Abstract
We previously showed that antimicrobial photodynamic inactivation (aPDI) of Gram-positive and Gram-negative bacteria mediated by the phenothiazinium dye, methylene blue (MB) was potentiated by addition of potassium thiocyanate (10mM). The mechanism was suggested to involve a singlet oxygen mediated reaction with SCN− to form sulfite and cyanide and then to produce sulfur trioxide radical anion. We now report that potassium selenocyanate KSeCN (concentrations up to 100 mM) can also potentiate (up to 6 logs of killing) aPDI mediated by a number of different photosensitizers: MB, Rose Bengal, and TPPS4 (as low as 200 nM). When a mixture of selenocyanate with these PS in solution was illuminated and then bacteria were added after the light, there was up to 6 logs killing (Gram-negative > Gram-positive) but the antibacterial species decayed rapidly (by 20 min). Our hypothesis to explain this antibacterial activity is the formation of selenocyanogen (SeCN)2 by reaction with singlet oxygen (1O2) as shown by quenching of 1O2 by SeCN− and increased photoconsumption of oxygen. The fact that lead tetra-acetate reacted with SeCN− (literature preparation of (SeCN)2) also produced a short-lived antibacterial species supports this hypothesis.
Keywords: antimicrobial photodynamic inactivation, Gram-positive and Gram-negative bacteria, potentiation by potassium selenocyanate, methylene blue, Rose Bengal, TPPS4, selenocyanogen, singlet oxygen
Introduction
Antimicrobial photodynamic inactivation (aPDI) is a new photochemical approach to kill pathogenic microorganisms [1], that has been developed in response to the ever-increasing threat of multi-antibiotic resistance, that has now spread to every corner of the world [2]. aPDI is not affected by the antibiotic resistance status of the bacteria [3], and is thought to be unlikely to cause any resistance to develop against itself [4]. The photosensitizer (PS) absorbs light of the correct wavelength boosting the PS molecule to the short-lived (nsec) first excited singlet state, whence it can undergo intersystem crossing to the excited triplet state, that has a sufficiently long lifetime (μsec) to carry out photochemistry [5]. This photochemistry can either take the form of energy transfer to ground state triplet oxygen to form the reactive oxygen species (ROS), singlet oxygen (1O2). Alternatively the triplet PS can undergo an electron transfer reaction to produce ROS that are radical species. Since both these ROS are highly reactive their lifetime is short and they cannot travel extended distances in biological media. Therefore the PS molecules should bind to the microbial cells to achieve efficient photoinactivation. In general Gram-positive bacteria are highly porous in nature, and many different PS structures are able to penetrate the cell wall so the ROS can efficiently kill them. On the other hand, most Gram-negative species are impermeable to many PS structures, unless the PS bears a pronounced cationic charge at biological pH, and are consequently less susceptible to aPDI killing. Fungal cells such the yeast Candida albicans, have an intermediate permeability barrier and in general are intermediate in their susceptibility to aPDI.
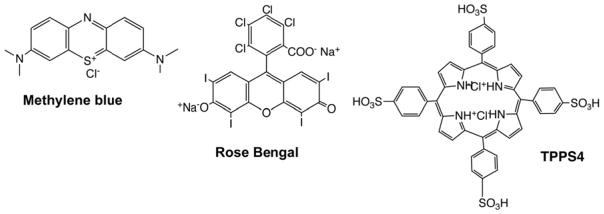
The photosensitizers that are chosen for aPDI should be inexpensive and generally regarded as safe. Two examples of dyes that meet this criterion are the phenothiazinium salt, methylene blue (MB) and the halogenated xanthene, Rose Bengal (RB) (Figure 1). While MB is cationic, it only has one positive charge on each molecule, and although it is able to inactivate Gram-positive bacteria, its activity is not particularly high. By contrast RB is anionic and its activity against Gram-positive bacteria is very high. However its activity against Gram-negative species is very low to non-existent. We also used a tetrasulfonated porphyrin (TPPS4) that was previously considered to be anionic in character, but which we recently showed also has some cationic character [6]. We have recently reported that a surprisingly wide variety of inorganic salts can potentiate the aPDI killing of microbial cells [7]. This strategy applied to Gram-positive bacteria, Gram-negative bacteria and also to fungi. A variety of salts can be employed, including potassium iodide [6–10], potassium bromide [11], sodium azide [12, 13], and sodium thiocyanate [14]. Potassium iodide, potassium bromide and sodium thiocyanate are non-toxic, while sodium azide has moderate toxicity. In the present study we report on a new salt that can potentiate aPDI, namely potassium selenocyanate. Although selenocyanate cannot be considered to be completely non-toxic, selenium is known to be an essential trace element for humans, and selenocyanate has been suggested to function as a low-toxicity inorganic selenium species that could be suitable for dietary supplementation [15]. Therefore we tested whether addition of KSeCN could potentiate aPDI in vitro.
Figure 1. Chemical structures of the three photosensitizers.
Methylene blue (MB); Rose Bengal (RB); 5,10,15,20-tetrakis(4-sulfonatophenyl)porphyrin dihydrochloride (TPPS4).
Materials and Methods
Compounds
We purchased methylene blue (MB), Rose Bengal (RB), 5,10,15,20-tetrakis(4-sulfonatophenyl)porphyrin dihydrochloride (TPPS4), potassium selenocyanate (KSeCN) and lead tetraacetate {Pb(OAc)4 from Sigma-Aldrich (St. Louis, MO, USA). Amplex red hydrogen peroxide/peroxidase assay kit was purchased from Invitrogen (Carlsbad, CA, USA). The singlet oxygen sensor green reagent (SOSG) and 3′-(p-hydroxyphenyl)-fluorescein (HPF) probes to detect singlet oxygen or hydroxyl radicals were purchased from Life Technologies (Grand Island, NY, USA). MB, RB, TPPS4 and KSeCN were prepared in distilled H2O (dH2O). Pb(OAc)4 was prepared in pH 7.4 distilled phosphate-buffered saline (PBS) and all PS stock solution (2.5 mM) were stored at 4°C in the dark for no more than 1 week prior to use. KSeCN, Pb(OAc)4, SOSG and HPF solutions were prepared freshly before each experiment.
Cells and culture conditions
In this study the bacterial strains were used as follow: Gram-positive bacteria: methicillin-resistant Staphylococcus aureus (MRSA) USA 300, Staphylococcus epidermidis ATCC27127 (S. epidermidis), Enterococcus faecalis ATCC29212 (E. faecalis), Bacillus subtilis ATCC6051 (B. subtilis), Bacillus atrophaeus ATCC 9372 (B. atrophaeus) and Bacillus megaterium ATCC14581 (B. megaterium); Gram-negative bacteria: Escherichia coli (E. coli) K-12 (ATCC 33780), Proteus mirabilis (P. mirabilis) ATCC 51393 (Xen 44), Pseudomonas aeruginosa (P. aeruginosa ) ATCC 19660 (Xen 5P), Klebsiella pneumoniae (K. pneumoniae ), Acinetobacter baumannii (A. baumannii) ATCC BAA 747 and E. coli UTI 89, UTI89 was a generous gift from Dr. Patrick Seed’s laboratory, which is a clinical cystitis isolate strain from humans previously described by Mulvey et al. [16].
A single colony of bacteria from an agar plate was suspended in 5 mL of brain heart infusion (BHI) broth and grown overnight in a shaker incubator (New Brunswick Scientific, Edison, NJ) at 120 rpm under aerobic conditions at 37 °C. An aliquot of 1 mL from an overnight bacterial suspension was refreshed in fresh BHI for 2–3 h at 37 °C to mid-log phase. Cell concentration was estimated by measuring the optical density (OD) at 600 nm (OD of 0.6 = 108 cells/mL). The bacterial suspension was centrifuged, washed, and resuspended in pH7.4 PBS to arrest microbial growth and used (108 cells/mL) for the experiments.
aPDI experiments
The initial studies used aPDI with suspensions of bacteria (108 cells/mL) and one of three different PS (10 μM MB, 200 nM RB and 200 nM TPPS4 for Gram-positive MRSA cells; 10 μM MB, 10 μM RB or 200 nM TPPS4 for Gram-negative E. coli cells) incubated with increasing concentrations of KSeCN between 0 to 100 mM in pH 7.4 PBS in the dark at room temperature for 30 min, cells irradiated with 10J/cm2 of 660nm for MB, 10J/cm2 of 540nm for RB and 10J/cm2 of 415nm for TPPS4. The light source we used for red and green light was a white lamp with a band-pass filter probe 660±15 nm or 540±15 nm (Lumacare, Newport Beach, CA, USA). For blue light we used an Omnilux Clear-U light-emitting diode (LED) array (Photo Therapeutics, Inc., Carlsbad, CA) at 415±15nm. The power densities were between 12–16 mW/cm2 measured with a power meter (Coherent, Santa Clara, California). The time to deliver 10 J/cm2 was 10–13 minutes. At the end of illumination CFUs were determined according to the method of Jett et al [17]. The aliquots were serially diluted tenfold in PBS to give dilutions of 10−1 to 10−5 times in addition to the original concentration and 10 μL aliquots of each of the dilutions were streaked horizontally on square BHI agar plates. Plates were incubated for 12–18 h at 37 °C in the dark to allow colony formation. Each experiment was performed at least three times.
For experiments where cells were added after light, we used 200 mM KSeCN plus a range of TPPS4 concentrations (0 to 10 μM) in pH 7.4 PBS irradiated with 10 J/cm2 415nm. After the end of illumination then immediately or at different time points we added an equal volume of E. coli or MRSA suspension in PBS (108 cells/mL) and mixed them thoroughly. After 30 min incubation, the aliquots were serially diluted as before. Each experiment was performed at least three times.
Suspensions of bacteria (Gram-positive species: MRSA, E. faecalis, S. epidermidis, B subtilis, B. atrophaeus and B. megaterium, or Gram-negative species: P. mirabilis, P. aeruginosa, K pneumoniae and A. baumannii) and 10 μM TPPS4 incubated with or without 100 mM of KSeCN in pH 7.4 PBS in the dark at room temperature for 30 min. Cells were irradiated with 10J/cm2 of 415nm light, the aliquots were serially diluted as before. For “cells after” 10 μM TPPS4 mixed with 100 mM of KSeCN in pH 7.4 PBS, was irradiated with10 J/cm2 415nm, then immediately was added an equal volume of bacterial suspension and mixed completely. After 30 min incubation in the dark at room temperature, the aliquots were serially diluted as before. Each experiment was performed at least three times.
A control group of cells treated with light alone (660, 540, 415 nm; no MB, RB or TPPS4 added) showed the same number of CFU as absolute control (data not shown). Survival fractions were routinely expressed as ratios of CFU of microbial cells treated with light and MB, RB or TPPS4 (or MB, RB or TPPS4 in the absence of light) to CFUs of microbes treated with neither.
Experiments with lead tetraacetate
Suspensions of bacteria (108 CFU/ml) were incubated at room temperature for 30 min with a range of concentration of Pb(OAc)4 dissolved in PBS with or without 100mM KSeCN. At the end of the incubation time, 100-μL aliquots were placed in 96 wells from each tube, serially diluted and streaked on BHI agar plates to determine CFU. 5 mM Pb(OAc)4 plus 10 mM KSeCN was mixed in PBS and after different times an equal volume of bacterial suspension was added. After 30 min incubation at room temperature, the aliquots were serially diluted as before. Each experiment was performed at least three times.
Mechanistic experiments
Activation of SOSG & HPF
Cell-free experiments were performed in 96-well black-sided plates. RB was used at 1μM in PBS, and SOSG or HPF (Molecular Probes, Invitrogen, USA) was added to each well at a final concentration of 10 μM. KSeCN solution (100 mM) was either added or not. Each experimental group contained four wells. All groups were illuminated simultaneously, and 540nm green light was delivered in sequential doses of 0–7 J/cm2. A microplate spectrophotometer (Spectra Max M5, Molecular Devices) was used for acquisition of fluorescence signals in the “slow kinetic” mode. The fluorescence excitation and emission at 504 and 525 nm for SOSG and 490 and 515 nm for HPF, respectively. Each time after an incremental fluence was delivered, the fluorescence was measured. Each experiment was performed at least three times.
Amplex red assay for hydrogen peroxide
Amplex red hydrogen peroxide/peroxidase assay kit was used to detect the production of H2O2 from TPPS4 with or without KSeCN plus light. The colorless probe Amplex red (10-acetyl-3, 7-dihydroxy-phenoxazine) reacts with H2O2 in the presence of peroxidase and forms resorufin (7-hydroxy-3H-phenoxazin-3-one). The detection process after TPPS4 with or without KSeCN mediated PDT was according to manufacturer’s instructions. The reaction systems contained 10 μM TPPS4 with or without 100 mM KSeCN were illuminated with increasing fluence of 415nm blue light and aliquots withdrawn and added to 50 μM Amplex Red reagent and 0.1 U/mL horseradish peroxidase (HRP) in Krebs–Ringer phosphate (consists of 145 mM NaCl, 5.7 mM Na3PO4, 4.86 mM KCl, 0.54 mM CaCl2, 1.22 mM MgSO4, 5.5 mM glucose, pH 7.35). After 30 min incubation, a fluorescence microplate reader (excitation 530 nm and emission ~590 nm) was used to measure incremental fluorescence after an incremental fluence of green light was delivered. Controls were (1) TPPS4 + light, (2) KSeCN + light, (3) Amplex red reagent alone. Each experiment was performed at least three times.
EPR-oximetry and EPR-spin trapping
The effect of selenocyanate on photoconsumption of oxygen mediated by the studied photosensitizers was examined by electron paramagnetic resonance (EPR)-oximetry, in a closed-chamber approach, using the water soluble oxygen-sensitive spin probe mHCTPO at concentration 0.1 mM, according to the method described elsewhere [18, 19]. EPR samples were run using microwave power 1.06 mW, modulation amplitude 0.006 mT, scan width 0.3 mT and scan time 5.2 s. The photosensitized generation of superoxide anion modulated by selenocyanate was determined by EPR-spin trapping using 100 mM DMPO as a spin trap, as described elsewhere [18, 19]. EPR samples were run using microwave power 10.6 mW, modulation amplitude 0.05 mT, center field 339.0 mT, scan width 8 mT and scan time 84 s. To increase the lifetime of the superoxide anion spin adduct 70–80% DMSO was used. The EPR system consisted of a Bruker EMX-AA 1579 EPR spectrometer (Bruker BioSpin, Germany) equipped with an optical resonant cavity, allowing irradiation of samples during ESR examination with green or red light, derived from a Cermax PE300CE-13FM 300W lamp in air-cooled housing (Perkin Elmer, USA) that was equipped with a combination of cut-off and band-pass filters. Data acquisition was facilitated by WIN EPR Bruker EMX software (version 4.33rev.05).
Direct time-resolved singlet oxygen measurements
The efficiency of selenocyanate to quench singlet oxygen was determined directly by measuring 1O2 lifetime in deuterium oxide (D2O) with and without KSeCN, as described elsewhere [14]. Time-resolved luminescence of 1O2 was measured at 1270 nm. Solutions of RB in 1-cm fluorescence cuvette (QA-1000, Helma Optik) were excited with 532 nm microjoule pulses (750 ps duration) generated by a microchip Nd:YAG laser (Pulselas-P-1064-FC, Alphalas GmbH, Goettingen, Germany) operating with 2–10 kHz repetition rate. To filter first and third harmonics of laser radiation, 50-cm water filter and dichroic mirrors (BK7series, Eksma Optics, Vilnius, Lithuania) were used. Near-infrared luminescence was measured perpendicularly to the excitation beam in a photon counting mode using a thermoelectric cooled NIR PMT module (Model H10330–45, Hamamatsu, Japan) equipped with 1100-nm cutoff filter and additional selected narrow-band filters (NB series, NDC Infrared Engineering LTD, Maldon, UK). A computer-mounted PCI-board multichannel scaler was used (NanoHarp 250, PicoQuant GmbH, Berlin, Germany) and data collection was synchronized with laser pulses using an ultrafast photodiode (UGP-300-SP, Alphalas GmbH, Goettingen, Germany) as a trigger. First-order luminescence decay fitting by the Levenberg-Marquardt algorithm was performed by custom-written software.
Statistics
Experiments were repeated three times and values are presented as means and error bars are standard deviations.
3. Results
We were able to demonstrate potentiation of aPDI mediated by three different PS each with a fundamentally different chemical structure as shown in Figure 1.
3.1 aPDI of E. coli and MRSA is potentiated by addition of selenocyanate
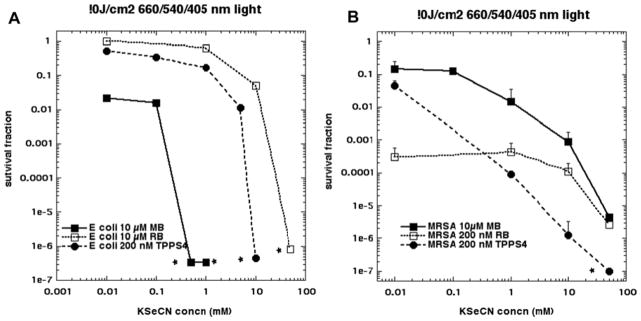
Figure 2A shows the results of adding increasing concentrations of KSeCN to a mixture of Gram-negative E. coli cells (10(8)/mL) and one of three different PS (10 μM, 10 μM RB or 200 nM TPPS4) that had been incubated for 30 min. Immediately after addition of the KSeCN, the light was turned on: 660 nm for MB; 540 nm for RB and 415 m for TPPS4 and 10 J/cm2 was delivered. There was only appreciable aPDI killing detected (1.5 logs) with MB alone (0.01 mM in the double log plot is equivalent in reality to zero KeSeCN); the other 2 non-cationic PS do not produce any aPDI effect against E. coli (at these reasonable concentrations such as 10 μM). When the KSeCN concentration reached 1 mM for MB at 10μM, 10 mM for TPPS4 at 200 nM, or 50 mM for RB at 10 μM, total eradication (> 6 logs) was achieved. In Figure 2B we show the analogous results for Gram-positive MRSA. We tried to select concentrations of PS that could produce some bacterial killing in the absence of KSeCN (equivalent to 0.01 mM), and these concentrations were 10 μM MB (1 log); 200 nM RB (3.5 logs) and 200 nM TPPS4 (1.5 logs). In each case there was marked potentiation of killing (3–5 logs more) by addition of KSeCN, especially when the concentration reached 50 mM.
Figure 2. Potentiation of aPDI by addition of KSeCN using three different PS.
(A) E. coli cells (108/mL) were incubated with the stated concentration of MB, RB or TPPS4 for 30 min, then the stated concentration of KSeCN was added and 10J/cm2 of the appropriate wavelength of light was delivered (660 nm for MB; 540 nm for RB; 415 nm for TPPS4); (B) Same as A but with MRSA cells (108/mL). * signifies eradication (zero CFU)
3.2 Stability of the antibacterial substance produced after illumination
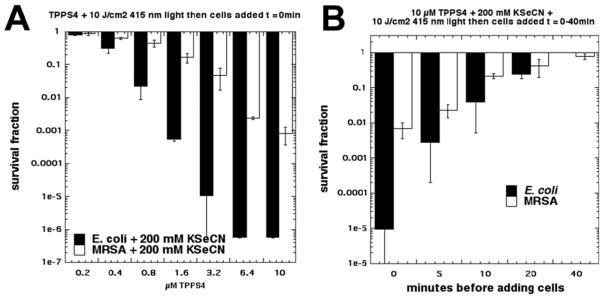
In order to detect whether the PDT action on KSeCN N produced an antibacterial substance, we delivered the light to a mixture of an increasing concentration of TPPS4 mixed with 200 mM KSeCN. The concentration of 200 mM KSeCN (rather than 100 mM) was chosen because an equal volume of bacterial suspension (10(8) cells/m mL) was added. TPPS4 was selected as the PS as it appeared to be the most active PS from data in Figure 2. Figure 3A shows that with E. coli and 200 nM TPPS4 plus KSeCN (a concentration which had been shown to eradicate E. coli when combined with KSeCN in the “cells in” format) did not produce any killing. However, when the concentration of TPPS4 was gradually increased, significant killing in the “cells after” format began to appear and eradication was achieved at 6.4 μM TPPS4. MRSA was clearly less susceptible to killing in the “cells after” format because even 10 0 μM TPPS4 only gave 3 logs of killing. We next asked how stable the antibacterial substance produced by PDT using 10 uM of TPPS4 plus 10 J/cm2 of 415 nm light and 200 mM KSeCN was, by adding the cells after an increasing delay time following cessation of the illumination period. Figure 3B shows that for both E. coli and MRSA the bactericidal action declined relatively rapidly and was totally eliminated by 40 min after the light had been switched off.
Figure 3. Effect of adding cells after light.
(A) A mixture of TPPS4 (different concentrations) and KSeCN (200 mM) was irradiated with 10J/cm2 415 nm light and immediately afterwards an equal volume of E. coli or MRSA cells was added, mixed and incubated for 30 min; (B) A mixture of TPPS4 (10μM) and KSeCN (200 mM) was irradiated with 10J/cm2 415 nm light and at different time points afterwards an equal volume of E. coli or MRSA cells was added, mixed and incubated for 30 min;
3.3 Testing other Gram-positive and Gram-negative bacteria
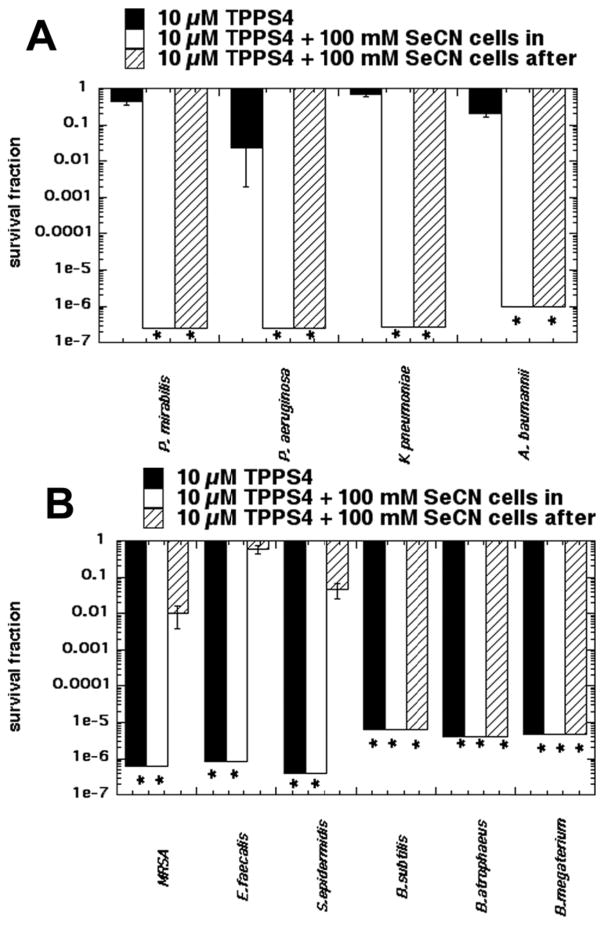
We carried out experiments to further investigate the difference in susceptibility we found between E. coli and MRSA. We first asked how generally applicable the killing of Gram-negative bacteria was by comparing four additional Gram-negative species namely P. mirabilis, P. aeruginosa, K pneumoniae and A. baumannii. Figure 4A shows the killing obtained with 10 uM TPPS4 excited with 10 J/cm2 415 nm light either alone, combined with 100 mM KSeCN in “cells in” format and also in “cells after” format. All four species were eradicated in both “cells in” and “cells after” formats while aPDI alone at most killed 1.5 logs.
Figure 4. Potentiation of aPDI (10 μM TPPS + 10 J/cm2 415 nm light) by added KSeCN (100mM) in other Gram-negative and Gram-positive species. Comparison of “cells in” and “cells after”.
(A) Gram-negatives: P. mirabilis, P. aeruginosa, K. pneumoniae, A. baumannii; (B) Gram-positives: MRSA, E. faecalis, S. epidermidis, B. subtilis, B. atrophaeus, B. megaterium. * signifies eradication (zero CFU)
Next we asked whether the relative resistance of MRSA bacteria (compared to Gram-negatives) was generally applicable to other Gram-positive species. We initially tested four Gram-positive species, MRSA, S. epidermidis, E. faecalis, and B subtilis. Since we found a large difference between B subtilis and the other Gram-positive species, we then extended the panel to include two additional Bacillus species, namely B. atrophaeus and B. megaterium. Since we used 10 μM TPPS4 and 10 J/cm2 of 415 nm which is a high dose of aPDI for Gram-positive bacteria, as expected all six species were eradicated in the aPDI alone format and also when combined with 100 mM KSeCN in the “cells in” format. The interesting comparison was seen in the aPDI plus KseCN and “cells after” format. MRSA and S. epidermidis showed about two logs killing, while E. faecalis showed no significant killing. On the other hand B. subtilis was completely eradicated, and when we tested two other Bacillus species, B. atrophaeus and B. megaterium these species were also eradicated.
3.4 Mechanistic studies
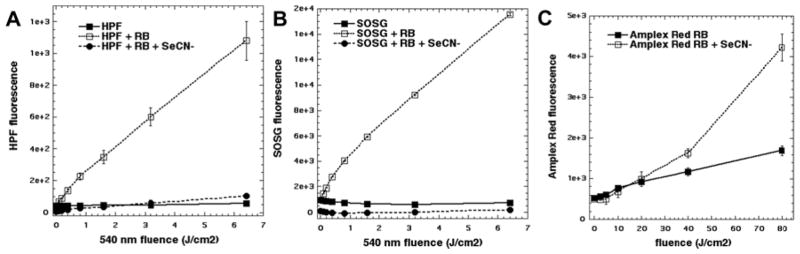
We asked whether singlet oxygen or hydroxyl radicals produced by the photodynamic reaction (RB + 540 nm light) were responsible for the oxidation of KSeCN to produce the bactericidal substance. In Figure 5A we see that the activation of the fluorescent probe HPF (specific for hydroxyl radicals) was completely quenched by addition of KSeCN. Likewise the activation of the fluorescent probe SOSG (specific for singlet oxygen) was completely quenched by KSeCN (Figure 5B). Since the two-electron oxidation of KSeCN by 1O2 would produce hydrogen peroxide, we then used the Amplex Red reagent to measure the production H2O2. Figure 5C shows that the addition of KSeCN produced more H2O2 from photoactivated RB, than was produced by RB alone.
Figure 5. Mechanistic studies.
(A) Reactions contained 10μM HPF probe; 1 μM RB; 100 mM KSeCN. Fluorescence was read after each aliquot of 540 nm light. (B) Same as (A) but with 10μM SOSG probe; (C) Reactions contained 10 μM RB and 100 mM KSeCN. At intervals after 540 nm light had been delivered, aliquots were withdrawn and measured using Amplex Red kit.
3.5 Oxygen photoconsumption and singlet oxygen quenching
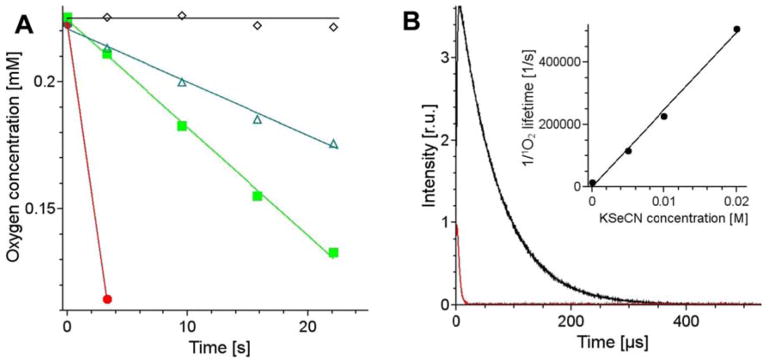
Selenocyanate at 10 mM dramatically accelerated the rate of photoconsumption of oxygen induced by excitation of samples containing 25 μM methylene blue (Fig 6A). Under the conditions used the EPR-oximetry allowed only to measure the first data point, which was acquired during several seconds of the sample irradiation. The observable rate of oxygen consumption photosensitized by MB in sample containing selenocyanate was partially inhibited by addition of 5 mM and 10 mM azide, indicating possible involvement of singlet oxygen. The interaction of selenocyanate with singlet oxygen has been confirmed by direct time-resolved measurements of singlet oxygen phosphorescence photogenerated by methylene blue in D2O (Fig 6B). It apparent that the lifetime of singlet oxygen is significantly shortened by 10 mM KSeCN. Measurements of the quenching effect of selenocynate at selected concentrations (inset in Fig 6B) yield second-order rate constant of the quenching, with the value 2.2×107 M−1s−1. Although this represents total quenching that consists of chemical and physical quenching, the data shown in Fig 6A suggest that reactive quenching of singlet oxygen by selenocyanate plays an important role. Singlet oxygen phosphorescence traces shown in Fig 6B indicate that selenocyanate not only quenches singlet oxygen but also reduces its intensity. The effect is probably due to several factors such as selenocyanate-induced photobleaching of methylene blue, accompanied by decomposition of KSeCN and the release of Se, which makes the sample turbid and less transparent, and possible quenching of the excited triplet state of methylene blue by selenocyanate.
Figure 6.
Changes in oxygen concentration induced by irradiation of 25 μM methylene blue in the absence (black empty diamonds) and presence of 10 mM KSeCN (red full circles), and after addition to selenocyanate-containing sample 10 mM azide (green full squares) or 5 mM azide (green empty triangles) (A). The effect of 10 mM selenocyanate (red trace) on singlet oxygen phosphorescence generated by photoexcitation of methylene blue in D2O (black trace) (B). Inset in (B) shows the reverse of the detected singlet oxygen lifetime as a function of KSeCN.
3.6 Comparison with the reaction between lead tetraacetate and selenocyanate
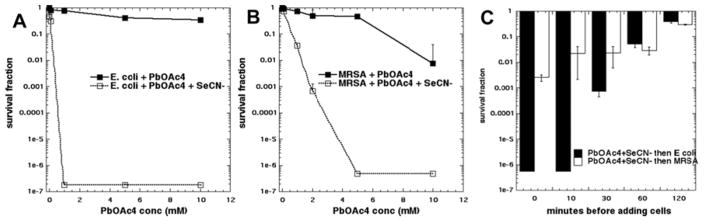
Since our leading hypothesis to explain the mechanism of the bactericidal potentiation effect was the photooxidation of KSeCN to produce the metastable pseudohalogen compound (SeCN)2, we asked whether there was another synthetic procedure we could use for comparison. Eventually we came upon the reaction of KSeCN with lead tetraacetate (see Figure 7). When these two salts were mixed there was an immediate precipitate of a red insoluble material. Up to 10 mM Pb(OAc)4 used alone did not kill any significant amount of E. coli (Figure 7A) while 10 mM Pb(OAc)4 alone (but not lower concentrations) killed two logs of MRSA (Figure 7B). When the reaction contained 100mM KSeCN there was a dramatic difference in killing observed. E. coli was eradicated with only 1 mM Pb(OAc)4 while MRSA was eradicated with 5 mM Pb(OAc)4. We then asked whether the antibacterial substance was unstable over the same timescale as the antibacterial substance produced from photoactivated TPPS4 and KSeCN. Figure 7C shows that the killing ability of the mixture did indeed decay over time and was totally eliminated after a 1-hour delay between producing the mixture and adding the bacterial cells.
Figure 7. Antibacterial effect of lead tetracetate is potentiated by SeCN−.
100 mM KSeCN was added to bacterial suspensions and increasing amounts of Pb(OAc)4 was added and incubated for 30 min. (A) E. coli; (B) MRSA; (C) 5 mM Pb(OAc)4 + 10 mM KSeCN were mixed and after different times an equal volume of bacterial suspension was added and incubated for 30 min.
Discussion
We have shown that aPDI killing of both Gram-positive bacteria and Gram-negative bacteria is strongly potentiated by addition of the inorganic salt KSeCN. Interestingly the potentiation was much stronger in the case of Gram-negative bacteria, than it was in the case of Gram-positive bacteria. This was somewhat unusual, as most published reports show that Gram-positive species are considerably more susceptible than Gram-negative species to almost all aPDI approaches. It is known that the Gram-negative double membrane barrier excludes non-cationic PS such as RB [10], and moreover that extracellularly produced singlet oxygen cannot easily diffuse into Gram-negative cells [20].
Moreover with Gram-negative E. coli, the concentration of KSeCN at which potentiation of killing was maximal, depended on the strength of binding of the PS to the bacteria. In Figure 2A it can be seen that with MB that binds to (and photoinactivates) E. coli, as low as 0.5 mM KSeCN was sufficient. In the case of TPPS4 which binds to (but does not photoinactivate E. coli [6]) it was necessary to use 10 mM KSeCN to achieve eradication. In the case of RB (which neither binds to nor photoinactivates E. coli [10]) it was necessary to use as high as 50 mM.
When we examined five different Gram-negative species with aPDI mediated by TPPS4 (a PS that has very limited PDT killing of Gram-negatives), we obtained eradication in every case whether the cells were present during light, or were added afterwards. When we repeated the experiments with a panel of Gram-positives (MRSA, S. epidermidis, E. faecalis and B. subtilis) we obtained eradication with TPPS4-PDT alone and also with TPPS4-PDT plus KSeCN in “cells in” format. However when we carried the “cells after” experiment the results were very different with only 2 logs of Staphylococcus species being killed, no killing of E. faecalis, and eradication was achieved only in the case of B. subtilis. This unexpected difference between Gram-positive strains prompted us to test two other species of Bacillus to see if Bacillus species in general were susceptible to the “cells after” approach of PDT + KSeCN. This indeed proved to be the case as both B. atrophaeus and B. megaterium were also eradicated. It is at present uncertain exactly why vegetative Bacillus cells are more susceptible to the bactericidal effects of the product of PDT and KSeCN (SeCN)2 while other Gram-positive species are not, and indeed why all Gram-negative species tested also seem to be susceptible. However, Bacillus species do have one thing in common with Gram-negative species, which is they are all rod-shaped, while other Gram-positive species are cocci with a spherical shape. Perhaps the greater surface area of rods compared to spheres accounts for better uptake or penetration of the bactericidal agent.
We recently reported that the addition of potassium iodide dramatically potentiated the aPDI killing of Gram-negative (and Gram-positive) bacteria by photoactivated MB [9], Photofrin [8], RB [10], and TPPS4 [6]. The singlet oxygen generated could oxidize iodide anion to produce enough iodine to eradicate both Gram-positive and Gram-negative bacteria. Interestingly TPPS4, which we initially assumed to be an anionic porphyrin, turned out to be cationic in character due to the two positively charged pyrrole nitrogen atoms and could bind to E. coli although it could not penetrate into the interior [6]. The mechanism of the potentiation by KSeCN appears to depend on the photochemical production of an antibacterial species formed in a reaction between the photoactivated PS and the added KSeCN. We found that there was more killing when the “cells in” format was employed compared to the “cells after format”. As shown in Figure 2A, 200 nM TPPS4 + 10 J/cm2 light + 10 mM KSeCN produced eradication of E. coli with “cells in” while 6.4 mM TPPS4 + 10 J/cm2 light + 100 mM KSeCN was necessary for eradication in the “cells after” format (see Figure 3A). There are two possible explanations for this major difference. Firstly it is possible that the putative (SeCN)2 is decomposing at the same time as it is being produced during the 30 minutes required for the illumination period. Therefore when the cells were added at the end of illumination (“cells after”), the amount of (SeCN)2 remaining was much less than the total amount of (SeCN)2 produced during the entire illumination time with “cells in”. Since we found that the antibacterial species is of very limited stability in water (up to 20 minutes) this is a distinct possibility. The second possible explanation is that there is a very short-lived reactive species produced (for instance the selenocyanate radical SeCN•) formed during the illumination, and this radical can efficiently inactivate bacteria. It is difficult to distinguish between these two possibilities
All the available evidence points to the mechanism involving the reaction of singlet oxygen with two molecules of SeCN− to produce the pseudohalogen species, selenocyanogen (SeCN)2. The involvement of singlet oxygen depends on the following lines of evidence. Activation of the fluorescent probes HPF and SOSG was completely quenched by addition of KSeCN (Figures 5A and 5B). Oxygen consumption was dramatically increased when RB was illuminated in the presence of KSeCN (Figure 6A). The 1270 nm luminescence signal from singlet oxygen was quenched in the presence of KSeCN and the corresponding lifetime was much shorter (Figure 6B). Singlet oxygen may carry out a 2-electron oxidation of SeCN− according to eq 1 to form (SeCN)2 plus hydrogen peroxide (H2O2). The increase in Amplex Red fluorescence when KSeCN was added to photoactivated RB (Figure 5C) supports this hypothesis.
| eq1 |
It is also possible that singlet oxygen could react with SeCN− by a 1-electron oxidation to produce two radicals as shown in eq2.
| eq2 |
The SeCN• radicals could then recombine to produce (SeCN)2 as shown in eq3
| eq3 |
However this mechanism may be less likely as shown by our failure to detect any superoxide formed in the reaction using the nitroblue tetrazolium reagent (data not shown).
(SeCN)2 was originally isolated by Birckenbach and Kellermann [21] in 1925 as an unstable yellow crystalline powder. Since then, there have been remarkably few papers discussing the chemical properties of (SeCN)2, except a review paper in Chemical Reviews by Walden and Auldrieth dating from 1928 [22]. We can trace no papers at all discussing the biological properties of (SeCN)2. It is known that (SeCN)2 adds to SeCN− anion to form a “tri-selenocyanate” (SeCN)3− anion (eq4) in the same way that iodide adds to iodine to form a triodide anion [23]. Since it is also known that tri-iodide anion is highly bactericidal [24], it would appear reasonable to hypothesize that (SeCN)3− could also be bactericidal.
| eq4 |
The literature methods for the preparation of (SeCN)2 involve oxidation of SeCN− by various reagents. Iodine pentafluoride was used to oxidize KSeCN to (SeCN)2 [25]. Meinke et al [26] used the reaction of silver selenocyanate with iodine in tetrahydrofuran at 0 °C to produce (SeCN)2. (SeCN)3− was produced during electrolysis (anodic oxidation) of KSeCN using platinum electrodes [27]. Kaufmann and Kogler [28] reported that the reaction between lead tetraacetate and KSeCN could produce the unstable (SeCN)2. This reaction was also mentioned by Challenger et al [29]. This convenient reaction allowed us to compare the product of aPDI reacting with KSeCN and the product of the reaction between Pb(OAc)4 and KSeCN in terms of relative antibacterial efficiency and the stability of the effect.
It is known that selenocyanogen is unstable in water. The decomposition reaction has been proposed to be as shown in eq5.
| eq5 |
The similar kinetics behavior of Pb(OAc)4 and aPDI in terms of the stability of the antibacterial activity produced by addition of KSeCN, provides additional evidence of the involvement of (SeCN)2.
We previously published that aPDI, mediated by MB and red light could be potentiated by addition of KSCN [14]. However the mechanism of KSCN appears to be different from the mechanisms in the present case of KSeCN. In the case of KSCN we were able to detect the sulfur trioxide radical anion that was proposed to arise by a 1-electron oxidation of sulfite anion that in turn was produced by decomposition of an addition product formed between singlet oxygen and SCN−. It should be noted that the strength of the potentiation of the antibacterial effect was much more pronounced in the case of KSeCN than it was in the case of KSCN.
The question of whether the potentiation of aPDI by KSeCN could have any clinical applications should be addressed. Undoubtedly SeCN− has more toxicity than SCN−, which is naturally present within the human body. It is known that SCN− is naturally present in saliva and bronchial secretions where it acts as an antimicrobial defense system in combination with endogenous peroxidase enzymes (lactoperoxidase) that can oxidize it to hypothiocyanite (OSCN−) in the presence of H2O2 [30, 31]. However Cupp-Sutton and Ashby [32] stated that the corresponding selenium species (hyposelenocyanite) “is not known”. Anan et al [33] showed using mass spectrometry, that SeCN− was produced in human hepatoma HepG2 cells that were exposed to sodium selenite. It was proposed that selenite was metabolized to SeCN− to temporarily reduce its toxicity, and that SeCN− could function as an intrinsic selenium pool in cultured cells. Clinically aPDI is carried out by topically applying the PS to the infected area followed by delivery of the light [34, 35]. In principle it would be possible to add KSeCN to the solution of the PS (for instance MB), and thereby increase the bacterial killing by many logs. Further studies are necessary to explore whether addition of KSeCN causes any (additional) damage to mammalian cells that are exposed to aPDI using the same condition as are routinely used to kill bacteria.
Acknowledgments
This work was supported by US-NIH grants R01AI050875 and R21AI121700. Research carried out at the Jagiellonian University (A.Z., A. K. and T.S) was supported by a grant from the National Science Centre (2011/03/B/NZ1/00007). Liyi Huang was supported by National Natural Science Foundation of China (81260239, 81472002), Guangxi Scientific and Technological Project (1355005-1-2), Guangxi Natural Science Foundation (2016GXNSFAA380312)
References
- 1.Hamblin MR. Curr Opin Microbiol. 2016;33:67–73. doi: 10.1016/j.mib.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Neill J. The Review on Antimicrobial Resistance Chaired by Jim O’Neill. 2015. [Google Scholar]
- 3.Tang HM, Hamblin MR, Yow CM. J Infect Chemother. 2007;13:87–91. doi: 10.1007/s10156-006-0501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kashef N, Hamblin MR. Drug Resist Updat. 2017;31:31–42. doi: 10.1016/j.drup.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abrahamse H, Hamblin MR. Biochem J. 2016;473:347–64. doi: 10.1042/BJ20150942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang L, El-Hussein A, Xuan W, Hamblin MR. J Photochem Photobiol B. 2017;178:277–286. doi: 10.1016/j.jphotobiol.2017.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamblin MR. Expert Rev Anti Infect Ther. 2017:1–11. doi: 10.1080/14787210.2017.1397512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang L, Szewczyk G, Sarna T, Hamblin MR. ACS Infect Dis. 2017;3:320–328. doi: 10.1021/acsinfecdis.7b00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vecchio D, Gupta A, Huang L, Landi G, Avci P, Rodas A, Hamblin MR. Antimicrob Agents Chemother. 2015;59:5203–12. doi: 10.1128/AAC.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wen X, Zhang X, Szewczyk G, El-Hussein A, Huang Y, Sarna T, Hamblin MR. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.00467-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, Huang YY, Kushida Y, Bhayana B, Hamblin MR. Free Radic Biol Med. 2016;95:74–81. doi: 10.1016/j.freeradbiomed.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang L, St Denis TG, Xuan Y, Huang YY, Tanaka M, Zadlo A, Sarna T, Hamblin MR. Free Radic Biol Med. 2012;53:2062–71. doi: 10.1016/j.freeradbiomed.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasimova KR, Sadasivam M, Landi G, Sarna T, Hamblin MR. Photochem Photobiol Sci. 2014;13:1541–8. doi: 10.1039/c4pp00021h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St Denis TG, Vecchio D, Zadlo A, Rineh A, Sadasivam M, Avci P, Huang L, Kozinska A, Chandran R, Sarna T, Hamblin MR. Free Radic Biol Med. 2013;65C:800–810. doi: 10.1016/j.freeradbiomed.2013.08.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi K, Suzuki N, Ogra Y. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulvey MA, Schilling JD, Hultgren SJ. Infect Immun. 2001;69:4572–9. doi: 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jett BD, Hatter KL, Huycke MM, Gilmore MS. Biotechniques. 1997;23:648–50. doi: 10.2144/97234bm22. [DOI] [PubMed] [Google Scholar]
- 18.Rozanowska M, Jarvis-Evans J, Korytowski W, Boulton ME, Burke JM, Sarna T. J Biol Chem. 1995;270:18825–30. doi: 10.1074/jbc.270.32.18825. [DOI] [PubMed] [Google Scholar]
- 19.Zadlo A, Burke JM, Sarna T. Photochem Photobiol Sci. 2009;8:830–7. doi: 10.1039/b901820d. [DOI] [PubMed] [Google Scholar]
- 20.Dahl TA, Midden WR, Hartman PE. J Bacteriol. 1989;171:2188–94. doi: 10.1128/jb.171.4.2188-2194.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birckenbach L, Kellermann K. Ber Dtsch Chem Ges. 1925;58:786. [Google Scholar]
- 22.Walden P, Audrieth IF. Chem Rev. 1928;5:339–359. [Google Scholar]
- 23.Martins ME, Castellano CE, Calandra AJ, Arvia AJ. Anal Chem. 1978;50:229–231. [Google Scholar]
- 24.Taylor SL, Fina LR, Lambert JL. Appl Microbiol. 1970;20:720–2. doi: 10.1128/am.20.5.720-722.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aynsley EE, Greenwood NN, Sprague MJ. J Chem Soc (Resumed) 1964:704–708. [Google Scholar]
- 26.Meinke PT, Krafft GA, Guram A. J Org Chem. 1988;53:3632–3634. [Google Scholar]
- 27.Martins ME, Castellano CE, Calandra AJ, Arvia AR. Anal Chem. 1978;50:229–231. [Google Scholar]
- 28.Kaufmann HP, Kögler F. Eur J Inorg Chem. 1926;59:178. [Google Scholar]
- 29.Challenger FC, Peters AT, Halevy J. J Chem Soc. 1926:1648–1655. [Google Scholar]
- 30.Gau J, Furtmuller PG, Obinger C, Arnhold J, Flemmig J. Biochem Biophys Rep. 2015;4:257–267. doi: 10.1016/j.bbrep.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashby MT. J Dent Res. 2008;87:900–14. doi: 10.1177/154405910808701003. [DOI] [PubMed] [Google Scholar]
- 32.Cupp-Sutton KA, Ashby MT. Antioxidants (Basel) 2016;5 doi: 10.3390/antiox5040042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anan Y, Kimura M, Hayashi M, Koike R, Ogra Y. Chem Res Toxicol. 2015;28:1803–14. doi: 10.1021/acs.chemrestox.5b00254. [DOI] [PubMed] [Google Scholar]
- 34.Kharkwal GB, Sharma SK, Huang YY, Dai T, Hamblin MR. Lasers Surg Med. 2011;43:755–67. doi: 10.1002/lsm.21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma SK, Mroz P, Dai T, Huang YY, St Denis TG, Hamblin MR. Isr J Chem. 2012;52:691–705. doi: 10.1002/ijch.201100062. [DOI] [PMC free article] [PubMed] [Google Scholar]