12 wells at a time, and performing the final OD600 measurements immediately after aspirating media from the wells. When working with potential antifungal compounds, be sure to visually inspect the plate for the appearance of unusual colors and/or crystallization in wells as this may affect the OD600 or OD492 measurements. If such problems arise, it may be necessary to reduce the concentration of the compound being tested and/or try dissolving the compound in different solvents. Finally, it is important to remember that the output from the Optical Density, XTT Reduction, or Crystal Violet Assays may not scale linearly between different strain backgrounds, mutants, or species. As such, interesting phenotypes should be validated using additional methods, such as confocal microscopy or the Microfluidics Assay.
Mixed Species Assays
When setting up mixed-species biofilms, it is important to know the numbers of C. albicans and bacterial cells being seeded for the biofilm (for dual-species biofilms, we often begin with 1:1 ratios of each species). We recommend that an accurate correlation of OD600 to cell density be established using a hemocytometer and/or the Colony-Forming Units Assay for each bacterial strain, as OD600 can be influenced by cell shape and size. It is essential that the assay include single species biofilms in the same media as the mixed-species biofilms, to aid in assessment of the mixed-species biofilms.
Silicone Square Assay
Including both positive and negative controls (e.g. wild-type and biofilm-defective mutant strains) as well as a blank control (uninoculated well) within each experiment is important to ensure proper experimental setup. Consistency in the treatment of samples is critical for reproducibility when performing this assay. It is necessary to note the top side of each silicone square, where the biofilm has been seeded; if the square gets flipped during manipulations, the biofilm will not form properly. Additional care must be taken when lifting the silicone squares during the wash step as well as when placing the silicone square into the fresh media plate. For all steps involved, we recommend handling the silicone square only from the edges using pointed forceps to prevent disturbing the adhered cells. When placing the washed silicone squares into the fresh media plate, be sure that the silicone squares are thoroughly submerged in the media. If bubbles are present at the surface of the media, push the silicone square below the bubbles to prevent the square from floating on top of the media.
Dry Weight Assay
There are a number of steps which can result in variability when performing the Dry Weight Assay. It is critical to recover all of the biofilm from the surface of the well when performing this assay. As such, second washes of the well may be necessary to collect biofilm fragments that escaped the first harvesting pass. Likewise, it is important to make sure that all of the biofilm is transferred from the pipette to the filtration membrane. Applying the biofilm to the center, rather than the edges, of the membrane is important as this reduces the chances of losing biofilm cells during later manipulations of the membrane. Finally, it is critical to dry all membranes thoroughly and uniformly in order to avoid variation due to residual moisture in the samples. Care should be taken to note any instances where dried biofilms have started to flake off from the membrane as this can affect dry weights if the biofilm fully detaches and is lost during weighing.
Cell Enumeration Assays
Colonies should only be counted once they have grown enough to be clearly visible to the naked eye and all plates should be counted at the same time if possible. As this assay is highly dependent on the homogenous disruption of biofilms and successful serial dilutions, there can be significant variability between samples. We recommend at least four replicates per strain or condition and to plate multiple dilutions from each replicate. If problems persist with the serial dilution method, we suggest visualizing the homogenized sample on a microscope to ensure that there are no cell clumps remaining. If cell clumps remain despite vigorous homogenization efforts, it may be best to use the live/dead fluorescence version of the assay to circumvent this problem. As with the other assays described in this chapter, if the effect of an antifungal compound is being tested, it is important to include controls for the solvent (without the compound) to ensure that the measured effect is due to the compound and not the solvent concentration in media. It is also recommended to use low retention filter tips when making serial dilutions, as C. albicans biofilms tend to stick to the walls of pipet tips, which may result in inaccuracies during serial dilutions. For the visualization of live/dead cells, it is important to include both a positive control (e.g. a known antifungal drug) and a negative control (e.g. media with no drug). The autofluorescent cells in the negative control can help in distinguishing live/dead cells in other samples as they provide a baseline of dead cell autofluorescence and the lack of fluorescence in live cells. When using this assay to distinguish between species in a mixed-species biofilm, it is important to make sure that only one species will grow on a given type of plate (e.g. C. albicans on plates containing antibiotics) or that it is possible to distinguish between colonies of the two species using a given media. The final number of cells from the species used can vary by at least an order of magnitude, as such it may be necessary to increase or decrease the number of serial dilutions accordingly.
Pheromone-Stimulated Assay
The reproducibility of this assay can be markedly improved by ensuring a uniform distribution of cells across the bottom of each well. To obtain this distribution, do not allow cells in the inoculum to settle. Gentle swirling of the plate immediately following addition of cells can aid in the even distribution of the cells across the surface of each well. The PBS wash step prior to measuring OD600 is the most critical and sensitive step in this protocol as entire biofilms can be inadvertently washed away. To minimize this, carefully pipet PBS down the sides of the well, while slowly rotating the plate (the media will tend to flow in evenly towards the center of the well). A layer of non-adhered cells may accumulate in the wells during the wash step; a very gentle agitation after the PBS has been added can help disperse these non-adhered cells. Wells will rapidly dry out after media has been aspirated, decreasing the effectiveness of the wash step. As such, we recommend aspirating media and washing one well at a time in order to prevent wells from drying out.
Microfluidics Assay
Including controls within each batch of experiments, such as an untreated wild-type strain and controls for any solvents used, is important to ensure proper experimental setup. Setting the direction of flow between inlet and outlet wells is a critical component of performing this assay. Cells must flow from the outlet wells towards the inlet wells during seeding of the biofilm, and sterile media must flow from the inlet wells towards the outlet wells during biofilm formation. This setup ensures that the sterile media does not get contaminated and that the biofilm is formed within the microfluidic channel rather than inside the inlet wells. If the cells flow too fast or for a longer duration than indicated in the protocol, the cells could contaminate the inlet wells. If this occurs, the inlet channel may become blocked due to biofilm growth and biofilm development will not properly take place within the microfluidic channel. Other key components of performing this assay are to ensure that the camera focus and exposure time are accurately set for each channel and that the apparatus is not disturbed throughout the experiment. Additionally, this assay must take place in a dark room. Any alterations in lighting or vibrations can lead to blurry images and videos. Finally, prewarming the media and cleaning the equipment are critical for preventing the development of air bubbles and for reducing issues during image acquisition.
Confocal Imaging
For CSLM, it is critical that the biofilms remain intact during the image acquisition process. The biofilms should be handled gently and kept in the dark until imaging is complete. Imaging should begin at the base of the biofilm and move towards the top. Care should be taken not to disturb the microscope or stage during the imaging process. We recommend at least two replicate wells per condition, and to obtain at least six Z-stack images per well from different areas of the well. When setting up Z-stack acquisition of images, care should be taken in obtaining the first image slice as the dye often cannot penetrate deep into the biofilm due to its thickness. It is important to ensure that the base of the biofilm, rather than the base of dye penetration, has been reached before starting image acquisition. This can be achieved by visualizing the substrate (polystyrene plate or silicone square), which will appear quite distinct from the biofilm, and moving upwards until the substrate is out of view. Once the substrate is just out of view, this is a good position to set for the first image slice.
Conditioned Media Harvesting for Proteomics
Although this assay is fairly robust in generating active protein mixtures, it is important to practice good sterile technique and to include at least one uninoculated well in each set of plates to act as a contamination control. As with all protein work, care should be taken to keep samples cold during the harvesting and processing steps to avoid the loss of protein activity due to enzymatic degradation and/or denaturation. Otherwise, the most important concern is to match growth times and media conditions as much as possible between different strains and conditions. Changes to the media may be necessary to improve biofilm formation for given strains; when making these changes, the effect on biofilm formation by all relevant strains should be noted. Likewise, care should be taken to avoid media or reagents with large amounts of peptides or full length proteins, which will increase the background baseline (noise) of the assay. If the yield from the initial two plate assays is insufficient, it may be necessary to scale up to four or even eight plates for harvesting.
When harvesting, be sure to note whether control wells have signs of contamination. If contamination is observed, we recommend discarding that batch of conditioned media and repeating the assay. Likewise, take care to note whether there are a noticeable number of free-floating planktonic cells in the well. The presence of at least some planktonic cells may be unavoidable, especially in the case of strains that form weaker biofilms. In these cases, the resulting profiling of the media may represent a mix of the biofilm and planktonic cell types.
Understanding Results
Culturing
With some exceptions, colonies of a strain should have consistent phenotypes when streaked onto a plate. Cultures should look visibly saturated after overnight growth and the uninoculated negative control cultures should remain clear. In most cases, there should not be visible flocculants (clumps of cells) in the overnight cultures. A normal cell density measured by OD600 for an overnight culture is around 10–12, although this can vary for certain mutant strains and based on the spectrophotometer (and path length) used to obtain the measurement.
Optical Density Assays
Plates should be examined visually for noticeable phenotypes and abnormal results in addition to the more quantitative OD600 and OD492 measurements from a plate reader. There should be a visible biofilm on the bottom of positive control wells while the negative control wells should remain clear (Figure 2). Contaminants in the negative control wells may appear as discreet spots on an otherwise clear well or as a cloudy haze throughout the well. Ideally, the density of biofilms should appear relatively uniform across a given well and between replicate wells of a given condition. Mutants or compounds that reduce biofilm formation should result in less dense or even clear wells. Wells where the addition of the compound of interest leaves a noticeable color, visible precipitation, or crystallization should be noted as this may affect OD600 or OD492 measurements. Position specific effects (e.g. noticeably lower density for all wells on one side of a plate) should be noted. Expected OD600 values will vary depending on the strain, media, duration of growth, and even the plate reader; wild-type densities in the 0.2–0.6 range are not unexpected.
Mixed Species Assays
The results from this assay are conceptually similar to those from the Optical Density Assay and its variants. It is possible that different bacterial species may not form robust single species biofilms in the media of choice (e.g. BHI-FBS) under aerobic conditions for a variety of reasons (e.g. nutritional or oxygen requirements). Despite this, the measurements from these single species control biofilms are important for contrasting against the dual-species biofilms formed by the same species in combination with C. albicans under the identical conditions. As the OD600 results do not distinguish cell density measurements between the two species, it will often be necessary to pair this assay with the Cell Enumeration Assays.
Silicone Square Assay
Silicone squares should be examined visually to ensure expected biofilm formation and expected turbidity of the surrounding medium. Positive control strains capable of forming normal biofilms should have clear surrounding medium with the silicone substrate completely covered by the biofilm (Figure 4). Negative control squares unable to form a biofilm should have dense turbidity of the surrounding medium with the silicone substrate devoid of cells. Blank (uninoculated) wells should be completely clear both in the medium as well as on the silicone square (Figure 4).
Dry Weight Assay
There should be a visible collection of biofilm cells on the dried membranes from the positive control wells and no visible cells on the dried membranes from the negative control wells. Although major biofilm defects will be clearly detectable using this assay, more minor biofilm defects will be less apparent or sometimes undetectable. In instances where a minor biofilm defect is anticipated, other biofilm assays should be used to assess the biofilm phenotype.
Cell Enumeration Assays
When counting colonies, the plates examined within 24 hr should contain single colonies (not lawns) and should be easily distinguished from one another. If no colonies are observed after 24 hr, plates should be examined daily for 48–72 hr, which will allow for the scoring of slow growing mutant strains. When using this assay with mixed-species biofilms, the results should be visually distinct colonies for each species or colonies of single species that only grow on specific selective media for that species.
Pheromone-Stimulated Assay
The results of this assay are conceptually similar to the Optical Density Assays. Wells that have not been treated with pheromone should appear mostly clear after the PBS wash, with a thin layer of yeast cells remaining on the bottom of the well. Wells treated with pheromone will form a fragile biofilm of varying thickness depending on strain background. Strains that respond strongly to pheromone will form a biofilm that coats the entire well; very few, if any, non-adherent cells should be washed away in those cases. Strains with a moderate response to pheromone will form a biofilm with upper layers that detach around the edges. Weak responders to pheromone will have an entire layer of non-adherent cells that comes off the well (oftentimes as a sheet) during the wash, with only the most adherent cells remaining. Strains that do not respond to pheromone will be washed off the well in small clumps of cells rather than sheets of biofilm.
Microfluidics Assay
At the completion of the experiment, the microfluidic plate can be examined visually. The inlet wells should have clear media and the outlet wells should have an obvious biofilm (in the case of a biofilm-forming strain). Additionally, most of the media should be in the outlet wells, with less than 100 μl present in the inlet wells. If any of the inlet wells still have noticeable amounts of media, it is possible that the channel became blocked during the experiment due to an air bubble. If the inlet wells have a biofilm, however, this indicates that the cells reached and contaminated the inlet well during the biofilm seeding step; the corresponding microfluidic channel should not be included in further analysis. The images generated at the end of the experiment can be analyzed using the Montage software. Images should not be blurry and the viewing field should not be obstructed by debris (Figure 7). During the generation of a time-lapse video using the Montage software, ensure that there were no air bubbles evident during the experiment. The video should begin by showing adhered yeast-form cells, followed by the formation of hyphae within 30 minutes (in the case of wild-type strains in biofilm-inducing media), followed by the formation of a thick biofilm covering most of the field of view at the end of the 12 hr experiment (Video S1).
Confocal Imaging
Wild-type C. albicans biofilms are highly structured, containing intercalating hyphae visible throughout the Z-stack of images (Figure 8). Usually yeast-form cells can be seen interspersed between the hyphae (Figure 8). Depending on the media and substrate used, the thickness of a typical wild-type biofilm after 24 hr can range from ~100–400 μm (and can be much thicker if grown for 48–60 hr). Mutant strains or strains exposed to antifungal agents may have a much thinner biofilm. When imaging mutant strains, it is important to note if particular cell types are evident in excess or in significantly reduced amounts relative to the wild-type strain.
Conditioned Media Harvesting for Proteomics
It is not possible to visually evaluate the harvested conditioned media at the end of the harvesting or processing steps. The exact concentration of the processed media will depend on the volume of media harvested and the final volume after the processing step; we typically get a yield of around 500 μl at 100–400 μg/ml from 50 ml of conditioned media. This material is consistently active in in vitro protease cleavage assays and at least several dozen distinct proteins are anticipated to be detected when shotgun proteomics are performed on a trypsin digest of the processed media.
Time Considerations
Culturing
The time required for streaking plates from frozen (−80°C) stocks, starting overnight cultures, and determining the densities of cultures will vary depending on the number of strains being used. Small numbers (less than 10) of strains will take only a few minutes for each of these steps.
Optical Density Assays
The most time-consuming steps of these assays are often the preparation of solutions (when screening compounds for anti-biofilm activity) and or the dilutions of strains (when screening different isolates or mutant strains). The length of these steps will depend greatly on the number of dilutions being made. Transferring these solutions to deep well 96- or 384-well plates can take between 10 min (for 96-well) and 30 min (for 384-well) for each plate. Once solutions and cells are in a suitable format or in cases where media and/or cells can be added from a sterile basin, starting a plate takes ~15 min. Washing and adding fresh media to the plate takes between 15 min (for 96-well) and 30 min (for 384-well) for each plate. Other than the dilution step, the overall setup process should be expected to take roughly 2.5 hr with active work required for less than half that time. Once familiar with this technique, it is possible to stagger the setup of two plates by offsetting them approximately 45 min and to setup multiple staggered sets of plates in a day. The disruption step, when applicable, will take between 30–45 min per plate. Aspirating the media and reading the density of wells will commonly take between 15 min (for 96-well) and 30 min (for 384-well) for each plate. Solutions for the XTT Reduction Assay will take ~15 min to prepare. Aspirating the existing media and adding the XTT solution to wells will take 15 min (for 96-well) to 30 min (for 384-well) plates. After the incubation step, which may vary in length (assume 30 min for initial assays), reading the plate will take ~5 min. The Crystal Violet Assay takes around 3–4 hr, although much of this time is taken up by the 45 min drying, staining, and destaining steps. Harvesting media for the Dispersal Assays takes ~10 min per plate. These times are approximate and are based on familiarity with the assay; steps may take considerably longer when performing the assay for the first time.
Mixed Species Assays
This assay will take longer at first for any new bacterial species due to the need to correlate the OD600 with actual cell numbers using a hemocytometer, which can take ~30 min per strain. Alternatively, the OD600-cell number correlation can be determined in advance using the Colony-Forming Units Assay. Once the OD600 to cell number correlation is established, this assay should take roughly the same amount of time as the Standard Optical Density Assay. Depending on the specific bacterial strain(s), however, there may be additional preparatory work required for culturing the specific species.
Silicone Square Assay
Determining the cell density, placing the silicone squares into the wells, and adding media and cells to wells takes ~20 min for one 12-well plate. Fresh 12-well plates with PBS and RPMI-1640 can be prepared during the 90-min adhesion step. The PBS wash step takes up the most time and care, about 1–3 min per silicone square or 12–36 min per 12-well plate.
Dry Weight Assay
It will take ~5–10 min to setup and ~10 min to wash each 6-well plate. Depending on the nature and robustness of the biofilms, it will take between 5–10 min to disrupt and homogenize each plate. It will take several minutes to set up the filter device (Figure 5), filter the biofilm, and remove the membrane for each sample. As such, processing times of at least 15 min can be expected for each 6-well plate.
Cell Enumeration Assays
The serial dilution/plate counting version of this assay takes significantly longer than the fluorescent dye based version. The most time-consuming steps of the former version of the assay are the serial dilutions and the plating of multiple dilutions for all replicates. A dilution series for four replicates of a given strain will generally take ~30–40 min. Once colonies have grown on the plates, it can take 1–5 min to count each plate depending on colony density. For the fluorescent dye-based protocol, the imaging step is the most time-intensive step of this assay. It takes ~15 min to acquire all representative images for each slide. Unlike the serial diluting/plate counting version, however, cell counting software, such as imageJ (NIH), make the analysis of each slide relatively fast (5–10 min per image).
Pheromone-Stimulated Assay
Most of the time requirements for this assay are similar to those for the related Standard Optical Density Assay. Seeding biofilms will take ~1 hr, which is longer than the Standard Optical Density Assay, due to the pheromone addition step. The final washing step can take ~45 min due to the care required while washing. Analysis with a spectrophotometer takes ~10 min for each plate.
Microfluidics Assay
This assay can be set up in less than 1 hr once the researcher is familiar with all the steps and the software. The experiment takes 12 hr to complete and the analysis, including the generation of time-lapse videos, can take 2–4 hr depending on the number of strains and conditions used in the experiment.
Confocal Imaging
This assay is time intensive, with the bulk of time spent on image acquisition using the confocal microscope. It takes ~15–20 min to obtain one set of Z-stack images, so each well can take ~3 hr to image.
Conditioned Media Harvesting for Proteomics
Determining cell density, adding media, and adding cells will take ~15–20 min for a set of four 6-well plates (24 wells total). The wash step takes ~5–10 min per plate, or ~30 min for a set of four 6-well plates. As such, the full setup process takes ~2.5 hr with active work for less than half of the time. Physically harvesting media from wells takes ~5–10 min per plate, or ~20–45 min for a set of four 6-well plates depending in part on the effort needed to disrupt the biofilm. Coupled with the spin and filtering steps, the full harvesting process will take between 30–60 min for a set of four 6-well plates. These steps become faster with practice; once familiar with the assay, it is possible to stagger the setup, washing, and processing of two sets of four 6-well plates. Subsequent processing steps will vary in length; the method we use takes ~2 hr for thawing and at least 3 hr for the concentration steps.
Supplementary Material
Biofilm formation of a C. albicans wild-type (WT) strain. The video shows the development of a WT biofilm in Spider medium using the Microfluidics Assay (Basic Protocol 10). Several yeast cells adhere to the channel, the cells form hyphae as time progresses, and a thick biofilm can be observed with intercalating hyphae and yeast cells as the biofilm matures. This biofilm was grown for 12 hr post-adherence using the BioFlux 1000Z under dynamic flow (0.5 dyne/cm2) at 37°C. The time-lapse video of biofilm formation is shown at 15 frames/sec.
Biofilm formation of a C. albicans biofilm-defective mutant strain. The video shows abnormal biofilm development of a known biofilm-defective mutant strain in Spider medium using the Microfluidics Assay (Basic Protocol 10). Fewer yeast cells adhere to the channel, hyphal formation is noticeably reduced, and the resulting biofilm is overall less dense relative to the wild-type (WT) strain shown in Video S1. This biofilm was grown for 12 hr post-adherence using the BioFlux 1000Z under dynamic flow (0.5 dyne/cm2) at 37°C. The time-lapse video of biofilm formation is shown at 15 frames/sec.
Significance Statement.
A significant virulence trait of the human fungal pathogen Candida albicans is its ability to form biofilms, communities of adhered cells encased in an extracellular matrix. C. albicans biofilms can develop on both biotic and abiotic surfaces, and are notoriously resistant to antimicrobial agents. Once formed, a biofilm acts as a resilient reservoir of cells that can lead to local and disseminated infections. We present several in vitro biofilm protocols, including protocols designed for high-throughput screening of mutant libraries and antifungal compounds as well as protocols designed to study specific aspects of biofilm development. These protocols allow for the detailed characterization of strains and antifungal compounds for effects on biofilm formation and serve as important precursors to in vivo studies.
Acknowledgments
We thank all members of the Nobile lab and BioSynesis team for helpful discussions on biofilm protocols and optimization procedures. This work was supported by National Institutes of Health (NIH) grants R21AI125801 (to C.J.N.), R35GM124594 (to C.J.N.), and R43AI131710 (to M.B.L.). C.L.E. was partially supported by a fellowship from the Sonoma County Mycological Association. D.L.R. was supported by a doctoral fellowship from the University of California Institute for Mexico and the United States (UC-MEXUS) and Consejo Nacional de Ciencia y Technologia (CONACYT).
LITERATURE CITED
- Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun. 2004;72(10):6023–6031. doi: 10.1128/IAI.72.10.6023-6031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie GS, Douglas LJ. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 1999;48(7):671–679. doi: 10.1099/00222615-48-7-671. [DOI] [PubMed] [Google Scholar]
- Bedell GW, Soll DR. Effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc-resistant and -sensitive pathways for mycelium formation. Infect Immun. 1979;26(1):348–354. doi: 10.1128/iai.26.1.348-354.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Uhl MA, Miller MG, Johnson AD. Identification and characterization of a Candida albicans mating pheromone. Mol Cell Biol. 2003;23(22):8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone RA, Fonzi WA. Virulence factors of Candida albicans. Trends Microbiol. 2001;9(7):327–335. doi: 10.1016/s0966-842x(01)02094-7. [DOI] [PubMed] [Google Scholar]
- Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001;183(18):5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels KJ, Srikantha T, Lockhart SR, Pujol C, Soll DR. Opaque cells signal white cells to form biofilms in Candida albicans. Embo J. 2006 doi: 10.1038/sj.emboj.7601099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas LJ. Medical importance of biofilms in Candida infections. Rev Iberoam Micol. 2002;19(3):139–143. [PubMed] [Google Scholar]
- Douglas LJ. Candida biofilms and their role in infection. Trends Microbiol. 2003;11(1):30–36. doi: 10.1016/s0966-842x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, Lanni F, Mitchell AP. Portrait of Candida albicans adherence regulators. PLoS Pathog. 2012;8(2):e1002525. doi: 10.1371/journal.ppat.1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox EP, Bui CK, Nett JE, Hartooni N, Mui MC, Andes DR, Nobile CJ, Johnson AD. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol Microbiol. 2015;96(6):1226–1239. doi: 10.1111/mmi.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox EP, Cowley ES, Nobile CJ, Hartooni N, Newman DK, Johnson AD. Anaerobic Bacteria Grow within Candida albicans Biofilms and Induce Biofilm Formation in Suspension Cultures. Curr Biol. 2014;24(20):2411–2416. doi: 10.1016/j.cub.2014.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati M, Ennis CL, Rodriguez DL, Nobile CJ. Visualization of Biofilm Formation in Candida albicans Using an Automated Microfluidic Device. J Vis Exp. 2017;(130) doi: 10.3791/56743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati M, Nobile CJ. Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes Infect. 2016;18(5):310–321. doi: 10.1016/j.micinf.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriott MM, Noverr MC. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Chemother. 2009;53(9):3914–3922. doi: 10.1128/AAC.00657-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriott MM, Noverr MC. Ability of Candida albicans mutants to induce Staphylococcus aureus vancomycin resistance during polymicrobial biofilm formation. Antimicrob Agents Chemother. 2010;54(9):3746–3755. doi: 10.1128/AAC.00573-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriott MM, Noverr MC. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 2011;19(11):557–563. doi: 10.1016/j.tim.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawser SP, Douglas LJ. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect Immun. 1994;62(3):915–921. doi: 10.1128/iai.62.3.915-921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Yip HK, Samaranayake YH, Yau JY, Samaranayake LP. Biofilm-forming ability of Candida albicans is unlikely to contribute to high levels of oral yeast carriage in cases of human immunodeficiency virus infection. J Clin Microbiol. 2003;41(7):2961–2967. doi: 10.1128/JCM.41.7.2961-2967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sudbery P. Candida albicans, a major human fungal pathogen. J Microbiol. 2011;49(2):171–177. doi: 10.1007/s12275-011-1064-7. [DOI] [PubMed] [Google Scholar]
- Krom BP, Cohen JB, McElhaney-Feser G, Busscher HJ, van der Mei HC, Cihlar RL. Conditions for optimal Candida biofilm development in microtiter plates. Methods Mol Biol. 2009;499:55–62. doi: 10.1007/978-1-60327-151-6_7. [DOI] [PubMed] [Google Scholar]
- Krom BP, Cohen JB, McElhaney Feser GE, Cihlar RL. Optimized candidal biofilm microtiter assay. J Microbiol Methods. 2007;68(2):421–423. doi: 10.1016/j.mimet.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Krom BP, Willems HM. In Vitro Models for Candida Biofilm Development. Methods Mol Biol. 2016;1356:95–105. doi: 10.1007/978-1-4939-3052-4_8. [DOI] [PubMed] [Google Scholar]
- Kuhn DM, Balkis M, Chandra J, Mukherjee PK, Ghannoum MA. Uses and limitations of the XTT assay in studies of Candida growth and metabolism. J Clin Microbiol. 2003;41(1):506–508. doi: 10.1128/JCM.41.1.506-508.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg BJ, Oude Lashof AM. Epidemiology of opportunistic invasive mycoses. Eur J Med Res. 2002;7(5):183–191. [PubMed] [Google Scholar]
- Kumamoto CA. Candida biofilms. Curr Opin Microbiol. 2002;5(6):608–611. doi: 10.1016/s1369-5274(02)00371-5. [DOI] [PubMed] [Google Scholar]
- LaFleur MD, Kumamoto CA, Lewis K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Chemother. 2006;50(11):3839–3846. doi: 10.1128/AAC.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFleur MD, Lucumi E, Napper AD, Diamond SL, Lewis K. Novel high-throughput screen against Candida albicans identifies antifungal potentiators and agents effective against biofilms. J Antimicrob Chemother. 2011;66(4):820–826. doi: 10.1093/jac/dkq530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafleur MD, Sun L, Lister I, Keating J, Nantel A, Long L, Ghannoum M, North J, Lee RE, Coleman K, Dahl T, Lewis K. Potentiation of azole antifungals by 2-adamantanamine. Antimicrob Agents Chemother. 2013;57(8):3585–3592. doi: 10.1128/AAC.00294-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Choi A, Bennett RJ. Defining pheromone-receptor signaling in Candida albicans and related asexual Candida species. Mol Biol Cell. 2011;22(24):4918–4930. doi: 10.1091/mbc.E11-09-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Kabrawala S, Fox EP, Nobile CJ, Johnson AD, Bennett RJ. Genetic control of conventional and pheromone-stimulated biofilm formation in Candida albicans. PLoS Pathog. 2013;9(4):e1003305. doi: 10.1371/journal.ppat.1003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Gulati M, Johnson AD, Nobile CJ. Development and regulation of single- and multi-species Candida albicans biofilms. Nat Rev Microbiol. 2018;16(1):19–31. doi: 10.1038/nrmicro.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Gulati M, Valle Arevalo A, Fishburn A, Johnson AD, Nobile CJ. Assessment and Optimizations of Candida albicans In Vitro Biofilm Assays. Antimicrob Agents Chemother. 2017;615 doi: 10.1128/AAC.02749-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Ribot JL. Candida albicans biofilms: more than filamentation. Curr Biol. 2005;15(12):R453–455. doi: 10.1016/j.cub.2005.06.020. [DOI] [PubMed] [Google Scholar]
- Morales DK, Hogan DA. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 2010;6(4):e1000886. doi: 10.1371/journal.ppat.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nailis H, Kucharikova S, Ricicova M, Van Dijck P, Deforce D, Nelis H, Coenye T. Real-time PCR expression profiling of genes encoding potential virulence factors in Candida albicans biofilms: identification of model-dependent and -independent gene expression. BMC Microbiol. 2010;10:114. doi: 10.1186/1471-2180-10-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett JE, Cain MT, Crawford K, Andes DR. Optimizing a Candida biofilm microtiter plate model for measurement of antifungal susceptibility by tetrazolium salt assay. J Clin Microbiol. 2011;49(4):1426–1433. doi: 10.1128/JCM.02273-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, Edwards JE, Filler SG, Mitchell AP. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2(7):e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Hartooni N, Mitchell KF, Hnisz D, Andes DR, Kuchler K, Johnson AD. A histone deacetylase complex mediates biofilm dispersal and drug resistance in Candida albicans. MBio. 2014;5(3):e01201–01214. doi: 10.1128/mBio.01201-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148(1–2):126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Johnson AD. Candida albicans Biofilms and Human Disease. Annual Reviews Microbiology. 2015;69:71–92. doi: 10.1146/annurev-micro-091014-104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol. 2005;15(12):1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Andes DR, Mitchell AP. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot Cell. 2006;5(10):1604–1610. doi: 10.1128/EC.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, Andes DR, Johnson AD, Mitchell AP. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 2009;7(6):e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norice CT, Smith FJ, Jr, Solis N, Filler SG, Mitchell AP. Requirement for Candida albicans Sun41 in biofilm formation and virulence. Eukaryot Cell. 2007;6(11):2046–2055. doi: 10.1128/EC.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue AJ, Eroy-Reveles AA, Knudsen GM, Ingram J, Zhou M, Statnekov JB, Greninger AL, Hostetter DR, Qu G, Maltby DA, Anderson MO, Derisi JL, McKerrow JH, Burlingame AL, Craik CS. Global identification of peptidase specificity by multiplex substrate profiling. Nat Methods. 2012;9(11):1095–1100. doi: 10.1038/nmeth.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce CG, Chaturvedi AK, Lazzell AL, Powell AT, Saville SP, McHardy SF, Lopez-Ribot JL. A Novel Small Molecule Inhibitor of Candida albicans Biofilm Formation, Filamentation and Virulence with Low Potential for the Development of Resistance. NPJ Biofilms Microbiomes. 2015;1 doi: 10.1038/npjbiofilms.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce CG, Saville SP, Lopez-Ribot JL. High-content phenotypic screenings to identify inhibitors of Candida albicans biofilm formation and filamentation. Pathog Dis. 2014;70(3):423–431. doi: 10.1111/2049-632X.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, Vande Walle K, Wickes BL, Lopez-Ribot JL. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Chemother. 2001;45(9):2475–2479. doi: 10.1128/AAC.45.9.2475-2479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard ML, Nobile CJ, Bruno VM, Mitchell AP. Candida albicans biofilm-defective mutants. Eukaryot Cell. 2005;4(8):1493–1502. doi: 10.1128/EC.4.8.1493-1502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll DR. The evolution of alternative biofilms in an opportunistic fungal pathogen: an explanation for how new signal transduction pathways may evolve. Infect Genet Evol. 2014;22:235–243. doi: 10.1016/j.meegid.2013.07.013. [DOI] [PubMed] [Google Scholar]
- Srikantha T, Daniels KJ, Pujol C, Sahni N, Yi S, Soll DR. Nonsex genes in the mating type locus of Candida albicans play roles in a/alpha biofilm formation, including impermeability and fluconazole resistance. PLoS Pathog. 2012;8(1):e1002476. doi: 10.1371/journal.ppat.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournu H, Van Dijck P. Candida biofilms and the host: models and new concepts for eradication. Int J Microbiol. 2012;2012:845352. doi: 10.1155/2012/845352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, Kohler JR, Kadosh D, Lopez-Ribot JL. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010;6(3):e1000828. doi: 10.1371/journal.ppat.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppuluri P, Pierce CG, Thomas DP, Bubeck SS, Saville SP, Lopez-Ribot JL. The transcriptional regulator Nrg1p controls Candida albicans biofilm formation and dispersion. Eukaryot Cell. 2010;9(10):1531–1537. doi: 10.1128/EC.00111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter MB, Salcedo EC, Lohse MB, Hartooni N, Gulati M, Sanchez H, Takagi J, Hube B, Andes DR, Johnson AD, Craik CS, Nobile CJ. Global Identification of Biofilm-Specific Proteolysis in Candida albicans. MBio. 2016;7(5) doi: 10.1128/mBio.01514-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Sahni N, Daniels KJ, Lu KL, Srikantha T, Huang G, Garnaas AM, Soll DR. Alternative mating type configurations (a/alpha versus a/a or alpha/alpha) of Candida albicans result in alternative biofilms regulated by different pathways. PLoS Biol. 2011;9(8):e1001117. doi: 10.1371/journal.pbio.1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Biofilm formation of a C. albicans wild-type (WT) strain. The video shows the development of a WT biofilm in Spider medium using the Microfluidics Assay (Basic Protocol 10). Several yeast cells adhere to the channel, the cells form hyphae as time progresses, and a thick biofilm can be observed with intercalating hyphae and yeast cells as the biofilm matures. This biofilm was grown for 12 hr post-adherence using the BioFlux 1000Z under dynamic flow (0.5 dyne/cm2) at 37°C. The time-lapse video of biofilm formation is shown at 15 frames/sec.
Biofilm formation of a C. albicans biofilm-defective mutant strain. The video shows abnormal biofilm development of a known biofilm-defective mutant strain in Spider medium using the Microfluidics Assay (Basic Protocol 10). Fewer yeast cells adhere to the channel, hyphal formation is noticeably reduced, and the resulting biofilm is overall less dense relative to the wild-type (WT) strain shown in Video S1. This biofilm was grown for 12 hr post-adherence using the BioFlux 1000Z under dynamic flow (0.5 dyne/cm2) at 37°C. The time-lapse video of biofilm formation is shown at 15 frames/sec.