Abstract
Notch receptor signaling is a highly conserved cell communication system in most multicellular organisms and plays a critical role at several junctures in animal development. In C. elegans, GLP-1/Notch signaling is essential for both germline stem cell maintenance and germ cell proliferation during gonad development. Here, we show that subunits (POLA-1, DIV-1, PRI-1, and PRI-2) of the DNA polymerase alpha-primase complex are required for germ cell proliferation in response to GLP-1/Notch signaling in different tissues at different developmental stages. Specifically, genetic and functional analyses demonstrated that 1) maternally contributed DIV-1 (regulatory subunit) is indispensable non-cell autonomously for GLP-1/Notch-mediated germ cell proliferation during early larval development, whereas POLA-1 (catalytic subunit) and two primase subunits, PRI-1 and PRI-2, do not appear to be essential; 2) germline POLA-1, PRI-1 and PRI-2 play a crucial role in GLP-1/Notch-mediated maintenance of proliferative cell fate during adulthood, while DIV-1 is dispensable; and 3) germline POLA-1, DIV-1, PRI-1, and PRI-2 function in tandem with PUF (Pumilio/FBF) RNA-binding proteins to maintain germline stem cells in the adult gonad. These findings suggest that the subunits of the DNA polymerase alpha-primase complex exhibit both discrete and shared functions in GLP-1/Notch or PUF-mediated germ cell dynamics in C. elegans. These findings link the biological functions of DNA replication machineries to signals that maintain a stem cell population, and may have further implications for Notch-dependent tumors.
Keywords: DNA pol α-primases, Notch signaling, PUF proteins, Germline, C. elegans
Graphical abstract
Self-renewal and differentiation of stem cells are tightly regulated by signal transduction pathways and intrinsic regulators. Using the nematode C. elegans germline as a model system, we show that subunits (POLA-1, DIV-1, PRI-1, and PRI-2) of the DNA polymerase alpha-primase complex promote germline stem cell proliferation in response to Notch signaling in different tissues at different developmental stages.
Introduction
Germline stem cells (GSCs) are characterized by their ability to self-renew and to give rise to either sperm or eggs (differentiation to gametes). A balance between self-renewal and differentiation of GSCs is strictly controlled by an intricate network encompassing both extrinsic pathways and intrinsic regulators [1]. Aberrant regulation of this network has impacts ranging from loss of the stem cell pool or a specific germ cell type to over-proliferation of undifferentiated germ cells. The latter has been implicated in germline tumors [1].
One of the key extrinsic cues for stem cell maintenance is Notch signaling [2, 3]. The resulting intercellular signal plays essential and varied roles in the regulation of many types of stem cells [2, 3]. In C. elegans, GLP-1/Notch signaling is required for GSC maintenance and germ cell proliferation during development [4] (Fig. 1A). Briefly, the Notch ligand, LAG-2, is expressed in stem cell niche (a function performed by “distal tip cells” [DTCs] in C. elegans) [5]. LAG-2 interacts with the GLP-1/Notch receptor, leading to its proteolytic cleavage. This is followed by the translocation of GLP-1/Notch intracellular domain (NICD) from the cell membrane to the nucleus, where it forms a ternary complex with LAG-1 (DNA-binding protein) and LAG-3/SEL-8 (transcription coactivator) to stimulate the expression of target genes [6, 7] (Fig. 1B). Among the target genes are fbf-2 [a member of the PUF (Pumilio/FBF) RNA-binding protein family] [8], lip-1 [a dual specificity phosphatase] [9, 10], lst-1 [lateral signaling target-1, no known homolog] [11, 12], sygl-1 [synthetic germline proliferation defect, no known homolog] [11]. Among them, lst-1 and sygl-1 function redundantly to promote germ cell proliferation and maintain GSCs in C. elegans [11]. For example, lst-1 and sygl-1 single mutant worms have self-fertile hermaphrodite germlines comparable in size and organization to wild-type worms [11]. In contrast, most lst-1 sygl-1 double mutant worms displayed a premature meiotic entry (defined as Glp [Germline proliferation defect]) phenotype [11]. Additionally, FBF-2 and LIP-1 proteins promote germ cell proliferation by inhibiting meiosis-promoting regulators such as GLD-1/Quaking and MPK-1/ERK [4, 9, 13] and cell cycle regulators like the cyclin E/CDK2 inhibitor, CKI-2 [14]). Therefore, reduced GLP-1/Notch activity inhibits germ cell proliferation. As an alternative fate, these cells differentiate into mature sperm [15]. Conversely, elevated GLP-1/Notch activity promotes germ cell proliferation and simultaneously inhibits their differentiation, resulting in germline tumors [16].
Figure 1. C. elegans germline and GLP-1/Notch signaling pathway.
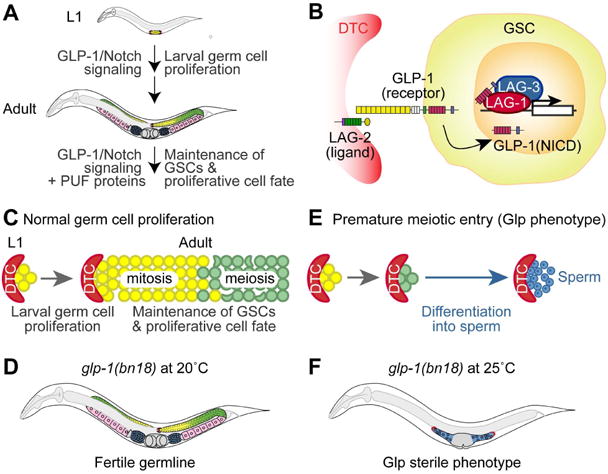
(A) Schematic of C. elegans germline development. GLP-1/Notch signaling pathway is required for germ cell proliferation during larval development. In coordination with PUF (Pumilio/FBF) proteins, GLP-1/Notch signaling is crucial for GSCs maintenance during adulthood. In the distal end, the somatic gonadal cells, called DTCs (distal tip cells), act as a GSC niche which is essential for GSC maintenance and germ cell proliferation. Germ cells (yellow) at the distal end of the germline divide mitotically. As germ cells move proximally, they enter meiosis (green) and differentiate into either sperm (blue) or oocytes (pink). GLP-1/Notch signaling is activated in the distal mitotic germline. (B) C. elegans GLP-1/Notch signaling pathway. The LAG-2 ligand, localized in the DTC, signals to both GLP-1/Notch receptor in GSCs and in mitotically dividing germ cells. Upon GLP-1 activation, the GLP-1/Notch intracellular domain (NICD), LAG-1 and LAG-3 form a ternary complex in the nucleus and activate the transcription of target genes. (C and D) Schematics of normal germ cell proliferation and the germline phenotype of glp-1(bn18) mutant at 20°C. In a normal germline, GLP-1/Notch signaling promotes GSC maintenance and germ cell proliferation. Once germ cells (yellow) move proximally, they enter meiosis (green). (E and F) Schematic of premature meiotic entry (also called Glp) phenotype and the germline phenotype of glp-1(bn18) mutant at 25°C. Germline with a Glp phenotype, occurring very early in larval development, has a complete depletion of proliferative cells and only ∼16 sperm being formed per gonad arm.
In this study, we explored the role of the DNA polymerase alpha-primase (henceforth termed DNA pol α-primase) complex in C. elegans GLP-1/Notch-mediated germ cell proliferation. Using a temperature sensitive glp-1(bn18) loss-of-function mutant worm, our focused RNAi screen has identified div-1 (a homolog of the human POLA2 regulatory subunit) as a positive regulator of GLP-1/Notch-mediated germ cell proliferation. Interestingly, DIV-1 is required non-cell autonomously for germ cell proliferation during early larval development. Conversely, other DNA pol α-primase subunits, POLA-1 (a homolog of mammalian POLA1 catalytic subunit), PRI-1 (a homolog of mammalian PRIM1 primase) and PRI-2 (a homolog of mammalian PRIM2 primase), control germ cell proliferation cell autonomously in the adult gonad. These findings suggest that each subunit may respond independently to GLP-1/Notch signaling to mediate germ cell proliferation at different developmental stages. The regulatory function of these subunits appears to be distinct from their initiation role in DNA replication. Furthermore, all four subunits have a shared cell autonomous function in PUF-mediated GSC maintenance. Therefore, our findings may provide a powerful model organism to investigate the connection between Notch signaling and DNA replication machineries during animal reproductive development.
Results
S phase arrest by HU enhances a Glp phenotype in glp-1(bn18) mutant worms
The cell cycle state of stem cells determines cell fate [17, 18]. Recent studies show that cyclin E and CDK1 (S phase entry factors) are critical for the maintenance of germline stem cells (GSCs) and proliferative cell fate in flies and worms [19-21]. Since GSCs have a short or absent G1 and a long S phase [20], it seems likely that events in early S phase involving this kinase complex might be essential to maintain their undifferentiated state. First, we asked whether S phase progression is functionally linked to GLP-1/Notch signaling that is essential for germ cell proliferation. We used a temperature sensitive glp-1(bn18) loss-of-function mutant worm that exhibits a sensitized background to determine the genetic connection of S phase progression to GLP-1/Notch signaling in vivo [20]. Proliferating germ cells in the glp-1(bn18) mutant worms resemble those of wild-type, albeit with a reduced germ cell number (∼50% of wild-type at young adult stage) at permissive temperature (20°C) (Fig. 1C, D) [22, 23]. At the restrictive temperature (25°C), however, a premature meiotic entry causes most glp-1(bn18) mutant worms to display the Glp (germline proliferation defect) phenotype, which produces just a few mature sperm cells and no oocytes (Fig. 1E, F) [15]. To determine the role of molecular events during S phase in GLP-1/Notch-mediated germ cell proliferation during early larval development, we treated wild-type and glp-1(bn18) mutants at L1 stage with 40 mM hydroxyurea (HU, a DNA synthesis inhibitor) at 20°C. After incubation for 18 hours in HU plate, worms were transferred to a normal NGM plate, and were allowed to grow to adults. The Glp phenotype was scored by staining whole worms with DAPI. Wild-type germlines did not show a Glp phenotype in the absence or presence of HU (Fig. 2A) and most glp-1(bn18) (-HU) mutants were normal and self-fertile (Fig. 2A, B). However, HU-treated glp-1(bn18) mutants exhibited a substantial increase in the Glp phenotype up to 93% (Fig. 2A, C). These results suggest that S phase arrest by HU impairs GLP-1/Notch-mediated germline proliferation during early larval development.
Figure 2. DIV-1 is required for GLP-1/Notch-mediated early germ cell proliferation.
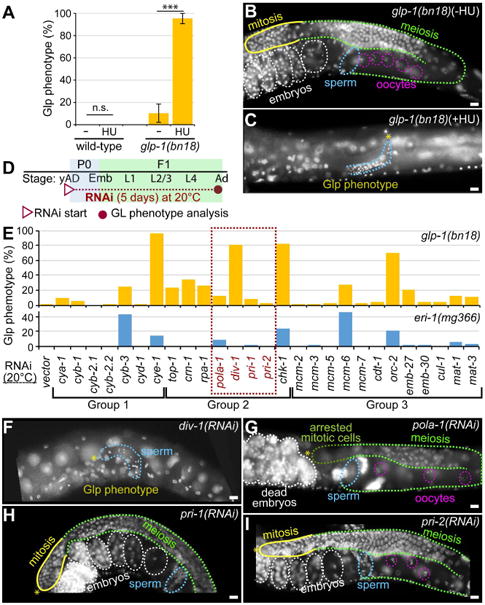
(A) The percentage of animals scored with a Glp phenotype following hydroxyl urea (HU) treatment. The Glp phenotype was strictly defined as no mitotic cells and only sperm by DAPI staining. Standard deviation bars were calculated from three independent experiments. n.s., not statistically significant; *** p<0.001. (B and C) DAPI-stained adult hermaphrodite germlines. (D) Schematic of RNAi experiment. (E) The percentage of animals scored with a Glp phenotype by RNAi of cell cycle regulator genes in glp-1(bn18) and eri-1(mg366) mutant worms at 20°C. The dotted red box contains genes that encode the subunits of the DNA pol α-primase complex. (F-I) DAPI-stained adult hermaphrodite germlines. (*) distal end; yellow lines, mitotic cells; broken green lines, meiotic cells; broken pink circles, oocyte nuclei; broken blue lines, sperm; broken white lines, embryos; broken dark green line in G, arrested mitotic cells. Scale bars: 10 μm.
DIV-1/POLA2 promotes GLP-1/Notch-mediated germ cell proliferation during early larval development
To further elucidate the role of molecular events during S phase in GLP-1/Notch-mediated germ cell proliferation during early larval development, we knocked down the expression of key regulator genes encoding Cyclins (Group 1), DNA replication processing proteins (Group 2), and DNA replication licensing proteins (Group 3), by feeding RNAi to glp-1(bn18) mutant worms at 20°C (Table 1). Specifically, young adult (yAd) glp-1(bn18) hermaphrodites (P0) were transferred to respective feeding RNAi plates to deplete the expression of both maternal and zygotic mRNAs (Fig. 2D). RNAi of these genes showed substantial embryonic lethality as previously reported by others [24, 25]. To bypass the requirement for GLP-1/Notch during embryogenesis and enable the monitoring of GLP-1/Notch-dependence during GSC expansion in larvae, RNAi knockdowns were performed in glp-1(bn18) mutant worms that have been upshifted to 20°C in early larval development. The germline phenotypes of the surviving F1 progeny were scored by DAPI staining fixed adult worms (3 days post L1, 5 days RNAi) (Fig. 2D). Consistent with the results of others [20], cye-1(RNAi) significantly enhanced the Glp phenotype of glp-1(bn18) mutant worms even at 20°C (Fig. 2E). Under the same conditions, we also examined the effects of S phase regulators on GLP-1/Notch-mediated germ cell proliferation. Interestingly, RNAi of some S phase genes, but not all, enhanced the Glp phenotype of glp-1(bn18) mutant worms with a wide range of penetrance (Fig. 2E, yellow bars). In particular, the Glp phenotype was considerably enhanced by inhibition of the div-1, chk-1, or orc-2 genes at 20°C (Fig. 2E, F). C. elegans div-1 (a homolog of the mammalian POLA2) encodes the regulatory subunit of the DNA pol α-primase complex and has an essential role in early embryogenesis [24]. The DIV-1 protein forms a complex with POLA-1 (catalytic subunit) and two primases, PRI-1 and PRI-2, to initiate DNA replication in proliferating cells. To test if other DNA pol α-primase complex subunits are required for GLP-1/Notch-mediated germ cell proliferation, we also depleted the expression of pola-1, pri-1, or pri-2 genes by RNAi in glp-1(bn18) mutant worms at 20°C. Intriguingly, RNAi of pola-1, pri-1, or pri-2 genes very slightly enhanced the Glp phenotype, suggesting a non-complex function for DNA pol α-primase (Fig. 2E, G-I). We also performed RNAi experiments in the same conditions on eri-1(mg366) mutant worms, which are hypersensitive to RNAi [26] (Fig. 2E, blue bars). Loss of cyb-3, pola-1, chk-1, mcm-6, or orc-2 expression partially enhanced the Glp phenotype in eri-1(mg366) mutant worms but at lower efficiencies, whereas no effect was seen with RNAi of div-1, pri-1, and pri-2 (Fig. 2E, blue bars). These results suggest that DIV-1 may have a specialized function in GLP-1/Notch-mediated germ cell proliferation during early larval development.
Table 1. Summary of RNAi genes.
| Gene | Molecular Identity | Mammalian Homolog |
|---|---|---|
| Group 1 (Genes encoding cyclins) | ||
| cya-1 | A-type cyclin | Cyclin A |
| cyb-1 | B-type cyclin | Cyclin B1 |
| cyb-2.1 | B-type cyclin | Cyclin B1 |
| cyb-2.2 | B-type cyclin | Cyclin B1 |
| cyb-3 | B-type cyclin | Cyclin B3 |
| cyd-1 | D-type cyclin | Cyclin D |
| cye-1 | E-type cyclin | Cyclin E |
| Group 2 (Genes encoding DNA replication processing proteins) | ||
| top-1 | Type 1 Topoisomerase | TOP-1 |
| crn-1 | 5′3′ exonuclease | Flap endonuclease 1 |
| rpa-1 | Replication protein A | RPA |
| pola-1 | DNA Polα catalytic subunit A | POLA1 |
| div-1 | DNA Polα regulatory subunit B | POLA2 |
| pri-1 | DNA primase subunit D | PRIM1 |
| pri-2 | DNA primase subunit C | PRIM2 |
| chk-1 | Serine Threonine protein kinase | CHK1 |
| Group 3 (Genes encoding DNA replication licensing proteins) | ||
| mcm-2 | DNA replication licensing factor | MCM2 |
| mcm-3 | DNA replication licensing factor | MCM2/3/5 |
| mcm-5 | DNA replication licensing factor | MCM5 |
| mcm-6 | DNA replication licensing factor | MCM6 |
| mcm-7 | DNA replication licensing factor | MCM7 |
| cdt-1 | DNA replication licensing factor | CDT1 |
| orc-2 | Origin recognition complex subunit | ORC2 |
| emb-27 | Anaphase-promoting complex (APC) subunit | CDC16 |
| emb-30 | APC/cyclosome component | ANAPC4 |
| cul-1 | Cullin | CUL1 |
| mat-1 | APC/cyclosome subunit | CDC27/APC3 |
| mat-3 | APC subunit | CDC23/APC8 |
Non-cell autonomous function of DIV-1 in GLP-1/Notch-mediated germ cell proliferation
The div-1(or148) loss-of-function mutant strain of worms displays embryonic lethality due to delayed embryonic cell division [24]. To avoid the lethality and characterize the function of DIV-1 in germ cell proliferation, we used a less penetrant approach instead of the div-1(or148) mutant worms. First, to test whether DIV-1 is necessary cell autonomously for early germ cell proliferation, we used an rrf-1(pk1417); glp-1(bn18) double mutant worm. The rrf-1(pk1417) mutation prevents RNAi effectiveness in the soma, but not in the germline [27]. Resulting phenotypes must therefore be due to gene function only in the germ cells themselves. We depleted both maternal and zygotic pools of div-1 and cye-1 mRNAs by RNAi in glp-1(bn18) and rrf-1; glp-1(bn18) mutant worms starting at the young adult (yAd) P0 stage, and subsequently analyzed their germline phenotypes in F1 progeny as worms reached adult stage (Fig. 3A). Because the expression of zygotic/somatic target genes is preserved in rrf-1; glp-1(bn18) mutant worms (Fig. 3B), reversion of phenotypes in this strain indicate that the target gene is affecting germ cells only through expression in neighboring somatic cells. Results showed that cye-1(RNAi) considerably enhanced the Glp phenotype of both glp-1(bn18) and rrf-1; glp-1(bn18) mutant worms even at 20°C (Fig. 3C). In contrast, div-1(RNAi) promoted the Glp phenotype in glp-1(bn18) mutant worms, but not in rrf-1; glp-1(bn18) double mutant worms at 20°C (Fig. 3C). Depletion of germline DIV-1 by div-1(RNAi) was confirmed by immunohistochemistry and western blot as well as by assessing the embryonic lethality of progeny (Data not shown). Therefore, CYE-1 functions within germ cells to promote proliferation, as one might expect. Unexpectedly, however, the DIV-1 functions in somatic cells to drive neighboring germ cells into proliferation. To confirm this result, we used a germline-RNAi incompetent ppw-1(pk1425); glp-1(bn18) double mutant. PPW-1 (a PAZ/PIWI domain-containing protein) belongs to the Argonaute family of proteins [28]. The ppw-1(pk1425) mutation prevents RNAi effectiveness in the germline, but not in the soma [28]. Resulting phenotypes must therefore be due to gene function only in the somatic cells themselves. Notably, div-1(RNAi) significantly enhanced the Glp phenotype in ppw-1; glp-1(bn18) double mutant worms at 20°C (Fig. 3C). These observations suggest that DIV-1 is required non-cell autonomously for GLP-1/Notch-mediated germ cell proliferation during early larval development. Because depletion of DIV-1 selectively from the soma (ppw-1 mutant background) enhances Glp phenotype, but its selective loss in germ cells (rrf-1 mutant background) has no effect on proliferation, DIV-1 must be acting from the somatic gonadal tissues to alter the fate of underlying germ cells. The path by which this occurs is not known.
Figure 3. Somatic DIV-1 promotes GLP-1/Notch-mediated germ cell proliferation during early larval development.
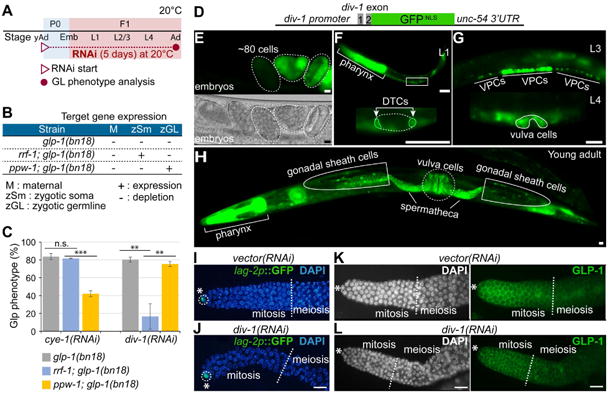
(A) Schematic of RNAi experiment. (B) RNAi in glp-1(bn18), rrf-1(pk1417); glp-1(bn18), and ppw-1(pk1425); glp-1(bn18) mutant worms. The expression of zygotic/somatic target genes is protected from RNAi in rrf-1(pk1417) and ppw-1(pk1425) mutant background. (C) The percentage of animals scored with a Glp phenotype. Standard deviation bars were calculated from three independent experiments. n.s., not statistically significant; ** p<0.01; *** p<0.001. (D) Design of div-1p∷GFP transgene. (E-H) The expression of div-1p∷GFP transgene in embryos (E), L1 (F), L3/L4 (G), and young adult (H). (I-L) The expression of LAG-2∷GFP and GLP-1. Young adult staged lag-2(promoter)∷GFP transgenic worms and wild-type worms were plated on div-1(RNAi) plates, and 5 days later, the expression levels of lag-2p∷GFP and GLP-1 were determined by staining dissected gonads with anti-GFP and anti-GLP-1 antibodies. Broken lines indicate the boundary between mitosis and meiosis. (*) distal end, Scale bars: 10 μm.
We then assessed the pattern of DIV-1 expression using a transgenic worm that expresses an extrachromosomal div-1(promoter)∷div-1(exon 1 and 2)∷GFP (henceforth referred to as div-1p∷GFP) transgene (Fig. 3D). The zygotic div-1p∷GFP was first detected at about the 80-cell stage in embryos and remained strongly expressed in dividing cells throughout the rest of embryogenesis (Fig. 3E). div-1p∷GFP was also highly expressed in the pharynx and DTCs in early larval stages (Fig. 3F), in vulva precursor cells in L3, and in vulva precursor cells in L4 (Fig. 3G). Most strikingly, div-1p∷GFP was strongly expressed in the somatic gonadal tissues, including gonadal sheath cells, spermatheca, and the vulva (Fig. 3H). It was previously reported that somatic sheath/spermatheca tissues interact with germ cells by induction events to control their proliferation and differentiation [29]. Therefore, we suggest that DIV-1 may function non-cell autonomously in somatic gonadal tissues to promote the GLP-1/Notch signaling that drives early germ cells to proliferate (see Fig. 6B).
Figure 6. Working Model.
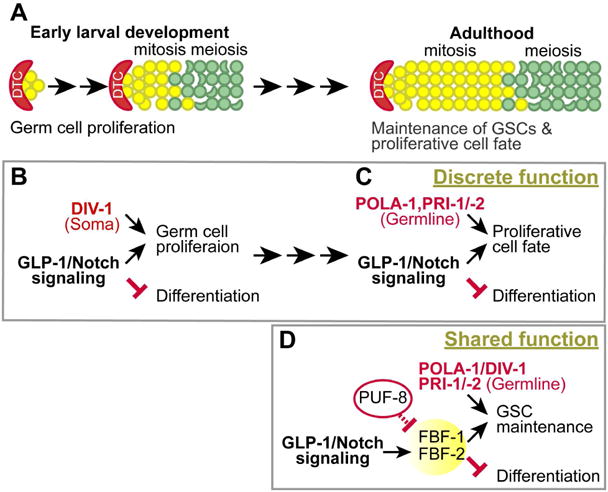
(A) Germline development during early larval stage and adulthood. (B and C) The discrete function of POLA-1, DIV-1, PRI-1, and PRI-2 in GLP-1/Notch-mediated germ cell proliferation either non-cell autonomously or cell autonomously during early larval development and adulthood, respectively. (D) The shared function of POLA-1, DIV-1, PRI-1, and PRI-2 in PUF-mediated GSC maintenance.
Next, we addressed what portion of soma-germ cell communication might require DIV-1. To do so, we tested whether div-1 knockdown disrupts somatic gonad development itself, or if it may affect the expression of LAG-2 and GLP-1, which are the ligand-receptor pair that signal maintenance of mitosis for germ cell proliferation. We performed div-1(RNAi) in lag-2(promoter)∷GFP transgenic and wild-type worms, and then immunostained dissected gonads with anti-GFP and anti-GLP-1 antibodies. Result showed that div-1(RNAi) did not affect somatic gonad development (data not shown) as the formation and morphology of the sheath was normal. Loss of DIV-1 did not reduce the expression of lag-2p∷GFP in the DTC (Fig. 3I, J), nor was there any substantial change in GLP-1 abundance in the membrane of germ cells (Fig. 3K, L). Thus, the primary signal for sustained mitosis of early germ cells is unperturbed by depletion of div-1. While it is still unclear how DIV-1 functions non-cell autonomously in somatic cells to influence germ cell proliferation, it is likely to modulate GLP-1/Notch signaling indirectly or its non-cell autonomous function may be independent of GLP-1/Notch signaling. Our findings do not rule out that div-1 knockdown may delay somatic development and affect soma-germline interaction by other means. Studying the function of DIV-1 in the somatic development remains a challenge for the future.
Once signaled, the maintenance of proliferative cell fate requires POLA-1, PRI-1, and PRI-2, but not DIV-1 during adulthood
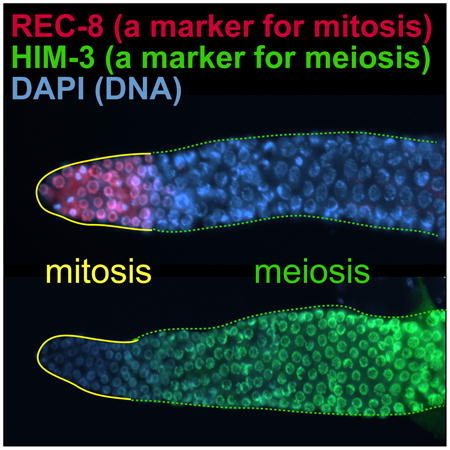
During adulthood, GLP-1/Notch signaling also maintains proliferative cell fate in the germline following the initial mitotic expansion in larvae. To examine whether div-1 is also required for maintenance of proliferative cell fate during adulthood, L4 staged glp-1(bn18) and rrf-1; glp-1(bn18) mutant worms were depleted of vector (negative control), cye-1 (positive control), pola-1, div-1, pri-1, or pri-2 by RNAi for 4 days at 20°C (Fig. 4A). Germline phenotypes were determined by staining dissected gonads with anti-REC-8 antibody, a marker of mitotic germ cells [30], and anti-HIM-3 antibody, a marker of meiotic germ cells [31]. In both glp-1(bn18) and rrf-1; glp-1(bn18) mutant worms, div-1 was not required for germ cell proliferation, whereas cye-1, pola-1, pri-1, and pri-2 were all found to be essential (Fig. 4B). Similar to wild-type, glp-1(bn18) mutant germlines maintain mitotically dividing cells, as evidenced by the REC-8(+)/HIM-3(-) region (yellow outlines) and transition to meiotic cells (REC-8(-)/HIM-3(+); broken green outlines) at 20°C (Fig. 4C). As previously described by others [20], cye-1(RNAi) dramatically enhanced the Glp phenotype of both glp-1(bn18) single and rrf-1; glp-1(bn18) double mutant worms, such that no mitotic germ cells are evident (Fig. 4B, D). RNAi of pola-1, pri-1, or pri-2 also enhanced the Glp phenotype of both mutant worms (Fig. 4B, E, G, H). However, div-1(RNAi) failed to enhance the Glp phenotype in both mutant worms (Fig. 4B, F). Since all these regulators are expressed in the C. elegans germline, we suggest that GLP-1/Notch-mediated maintenance of proliferative cell fate requires germline POLA-1, PRI-1, and PRI-2, but not DIV-1 during adulthood. To confirm this result, we performed somatic RNAi in ppw-1; glp-1(bn18) double mutant worms, in which RNAi is largely defective in the germline [28]. Their germline phenotypes were determined by staining dissected gonads with anti-REC-8 and anti-HIM-3 antibodies (Fig. 4I-N). Although the percentage of Glp phenotype by cye-1(RNAi) was slightly decreased in the ppw-1; glp-1(bn18) mutant worms in comparison to rrf-1; glp-1(bn18) mutant worms, cye-1(RNAi) significantly enhanced the Glp phenotype in both rrf-1; glp-1(bn18) and ppw-1; glp-1(bn18) mutant worms (Fig. 4B, J). This result indicates that CYE-1 appears to be critical for GLP-1/Notch-mediated germ cell proliferation in both somatic and germline tissues. However, the percentages of Glp phenotype by RNAi of pola-1, pri-1, or pri-2 were significantly decreased in the ppw-1; glp-1(bn18) mutant worms (Fig. 4B, K-N). These results suggest that GLP-1/Notch-mediated maintenance of proliferative cell fate requires cell autonomously POLA-1, PRI-1, and PRI-2, but not DIV-1 during adulthood.
Figure 4. POLA-1, PRI-1 and PRI-2 are critical cell autonomously for GLP-1/Notch-mediated maintenance of proliferative cell fate during adulthood.
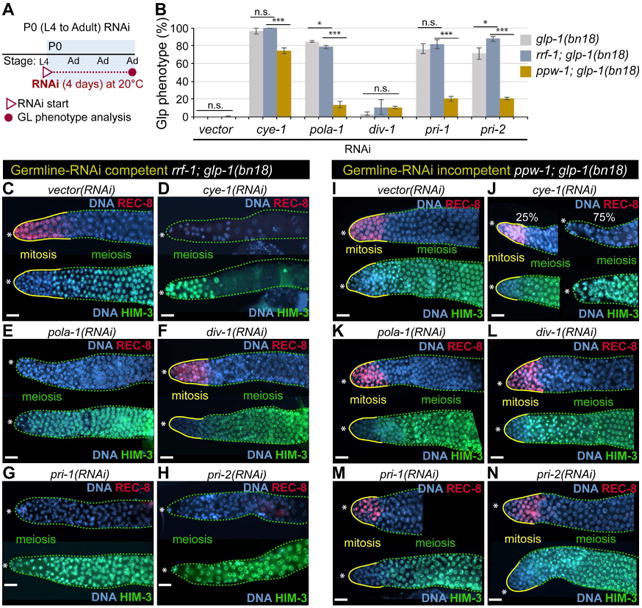
(A) Schematic of RNAi experiment. (B) The percentage of worms exhibiting a Glp phenotype at 20°C. Standard deviation bars were calculated from three independent experiments. n.s., not statistically significant; * p<0.05; ** p<0.01; *** p<0.001. (C-N) Germline staining with anti-REC-8 and anti-HIM-3 antibodies. Solid yellow lines, mitotic cells (REC-8(+)/HIM-3(-)); broken green lines, meiotic cells (REC-8(-)//HIM-3(+)). (*) distal end. Scale bars: 10 μm.
Fox et al. previously reported that a 2-day pri-1(RNAi) silencing failed to enhance the Glp phenotype of rrf-1; glp-1(bn18) mutant worms [20]. However, our study shows that 4-day RNAi silencing of pola-1, pri-1 or pri-2 dramatically enhances the Glp phenotype of rrf-1; glp-1(bn18) mutant worms (Fig. 4B). We therefore reevaluated the roles of both primase (PRI-1 and PRI-2) on GLP-1/Notch-mediated maintenance of proliferative cell fate. In this study, we did not test the effect of POLA-1 potential redundancy with PRI-1 and PRI-2 due to genetic complexity/efficacy of such triple depletions. As previously observed by others [20], most pri-1(RNAi) and pri-2(RNAi) germlines expressed REC-8 after a 2-day depletion. However, after 4-days of silencing, adult germlines exhibited a pronounced Glp phenotype, indicated by a lack of REC-8 and positive HIM-3 staining beginning at the DTC (presumptive niche; Fig. 4B, G, H). This result suggested that germlines with a larval mitotic germ cell population progressively lost all those proliferative cells after 4-days of pri-1(RNAi) or pri-2(RNAi) silencing. To test whether the delay in Glp phenotype development was due to redundancy, we performed double RNAi of pri-1 and pri-2 in rrf-1; glp-1(bn18) for 2 days at 20°C and scored the percentage of worms with the Glp phenotype, again by staining gonads with anti-REC-8 and anti-HIM-3 antibodies. Interestingly, the percentage of pri-1/-2(RNAi) worms with the Glp phenotype was significantly increased (52% ± 3, n=98) even at just 2 days. These results suggest that the two primase subunits (PRI-1 and PRI-2) of the DNA pol α-primase complex must be present in adult germ cells for the GLP-1/Notch-mediated signal that maintains a proliferating population (see Fig. 6C).
DIV-1 may work with FBF/PUF RNA-binding proteins to maintain GSCs
In addition to GLP-1/Notch signaling, a battery of mRNA regulators function to maintain the balance between proliferation and differentiation (Fig. 5A). A well-conserved mRNA translational regulator is the PUF (Pumilio/FBF) RNA-binding protein. PUF proteins control various physiological processes, including GSC maintenance and cell fate specification, by interacting with 3′ untranslated regions (3′UTRs) and modulating mRNA translation in a wide variety of eukaryotes [32]. Translational control is prevalent in the germlines of all organisms in order to promote gamete development and circumvent meiotic suppression of gene transcription [33-35]. C. elegans has multiple PUF genes with individualized roles. In particular, FBF-1 and FBF-2 proteins play an essential role in GSC maintenance and germ cell proliferation [36]. FBF-1 and FBF-2 proteins are 96% identical (henceforth called FBF), and are largely redundant: fbf-1 and fbf-2 single mutant worms are both self-fertile with germlines morphologically identical to wild-type [36]. In contrast, fbf-1 fbf-2 double mutant worms maintain their GSCs until the L4 stage, but then most GSCs leave mitotic cell cycle, enter meiosis, and the entire (small) population eventually differentiates into sperm at 20°C, leaving no GSC population (Glp phenotype) [36] (Fig. 5B, C). Remarkably, the Glp phenotype of fbf-1 fbf-2 mutant worms was suppressed by loss of PUF-8 as evidenced by the restoration of mitotically dividing cells (EdU-positive) in the germlines of puf-8 fbf-1 fbf-2 triple mutant worms at 20°C (Fig. 5B, D). These results indicate that PUF-8 antagonizes FBF-mediated GSC maintenance. Finally, we sought to establish whether the DNA pol α-primase subunits are required to act in germ cells for PUF-mediated GCS maintenance. We again performed RNAi of pola-1, div-1, pri-1, or pri-2 this time in rrf-1; puf-8 fbf-1 fbf-2 quadruple mutant worms at 20°C. Again, the rrf-1(pk1417) mutation abrogates RNAi in the soma, but not in the germline [27]. Most vector(RNAi); rrf-1; puf-8 fbf-1 fbf-2 germlines were positive for EdU-labeling at 20°C (Fig. 5E, F). However, the RNAi of pola-1, pri-1, pri-2, or div-1 genes dramatically subverted the restoration of mitotically dividing germ cells in rrf-1; puf-8 fbf-1 fbf-2 mutant worms, which were all negative for EdU-labeling (Fig. 5E, G-J). These results suggest that germline POLA-1, DIV-1, PRI-1, and PRI-2 may have a shared function in the PUF-mediated contribution to GSC maintenance and mitotic cell cycle progression that is germ cell autonomous (see Fig. 6D).
Figure 5. POLA-1, DIV-1, PRI-1 and PRI-2 are necessary cell autonomously for PUF-mediated GSC maintenance.
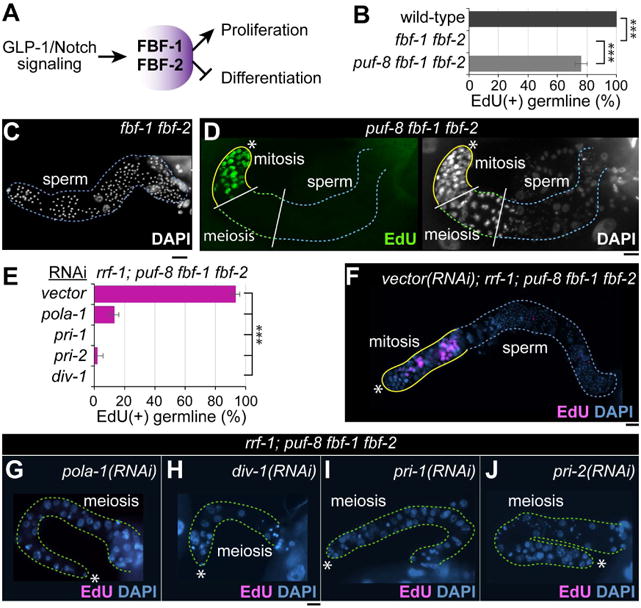
(A) A simplified network controlling the germ cell proliferation/differentiation decision. (B) The percentage of worms with EdU-positive germ cells. Standard deviation bars were calculated from three independent experiments. *** p<0.001. (C) DAPI staining of dissected adult fbf-1 fbf-2 gonads. (D) EdU staining of dissected adult puf-8 fbf-1 fbf-2 mutant gonads. (E) The percentage of worms with EdU-positive germ cells. Standard deviation bars were calculated from three independent experiments. *** p<0.001. (F-I) EdU staining of dissected adult gonads. Solid yellow lines, a region with EdU(+) cells; broken green lines, a region differentiating cells (meiotic cells and sperm); (*) distal end. Scale bars: 10 μm.
Discussion
In eukaryotes, intercellular signaling through Notch receptors regulates growth and differentiation in several contexts during animal development [37]. Moreover, aberrant regulation of the Notch signaling pathway is highly correlated to human disease, including cancers [38-40]. In C. elegans, the mechanisms of GSC maintenance and proliferation largely rely on GLP-1/Notch signaling [41] and a battery of mRNA regulators (e.g., PUF proteins) [4]. Here, we report on new, tenable links between DNA pol α-primase subunits, GLP-1/Notch signaling, and PUF proteins for those germ cell proliferation and maintenance functions (Fig. 6A-C). Specifically, DIV-1 is required non-cell autonomously for GLP-1/Notch-mediated germ cell proliferation during early larval development, whereas other subunits do not appear to be essential (Fig. 6B). During adulthood, GLP-1/Notch-mediated maintenance of proliferative cell fate requires POLA-1, PRI-1, and PRI-2, but not DIV-1 (Fig. 6C). However, all these subunits work with FBF/PUF RNA-binding proteins to maintain GSCs (Fig. 6D). How is it that DIV-1 is not required for GSC maintenance/proliferation in the germline during adulthood, but becomes important when PUF proteins are missing? While it still eludes us, we speculate that DIV-1 may regulate GSC proliferation within the germline, but it is largely compensated when PUF proteins are present. Understanding how this works at the molecular level will be an important challenge for the future.
The major role of the DNA pol α-primase complex is in the initiation of DNA replication at chromosomal origins. However, this complex has also been implicated in a variety of cellular processes including genome integrity, DNA metabolism, and telomere maintenance. For example, mutations in the subunits of the DNA pol α-primase complex have been demonstrated to highly influence the checkpoint pathway. In Saccharomyces cerevisiae, the pri1-M4 mutation in the gene coding for the p48 primase subunit was associated with an accelerated progression of the S phase as well as hypersensitivity of DNA to damage [42]. Also, a missense mutation in the small subunit of DNA primase (Prim1) led to a profound apoptotic activity in retinal neurons in zebrafish [43]. Interestingly, apoptosis induced by the Prim1 mutation was also dependent on activation of ataxia telangiectasia mutated (ATM), Chk2, and p53, pointing at a possible role of Prim1 in genomic surveillance and response to DNA damage [43]. Similarly, differences in the requirement for phosphorylation by kinases provided supportive evidence for the interplay between replication proteins and the checkpoint pathway. Pellicioli et al. reported that Rad53, the S. cereveisiae homolog of Chk2, regulates phosphorylation and activity of the DNA pol α-prim B subunit under DNA stress [44]. Several studies have demonstrated the role played by the DNA pol α-primase complex in the maintenance of chromosomal stability. Cdc13, a telomere-binding protein, was shown to interact with DNA pol α and the Cdc13-Stn1-Ten1 (CST) complex, required for telomere protection, was proposed to regulate DNA pol α-primase activity [45, 46]. Moreover, increasing evidence suggests that DNA pol α is also involved in the epigenetic control of chromatin assembly and gene expression. Nakayama et al. demonstrated that a mutated swi7 gene, which encodes the catalytic subunit of DNA pol α-primase complex, results in altered localization of the chromodomain protein Swi6, which also directly interacts with DNA pol α in vitro [47].
During S phase POLA2 is recruited to DNA at the replicative forks. A POLA2 (G583R) mutation leads to mislocalization of the subunit to the cytoplasm instead of the nucleus. This not only inhibits DNA replication in cancer cells [e.g., non-small cell lung cancer (NSCLC)], it also sensitizes them to chemotherapy (e.g., Gemcitabine) [48]. In addition, the synthetic retinoid 6-[3-(1-adamantyl)-4-hydroxyphenyl]-2-naphthalene carboxylic acid (CD437) is a potent inhibitor of POLA1 and was shown to selectively induce apoptosis in human lung cancer cells [49]. Inhibition of DNA replication by CD437 elicits different responses in cancer cells. Whereas CD437-mediated inhibition of POLA1 leads to cell death in cancer cells, it merely induces cell cycle arrest in normal epithelial cells [49]. Notably, Notch signaling is altered in approximately one third of NSCLCs, which are the leading cause of cancer-related deaths [50]. Although the role of POLA1/2 in NSCLCs with elevated Notch activity has not yet been studied, the fundamental mechanism that we observe in GSC maintenance may be conserved with that in tumorigenesis. At the molecular level, it is intriguing that DNA primases play a key role in the initiation of DNA replication and in the synthesis of Okazaki fragments on the lagging strand. Primases are also involved in checkpoint pathways, coupling DNA replication to DNA repair [43, 51, 52]. Importantly, mammalian DNA primases, PRIM-1 and PRIM-2 are highly associated with human malignancies [51, 53, 54]. In this study, we also found that the subunits of the DNA pol α-primase complex promote GLP-1/Notch-mediated proliferation with discrete and shared function in C. elegans germline. To date, this novel role has not yet been reported in any other organisms. Therefore, understanding the physiological processes that link Notch signaling to the role of the DNA pol α-primase subunits may hold promise for the development of antineoplastic molecular and cellular therapies.
Recently, it was reported that Notch signaling is tightly linked to cell cycle progression during vulval development in C. elegans [55]. Specifically, CYD-1/Cyclin D and CYE-1/Cyclin E stabilize LIN-12/Notch receptor in vulva precursor cells (VPCs) [55]. In C. elegans germline, CYE-1, but not CYD-1, is essential for GLP-1/Notch-mediated proliferation [20]. Also, C. elegans CYD-1 and CYE-1 induce distinct cell cycle re-entry programs in differentiated muscle cells [56]. These studies suggest that cell fate determination in very varied tissues (e.g., germline and soma) and at disparate times in development (e.g., early lineage establishment and late terminal differentiation), may be regulated by different cell cycle progression events and regulators, including Notch. In this study, we also showed that depletion of individual DNA pol α-primase subunits (POLA-1, DIV-1, PRI-1, or PRI-2) affected GLP-1/Notch-mediated germ cell proliferation differently from somatic or germline sources at different developmental stages (see Fig. 6A-D). Specifically, somatic DIV-1 promotes Notch-mediated germ cell proliferation non-cell autonomously during early larval development. Although it remains elusive how cell cycle signature or regulators in somatic tissues influence the proliferation of their neighboring germ cells, previous studies showed that somatic gonadal tissues such as DTC and sheath cells establish the both germ cell proliferation and differentiation patterns [57]. Our study points to a somatic molecular pathway that may be responsible for such induction events. Understanding the mechanism of how a somatic protein with a DNA replication function, DIV-1, influences Notch signaling for germ cell proliferation non-cell autonomously is still a major challenge for future studies.
Materials and Methods
Nematode Strains
All strains were derived from Bristol strain N2 and maintained at 20°C as described unless otherwise noted [58]. Mutations and balancers used in this work include: LG I: rrf-1(pk1417), ppw-1(pk1426); LG II: fbf-1(ok91), fbf-2(q738), puf-8(q725); mIn1[mIs14 dpy-10(e128)]; LG III: glp-1(bn18). MHL52 [div-1(promoter)∷div-1(exon1 and 2)∷GFP], JK2868 [lag-2p∷GFP+unc-119(+)], TG1755 [unc-119(ed3); Itls37 ((pAA64) pie-1p∷mCherry∷his-58 + unc-119(+); gtIs66 (pie-1p∷GFP(lap)∷div-1 + unc-119(+)].
Hydroxyurea (HU) Treatment
L1 staged wild-type and glp-1(bn18) mutant worms were transferred to NGM (Nematode Growth Media) agar plates containing 40 mM HU (Sigma-Aldrich, St. Louis, MO) and incubated at 20°C. After 18 h with HU plate incubation, worms were transferred to a normal NGM plate and allowed to grow to adults.
pola-1∷L4440 feeding RNAi vector
To construct a pola-1∷L4440 feeding RNAi vector, pola-1 genomic fragments (958 nt) were amplified from wild-type (N2) DNA by PCR using two primers [pola1(F): 5′-CCAGCGGTAACGCCTGGGAG-3′ and pola1(R): 5′-GCTGTGAGCTCCCATTGGCC-3′]. PCR products were purified by QIAquick PCR purification kit (QIAGEN, Chatsworth, CA; Cat # 28106). Purified pola-1 PCR products and feeding RNAi vector (L4440) (Addgene, Plasmid #1654) were double digested by Xba1 and Pst1 (New England Biolabs, Ipswich, MA, USA) for 4 hours at 37°C. The digested DNA fragments were eluted by QIAquick Gel Extraction kit (QIAGEN, Cat # 28704). Eluted pola-1 DNA fragments were introduced into the eluted feeding RNAi vector L4440 by a ligation enzyme reaction. Finally, cloned pola-1∷L4440 plasmids were transformed into HT115 (DE3), an RNase III-deficient E. coli strain with IPTG-inducible T7 polymerase activity.
Feeding RNAi
RNAi constructs were obtained from the C. elegans ORF RNAi Library (Thermo Fisher Scientific, San Jose, CA, USA). For yAd(P0)➔Ad(F1) RNAi (see Fig. 2D, 3A), young adult (yAd) staged worms (P0) were placed on feeding RNAi plates seeded bacteria expressing double stranded RNAs (dsRNAs) corresponding to the gene of interest as previously described [59-61]. The germline phenotypes were analyzed by DAPI staining when F1 progeny reached adult stage at 20°C (3 days from L1). For yAd(P0)➔Ad(P0) RNAi (see Fig. 4A), synchronized L4 staged worms were placed on feeding RNAi plates and incubated at 20°C and the germline phenotypes were analyzed by DAPI staining 4 days later.
Germline Antibody Staining
Germline antibody staining was performed as previously described [62]. Briefly, dissected gonads were fixed in 3% paraformaldehyde/0.1M K2HPO4 (pH 7.2) solution for 10-20 min, and then post-fixed with cold 100% methanol for 5 min. After blocking for 30 min in 1× PTW (1×PBS + 0.1% Tween20)/0.5% BSA (Bovine Serum Albumin) solution, primary antibody was added, followed by incubation for 2 h at 20°C. The dissected gonads were washed three times for at least 30 min with 1× PTW/0.5% BSA solution and incubated in the same solution containing the fluorescence-conjugated secondary antibodies for 1-2 h at 20°C. After washing three times in 1× PTW/0.5% BSA solution for at least 30 min, the dissected gonads were stained with 100 ng/mL DAPI solution for 10 min at 20°C and were again washed in 1× PTW/0.5% BSA solution three times. The antibody staining was observed using fluorescence microscopy. Antibodies used in this study include anti-REC-8 (1:200 dilution, kindly provided by Dr. Josef Loidl's lab, University of Vienna, Austria), anti-HIM-3 (1:400 dilution, Novus Biologicals, Littleton, CO, USA), anti-GFP (1:400 dilution, Abcam, Cambridge, MA, USA), and anti-GLP-1 (1:200 dilution, kindly provided by Dr. Judith Kimble's lab, University of Wisconsin-Madison, WI, USA).
Generation of div-1p∷GFP Transgenic Worms
To make a div-1p∷GFP transgene, the 5′ upstream sequences (1,940 bp), exon1, intron 1, and exon 2 of the div-1 gene were amplified from wild-type (N2) genomic DNA using two primers [div-1(Sph1)F1: 5′-AGGTTAGCATGCATCCGGAATCACCCACACTTTAC-3′ and div-1(BamHI)R1: 5′-CGCGGATCCGGTTTTTTCACAGATCTCGGCGTG-3′]. Purified PCR products were digested by Sph1 and BamH1 (New England Biolabs, Ipswich, MA, USA), and inserted into the pPD95.67 vector upstream and in frame with a C. elegans optimized gfp gene. Wild-type worms were injected with the div-1p∷GFP transgene (50 ng/ul) and a pCFJ90 co-injection marker (myo-2p∷mCherry). A total of three transgenic lines expressing both div-1p∷GFP and myo-2p∷mCherry were generated. pPD95.67 and pCFJ90 were gifts from Andrew Fire (Stanford University; Addgene plasmid #1490) and Erik Jorgensen (University of Utah; Addgene, plasmid #19327), respectively.
EdU (5-ethynyl-2′-deoxyuridine) Labeling
For metabolic labeling of DNA synthesis, animals were incubated with rocking in M9/0.1% Tween 20/1 mM EdU for 30 min at 20°C. Gonads were dissected and fixed in 3% paraformaldehyde/0.1M K2HPO4 (pH 7.2) solution for 10-20 min, followed by -20°C methanol fixation for 10 min. Dissected gonads were blocked in 1× PTW, 0.5% BSA solution for 30 min at 20°C. EdU labeling was detected using the Click-iT EdU Alexa Fluor 488 Imaging Kit (Invitrogen, San Jose, CA, USA, #C10337), according to the manufacturer's instructions. After washing three times with 1× PTW/0.5% BSA solution for at least 30 min at 10-min intervals, the dissected gonads were stained with DAPI solution (100 ng/mL) for 10 min at 20°C and were finally washed with 1× PTW, 0.5% BSA solution three times. The EdU labeling was observed by fluorescence microscopy.
Western Blot
For the preparation of whole protein samples, collected C. elegans worms were lysed in Laemmli sample buffer (Bio-Rad Laboratories, Inc., Hercules, CA, USA) by boiling for 10 minutes. The lysed proteins analyzed by 10% sodium-dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Bio-Rad Laboratories, Inc.). Transferred membranes were blocked with 5% nonfat dry milk (Biotium, Inc., Fremont, CA, USA) in Tris-buffered saline (TBS) with 0.5% Tween 20 (TBST) and anti-GFP antibodies (1:1000 dilution, Abcam) at 4°C. The blot was probed with HRP-conjugated anti-rabbit secondary antibodies in 5% (w/v) nonfat dry milk in TBST for 1 hour at room temperature. Western blot images were obtained using the LI-COR C-DiGit Chemiluminescence Western Blot Scanner and Image software (LI-COR, Lincoln, Lebraska, USA).
Data Analysis
Statistical significance was analyzed using the two-tailed student's t-test. The error bars reflect respective standard deviation values.
Acknowledgments
We are grateful to Dr. Eleanor Maine (Syracuse University), Dr. Elizabeth T. Ables (ECU), and Dr. Chris Geyer (ECU) for advice and discussion during this work. We thank Dr. Judith Kimble (University of Wisconsin-Madison) for C. elegans mutant strains and GLP-1 antibody and Dr. Josef Loidl (University of Vienna) for REC-8 antibody. This work was supported in part by the Vidant Medical Center-Cancer Research and Education Fund, Brody Brother's Grant (BBE 213152), BSOM Seed/Bridge Grant, NIH (1R15GM112174-01A1) to M-H.L, and NSF (grant MCB1714264) to B.D.K and M-H.L. The Caenorhabditis Genetics Center (CGC) is supported by the National Institutes of Health – Office of Research Infrastructure Programs (P40 OD010440).
Abbreviations
- C. elegans
Caenorhabditis elegans
- DNA pol α-primases
DNA polymerase α-primases
- GLP
Germline proliferation defect
- PUF
PUMILIO/FBF
- GSC
Germline stem cell
- NICD
Notch intracellular domain
- HU
Hydroxyl urea
- RNAi
RNA interference
- GFP
Green fluorescent protein
- DTC
Distal tip cell
- 3′UTR
3′ untranslated region
- EdU
5-ethynyl-2′-deoxyuridine
Footnotes
Author Contributions: DSY, DSC, MAA, and MHL conceived and designed the experiments. DSY, DSC, MAA, BDK and MHL analyzed the data, and wrote the paper. DSY, DSC, and MHL performed the experiments.
Conflict of Interest: The authors declare that there is no conflict of interest.
References
- 1.Wong MD, Jin Z, Xie T. Molecular mechanisms of germline stem cell regulation. Annual review of genetics. 2005;39:173–95. doi: 10.1146/annurev.genet.39.073003.105855. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Sato C, Cerletti M, Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Current topics in developmental biology. 2010;92:367–409. doi: 10.1016/S0070-2153(10)92012-7. [DOI] [PubMed] [Google Scholar]
- 3.Greenwald I, Kovall R. Notch signaling: genetics and structure. WormBook : the online review of C elegans biology. 2013:1–28. doi: 10.1895/wormbook.1.10.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annual review of cell and developmental biology. 2007;23:405–33. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- 5.Henderson ST, Gao D, Lambie EJ, Kimble J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development. 1994;120:2913–24. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- 6.Petcherski AG, Kimble J. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature. 2000;405:364–8. doi: 10.1038/35012645. [DOI] [PubMed] [Google Scholar]
- 7.Doyle TG, Wen C, Greenwald I. SEL-8, a nuclear protein required for LIN-12 and GLP-1 signaling in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7877–81. doi: 10.1073/pnas.97.14.7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamont LB, Crittenden SL, Bernstein D, Wickens M, Kimble J. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Developmental cell. 2004;7:697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 9.Lee MH, Hook B, Lamont LB, Wickens M, Kimble J. LIP-1 phosphatase controls the extent of germline proliferation in Caenorhabditis elegans. The EMBO journal. 2006;25:88–96. doi: 10.1038/sj.emboj.7600901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hajnal A, Berset T. The C.elegans MAPK phosphatase LIP-1 is required for the G(2)/M meiotic arrest of developing oocytes. The EMBO journal. 2002;21:4317–26. doi: 10.1093/emboj/cdf430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kershner AM, Shin H, Hansen TJ, Kimble J. Discovery of two GLP-1/Notch target genes that account for the role of GLP-1/Notch signaling in stem cell maintenance. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3739–44. doi: 10.1073/pnas.1401861111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoo AS, Bais C, Greenwald I. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science. 2004;303:663–6. doi: 10.1126/science.1091639. [DOI] [PubMed] [Google Scholar]
- 13.Yoon DS, Alfhili MA, Friend K, Lee MH. MPK-1/ERK regulatory network controls the number of sperm by regulating timing of sperm-oocyte switch in C. elegans germline. Biochemical and biophysical research communications. 2017;491:1077–1082. doi: 10.1016/j.bbrc.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalchhauser I, Farley BM, Pauli S, Ryder SP, Ciosk R. FBF represses the Cip/Kip cell-cycle inhibitor CKI-2 to promote self-renewal of germline stem cells in C. elegans. The EMBO journal. 2011;30:3823–9. doi: 10.1038/emboj.2011.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kodoyianni V, Maine EM, Kimble J. Molecular basis of loss-of-function mutations in the glp-1 gene of Caenorhabditis elegans. Molecular biology of the cell. 1992;3:1199–213. doi: 10.1091/mbc.3.11.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berry LW, Westlund B, Schedl T. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development. 1997;124:925–36. doi: 10.1242/dev.124.4.925. [DOI] [PubMed] [Google Scholar]
- 17.Pauklin S, Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–47. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seidel HS, Kimble J. Cell-cycle quiescence maintains Caenorhabditis elegans germline stem cells independent of GLP-1/Notch. eLife. 2015;4 doi: 10.7554/eLife.10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ables ET, Drummond-Barbosa D. Cyclin E controls Drosophila female germline stem cell maintenance independently of its role in proliferation by modulating responsiveness to niche signals. Development. 2013;140:530–40. doi: 10.1242/dev.088583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fox PM, Vought VE, Hanazawa M, Lee MH, Maine EM, Schedl T. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development. 2011;138:2223–34. doi: 10.1242/dev.059535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeong J, Verheyden JM, Kimble J. Cyclin E and Cdk2 control GLD-1, the mitosis/meiosis decision, and germline stem cells in Caenorhabditis elegans. PLoS genetics. 2011;7:e1001348. doi: 10.1371/journal.pgen.1001348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiao L, Lissemore JL, Shu P, Smardon A, Gelber MB, Maine EM. Enhancers of glp-1, a gene required for cell-signaling in Caenorhabditis elegans, define a set of genes required for germline development. Genetics. 1995;141:551–69. doi: 10.1093/genetics/141.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maine EM, Hansen D, Springer D, Vought VE. Caenorhabditis elegans atx-2 promotes germline proliferation and the oocyte fate. Genetics. 2004;168:817–30. doi: 10.1534/genetics.104.029355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Encalada SE, Martin PR, Phillips JB, Lyczak R, Hamill DR, Swan KA, Bowerman B. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Developmental biology. 2000;228:225–38. doi: 10.1006/dbio.2000.9965. [DOI] [PubMed] [Google Scholar]
- 25.Brauchle M, Baumer K, Gonczy P. Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Current biology : CB. 2003;13:819–27. doi: 10.1016/s0960-9822(03)00295-1. [DOI] [PubMed] [Google Scholar]
- 26.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–9. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 27.Sijen T, Plasterk RH. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–4. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- 28.Tijsterman M, Okihara KL, Thijssen K, Plasterk RH. PPW-1, a PAZ/PIWI protein required for efficient germline RNAi, is defective in a natural isolate of C. elegans. Curr Biol. 2002;12:1535–40. doi: 10.1016/s0960-9822(02)01110-7. [DOI] [PubMed] [Google Scholar]
- 29.Killian DJ, Hubbard EJ. C. elegans pro-1 activity is required for soma/germline interactions that influence proliferation and differentiation in the germ line. Development. 2004;131:1267–78. doi: 10.1242/dev.01002. [DOI] [PubMed] [Google Scholar]
- 30.Hansen D, Wilson-Berry L, Dang T, Schedl T. Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development. 2004;131:93–104. doi: 10.1242/dev.00916. [DOI] [PubMed] [Google Scholar]
- 31.Zetka MC, Kawasaki I, Strome S, Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes & development. 1999;13:2258–70. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends in genetics : TIG. 2002;18:150–7. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 33.Lee MH, Mamillapalli SS, Keiper BD, Cha DS. A systematic mRNA control mechanism for germline stem cell homeostasis and cell fate specification. BMB reports. 2016;49:93–8. doi: 10.5483/BMBRep.2016.49.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friday AJ, Keiper BD. Positive mRNA Translational Control in Germ Cells by Initiation Factor Selectivity. BioMed research international. 2015;2015:327963. doi: 10.1155/2015/327963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nousch M, Eckmann CR. Translational control in the Caenorhabditis elegans germ line. Advances in experimental medicine and biology. 2013;757:205–47. doi: 10.1007/978-1-4614-4015-4_8. [DOI] [PubMed] [Google Scholar]
- 36.Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–3. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 37.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–73. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 38.Allenspach EJ, Maillard I, Aster JC, Pear WS. Notch signaling in cancer. Cancer biology & therapy. 2002;1:466–76. doi: 10.4161/cbt.1.5.159. [DOI] [PubMed] [Google Scholar]
- 39.Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27:5124–31. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
- 40.Yuan X, Wu H, Xu H, Xiong H, Chu Q, Yu S, Wu GS, Wu K. Notch signaling: an emerging therapeutic target for cancer treatment. Cancer letters. 2015;369:20–7. doi: 10.1016/j.canlet.2015.07.048. [DOI] [PubMed] [Google Scholar]
- 41.Austin J, Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51:589–99. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- 42.Marini F, Pellicioli A, Paciotti V, Lucchini G, Plevani P, Stern DF, Foiani M. A role for DNA primase in coupling DNA replication to DNA damage response. EMBO J. 1997;16:639–50. doi: 10.1093/emboj/16.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi M, Fujimori-Tonou N, Yoshimura Y, Kishi T, Okamoto H, Masai I. Mutation of DNA primase causes extensive apoptosis of retinal neurons through the activation of DNA damage checkpoint and tumor suppressor p53. Development. 2008;135:1247–57. doi: 10.1242/dev.011015. [DOI] [PubMed] [Google Scholar]
- 44.Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–72. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–88. [PMC free article] [PubMed] [Google Scholar]
- 46.Lue NF, Chan J, Wright WE, Hurwitz J. The CDC13-STN1-TEN1 complex stimulates Pol alpha activity by promoting RNA priming and primase-to-polymerase switch. Nat Commun. 2014;5:5762. doi: 10.1038/ncomms6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakayama J, Allshire RC, Klar AJ, Grewal SI. A role for DNA polymerase alpha in epigenetic control of transcriptional silencing in fission yeast. EMBO J. 2001;20:2857–66. doi: 10.1093/emboj/20.11.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mah TL, Yap XN, Limviphuvadh V, Li N, Sridharan S, Kuralmani V, Feng M, Liem N, Adhikari S, Yong WP, Soo RA, Maurer-Stroh S, Eisenhaber F, Tong JC. Novel SNP improves differential survivability and mortality in non-small cell lung cancer patients. BMC genomics. 2014;15(9):S20. doi: 10.1186/1471-2164-15-S9-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun SY, Yue P, Chen X, Hong WK, Lotan R. The synthetic retinoid CD437 selectively induces apoptosis in human lung cancer cells while sparing normal human lung epithelial cells. Cancer research. 2002;62:2430–6. [PubMed] [Google Scholar]
- 50.Westhoff B, Colaluca IN, D'Ario G, Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G, Viale G, Pece S, Di Fiore PP. Alterations of the Notch pathway in lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22293–8. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yatsula B, Galvao C, McCrann M, Perkins AS. Assessment of F-MuLV-induced tumorigenesis reveals new candidate tumor genes including Pecam1, St7, and Prim2. Leukemia. 2006;20:162–5. doi: 10.1038/sj.leu.2404034. [DOI] [PubMed] [Google Scholar]
- 52.Michael WM, Ott R, Fanning E, Newport J. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science. 2000;289:2133–7. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- 53.Petrocca F, Lieberman J. Promise and challenge of RNA interference-based therapy for cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:747–54. doi: 10.1200/JCO.2009.27.6287. [DOI] [PubMed] [Google Scholar]
- 54.Yotov WV, Hamel H, Rivard GE, Champagne MA, Russo PA, Leclerc JM, Bernstein ML, Levy E. Amplifications of DNA primase 1 (PRIM1) in human osteosarcoma. Genes, chromosomes & cancer. 1999;26:62–9. doi: 10.1002/(sici)1098-2264(199909)26:1<62::aid-gcc9>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 55.Nusser-Stein S, Beyer A, Rimann I, Adamczyk M, Piterman N, Hajnal A, Fisher J. Cell-cycle regulation of NOTCH signaling during C. elegans vulval development. Molecular systems biology. 2012;8:618. doi: 10.1038/msb.2012.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korzelius J, The I, Ruijtenberg S, Prinsen MB, Portegijs V, Middelkoop TC, Groot Koerkamp MJ, Holstege FC, Boxem M, van den Heuvel S. Caenorhabditis elegans cyclin D/CDK4 and cyclin E/CDK2 induce distinct cell cycle re-entry programs in differentiated muscle cells. PLoS genetics. 2011;7:e1002362. doi: 10.1371/journal.pgen.1002362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korta DZ, Hubbard EJ. Soma-germline interactions that influence germline proliferation in Caenorhabditis elegans. Developmental dynamics : an official publication of the American Association of Anatomists. 2010;239:1449–59. doi: 10.1002/dvdy.22268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–72. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 60.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–12. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 62.Yoon DS, Pendergrass DL, Lee MH. A simple and rapid method for combining fluorescent in situ RNA hybridization (FISH) and immunofluorescence in the C. elegans germline. MethodsX. 2016;3:378–85. doi: 10.1016/j.mex.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


