Abstract
Proton magnetic resonance spectroscopy measurements of glutamate and GABA are important in neuropsychiatric research. Some study designs require simultaneous measurement of both metabolites. GABA measurement requires specialized pulse sequences, the most common approach being J-difference spectral editing with MEGA-PRESS. This method enables two different strategies for concurrently measuring glutamate - from either off-resonance or difference spectra. However, it is uncertain how either strategy compares to conventional glutamate measurements. Here we compared these approaches in 49 subjects (28 healthy volunteers and 21 first-episode psychosis patients), in whom both PRESS (TE 80) and MEGA-PRESS (TE 68) spectra were obtained from dorsolateral prefrontal cortex. Glutamate and glx estimates from MEGA-PRESS difference and off-resonance spectra were compared to glutamate and glx estimates from PRESS spectra using correlational analyses. In healthy volunteers, correlations between PRESS and MEGA-PRESS off-resonance values were r ≥ 0.88 and were significantly higher than correlations between PRESS and MEGA-PRESS difference spectrum values (r ≤ 0.36). Patients showed a similar pattern. Lower correlations with difference spectrum values may reflect a disproportionate impact of field instabilities on co-edited glutamate signals. The results suggest that MEGA-PRESS off-resonance spectra can substitute for separately-acquired PRESS spectra in studies requiring simultaneous glutamate and GABA measurements.
Keywords: glutamate, GABA, MRS, simultaneous, brain, metabolites, measurement
Graphical abstract
1. Introduction1
Proton magnetic resonance spectroscopy (1H-MRS) is a technique for the non-invasive measurement of neurometabolites in defined regions of the human brain. Using optimized acquisition sequences, 10 or more different brain metabolites may be present in sufficiently high concentration to be measurable with clinical scanners. Among these metabolites, glutamate and GABA are of particular interest, as their functions include serving as the principal excitatory and inhibitory neurotransmitters, respectively. Disruption of glutamatergic and GABAergic systems has been implicated in a variety of neuropsychiatric disorders, including psychotic disorders, seizure disorders, and mood disorders (Jun et al., 2014; Luykx et al., 2012; Maddock and Buonocore, 2012; Marsman et al., 2013; Schur et al., 2016; van Veenendaal et al., 2015). Aspects of brain metabolism or neurotransmission involving glutamate and GABA may be promising targets for the development of novel treatments for these disorders (Poels et al., 2014; Waschkies et al., 2014). Thus, 1H-MRS may have a key role in testing the engagement of targeted mechanisms by such novel treatments. While many clinical investigators may wish to measure glutamate and GABA simultaneously, it is not yet clear to what extent this can be adequately done using a single 1H-MRS acquisition sequence. Glutamate is most often measured using conventional PRESS or STEAM sequences, while measuring GABA on a clinical scanner requires use of a specialized acquisition sequence (Wijtenburg et al., 2015; Harris et al., 2017), the most widely used of which is the MEGA-PRESS sequence (Mescher et al., 1998). MEGA-PRESS sequences used to measure GABA can also provide nominal measures of glutamate (Mescher et al., 1998), but it is not yet known to what extent such glutamate measurements are comparable to those acquired using PRESS or STEAM. The goal of the current study is to compare glutamate measurements obtained using a conventional PRESS sequence with those acquired using a widely available, GABA-optimized, MEGA-PRESS sequence.
A key challenge in measuring glutamate is distinguishing its resonances from nearby resonances arising from glutamine, GABA and glutathione. When using conventional pulse sequences at field strengths below 3T, spectral resolution is not sufficient for curve-fitting algorithms to adequately isolate the glutamate resonances. Under such conditions, the total signal near the glutamate resonances is referred to as glx, which indicates the combined signal from glutamate and some of these other resonances, in proportions that vary with different scanning parameters and basis sets. Even at field strengths of 3T or higher, separation from other metabolites (especially glutamine) is sometimes a problem for the measurement of glutamate with conventional pulse sequences, and the degree of overlap varies with echo time (Schubert et al., 2004). In addition, glutamate measurement may be hampered by overlap with macromolecules at short echo times and by the J-evolution of its complex multiplet resonances at longer echo times. In an effort to find an optimal echo time for measuring glutamate at 3T, one group (Schubert et al., 2004) showed that a PRESS sequence using TE= 80 ms produced both a strong signal for the 2.34 and 3.74 ppm glutamate resonances and good separation of the glutamate resonance at 2.34 ppm from macromolecules, glutamine, and other resonances. These investigators also showed good correspondence between glutamate values obtained with this method and those obtained using a specialized multiple quantum coherence filter sequence designed to isolate glutamate. Subsequently, this method has been used in a variety of clinical investigations, including two independent studies demonstrating elevated hippocampal glutamate in patients with schizophrenia (Gallinat et al., 2016; Kraguljac et al., 2013).
The 1H-MRS spectrum of GABA contains three multiplet resonances. When acquired with conventional acquisition methods, all three resonances are almost completely obscured by overlapping signals from more highly concentrated brain metabolites. Thus, special acquisition techniques are needed (Wijtenburg et al., 2015; Harris et al., 2017). The most widely used method for identifying and measuring GABA is J-difference editing, typically using a MEGA-PRESS sequence with a TE of ~68 msec (Mescher et al., 1998; Mullins et al., 2014). This method takes advantage of the fact that the C2 methylene resonance of GABA at 3.01 ppm is J-coupled to the C3 methylene resonance of GABA at 1.89 ppm (Govind et al. 2015). The J-evolution associated with this coupling causes inversion of the outer triplet peaks of GABA at 3.01 ppm at echo times near 68 msec. The on-resonance editing pulse selectively inverts the resonance at 1.89 ppm, which reverses the J-modulation of the coupled peak at 3 ppm, such that the magnetization of this signal fully refocuses at echo times near 68 msec. The GABA-optimized MEGA-PRESS sequence interleaves an on-resonance editing pulse with an off-resonance editing pulse. In post-processing, the off-resonance acquisition is subtracted from the on-resonance acquisition, yielding an upright GABA resonance at 3.0 ppm, while resonances that lack a coupling partner near 1.89 ppm are subtracted away (Mescher et al., 1998). Thus, the MEGA-PRESS difference spectrum contains a quantifiable GABA signal that is relatively isolated from most other resonances. In addition, resonances from glutamate, glutamine and glutathione have coupling partners near 2.1 ppm, and these signals often co-edit with GABA. Many MEGA-PRESS sequences use an editing pulse bandwidth that is sufficiently broad to at least partially refocus the J-evolution of these 2.1 ppm coupling partners. When this is the case, glutamate resonances at 3.74 and 2.34 ppm, glutamine resonances at 3.75 and 2.45 ppm, and glutathione resonances at 3.77 and 2.53 ppm co-edit with GABA and are retained in the difference spectrum. Many investigators have used co-edited signals in the difference spectra to quantify glutamate or glx (Kegeles et al., 2012; Milak et al., 2016; Nezhad et al. 2018; Yoon et al., 2010). Alternatively, the off-resonance spectrum from a GABA-optimized MEGA-PRESS sequence will be very similar to a conventional PRESS sequence when the frequency selectivity of the off-resonance editing pulse is outside the range of most metabolites (e.g. 7.5 ppm). Many investigators have used these off-resonance spectra to measure glutamate (Maddock et al., 2016; Ongur et al., 2011; Stan et al., 2015). Because of the similarity to a PRESS sequence, glutamate measurements from the off-resonance spectra acquired with a MEGA-PRESS sequence should correspond closely those from a PRESS sequence acquired with a similar echo time. In contrast, the difference spectra from a MEGA-PRESS sequence are quite different from PRESS spectra. Although glutamate signal is present in both types of spectra, it is not easy to predict how closely the glutamate values will correspond between the two acquisition strategies. This study is designed to directly compare these different approaches to measuring glutamate. The ability to measure glutamate and GABA simultaneously is particularly important when the time course of activity-dependent changes in these metabolites is the focus of study. If glutamate values measured from a MEGA-PRESS scan are found to be substantially equivalent to those measured from a conventional PRESS scan, then investigators can have confidence in using the MEGA-PRESS glutamate values as a proxy for glutamate measured from a separately acquired PRESS scan.
2. Methods
2.1 Participants
Participants were 28 healthy volunteers (24 male, aged 19 to 37, mean age 26) and 21 patients receiving treatment for first-episode psychosis (15 male, aged 18 to 32, mean age 23), all of whom were recruited as part of a larger study of cognition and brain function in first-episode psychosis. All participants were medically and psychiatrically assessed prior to brain imaging, and were free of significant medical problems. Healthy volunteers were free of current or past psychiatric diagnoses. All participants provided informed consent following a protocol approved by the IRB of the University of California, Davis and were screened for magnetic resonance imaging contraindications.
2.2 Magnetic resonance imaging and spectroscopy acquisitions
MR data were acquired using a 3 Tesla Siemens TIM/trio scanner (Berlin/Munich, Germany) with a 32-channel radiofrequency head coil. The single scanning session began with a T1-weighted structural scan (MPRAGE, TR/TE = 2500/4 ms, 1100 ms TI, flip angle = 7°, FoV 256 * 256, 0.95 mm3 voxel size, acceleration factor of two) that was used to guide voxel placement. A 30 * 15 * 35 mm voxel was centered on the left middle frontal gyrus (dlPFC), with the voxel orientation rotated to maximize the amount of cortical grey matter, while accommodating each individual participant's anatomy. The outer surface of the voxel was positioned a few millimeters inside the cortical surface to avoid inclusion of meninges in the measurements. Figure 1 demonstrates typical voxel placement. A combination of advanced automated and manual voxel shimming was used to minimize line width. A series of four MEGA-PRESS subscans were then acquired from the dlPFC voxel. MEGA-PRESS scanning parameters were TR/TE = 1500/68 ms, edit pulse frequencies = 1.9 ppm (on) and 7.5 ppm (off), Gaussian edit pulse bandwidth = 45 Hz, delta frequency = −1.7 ppm relative to water (optimized for signal detection at 3.0 ppm), water suppression bandwidth = 50 Hz, NEX = 72 each for on- and off- resonance acquisitions, duration = 3.6 min). Dividing the acquisition into four short subscans allowed for a quick updating and re-centering of the water frequency between scans. This was intended to reduce the effects of frequency offset (drift) on the editing pulse and required only a few seconds. Thus, the four subscans were acquired in rapid succession. The subscans were later combined off-line for analysis. Total data acquisition time for all four MEGA-PRESS scans was 14.4 minutes and included 288 excitations each for the on- and off- resonance acquisitions. Next, a single PRESS scan was acquired from the same dlPFC voxel. PRESS scanning parameters were TR/TE = 1500/80 ms, delta frequency =− 1.7 ppm, water suppression bandwidth = 50 Hz, NEX = 160, duration = 4.0 min. Most subjects subsequently had MEGA-PRESS, but not PRESS, scans acquired from a second brain region. These data will be reported separately.
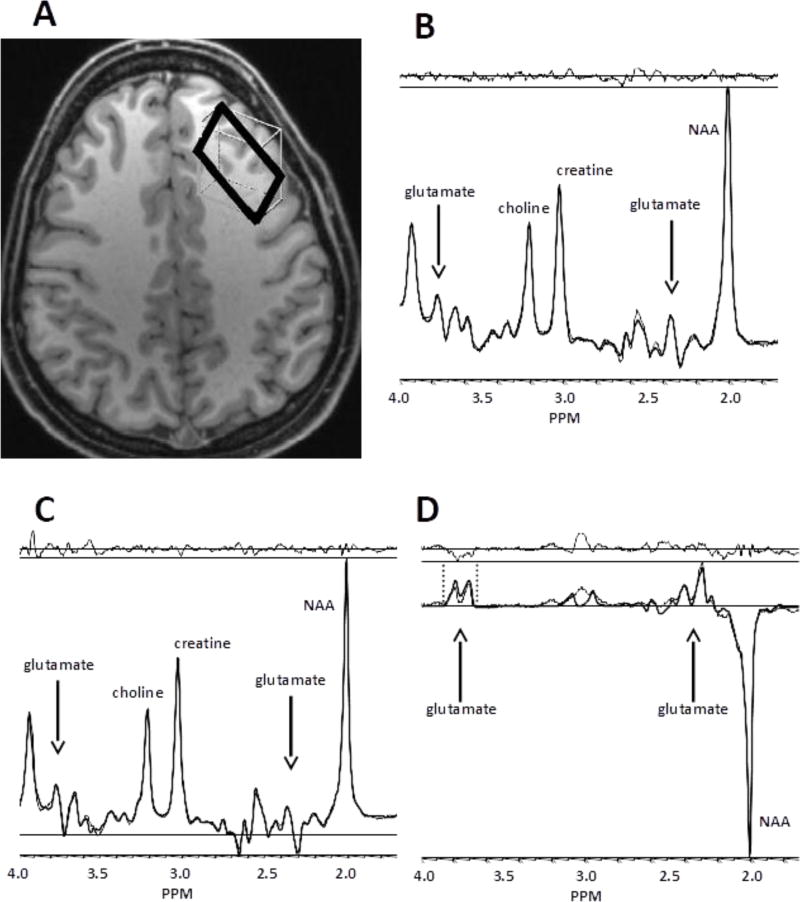
Figure 1. Voxel location and typical spectra.
Panel A depicts an axial slice showing the dlPFC voxel. Panels B, C, and D show examples of PRESS, MEGA-PRESS off-resonance, and MEGA-PRESS difference spectra, respectively. For B through D, the model fit (heavy line) is superimposed on the raw spectral data (fine line). The residual (fit minus raw data) is shown at the top of each panel. Arrows indicate locations of visible glutamate resonances. Note that glutamine and other resonances may overlap visually with glutamate resonances. Vertical dashed lines in D indicate the ppm range used for peak integration of the glx resonances centered at 3.74 ppm.
2.3 Data processing and analysis
Glutamate and other metabolite signals were quantified from PRESS spectra, MEGA-PRESS difference spectra (MP-Diff), and MEGA-PRESS off-resonance spectra (MP-Off) using either model fitting with LCModel v6.3 1-L (Provencher, 1993) or a peak integration procedure.
2.3.1 Model fitting of PRESS spectra
For PRESS spectra, metabolite values were fit using an analysis window from 4.0 to 1.7 ppm and a simulated basis set provided with LCModel that included the following metabolites: glutamate, glutamine, glutathione, creatine, phosphocreatine, n-acetylaspartate (NAA), n-acetylaspartylglutamate (NAAG), phosphocholine, glycerophosphorylcholine, myo-inositol, scyllo-inositol, aspartate, taurine, GABA, and glucose. Glx for this and for all model fitting analyses in this study is defined as the fit estimate for glutamate plus the fit estimate for glutamine. Glutamate and glx from PRESS acquisitions were normalized to Creatine (calculated as creatine + phosphocreatine) for statistical comparisons.
2.3.2 Model fitting of MEGA-PRESS off-resonance spectra and difference spectra
For both the difference and the off-resonance MEGA-PRESS spectra, the first processing step consisted of phase and frequency correction of the averaged data from each of the four 3.6-minute subscans using LCModel. The four aligned spectra were then combined in the time domain for quantification of metabolite values with LCModel. The basis sets used for both the difference and the off-resonance spectra were based on published coupling constants and chemical shifts (Govindaraju et al. 2000; Govind et al. 2015; Kaiser et al., 2008) and simulated with custom software (Murdoch, 1982). The simulations were designed for use with the Siemens MEGA-PRESS sequence and are available for download at: http://purcell.healthsciences.purdue.edu/mrslab/basis_sets.html. For the MP-Off spectra, metabolite values were fit in LCModel using an analysis window from 4.0 to 1.7 ppm. The basis set included glutamate, glutamine, glutathione, creatine, NAA, NAAG, glycerophosphorylcholine, myo-inositol, scyllo-inositol, aspartate, taurine, and GABA (Long et al., 2014). Glutamate and glx from the MP-Off spectra were normalized to creatine. For the difference spectra, metabolite values were fit using an analysis window from 4.2 to 1.95 ppm, as recommended in the LCModel manual. The basis set included GABA, glutamate, glutamine, glutathione, NAA, and NAAG (Long et al., 2014). Glutamate and glx from the difference spectra were normalized to creatine as measured in the MP-Off spectra.
2.3.3 Peak integration of glx in the MEGA-PRESS difference spectra
The peak integration method cannot separate overlapping signals. Thus a measurement of glutamate alone was not considered feasible with peak integration. Our estimate of glx by peak integration included combined signal from co-edited glutamate and glutamine, plus a small contribution from co-edited glutathione. This was quantified using jMRUI software (version 5.2) (Stefan et al., 2009) and Excel to calculate the area under the curve of the pseudo-doublet resonance centered at 3.74 (±0.10) ppm in the MP-Diff spectra, as previously described (Greenhouse et al., 2016; Yoon et al., 2010). This glx estimate was normalized to creatine, which was quantified by integration of the peak at 3.02 (±0.09) ppm in the summed (on- plus off- resonance) MEGA-PRESS spectra.
2.3.4 Statistical analyses
Each of the MEGA-PRESS-based methods for measuring glutamate or glx were compared to the PRESS-based measurement acquired from the same voxel in each subject. Pearson’s correlation (r) with the PRESS measure and the corresponding r2 value (reflecting the proportion of variance shared by the two variables) in the sample of healthy volunteers were considered the primary outcome measures for assessing similarity to the PRESS values. Statistical significance of differences between these r values was calculated by the method of Steiger (Steiger, 1980), which is appropriate when the two correlations share one variable in common as in this study (i.e. comparing the correlation between PRESS and MP-Off values to the correlation between PRESS and MP-Diff values). Alpha for significance differences between correlations was set at .05, two-tailed.
In addition, the coefficient of variation (CoV) across the sample of normal volunteers was calculated and compared for each measure of glutamate and glx. This comparison was intended to provide an estimate of the nuisance variance (noise) introduced by the measurement strategy. Since the underlying true sample variance should be similar for all measurement methods, we reasoned that methods resulting in higher CoVs would have added more nuisance variance than methods associated with lower CoVs. Calculation of the significance of differences between the CoVs was based on the t-distribution (Zaiontz, 2017). In addition to the primary analyses conducted in the sample of healthy volunteers, Pearson’s r, r2, and CoV values were also calculated for the sample of patients and for the combined sample of patients and controls.
Differences in dlPFC glutamate values between the patient and control groups were also examined. As the PRESS acquisitions with TE = 80 were considered a priori our best measure of glutamate, these data were used in testing for group effects, which were examined by analysis of variance with group as a factor. Alpha was set at .05, two-tailed.
3. Results
3.1 Quality of MRS data
All spectra were visually inspected for significant artifact due to excess water, excess lipid, or motion (Ernst and Chang, 1996) resulting in 6 of the 49 subjects being excluded from the analysis. Reasons for exclusion were: lipid contamination of MP-Off (2 patients) or PRESS spectra (1 control), water contamination of MP-Off spectra (1 patient and 1 control), and excessive within-scan motion (1 control). An additional patient was excluded when her history of an old ischemic stroke was discovered. Thus, the analysis was based on complete data from 25 healthy controls and 17 patients. Spectral quality indices for included data were as follows. Median LCModel estimates of signal-to-noise and full-width half-maximum for PRESS and MP-OFF spectra were 36 (range 24 – 43) and .036 ppm (range .024 – .048), respectively. The corresponding values for the MP-Diff spectra were 23 (range 15 – 27) and .040 ppm (range .028 – .055). Cramer-Rao lower bounds (CRLBs) in PRESS and MP-Off spectra were all ≤ 7 for glutamate and ≤ 8 for glx. CRLBs in MP-Diff spectra were all ≤ 7 for glutamate and ≤ 5 for glx.
3.2 Correlation analyses
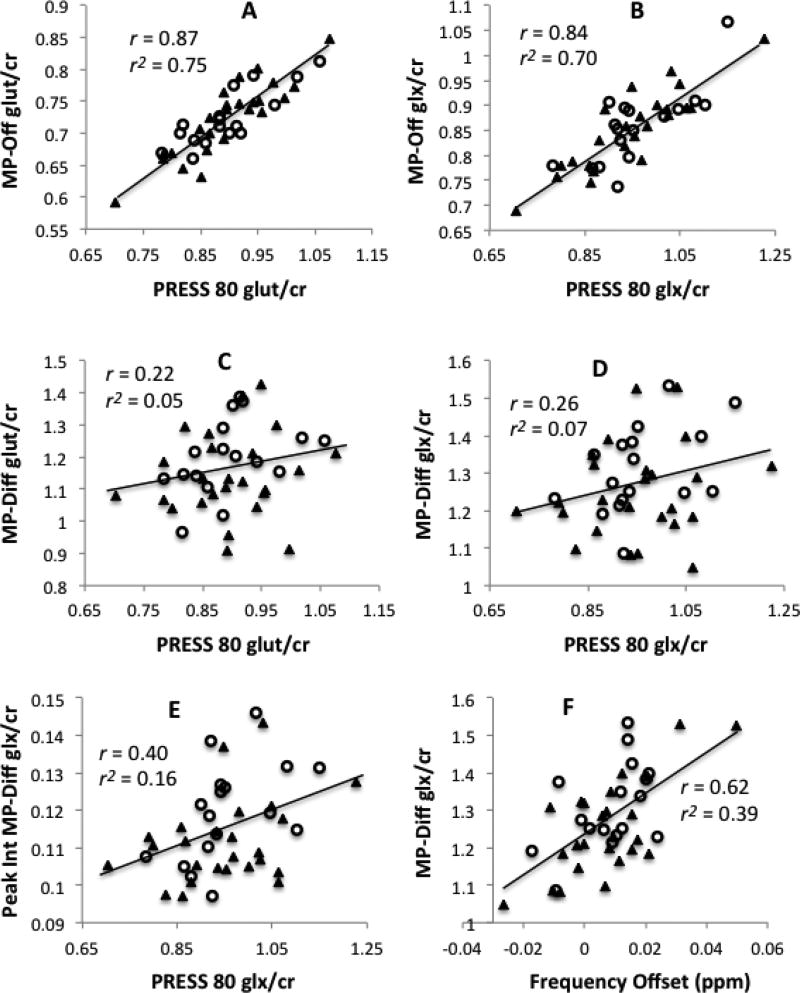
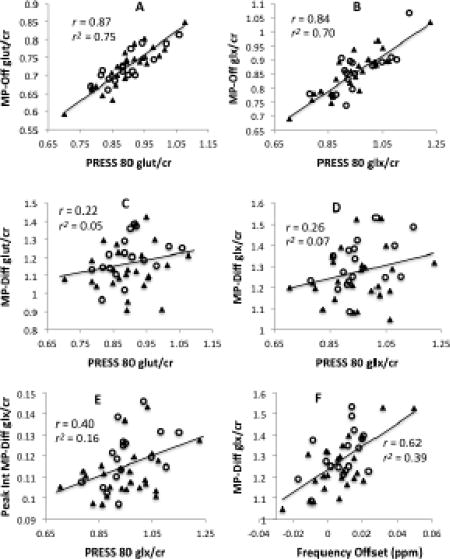
Tables 1 and 2 show Pearson correlations (r) and r2 between the different MEGA-PRESS-derived measures and the PRESS-derived measure of glutamate (Table 1) and glx (Table 2). Additional correlations are shown in the Supplemental Tables. For both glutamate and glx in the healthy volunteers, measures derived from the MP-Off spectra demonstrated strong correlations and a high percentage of shared variance (r2 ≥ .77) with the PRESS-based values. In contrast, measures derived from the MP-Diff spectra, whether by LCModel fits or by peak integration, evidenced little shared variance with the PRESS-based values (all r2 ≤ .13 in healthy volunteers). For both glutamate and glx in the sample of 25 healthy volunteers, the correlations between MP-Off and PRESS were significantly greater than the correlations between MP-Diff or MP-Diff-PI and PRESS (all p < .0001). Similar significant differences were observed across the combined sample of volunteers and patients (all p < .0001). In the smaller sample of patients alone, these differences were in the same direction, and they were significant for glutamate but not for glx. Figure 2 shows scatterplots of glutamate and glx values obtained from the PRESS spectra versus the three methods based on the MEGA-PRESS-derived spectra in the combined sample of healthy volunteers and patients.
Table 1.
Correlations between PRESS and two MEGA-PRESS measures of glutamate
| Controls (n=25) | Patients (n=17) | All (n=42) | ||||
|---|---|---|---|---|---|---|
| r (r2) with PRESS |
CoV (%) |
r (r2) with PRESS |
CoV (%) |
r (r2) with PRESS |
CoV (%) |
|
| PRESS | --- | 9.2 | --- | 8.2 | --- | 8.7 |
| MP-Off | 0.89 (0.79) | 8.2 | 0.83 (0.69) | 6.1 | 0.87 (0.75) | 7.3 |
| MP-Diff | 0.11 (0.01)** | 11.5 | 0.42 (0.18)* | 9.7 | 0.22 (0.05)** | 11.0 |
CoV = coefficient of variation; PRESS = PRESS with 80 ms TE; MP-Off = MEGA-PRESS off resonance spectra; MP-Diff = MEGA-PRESS difference spectra; all correlations shown are between glutamate/creatine ratios from LCModel fits.
lower than PRESS vs MP-Off correlation, p < .0001
lower than PRESS vs MP-Off correlation, p < .05
Table 2.
Correlations between PRESS and three MEGA-PRESS measures of glx
| Controls (n=25) | Patients (n=17) | All (n=42) | ||||
|---|---|---|---|---|---|---|
| r (r2) with PRESS |
CoV (%) |
r (r2) with PRESS |
CoV (%) |
r (r2) with PRESS |
CoV (%) |
|
| PRESS | --- | 11.9 | --- | 9.8 | --- | 10.9 |
| MP-Off | 0.88 (0.77) | 9.2 | 0.76 (0.58) | 8.9 | 0.84 (.70) | 9.0 |
| MP-Diff | 0.16 (0.03)** | 10.1 | 0.44 (0.20)a | 8.8 | 0.26 (.07)** | 9.7 |
| MP-Diff-PI | 0.36 (0.13)** | 10.2 | 0.49 (0.24)a | 11.0 | 0.40 (.16)** | 11.0 |
Abbreviations as in Table 1; MP-Diff-PI = MEGA-PRESS difference spectra with glx/creatine quantified by peak integration, all others are glx/creatine from LCModel fits. Note that since peak integration cannot estimate glutamate separately from glutamine, peak integration values are only shown in Table 2.
lower than PRESS vs MP-Off correlation, p < .0001
lower than PRESS vs MP-Off correlation, p < .05
NS, p ≤ .14
Figure 2. Values from PRESS versus MEGA-PRESS acquisitions.
Scatterplots of glutamate estimated from PRESS spectra (PRESS 80) versus MEGA-PRESS off-resonance spectra (MP-Off) (panel A); glx estimated from PRESS 80 versus MP-OFF (panel B); glutamate estimated from PRESS 80 versus MEGA-PRESS difference spectra (MP-Diff) (panel C); glx estimated from PRESS 80 versus MP-Diff (panel D); and glx estimated from PRESS 80 versus MP-Diff by peak integration (Peak Int MP-Diff) (panel E). MP-Off values correlate strongly with PRESS 80 values but MP-Diff values do not. Panel F illustrates the strong relationship between MP-Diff glx values and downfield frequency offset occurring during the MEGA-PRESS acquisition. All data are from the combined sample of control subjects and patients (N=42), with closed triangles (▲) indicating healthy volunteers and open circles (○) indicating patients.
3.3 Coefficients of variation
The coefficients of variation for glutamate and glx values are shown in Table 1 and Table 2, respectively. In the healthy volunteers, Glutamate and glx CoVs from the MP-Off or the MP-Diff spectra did not differ significantly from CoVs from the PRESS spectra. In the combined sample, glutamate CoV, but not glx CoV, was significantly higher in MP-Diff than in MP-Off spectra (z = 2.69, n = 42, p < .01).
3.4 Group comparisons
Analysis of variance of PRESS glutamate/creatine ratios with group as a factor showed no differences between the patient and control groups (mean = .90, s.d. = .07; mean = .90, s.d. = .08, respectively; F = 0.00, df = 1, 40, NS). Similarly, no group difference was seen for glx/creatine (mean = .96, s.d. = .09; mean = .95, s.d. = .11, respectively; F = 0.06, df = 1, 40, NS).
4. Discussion
4.1 Glutamate from off-resonance, but not difference spectra, corresponds to PRESS values
As expected, glutamate and glx measurements generated from the off-resonance spectra of a GABA-optimized MEGA-PRESS sequence were highly correlated with glutamate and glx measurements derived from a conventional PRESS scan acquired with a TE of 80 msec. In contrast, glutamate and glx measurements generated from the difference spectra of a GABA-optimized MEGA-PRESS sequence had little shared variance (quantified as r2) with these same measurements derived from a PRESS scan. In a sample of young healthy volunteers, the difference in the correlations between these paired measurements was highly significant. A similar pattern of results was observed in a smaller sample of medicated, first episode psychosis patients, and in the larger sample combining both patients and healthy volunteers. It is worth noting that similar results were observed whether glutamate was quantified alone or whether the combined signals from glutamate and glutamine (glx) were quantified together. In both the control subjects and in the combined sample, we observed no significant differences in how well glutamate versus glx values from the MP-Off or MP-Diff spectra correlated with PRESS spectra values. The results provide empirical support for using the off-resonance spectra rather than the difference spectra from a GABA-optimized MEGA-PRESS sequence as a proxy for a conventional PRESS sequence. This strategy may be particularly useful when investigators are interested in obtaining simultaneous measurements of GABA and glutamate from the same voxel.
The coefficients of variation for glutamate and glx values did not differ significantly between the PRESS and the off-resonance MEGA-PRESS acquisitions. However, CoVs were slightly lower in every comparison for the latter (Tables 1 and 2). This apparent, slight reduction in noise variance may partly stem from the MEGA-PRESS off-resonance spectra being acquired for a total of 7.2 minutes (288 reps, or one-half of the total 14.4 minute MEGA-PRESS acquisition time), while the PRESS spectra were acquired for only 4 minutes (160 reps). Since reliable GABA measurements using MEGA-PRESS require longer scan times than conventional PRESS scans (Brix et al., 2017), this potential advantage for the off- resonance glutamate measurements is inherent in the design of most GABA studies. In any case, our results provide no evidence that glutamate or glx values from off-resonance MEGA-PRESS acquisitions are noisier than those from PRESS acquisitions. However, in the combined sample, glutamate estimates from the MEGA-PRESS difference spectra had significantly higher CoVs than glutamate estimates from the off-resonance spectra, suggesting the presence of additional noise in the difference spectrum estimates of glutamate.
Our results suggest that glutamate estimates from PRESS and MP-Off spectra are measuring similar underlying quantities, and that these quantities differ from what is being measured by the glutamate estimates from MP-Diff spectra. Both the method of generating the spectra and the basis sets for quantifying glutamate from them are highly similar for the PRESS and the MP-Off spectra, and dissimilar for the MP-Diff spectra. Thus, it was expected that PRESS and MP-Off spectra would provide similar measures of glutamate. Many factors may have contributed to the poorer correspondence between MP-Diff and PRESS measures of glutamate and glx. Our quality metrics suggest that low quality spectra are not the problem. In particular, our difference spectra all comfortably exceed the quality thresholds recently recommended by Nezhad et al. (2018) for glutamate measurements in difference spectra (difference spectra from the 25 healthy volunteers are shown in Supplemental Figure 2). However, editing parameters that are suboptimal for glutamate may play an important role. The editing pulse frequency (1.9 ppm) used in our MEGA-PRESS editing sequence used was optimized for editing GABA, but not for editing glutamate (for which 2.1 ppm is optimal). Furthermore, the GABA-optimized echo time may have been less than optimal for editing the 2.34 ppm and 3.74 ppm resonances of glutamate. In addition, the basis set used may have not been optimal for quantifying glutamate in the difference spectra. These factors may have reduced the efficiency and increased the variability of the measurement of co-edited glutamate in our subjects, and thus contributed to the dissimilarity between MP-Diff and PRESS values. We explore in more detail below some of these possibilities.
4.2 Possible role of frequency offset during the MEGA-PRESS acquisition
Frequency offset in an MRS acquisition can result from imprecision in the prescan frequency calibration or from frequency changes over time due to several factors, including magnetic field drift and subject motion during the scan. Metabolite measurements from MEGA-PRESS difference spectra can be particularly vulnerable to the confounding effects of frequency offset. In the ideal implementation of our MEGA-PRESS sequence, the on-resonance editing pulse will remain centered at 1.9 ppm throughout the entire acquisition. With a bandwidth of 0.365 ppm (45 Hz), our Gaussian editing pulse has only an off-center interaction with the glutamate resonance at 2.1 ppm (Supplemental Figure 1). Frequency offset during the scan changes the location of the editing pulse in the frequency domain and alters its interactions with the 2.1 ppm glutamate resonance, thus altering its efficiency for editing glutamate. Importantly, the longer scan times used for GABA editing increase the likelihood of significant field drift or subject motion during the acquisition. Van der Veen et al. (2017) recently demonstrated the effects of frequency offset on metabolite values measured in the difference spectra of a GABA-optimized MEGA-PRESS acquisition at 3T. Among their findings was the observation that the glutamate/creatine ratio in the difference spectra was significantly and positively correlated with the average downfield frequency offset (i.e. toward higher ppm values) during the MEGA-PRESS scan (r = 0.48 and r = 0.51 in two frontal voxels). This most likely reflects increased editing efficiency for glutamate caused by the downfield frequency offset. With downfield offset, the center of the editing pulse moves closer to the glutamate coupling partner at 2.1 ppm and generates a larger glutamate signal in the difference spectrum.
In light of these considerations, we estimated the average frequency offset during the MEGA-PRESS acquisition to see if it could account for the low correlation between PRESS and MP-Diff values for glutamate and glx. We used the observed frequency of the creatine peak prior to frequency alignment to estimate the frequency offset in each of the four subscans of our MEGA-PRESS acquisition. Across our full sample of 42 subjects, the average downfield frequency offset ranged from -.026 ppm to +.050 ppm (mean = +.0075 ppm). Similar to van der Veen et al., we found that frequency offset was significantly and positively correlated with LCModel fits for glutamate/creatine and glx/creatine in the difference spectra (r = 0.36, p = .02 and r = 0.62, p < .0001; respectively, both N = 42) (Figure 2, panel F). Glx/creatine quantified by peak integration was similarly positively correlated with frequency offset (r = 0.60, p < .0001, N = 42). In every case, the frequency offset estimate accounts for a greater proportion of the variance in the MP-Diff values than is accounted for by the PRESS values. Frequency offset did not correlate significantly with glutamate or glx values from MP-Off or PRESS spectra, suggesting that the difference spectra were selectively vulnerable to this confounding effect (Supplemental Table). We then used partial correlation analysis on the full sample to see if controlling for the effect of frequency offset would improve the correlations between MP-Diff and PRESS values for glutamate and glx. Controlling for frequency offset led to either no change or modest increases in these correlations, yielding r = 0.22, r = 0.33, and r = 0.50 for glutamate by LCModel, glx by LCModel, and glx by peak integration, respectively (all N = 42). In all cases, these partial correlations (controlled for frequency offset) were still significantly lower than the corresponding correlations between PRESS and MP-Off values (Supplemental Table).
By selectively influencing difference spectra values, frequency offset contributed to the poor correspondence between glutamate and glx values from PRESS and those from MP-difference spectra. However, controlling for frequency offset did not eliminate the discrepancy between the two methods. Thus, additional factors must also contribute to this poor correspondence. For example, more transient frequency offsets could have affected the difference spectra values, but would not have been detected by our method of estimation from the 3.6 minute averaged subscans. In addition, non-linear effects of frequency offset and other sources of instability differentially affecting glutamate signals in the difference spectra could also contribute to their poor correspondence with PRESS values.
4.3 Possible role of the MEGA-PRESS difference spectrum basis set
Bhogal et al. (2017) recently reported on the effects of different processing parameters on the quantification of identical 7T 1H-MRS scans. In their study, four different research labs analyzed the same 60 scans with LCModel software using their preferred (but different) processing pipelines. They found that, unlike NAA fits, glutamate fits showed considerable variation between processing pipelines (inter-lab r values ranged from 0.66 to 0.94). The authors considered the different basis sets used by each lab to be one of the parameters responsible for the varying glutamate values. Other potential factors identified included the approach to modeling the baseline, the analysis window, and prior information on phasing. For various reasons, these latter parameters seemed unlikely to account for the discrepant results we observed with our difference spectra values. In difference spectra, baseline fluctuations are largely subtracted away. The analysis window used for the difference spectra was explicitly prescribed by the LCModel manual. We used the same prior information on phasing in the analysis of all of our spectra. To test the possibility that a shortcoming in our basis set could account for the poor correlations observed with the difference spectra, we re-analyzed these spectra using the MEGA-PRESS difference spectrum basis set provided by LCModel. This is a widely-used alternative that appears to incorporate slightly different chemical shift and coupling values. Results of this re-analysis supported Bhogal et al.’s observation that the choice of basis set influences the glutamate estimates. The correlation between the glutamate values estimated by the two basis sets was r = 0.67, suggesting considerable variability is introduced by the basis sets. In contrast, the glx values were very similar for the two basis sets (r = 0.90). Importantly, use of the alternative basis set led to only a very slight improvement in the correlation between the PRESS and the MP-Diff glutamate values (r = 0.35 vs. r = 0.32). There was no change in the corresponding correlation for the glx values (r = 0.27 for both basis sets). This re-analysis does not support the idea that the poor correlations between PRESS and MP-Diff values are due to an idiosyncratic shortcoming of the basis set used for the difference spectra.
It is possible that basis sets simulated from more complete modeling of the localization pulses and of the frequency offset effects on the difference spectra could improve the correspondence between difference spectra and PRESS spectra values for glutamate and glx. Many widely available basis sets, including all basis sets used in this study, treat the localization pulses as hard pulses, and do not model the four-compartment effect or imperfect RF profiles that reduce the observable signal from coupled spins (Kaiser et al., 2008; Mullins et al., 2014). These effects can be addressed by simulation of basis sets that account for them or by use of acquisition parameters that minimize them (Kaiser et al., 2008). In addition, van der Veen et al. (2017) have recently shown that basis sets can be tailored to account for the unique patterns of frequency offset occurring in individual subjects. It is possible that more precisely simulated basis sets or improved acquisition parameters would have reduced the discrepancy we observed between PRESS and MP-Diff values for glutamate and glx.
4.4 Interpreting correlations between normalized metabolite values from the same voxel
It is worth noting that we have not reported the statistical significance (p values) of correlations between pairs of glutamate or glx measurements. The necessity to normalize measured metabolite values (typically to creatine or water) creates difficulties in assigning significance to correlations between metabolites from the same voxel. This is due to shared variance in the denominators (e.g. creatine or water). In essence, the r value observed when there is no true correlation between the metabolite values in the numerator (null hypothesis) is not zero but some unknown positive value (Maddock, 2014; Pearson, 1897). Across many studies in our lab, the median correlation among all pairs of well-fit metabolites measured in the same voxel and normalized to the same creatine value has ranged from r = 0.2 to r = 0.4. This may serve as a rough estimate of the expected r value between creatine-normalized metabolites when there is no true correlation between the metabolites in the numerators, as may be the case for glutamate measured in the PRESS and MP-Diff spectra here. However, assessing the statistical significance of differences between two such correlations is largely free of this problem and can be estimated using conventional methods (Maddock, 2014). Thus, our conclusion that glutamate and glx measurements from PRESS spectra correlated more strongly with the same measurements from MEGA-PRESS off-resonance spectra than with those from MEGA-PRESS difference spectra is not compromised by the use of creatine-normalized metabolite values.
4.5 Limitations
Our study has limitations. The individual transients of the MEGA-PRESS acquisition were not archived for post-processing. Thus, phase and frequency alignment was performed on the 3.6 minute subscans rather than on each transient. Alignment of individual transients improves the quality of the final difference spectra used for quantification of metabolites. This limitation in our processing pipeline may have contributed to the poor performance of the difference spectra values for glutamate and glx. However, alignment of each transient during post-processing does not correct for the changes in MEGA-PRESS editing efficiency due to frequency offset during acquisition (van der Veen et al., 2017). In addition, we used a single set of conventional scanning parameters (a PRESS sequence with TE = 80 on a 3T system) to examine correlations with MEGA-PRESS-derived measures of glutamate and glx. Replication in an independent sample using other scanning parameters for comparison with MEGA-PRESS would increase confidence in the generalizability of the current results. In this regard, a recent re-examination of archival data showed a pattern similar to our findings. On a 4T system, Meyerhoff and colleagues (Mon et al., 2012) used a STEAM sequence (TE = 12 msec) to measure glutamate and a MEGA-PRESS sequence (TE = 71 msec) to measure GABA and glx in three cortical voxels (anterior cingulate, dorsolateral prefrontal and parieto-occipital). In their combined sample of 82 subjects (recovering alcoholics and age-matched controls), correlation coefficients between glutamate values from STEAM and glx values from MEGA-PRESS difference spectra of the same voxels from the same subjects were low and similar to our findings (all r < 0.23 and all r2 ≤ 0.05) (Meyerhoff, personal communication). Taken together, these data are consistent with the view that glx measures derived from MEGA-PRESS difference spectra have little correspondence with conventional glutamate measurements across different echo times, acquisition sequences (PRESS and STEAM) and field strengths (3T and 4T). Further studies are needed to ascertain whether our finding of a close correspondence between glutamate and glx values derived from MEGA-PRESS off-resonance spectra and conventional spectra is similarly robust to differences in acquisition parameters. While many MEGA-PRESS investigations of GABA direct the off-resonance editing pulse to approximately 7.5 ppm as was done here (Mullins et al., 2014), some investigators direct the off-resonance pulse to 1.5 ppm in order to null co-editing macromolecular resonances (Harris et al., 2015). An editing pulse in this part of the spectrum would not be expected to alter the glutamate resonances in the off-resonance spectrum. However, the current data cannot address how well glutamate measures from the off-resonance spectra of this type of MEGA-PRESS sequence would correlate with a conventional PRESS acquisition.
An important limitation of our study is that it cannot assess the relative utility of glutamate values from PRESS, MEGA-PRESS off resonance, and MEGA-PRESS difference spectra in testing hypotheses about brain glutamate. Under ideal conditions (phantom studies), all three approaches are sensitive to glutamate concentration (Henry et al., 2011; Nezhad et al. 2018). However, in vivo conditions include subject motion, longer scan times, and more complex baselines. On one hand, the relatively flat baselines of MEGA-PRESS difference spectra offer an advantage over PRESS spectra for measuring glutamate. On the other hand, subject motion and frequency offset over long acquisition times add more nuisance variance to glutamate measurements from MEGA-PRESS difference spectra than to those from PRESS spectra. In the current study, we replicated the finding of Van der Veen et al. (2017) that frequency offset during the MEGA PRESS acquisition significantly confounded glutamate measurements from the difference spectra. There is a large literature based on PRESS measurements of glutamate, and it is conventionally considered a useful method. Our results demonstrate that MEGA-PRESS off resonance glutamate values can serve as a proxy for PRESS values. However, it remains an open question whether glutamate values from PRESS spectra are more or less valid than glutamate values from MEGA-PRESS difference spectra in in vivo studies.
4.6 Conclusions
In summary, we demonstrate that glutamate and glx measurements generated from good quality off-resonance spectra of a GABA-optimized MEGA-PRESS sequence are highly correlated with glutamate and glx measurements derived from a subsequently acquired conventional PRESS scan (TE = 80). In contrast, glutamate and glx estimates derived from the difference spectra of a GABA-optimized MEGA-PRESS sequence correspond poorly with such measurements from a PRESS scan. For many experimental questions, it may be advantageous to measure glutamate and GABA simultaneously in the same voxel with the same pulse sequence. In this circumstance, our results suggest that glutamate estimates from the off-resonance spectra of a GABA-optimized MEGA-PRESS sequence can be used as a proxy for glutamate values from separately acquired PRESS sequence.
Supplementary Material
Highlights.
Glutamate can be measured two ways during a GABA-optimized MEGA-PRESS scan.
These glutamate measurements have not been directly compared to conventional methods.
MEGA-PRESS off-resonance glutamate values correspond well to PRESS values.
MEGA-PRESS difference spectra glutamate values correspond poorly to PRESS values.
MEGA-PRESS off-resonance glutamate values can serve as a proxy for PRESS values.
Acknowledgments
We thank Costin Tanase, PhD for assistance with MRS protocol development; Jovian Lam, B.S. for assistance with data collection; and study volunteers and their families for participation in research studies.
Funding: This work was supported by the National Institutes of Health R01MH105411 and P50-MH106438.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
1H-MRS: proton magnetic resonance spectroscopy; dlPFC: dorsolateral prefrontal cortex; CoV: coefficient of variation: CRLB: Cramer-Rao lower bounds; FoV: field of view; glx: glutamate+glutamine; MEGA-PRESS: Mescher-Garwood point-resolved spectroscopy; MP-Diff: MEGA-PRESS difference spectrum; MP-Off: MEGA-PRESS off-resonance spectrum; NAA: n-acetylaspartate; NAAG: n-acetylaspartylglutamate; NEX: number of excitations; PRESS: point-resolved spectroscopy; RF: radio frequency; STEAM: stimulated echo acquisition mode; TE: echo time; TR: repetition time.
Conflicts of Interest
The authors have no conflicts of interest to report.
References
- Brix MK, Ersland L, Hugdahl K, Dwyer GE, Gruner R, Noeske R, et al. Within- and between-session reproducibility of GABA measurements with MR spectroscopy. J. Magn. Reson. Imaging. 2017 doi: 10.1002/jmri.25588. [DOI] [PubMed] [Google Scholar]
- Bhogal AA, Schür RR, Houtepen LC, van de Bank B, Boer VO, Marsman A, et al. 1H–MRS processing parameters affect metabolite quantification: The urgent need for uniform and transparent standardization. NMR Biomed. 2017;30:e3804. doi: 10.1002/nbm.3804. [DOI] [PubMed] [Google Scholar]
- Dydak U, Jiang YM, Long LL, Zhu H, Chen J, Li WM, et al. In vivo measurement of brain GABA concentrations by magnetic resonance spectroscopy in smelters occupationally exposed to manganese. Environ. Health Perspect. 2011;119:219–224. doi: 10.1289/ehp.1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Chang L. Elimination of artifacts in short echo time H MR spectroscopy of the frontal lobe. Magn. Reson. Med. 1996;36:462–468. doi: 10.1002/mrm.1910360320. [DOI] [PubMed] [Google Scholar]
- Gallinat J, McMahon K, Kuhn S, Schubert F, Schaefer M. Cross-sectional study of glutamate in the anterior cingulate and hippocampus in schizophrenia. Schizophr. Bull. 2016;42:425–433. doi: 10.1093/schbul/sbv124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13:129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Govind V, Young K, Maudsley AA. Corrigendum: Proton NMR chemical shifts and coupling constants for brain metabolites. Govindaraju V, Young K, Maudsley AA, NMR Biomed. 2000; 13:129-153. NMR Biomed. 2015;28:923–924. doi: 10.1002/nbm.3336. [DOI] [PubMed] [Google Scholar]
- Greenhouse I, Noah S, Maddock RJ, Ivry RB. Individual differences in GABA content are reliable but are not uniform across the human cortex. Neuroimage. 2016;139:1–7. doi: 10.1016/j.neuroimage.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AD, Puts NA, Barker PB, Edden RA. Spectral-editing measurements of GABA in the human brain with and without macromolecule suppression. Magn. Reson. Med. 2015;74:1523–1529. doi: 10.1002/mrm.25549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AD, Saleh MG, Edden RA. Edited 1 H magnetic resonance spectroscopy in vivo: Methods and metabolites. Magn. Reson. Med. 2017;77:1377–1389. doi: 10.1002/mrm.26619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry ME, Lauriat TL, Shanahan M, Renshaw PF, Jensen JE. Accuracy and stability of measuring GABA, glutamate, and glutamine by proton magnetic resonance spectroscopy: A phantom study at 4 Tesla. J. Magn. Reson. 2011;208:210–218. doi: 10.1016/j.jmr.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun C, Choi Y, Lim SM, Bae S, Hong YS, Kim JE, et al. Disturbance of the glutamatergic system in mood disorders. Exp. Neurobiol. 2014;23:28–35. doi: 10.5607/en.2014.23.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser LG, Young K, Meyerhoff DJ, Mueller SG, Matson GB. A detailed analysis of localized J-difference GABA editing: theoretical and experimental study at 4T. NMR Biomed. 2008;21:22–32. doi: 10.1002/nbm.1150. [DOI] [PubMed] [Google Scholar]
- Kegeles LS, Mao X, Stanford AD, Girgis R, Ojeil N, Xu X, et al. Elevated prefrontal cortex gamma-aminobutyric acid and glutamate-glutamine levels in schizophrenia measured in vivo with proton magnetic resonance spectroscopy. Arch. Gen. Psychiatry. 2012;69:449–459. doi: 10.1001/archgenpsychiatry.2011.1519. [DOI] [PubMed] [Google Scholar]
- Kraguljac NV, White DM, Reid MA, Lahti AC. Increased hippocampal glutamate and volumetric deficits in unmedicated patients with schizophrenia. JAMA Psychiatry. 2013;70:1294–1302. doi: 10.1001/jamapsychiatry.2013.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Z, Li XR, Xu J, Edden RA, Qin WP, Long LL, et al. Thalamic GABA predicts fine motor performance in manganese-exposed smelter workers. PLoS ONE. 2014;9(2):e88220. doi: 10.1371/journal.pone.0088220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luykx JJ, Laban KG, van den Heuvel MP, Boks MP, Mandl RC, Kahn RS, et al. Region and state specific glutamate downregulation in major depressive disorder: a meta-analysis of (1)H-MRS findings. Neurosci. Biobehav. Rev. 2012;36:198–205. doi: 10.1016/j.neubiorev.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Maddock R. The problem of spurious correlations between pairs of brain metabolite values measured in the same voxel with magnetic resonance spectroscopy. JAMA Psychiatry. 2014;71:338–339. doi: 10.1001/jamapsychiatry.2013.4343. [DOI] [PubMed] [Google Scholar]
- Maddock RJ, Buonocore MH. MR spectroscopic studies of the brain in psychiatric disorders. Curr. Top. Behav. Neurosci. 2012;11:199–251. doi: 10.1007/7854_2011_197. [DOI] [PubMed] [Google Scholar]
- Maddock RJ, Casazza GA, Fernandez DH, Maddock MI. Acute modulation of cortical glutamate and GABA content by physical activity. J. Neurosci. 2016;36:2449–2457. doi: 10.1523/jneurosci.3455-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsman A, van den Heuvel MP, Klomp DW, Kahn RS, Luijten PR, Hulshoff Pol HE. Glutamate in schizophrenia: a focused review and meta-analysis of (1)H-MRS studies. Schizophr. Bull. 2013;39:120–129. doi: 10.1093/schbul/sbr069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11:266–272. doi: 10.1002/(sici)1099-1492(199810)11:6<266::aid-nbm530>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Milak MS, Proper CJ, Mulhern ST, Parter AL, Kegeles LS, Ogden RT, et al. A pilot in vivo proton magnetic resonance spectroscopy study of amino acid neurotransmitter response to ketamine treatment of major depressive disorder. Mol. Psychiatry. 2016;21:320–327. doi: 10.1038/mp.2015.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mon A, Durazzo TC, Meyerhoff DJ. Glutamate, GABA, and other cortical metabolite concentrations during early abstinence from alcohol and their associations with neurocognitive changes. Drug Alcohol Depend. 2012;125:27–36. doi: 10.1016/j.drugalcdep.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins PG, McGonigle DJ, O'Gorman RL, Puts NA, Vidyasagar R, Evans CJ, et al. Current practice in the use of MEGA-PRESS spectroscopy for the detection of GABA. Neuroimage. 2014;86:43–52. doi: 10.1016/j.neuroimage.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch JB. Computer Studies of Multiple-Quantum Spin Dynamics,” Ph.D. thesis. University of California, Berkeley; 1982. (published as Lawrence Berkeley Laboratory report LBL-15254). [Google Scholar]
- Nezhad FS, Anton A, Michou E, Jung J, Parkes LM, Williams SR. Quantification of GABB , glutamate and glutamine in a single measurement at 3 T using GABA-edited MEGA - PRESS. NMR Biomed. 2018;31:e3847. doi: 10.1002/nbm.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongur D, Haddad S, Prescot AP, Jensen JE, Siburian R, Cohen BM, et al. Relationship between genetic variation in the glutaminase gene GLS1 and brain glutamine/glutamate ratio measured in vivo. Biol. Psychiatry. 2011;70:169–174. doi: 10.1016/j.biopsych.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson K. On a form of spurious correlation which may arise when indices are used in the measurement of organs. Proc. R. Soc. Lond. 1897;60:489–498. [Google Scholar]
- Poels EM, Kegeles LS, Kantrowitz JT, Slifstein M, Javitt DC, Lieberman JA, et al. Imaging glutamate in schizophrenia: review of findings and implications for drug discovery. Mol. Psychiatry. 2014;19:20–29. doi: 10.1038/mp.2013.136. [DOI] [PubMed] [Google Scholar]
- Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn. Reson. Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- Schubert F, Gallinat J, Seifert F, Rinneberg H. Glutamate concentrations in human brain using single voxel proton magnetic resonance spectroscopy at 3 Tesla. Neuroimage. 2004;21:1762–1771. doi: 10.1016/j.neuroimage.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Schur RR, Draisma LW, Wijnen JP, Boks MP, Koevoets MG, Joels M, et al. Brain GABA levels across psychiatric disorders: A systematic literature review and meta-analysis of (1) H-MRS studies. Hum. Brain Mapp. 2016;37:3337–3352. doi: 10.1002/hbm.23244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan AD, Ghose S, Zhao C, Hulsey K, Mihalakos P, Yanagi M, et al. Magnetic resonance spectroscopy and tissue protein concentrations together suggest lower glutamate signaling in dentate gyrus in schizophrenia. Mol. Psychiatry. 2015;20:433–439. doi: 10.1038/mp.2014.54. [DOI] [PubMed] [Google Scholar]
- Stefan D, Di Cesare F, Andrasescu A, Popa E, Lazariev A, Vescovo E, et al. Quantitation of magnetic resonance spectroscopy signals: The jMRUI software package. Meas. Sci. Technol. 2009;20:104035. doi: 10.1088/0957-0233/20/10/104035. (9 pp) [DOI] [Google Scholar]
- Steiger JH. Tests for comparing elements of a correlation matrix. Psychol. Bull. 1980;87:245–251. [Google Scholar]
- van der Veen JW, Marenco S, Berman KF, Shen J. Retrospective correction of frequency drift in spectral editing: The GABA editing example. NMR Biomed. 2017;30:e3725. doi: 10.1002/nbm.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veenendaal TM, IJff DM, Aldenkamp AP, Hofman PA, Vlooswijk MC, Rouhl RP, et al. Metabolic and functional MR biomarkers of antiepileptic drug effectiveness: A review. Neurosci. Biobehav. Rev. 2015;59:92–99. doi: 10.1016/j.neubiorev.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Waschkies CF, Bruns A, Muller S, Kapps M, Borroni E, von Kienlin M, et al. Neuropharmacological and neurobiological relevance of in vivo (1)H-MRS of GABA and glutamate for preclinical drug discovery in mental disorders. Neuropsychopharmacology. 2014;39:2331–2339. doi: 10.1038/npp.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijtenburg SA, Yang S, Fischer BA, Rowland LM. In vivo assessment of neurotransmitters and modulators with magnetic resonance spectroscopy: Application to schizophrenia. Neurosci. Biobehav. Rev. 2015;51:276–295. doi: 10.1016/j.neubiorev.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Maddock RJ, Rokem A, Silver MA, Minzenberg MJ, Ragland JD, et al. GABA concentration is reduced in visual cortex in schizophrenia and correlates with orientation-specific surround suppression. J. Neurosci. 2010;30:3777–3781. doi: 10.1523/jneurosci.6158-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiontz C. [Accessed May 21, 2017];Real statistics using excel. 2017 www.real-statistics.com/students-t-distribution/coefficient-of-variation-testing/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.