Abstract
Introduction
GNE myopathy is an adult-onset muscle disorder characterized by impaired sialylation of (muscle) glycans, detectable by lectin histochemistry. We describe a standardized method to quantify (lectin-)fluorescence in muscle sections, applicable for diagnosis and response to therapy for GNE myopathy.
Methods
Muscle sections were fluorescently labeled with the sialic acid-binding Sambucus nigra agglutinin (SNA) lectin and antibodies to sarcolemma residence protein Caveolin-3 (CAV-3). Entire tissue sections were imaged in tiles and fluorescence was quantified.
Results
SNA fluorescence co-localizing with CAV-3 was ~50% decreased in GNE myopathy biopsies compared to muscle-matched controls, confirming previous qualitative results.
Discussion
This quantitative fluorescence method can accurately determine sialylation status of GNE myopathy muscle biopsies. This method is adaptable for expression of other membrane-associated muscle proteins, and may be of benefit for disorders in which therapeutic changes in expression are subtle and difficult to assess by other methods.
Keywords: Caveolin-3, N-acetylmannosamine (ManNAc), sarcolemma, sialylation, SNA lectin
INTRODUCTION
GNE myopathy is an autosomal recessive disorder characterized by adult-onset, slowly progressive, distal and proximal myopathy that typically leaves patients wheelchair dependent 10–20 years after onset. Histologically, it is associated with muscle fiber degeneration and formation of tubulofilamentous vacuoles in muscle tissue.1–5 GNE myopathy is caused by biallelic, mostly missense mutations in the GNE gene, encoding the rate-limiting enzyme of sialic acid biosynthesis, UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE).6–9 Sialic acids are the most abundant terminal sugar residues on glycans (glycoproteins and glycolipids), where they regulate several biologic functions, including cellular interactions, adhesions and signaling.10,11
The exact pathophysiology of GNE myopathy remains unknown, but the partial dysfunction in GNE enzyme activities due to missense mutations suggests involvement of impaired sialylation of muscle glycans.12,13 Such impaired sialylation was identified in a select group of (sialo-) glycans in GNE myopathy patients;14–18 some of the glycans may aid in diagnosing GNE myopathy.17,18 There are no robust biomarkers developed for diagnosis or for demonstrating intracellular response to therapy.
Lectins are sugar-binding proteins with ligand specificities for defined carbohydrate sequences.19 Staining of GNE myopathy human and mouse muscle sections with sialic acid binding lectins or lectins binding to desialylated sugar moieties demonstrated hyposialylation of sarcolemmal membranes.4,12,15,18,20,21 In particular, use of the lectin SNA (Sambucus nigra agglutinin) that predominantly recognizes terminal sialic acid (Neu5Ac) in an α(2,6)-linkage with either galactose or with N-acetylgalactosamine (GalNAc),24,25,26 was previously proven informative for GNE myopathy muscle sialylation status.4,15,18
Lectin histochemistry demonstrated that sialic acid residues in a α(2,6)-linkage with either galactose or with N-acetylgalactosamine (GalNAc), present on sarcolemmal glycans appeared to be absent or decreased in human and mouse GNE myopathy muscle sections (Supplemental Figure S1).4,15,18 In addition, GNE myopathy mice receiving 12 weeks of oral therapy with the sialic acid precursor N-acetylmannosamine (ManNAc) showed re-sialylation of sarcolemmal membranes by lectin histochemistry (Supplemental Figure S1C),21 and ManNAc therapy ameliorated the myopathic phenotype in GNE myopathy mice.22 These encouraging murine results suggest that lectin staining of muscle biopsies not only serves as a biomarker aiding diagnosis of GNE myopathy,4,18 but may also to demonstrate intracellular response to sialylation-increasing therapies. Clinical studies of oral ManNAc therapy in GNE myopathy subjects are currently ongoing (clinicaltrials.gov Identifier NCT02346461)4,23 and a robust biomarker of intracellular response to therapy of skeletal muscle, the only affected tissue in GNE myopathy, is pivotal for demonstration of biochemical efficacy.
Therefore, we sought to develop SNA lectin staining of skeletal muscle biopsies as a biomarker for GNE myopathy. GNE myopathy patients’ muscle biopsies are available, since they are often acquired as part of the diagnostic evaluation.1,2,4,5 In previous lectin studies for human GNE myopathy muscle biopsy sections, either paraffin embedded4,18 or frozen,12,15 were imaged and presented in a qualitative way, with the investigator determining the microscope settings and the muscle region in the biopsy appropriate for imaging. Here we present a standardized, reproducible method to image and quantify fluorescent lectin binding to muscle membranes (marked by sarcolemma residence protein Caveolin-3) in entire muscle biopsy slides.
MATERIALS and METHODS
Subjects and Muscle Biopsies
Frozen human control muscle biopsy slides (n=4) and human GNE myopathy muscle biopsy slides (n=6) (Table 1) were acquired from the Department of Neuromuscular Research, National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP), Tokyo, Japan. Muscle tissue was obtained through open biopsies on control individuals and patients participating in clinical studies approved by the NCNP Institutional Review Board; written informed consent was obtained from all muscle biopsy donors. Biopsies were frozen in 2-methylbutane, cooled in liquid nitrogen and cut into 8mm cross sections.
Table 1.
Demographics, GNE mutations, muscle type and fluorescence quantitation per biopsy
| Donor | Age (y) |
Sex | GNE Mutations * |
Muscle Biopsied † |
No. Cells in Biopsy ‡ |
Avg cell length § |
SNA Area/Cav-3 Area ‖ (per avg cell length) ¶ |
SNA/CAV-3 Total Intensity ‖ (per avg cell length) ¶ |
|---|---|---|---|---|---|---|---|---|
| Control Group: | ||||||||
| C1 | 27 | Male | Normal | biceps branchii (L) | 5,338 | 23.26 | 4,988,767 (214,505) | 2,141,539,604 (92,080,962) |
| C2 | 42 | Female | Normal | biceps branchii (L) | 6,146 | 18.44 | 4,585,638 (248,657) | 1,298,005,973 (70,384,594) |
| C3 | 32 | Female | Normal | biceps branchii (L) | 3,137 | 16.80 | 2,791,129 (166,133) | 882,982,901 (52,556,721) |
| C4 | 31 | Male | Normal | biceps branchii (L) | 13,304 | 17.94 | 10,202,823 (568,718) | 3,138,608,250 (174,949,970) |
|
| ||||||||
| Average: | 6,981 | 19.11 | 5,642,089 (299,503) | 1,865,284,182 (97,493,062) | ||||
|
| ||||||||
| GNE Myopathy Group: | ||||||||
| GNE-M1 | 27 | Male | p.V603L/ p.V603L | biceps branchii (L) | 1,393 | 11.53 | 816,286 (70,798) | 367,144,808 (31,843,163) |
| GNE-M2 | 29 | Male | p.V603L/p.V603L | biceps branchii (L) | 7,332 | 17.46 | 5,328,311 (305,148) | 1,391,871,694 (79,711,274) |
| GNE-M3 | 24 | Male | p.V603L/p.A622T | biceps branchii (L) | 3,114 | 16.14 | 1,936,055 (119,976) | 816,679,955 (50,608,913) |
| GNE-M4 | 55 | Male | p.D207V/pV603L | triceps branchii (L) | 1,223 | 23.14 | 1,404,210 (60,683) | 540,816,086 (23,371,399) |
| GNE-M5 | 40 | Female | p.D207V/p.K458N # | hamstring (R) | 3,318 | 23.81 | 3,831,255 (160,939) | 1,529,534,462 (64,251,094) |
| GNE-M6 | 39 | Female | p.D207V/p.V603L | tibialis anterior (R) | 2,600 | 21.06 | 2,279,575 (108,224) | 819,170,403 (38,890,517) |
|
| ||||||||
| Average: | 3,163 | 18.86 | 2,599,282 (137,628) | 910,869,568 (48,112,727) | ||||
Mutation nomenclature according to GNE2 isoform GenBank NP_001121699.
L, left; R, right.
Manually determined, using ImageJ software.
Avg = average. As calculated by Image J software, see text for details.
Area or Intensity of fluorescence (in pixels) determined by Definiens Architect software.
Plotted in Figure 2.
p.K458N is a novelGNE variant (disease causing by MutationTaster, deleterious by SIFT, benign by polyphen, CADD_phred score of 25).
Sectioning and Fluorescence Staining
Muscle sections were double stained with Caveolin-3 (CAV-3) antibody (R&D Systems, Minneapolis, MN) and SNA lectin (Vector Laboratories). The CAV-3 protein is not glycosylated and is found in plasma membranes of most cell types. CAV-3 antibodies are commonly used as sarcolemma markers in muscle imaging studies.27,28 The lectin SNA predominantly recognizes terminal sialic acid (Neu5Ac) in an α(2,6)-linkage with either galactose or with N-acetylgalactosamine (GalNAc).24,25,26
Slides containing frozen muscle sections were transferred from −80°C storage into pre-chilled glass dishes and thawed to room temperature for 30 min. Staining for all 9 slides (GNE myopathy and controls) was performed in 1 batch under the same conditions. Thawed muscle sections were fixed in acetone for 20 min, followed by 1 min of air drying and washing with phosphate-buffered saline (PBS) for 10 min. The fixed muscle sections were then blocked at room temperature with 1X Carbo-Free Blocking solution (Vector Laboratories, Burlingame, CA) for 1 h, followed by overnight incubation at 4°C in a mixture of 8 µg/mL Caveolin-3 (CAV-3) antibody (R&D Systems, Minneapolis, MN) and 5 µg/mL fluorescein isothiocyanate (FITC)-labeled SNA lectin (Vector Laboratories) in Carbo-Free Blocking solution. Next, the sections were washed at room temperature 3 times 5 min with 0.1% Tween/PBS and 5 min with PBS. The slides were then incubated at room temperature for 1 h with secondary antibodies (to visualize CAV-3 antibody staining) donkey anti-mouse AlexaFluor555 (A-31570, Invitrogen/Thermo Fisher Scientific, Rockville, MD) in Carbo-Free Blocking solution. After washing again 3 times 5 min with 0.1% Tween/PBS and 5 min with 1X PBS, the slides were mounted with Vectashield containing the nuclear dye DAPI (Vector Laboratories).
Image Acquisition Procedure by Inverted Microscopy
Imaging was performed on an Axio Observer Z1 inverted microscope (Zeiss Microimaging Inc, Thornwood, NY) with a 20× DIC objective, using an Axiocam MRm Rev 3 digital camera with Colibri LED illumination (Zeiss Microimaging Inc). Image settings were established on a control stained muscle slide. Entire muscle sections on each slide were digitally imaged using Tile-imaging (with local focus surface support points, 10% Tile Overlap) and Z-stack imaging (5 Z-slices per slide; 8 µm range, 2 µm interval slices) using ZEN2 Pro software (Zeiss Microimaging Inc.). All slides were imaged in one session with the same microscope settings. Image processing was performed with ZEN2 Pro software, the focus planes of the entire Z-Stack image was calculated to one extended depth of focus (EDF) image with Maximum Projection parameters. All tiles were stitched together into one image file by stitching processing.
Selected Area Imaging by Confocal Microscopy
After finalizing the Tile-imaging for each slide, 4–5 selected areas in each slide were also digitally imaged as a Z-stack with a Zeiss LSM 510 META confocal laser-scanning microscope (Zeiss Microimaging Inc). Images were acquired using a Plan-Apochromat 20X DIC objective. All confocal images are displayed as 1D projections of confocal Z-stacks (Supplemental Figure S1).
Membrane Fluorescence Quantitation with Image Analysis Software
Image analysis of SNA/CAV-3 fluorescent image files was performed using Definiens Architect Software (Definiens, Cambridge, MA) with specialized algorithms and applications. Entire image files (Figure 1A, 1B) were imported in the software. Nonspecific artifacts/fluorescence, scrambled biopsy edges and non-muscle or non-intact areas (i.e., mostly fibrous tissue areas) (Figure 1C) were manually annotated in each image file to be excluded from analysis (Figure 1D). Images were analyzed automatically in one batch. For image analysis, the CAV-3 fluorescent area was identified by the software and set as a mesh for sarcolemma membrane area (Figure 1E). The software then determined the SNA fluorescent signal co-localization (intensity (pixels), area of fluorescence) on the CAV-3 mesh (Figure 1F). The software provided fluorescence quantitation per image file in the following outputs:
Area of SNA fluorescence co-localized on CAV-3 mesh (area is calculated in pixels)
Total intensity of SNA fluorescence co-localized on CAV-3 mesh (intensity is calculated by sum of intensity and # of pixels).
In addition, the number of cells per biopsy was manually determined, using ImageJ software (NIH, Bethesda, MD). The diameter per cell was manually annotated with Image J software (annotated as narrowest distance at/near center of each cell), which was used to determine membrane length per cell and average membrane length per cell for each biopsy (calculated with Image J software).
FIGURE 1. Representative Projections of Entire Muscle Section Image Files.
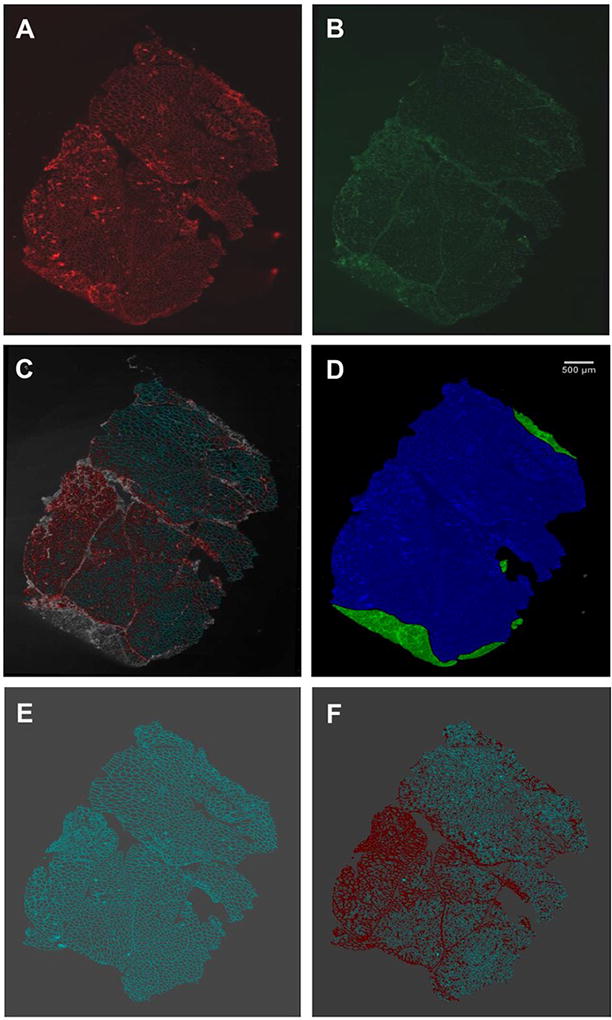
Projections of entire image files of biopsy GNE-M3 are shown to illustrate the fluorescence quantitation process. Biopsy slides were co-stained with membrane marker CAV-3 (secondary antibody AlexaFluor555-labeled) (A) and sialic-acid binding lectin SNA (FITC-labeled) (B). CAV-3/SNA fluorescence overlay image (C). Manual annotation to exclude non-specific areas (green) from analysis (D). Caveolin fluorescence set as a mesh for muscle membrane area (E). SNA fluorescence co-localizing on CAV-3 mesh (F). Fluorescence overlay images of all 9 biopsies are displayed in Supplemental Figure S1.
Statistical Analysis
Statistical analysis of fluorescence quantitation was analyzed with the Mann-Whitney unpaired t-test.
RESULTS
The biopsy donors in both the control and GNE myopathy groups were comparable in male/female distribution, range of age at biopsy and variation in GNE gene mutations, and most biopsies were acquired from the biceps branchii (Table 1). Each biopsy was of adequate quality with a large number of intact muscle cells per biopsy slide (range 1,223–13,304 total cells) (Table 1, Supplemental Figure S2). The average cell membrane length varied from 11.53–23.81, reflecting the cut of each biopsy (pure cross-section or more longitudinal cut resulting in larger cells).
Each biopsy slide was double-stained with FITC-labeled SNA lectin and CAV-3 antibodies (visualized with AlexaFluor555-labeled secondary antibodies). Initial qualitative imaging on muscle biopsy sections showed that in frozen sections, the SNA fluorescent signal is less distinctive than in paraffin embedded sections (Supplemental Figure S1A, S1C), which called for quantitation of the SNA fluorescent signal.
To standardize quantitative lectin analysis, staining methods and imaging procedures were optimized using control and GNE myopathy muscle slides. All biopsies were stained with SNA and CAV-3 in one batch, followed by imaging entire tissue sections on each slide in one batch by Tile and Z-stack imaging in automated fashion with similar microscope settings by the same investigator. One image file was created for each biopsy, with annotations for regions to exclude from analysis (Figure 1, Supplemental Figure S2), followed by quantitative analysis of fluorescent area and intensity in each image file (Table 1).
Because the size and quality of each biopsy varied, we used the relative measures SNA area and SNA intensity colocalized with CAV-3 per biopsy as outcome parameters, and divided them by the average length of cells per biopsy to correct for a different cut (cross-section cut versus more longitudinal cut) of each biopsy. These values were averaged for each group (control and GNE myopathy) of biopsies. Both the SNA area (299.5*103 in control group versus 137.6 *103 in GNE Myopathy group) and SNA intensity (97.5*106 in control group versus 48.1*106 in GNE Myopathy group) co-localized with CAV-3 per the average cell length showed a ~ 50% decrease in the GNE myopathy group compared to the control group (Table 1). Although substantial, this difference was not statistically significant (unpaired t-test; P = 0.0667, Figure 2).
FIGURE 2. Dot Plots of Fluorescence Quantitation Results.
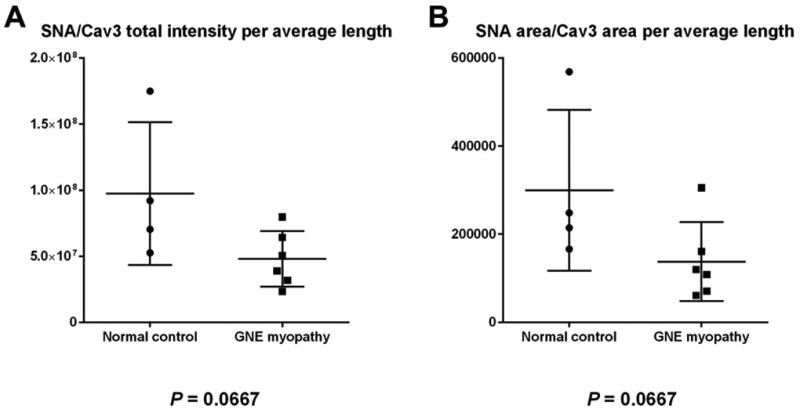
Dot plots of SNA/Cav-3 area (A) and SNA/CAV-3 intensity (B) results, comparing control muscle biopsies (n=4) with GNE myopathy muscle biopsies (n=6). A difference in fluorescence expression between groups is present with each output parameter, although not statistically significant (Mann-Whitney unpaired t-test P value 0.0667 for both parameters).
DISCUSSION
Fluorescent stained muscle biopsy slides often show varied fluorescent intensity in different areas of the tissue; this is related to differences in size, cut, and quality (fibrous tissue vs muscle cells) of each biopsy. It is often at the discretion of the investigator performing the imaging to capture representative fields of the tissue, determine microscope settings and qualitatively analyze the results, i.e., score the fields by eye. While this is an accepted practice,12,14,15,17,18 quantitation of the fluorescence signal in the entire biopsy may be more suitable if diagnosis/outcome or clinical trial decisions are based on such results. A few other studies have explored quantitation of fluorescence signals in muscle biopsy slides, mostly for dystrophin-expression related studies.29,30,31
Here we assessed quantitative analysis of SNA fluorescent signal in entire muscle biopsy slides as a biomarker for GNE myopathy, which is associated with impaired sarcolemma sialylation. The quantitative analysis method described in this manuscript includes standardized fluorescent staining and imaging of entire cross-section biopsy slides, with unbiased software analysis of the image files. This software program has previously been used in muscle image analysis for other purposes.31 Previous imaging demonstrated that SNA-reactive glycans are expressed at muscle membranes and all muscle fibers appear to express SNA-reactive glycans equally,4,15,18,21 Therefore, we imaged entire muscle sections and included each intact muscle cell (stained by caveolin) in our analysis.
Even with the small number of biopsies per group, we demonstrated that average areas and intensities of SNA fluorescence per CAV-3 area was ~50% (p = 0.0667) reduced in GNE myopathy biopsies compared to controls. Statistical significance was absent likely to the limited number of biopsies in each group. These findings are in line with previously published, sometimes subtle, qualitative GNE myopathy lectin imaging results.12,15,18
The use of the non-glycosylated protein, CAV-3, as sarcolemma marker proved appropriate, because CAV-3 is evenly distributed among sarcolemma membranes and muscle fiber types. The use of a software program for muscle membrane analysis using CAV-3 as a mesh, as established in this manuscript, is easily adaptable for any other purpose involving expression analysis of muscle membrane components, and may be of benefit for disorders in which therapeutic changes in expression are subtle and difficult to assess by other methods.
For future use of this method for diagnosing GNE myopathy, ranges of normal and disease fluorescence intensities/areas need to be established in a larger group of control and GNE myopathy muscle biopsies. A suspected GNE myopathy diagnosis based on muscle biopsy lectin analysis needs to be confirmed by genetic analysis identifying biallelic mutations in the GNE gene, because it is possible that other muscle disorders (e.g., dystroglycanopathies) show decreased sialylation/ glycosylation of sarcolemma membranes.32
Our established quantitation method is well-suited to demonstrate response to therapy, particularly in muscle biopsies of the same muscles in the same individuals acquired before and after therapy. We have now established and validated this method for GNE myopathy muscle biopsies, and intend to apply it to muscle biopsies acquired in our ongoing Phase 2 clinical trial of ManNAc for GNE myopathy (clinicaltrials.gov Identifier NCT02346461).
Supplementary Material
Acknowledgments
The authors thank the participating muscle biopsy donors. We thank Megumu Ogawa (NIN, NCNP, Japan) for her technical expertise in preparing the muscle biopsy specimen, Carla Ciccone (NHGRI, NIH, USA), Heidi Dorward (NHGRI, NIH, USA) and Steven Titus (NCATS, NIH, USA) for expert laboratory assistance and training, Andrzej Cholewinski (Definiens Inc., USA) for assistance with image parameters and analysis using the Definiens software, and the UDP Translational Laboratory (UDP, NIH, USA) for microscope use.
This study was supported by the Intramural Research Programs of the National Human Genome Research Institute and the National Center for Advancing Translational Sciences, National Institutes of Health, Bethesda, Maryland, USA. This study was partially funded by Escala Therapeutics (New York, USA) through a Cooperative Research and Development Agreement.
ABBREVIATIONS
- CAV-3
caveolin-3
- DAPI
4',6-diamidine-2'-phenylindole dihydrochloride
- EDF
extended depth of focus
- FITC
fluorescein isothiocyanate
- GalNAc
N-acetylgalactosamine
- GNE
UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase
- HPA
Helix pomatia agglutinin (lectin)
- ManNAc
N-acetylmannosamine
- MMA
Maackia amurensis agglutinin (lectin)
- NIH
National Institutes of Health
- PBS
phosphate-buffered saline
- PNA
Peanut agglutinin (lectin)
- SBA
Soybean agglutinin (lectin)
- SNA
Sambucus nigra agglutinin (lectin)
- VVA
Vicia villosa agglutinin (lectin)
- WGA
Wheat germ agglutinin (lectin)
Footnotes
Ethical Publication Statement:
The authors confirm that they have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Conflict of Interest Disclosure:
None of the authors has any conflict of interest to disclose.
Part of the material presented in this manuscript is contained within poster presentations made at the 22nd International Congress of the World Muscle Society (Oct 3–7, 2017; St. Malo, France) and at the Society for Glycobiology 2017 Annual meeting (Nov 5–8, 2017; Portland, OR).
References
- 1.Nonaka I, Sunohara N, Ishiura S, Satoyoshi E. Familial distal myopathy with rimmed vacuole and lamellar (myeloid) body formation. J Neurol Sci. 1981;51:141–155. doi: 10.1016/0022-510x(81)90067-8. [DOI] [PubMed] [Google Scholar]
- 2.Argov Z, Yarom R. Rimmed vacuole myopathy’ sparing the quadriceps: a unique disorder in Iranian Jews. J Neurol Sci. 1984;64:33–43. doi: 10.1016/0022-510x(84)90053-4. [DOI] [PubMed] [Google Scholar]
- 3.Argov Z, Mitrani-Rosenbaum S. The hereditary inclusion body myopathy enigma and its future therapy. Neurotherapeutics. 2008;5:633–637. doi: 10.1016/j.nurt.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huizing M, Malicdan MV, Krasnewich DM, Manoli I, Carrillo-Carrasco N. GNE Myopathy. In: Valle D, Beaud AL, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A, et al., editors. Online Metabolic and Molecular Bases of Inherited Disease. New York, NY: McGraw-Hill; 2014. http://ommbid.mhmedical.com/book.aspx+bookID+474. [Google Scholar]
- 5.Nishino I, Carrillo-Carrasco N, Argov Z. GNE myopathy: current update and future therapy. J Neurol Neurosurg Psychiatry. 2015;86:385–392. doi: 10.1136/jnnp-2013-307051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinderlich S, Stasche R, Zeitler R, Reutter W. A bifunctional enzyme catalyzes the first two steps in Nacetylneuraminic acid biosynthesis of rat liver. Purification and characterization of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. J Biol Chem. 1997;272:24313–24318. doi: 10.1074/jbc.272.39.24313. [DOI] [PubMed] [Google Scholar]
- 7.Keppler OT, Hinderlich S, Langner J, Schwartz-Albiez R, Reutter W, Pawlita M. UDP-GlcNAc 2-epimerase: a regulator of cell surface sialylation. Science. 1999;284:1372–1376. doi: 10.1126/science.284.5418.1372. [DOI] [PubMed] [Google Scholar]
- 8.Eisenberg I, Avidan N, Potikha T, Hochner H, Chen M, Olender T, et al. The UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase gene is mutated in recessive hereditary inclusion body myopathy. Nat Genet. 2001;29:83–87. doi: 10.1038/ng718. [DOI] [PubMed] [Google Scholar]
- 9.Celeste F, Vilboux T, Ciccone C, de Dios JK, Malicdan MC, Leoyklang P, et al. Mutation Update for GNE Gene Variants Associated with GNE Myopathy. Hum Mutat. 2014;35:915–926. doi: 10.1002/humu.22583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schauer R. Sialic acids as regulators of molecular and cellular interactions. Curr Opin Struct Biol. 2009;19:507–514. doi: 10.1016/j.sbi.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varki A. Sialic acids in human health and disease. Trends Mol Med. 2008;14:351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noguchi S, Keira Y, Murayama K, Ogawa M, Fujita M, Kawahara G, et al. Reduction of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase activity and sialylation in distal myopathy with rimmed vacuoles. J Biol Chem. 2004;279:11402–11407. doi: 10.1074/jbc.M313171200. [DOI] [PubMed] [Google Scholar]
- 13.Sparks SE, Ciccone C, Lalor M, Orvisky E, Klootwijk R, Savelkoul PJ, et al. Use of a cell free system to determine UDP-N-acetylglucosamine 2-epimerase and N-acetylmannosamine kinase activities in human hereditary inclusion body myopathy. Glycobiology. 2005;15:1102–1110. doi: 10.1093/glycob/cwi100. [DOI] [PubMed] [Google Scholar]
- 14.Huizing M, Rakocevic G, Sparks SE, Mamali I, Shatunov A, Goldfarb L, et al. Hypoglycosylation of alpha-dystroglycan in patients with hereditary IBM due to GNE mutations. Mol Genet Metab. 2004;81:196–202. doi: 10.1016/j.ymgme.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Tajima Y, Uyama E, Go S, Sato C, Tao N, Kotani M, et al. Distal myopathy with rimmed vacuoles: impaired O-glycan formation in muscular glycoproteins. Am J Pathol. 2005;166:1121–1130. doi: 10.1016/S0002-9440(10)62332-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paccalet T, Coulombe Z, Tremblay JP. Ganglioside GM3 levels are altered in a mouse model of HIBM: GM3 as a cellular marker of the disease. PLoS One. 2010;5:e10055. doi: 10.1371/journal.pone.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ricci E, Broccolini A, Gidaro T, Morosetti R, Gliubizzi C, Frusciante R, et al. NCAM is hyposialylated in hereditary inclusion body myopathy due to GNE mutations. Neurology. 2006;66:755–758. doi: 10.1212/01.wnl.0000200956.76449.3f. [DOI] [PubMed] [Google Scholar]
- 18.Leoyklang P, Malicdan MC, Yardeni T, Celeste F, Ciccone C, Li X, et al. Sialylation of Thomsen-Friedenreich antigen is a noninvasive blood-based biomarker for GNE myopathy. Biomark Med. 2014;8:641–652. doi: 10.2217/bmm.14.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharon N. Lectins: carbohydrate-specific reagents and biological recognition molecules. J Biol Chem. 2007;282:2753–2764. doi: 10.1074/jbc.X600004200. [DOI] [PubMed] [Google Scholar]
- 20.Voermans NC, Guillard M, Doedée R, Lammens M, Huizing M, Padberg GW, et al. Clinical features, lectin staining, and a novel GNE frameshift mutation Hereditary Inclusion Body Myopathy. Clin Neuropathol. 2010;29:71–77. [PMC free article] [PubMed] [Google Scholar]
- 21.Niethamer TK, Yardeni T, Leoyklang P, Ciccone C, Astiz-Martinez A, Jacobs K, et al. Oral monosaccharide therapies to reverse renal and muscle hyposialylation in a mouse model of GNE myopathy. Mol Genet Metab. 2012;107:748–755. doi: 10.1016/j.ymgme.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malicdan MC, Noguchi S, Hayashi YK, Nonaka I, Nishino I. Prophylactic treatment with sialic acid metabolites precludes the development of the myopathic phenotype in the DMRV-hIBM mouse model. Nat Med. 2009;15:690–695. doi: 10.1038/nm.1956. [DOI] [PubMed] [Google Scholar]
- 23.Xu X, Wang AQ, Latham LL, Celeste F, Ciccone C, Malicdan MC, et al. Safety, pharmacokinetics and sialic acid production after oral administration of N-acetylmannosamine (ManNAc) to subjects with GNE myopathy. Mol Genet Metab. 2017;122:126–134. doi: 10.1016/j.ymgme.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iskratsch T, Braun A, Paschinger K, Wilson IB. Specificity analysis of lectins and antibodies using remodeled glycoproteins. Anal Biochem. 2009;386:133–146. doi: 10.1016/j.ab.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Shibuya N, Goldstein IJ, Broekaert WF, Nsimba-Lubaki M, Peeters B, Peumans WJ. The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(alpha 2–6)Gal/GalNAc sequence. J Biol Chem. 1987;262:1596–1601. [PubMed] [Google Scholar]
- 26.Fischer E, Brossmer R. Sialic acid-binding lectins: submolecular specificity and interaction with sialoglycoproteins and tumour cells. Glycoconj J. 1995;12:707–713. doi: 10.1007/BF00731268. [DOI] [PubMed] [Google Scholar]
- 27.McNally EM, de Sá Moreira E, Duggan DJ, Bönnemann CG, Lisanti MP, Lidov HG, et al. Caveolin-3 in muscular dystrophy. Hum Mol Genet. 1998;7:871–877. doi: 10.1093/hmg/7.5.871. [DOI] [PubMed] [Google Scholar]
- 28.Minetti C, Sotgia F, Bruno C, Scartezzini P, Broda P, Bado M, et al. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat Genet. 1998;18:365–368. doi: 10.1038/ng0498-365. [DOI] [PubMed] [Google Scholar]
- 29.Arechavala-Gomeza V, Kinali M, Feng L, Brown SC, Sewry C, Morgan JE, et al. Immunohistological intensity measurements as a tool to assess sarcolemma associated protein expression. Neuropathol Appl Neurobiol. 2010;36:265–2674. doi: 10.1111/j.1365-2990.2009.01056.x. [DOI] [PubMed] [Google Scholar]
- 30.Taylor LE, Kaminoh YJ, Rodesch CK, Flanigan KM. Quantification of dystrophin immunofluorescence in dystrophinopathy muscle specimens. Neuropathol Appl Neurobiol. 2012;38:591–601. doi: 10.1111/j.1365-2990.2012.01250.x. [DOI] [PubMed] [Google Scholar]
- 31.Beekman C, Sipkens JA, Testerink J, Giannakopoulos S, Kreuger D, van Deutekom JC, et al. A sensitive, reproducible and objective immunofluorescence analysis method of dystrophin in individual fibers in samples from patients with duchenne muscular dystrophy. PLoS One. 2014;9:e107494. doi: 10.1371/journal.pone.0107494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muntoni F, Torelli S, Wells DJ, Brown SC. Muscular dystrophies due to glycosylation defects: diagnosis and therapeutic strategies. Curr Opin Neurol. 2011;24:437–442. doi: 10.1097/WCO.0b013e32834a95e3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


