Abstract
Shigella is an enteroinvasive human pathogen that infects the colonic epithelium and causes Shigellosis, an infectious diarrheal disease. There is no vaccine for the prevention or treatment of Shigellosis and antibiotic resistant strains of Shigella are increasing, emphasizing the need for deeper understanding of Shigella pathogenesis in order to design effective antimicrobial therapies. Small animal models do not recapitulate Shigellosis, therefore tissue cultured cells have served as model systems to study Shigella pathogenesis. Here, we describe protocols to enumerate Shigella invasion, cell-cell spread, and plaque formation in the tissue cultured cell lines Henle-407 and CoN-841. Additionally, we describe a new method to study Shigella invasion in primary intestinal enteroids. These protocols can be used to examine different aspects of Shigella virulence.
Keywords: Shigella, tissue culture, invasion, plaque, enteroid
INTRODUCTION
Shigella is a human-specific gastrointestinal pathogen that causes the diarrheal disease Shigellosis. After Shigella is ingested and traverses the gastrointestinal tract, it is endocytosed by colonic M-cells. Here, it invades the colon epithelium, and replicates within the host cell cytosol. Shigella then spreads to adjacent cells by exploiting host actin polymerization. This process results in colonic lesions, and a robust inflammatory response in the host. There is no small animal model that faithfully recapitulates Shigella pathogenesis, thus tissue culture models have been extensively applied to study the different aspects of Shigella virulence. The protocols here describe tissue culture assays to study different aspects of Shigella pathogenesis in epithelial cells. These protocols have been optimized with the laboratory model strain S. flexneri 2457T (serotype 2A) but can be applied to other Shigella strains as well. Likewise, different tissue culture cell lines can be used to answer specific questions regarding Shigella pathogenesis. We describe here how to enumerate Shigella host invasion, cell-cell spread, growth rate, and plaque formation. Finally, we describe a protocol to perform Shigella infections of human intestinal enteroids.
STRATEGIC PLANNING
It is important to choose the tissue culture model to best address your experimental question. Henle-407 is a HeLa cell contaminant line with a STR profile identical to HeLa, and both cell lines respond similarly to Shigella infection. Henle-407 or HeLa cells are quick growing and easy to manipulate, but have altered immune signaling pathways and are subject to the Warburg effect, a documented phenomenon where cancer-derived cell lines have elevated glycolysis and lactic acid fermentation (Hsu and Sabatini, 2008). CoN-841 cells are an untransformed epithelial cell line; these cells grow slower than Henle-407 or HeLa cells, but are not subject to the Warburg effect. The most physiologically relevant tissue culture currently available are intestinal enteroids (Sato et al., 2011). Intestinal enteroids are derived from stem cells acquired from human biopsies. These enteroids can be cultivated and differentiated into 3-dimensional spheres grown in matrigel or polarized monolayers grown on transwells. While intestinal enteroids are difficult to culture and expensive to maintain, they are ideal for experiments requiring a primary, untransformed cell line.
Shigella spp. are highly infectious, with studies estimating the infectious dose as low as 10 Shigella cells (FDA, 2012); therefore, it is important to adhere to BSL-2 protocols when handling Shigella cultures. Henle-407 cells contain human papilloma virus, and thus require BSL-2 protocols for handling. Human intestinal enteroids are typically derived from anonymized patient biopsy samples that could contain human pathogens, requiring BSL-2 protocols for handling.
BASIC PROTOCOL 1. SHIGELLA HOST CELL INVASION
Shigella is an intracellular pathogen which primarily replicates within the cytosol of a host colon cell; cytosolic invasion is dependent on a Shigella contact-dependent type III secretion system (T3SS) which delivers effectors into a host cell to induce endocytosis. Many Shigella genes are required for host cell invasion, and different environmental conditions also influence Shigella-host invasion efficiency. For example, supplementing sodium deoxycholate to Shigella cultures prior to invasion can significantly increase invasion efficiency (Pope et al., 1995). This protocol describes how to quantify Shigella invasion frequency. Host cells are cultured in 6-well plates, and Shigella is added to the host cells at a relatively high multiplicity of infection (MOI) for a short period. Gentamicin is used to inhibit extracellular Shigella growth, since it inhibits Shigella growth but cannot penetrate into an infected host cell. After infection, invaded host cells are enumerated using light microscopy. This protocol has been optimized for both Henle-407 cells and CoN-841 cells. Appropriate Shigella genetic controls to include could be a Shigella strain lacking its virulence plasmid or a Shigella T3SS genetic mutant.
Materials
Shigella glycerol freezer stock
Tryptic soy agar plates with 0.01% w/v congo red (TSA + CR)
37 °C incubated shaker
-
Luria-Bertani broth (LB)
Optional: Adding sodium deoxycholate to LB will increase Shigella invasion.
Some LB recipes call for glucose, this can alter Shigella aggregation in combination with sodium deoxycholate (Nickerson et al., 2017).
MEM (see recipe)
Henle-407 (ATCC CCL6) cultured in a T25 flask or CoN-841 (ATCC CRL-1790) cells cultured in a T75 flask
37 °C humidified incubator with 5% CO2
6-well tissue-culture treated plate (Corning #3516)
-
Trypsin
0.1% or 0.05% final concentration
Spectrophotometer
Centrifuge with a swinging-bucket rotor, capable of spinning plates
Phosphate buffered saline (PBS)
-
Gentamicin sulfate
20 μg / mL or 4 μg / mL final concentration
Camco Quik Stain® II (Wright - Giemsa)
Microscope with 100x oil objective
Stage 1: Preparing Shigella cultures
Note: Shigella that has retained its virulence plasmid will present as red colonies when streaked on TSA + CR, whereas Shigella that has lost its virulence plasmid will present as white colonies.
-
1
Streak the desired Shigella strain(s) from a freezer stock on TSA + CR.
-
2
Incubate plate overnight at 37 °C.
Shigella streaked on TSA + CR agar plates can be used up to one week if the plates are sealed and stored at 4 °C.
-
3
One day prior to infection, pick 3 similarly sized red colonies and separately inoculate 3 separate test tubes with 5 mL LB medium.
-
4
Incubate the tubes at 30 °C with shaking overnight.
Shigella overnight cultures are grown at a lower temperature to maintain the virulence plasmid.
Stage 2: Preparing host cells
Note: Prepare 3 wells per strain or condition to be tested.
-
5
One day prior to infection, trypsinize a confluent monolayer of Henle-407 cells or CoN-841 cells.
Using a higher trypsin concentration improves recovery of CoN-841 cells. Use 0.05% trypsin for Henle-407 cells and 0.1% trypsin for CoN-841 cells.
-
6
Dilute Henle-407 cells 1:10 or CoN-841 cells 1:5 in 12 mL MEM.
-
7
Dispense 2 mL cells per well of a 6-well plate.
-
8
Incubate the plate overnight at 37 °C at 5% CO2.
Stage 3: Infecting host cells
-
9
Inoculate new tubes with 5 mL LB with 50 μL overnight Shigella culture.
Sodium deoxycholate can be supplemented to the Shigella cultures at a final concentration of 0.1% w/v to increase Shigella invasion.
-
10
Incubate the Shigella cultures at 37 °C with shaking (200 rpm) until the cultures are mid-exponential phase, approximately 3–3.5 hours.
Cultures are at mid-exponential phase between an OD650 (10 mm lightpath) of 0.6–0.9
-
11
Read the OD650 of the Shigella cultures and dilute or concentrate them to 2 x 109 cfu / mL in sterile saline or PBS.
An OD650 of 1.0 (10 mm lightpath) is equivalent to 8 x 108 cfu / mL.
Shigella can be concentrated by centrifuging cultures in 1.5 mL Eppendorf tubes at 16,000 x g for 2 minutes, then aspirating away the media and resuspending the pellet in the appropriate volume of saline.
-
12
Add 100 μL Shigella to each well of the 6-well plate. Swirl the plate to distribute the Shigella in the plate well.
-
13
Centrifuge the plate at 200 x g for 10 minutes.
-
14
Incubate the plate for 30 minutes at 37 °C at 5% CO2.
-
15
Remove the media by aspiration and wash the cells 4 times with 2 mL sterile PBS warmed to 37 °C.
-
16
Add MEM containing gentamicin (20 μg / mL for Henle-407, 4 μg / mL for CoN-841).
-
17
Incubate the plate for 90 minutes at 37 °C at 5% CO2.
-
18
Remove the media by aspiration and wash the cells 2 times with 2 mL sterile PBS warmed to 37 °C.
-
19
Add 500 μL Wright-Giemsa to each well and let the plate stain for 2 minutes.
-
20
Add 2 mL distilled water to each well then remove all the liquid by aspiration.
If dye precipitant remains on the plate, an additional wash with water will remove this precipitant.
-
21
Let the plates dry for at least 30 minutes.
-
22
Examine the Henle-407 or CoN-841 cells using brightfield microscopy using a 100x oil objective (see Figure 1 for example).
-
23
Assess 300 Henle-407 or CoN-841 cells per well and score a cell as positively invaded if it contains 3 or more intracellular Shigella.
-
24
Calculate the percent of invaded cells by dividing the number of invaded Henle-407 or CoN-841 cells by the number of total cells counted.
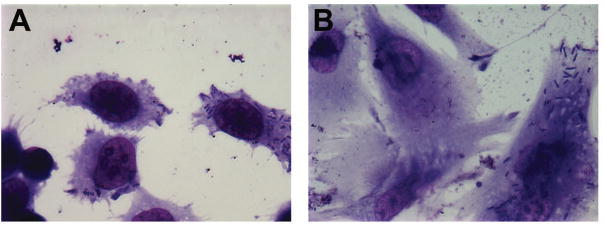
Figure 1.
(A) Henle-407 invaded by the S. flexneri WT strain. Shigella cells present as small dark purple rods in the host cytosol. (B) CoN-841 cells invaded by the S. flexneri WT strain.
ALTERNATE PROTOCOL 1. SHIGELLA CELL-CELL SPREAD
After invading a host cell in tissue culture, Shigella exploits host actin polymerization to spread to adjacent cells within a monolayer. Basic Protocol 1 can be modified to quantify the frequency of Shigella cell-cell spread. Host cells are infected with Shigella at a relatively low MOI for a longer time period, and light microscopy is used to enumerate instances where Shigella has spread to adjacent cells. A control to include is a Shigella mutant defective in cell-cell spread, such as a Shigella ΔicsA strain.
Stage 1: Preparing Shigella and host cells
-
1
Prepare Shigella cultures as detailed in Basic Protocol 1; Stage 1.
-
2
Two days prior to infection, trypsinize a confluent monolayer of Henle-407 cells.
-
3
Dilute Henle-407 cells 1:10 12 mL MEM.
-
4
Dispense 2 mL cells per well of a 6-well plate.
-
5
Incubate the plate two days at 37 °C at 5% CO2.
Stage 2: Infecting host cells
-
6
Inoculate new tubes with 5 mL LB with 50 μL overnight Shigella culture.
-
7
Incubate the Shigella cultures at 37 °C with shaking at 200 rpm until the cultures are mid-exponential phase, approximately 3–3.5 hours.
Cultures are at mid-exponential phase between an OD650 (10 mm lightpath) of 0.6–0.9
-
8
Read the OD650 of the Shigella cultures and dilute or concentrate them to 1 x 107 cfu / mL in sterile saline or PBS.
An OD650 of 1.0 (10 mm lightpath) is equivalent to 8 x 108 cfu / mL.
Shigella can be concentrated by centrifuging cultures in 1.5 mL Eppendorf tubes at 16,000 x g for 2 minutes, then aspirating away the media and resuspending the pellet in the appropriate volume of saline.
-
9
Add 100 μL Shigella to each well of the 6-well plate.
-
10
Centrifuge the plate at 200 x g for 10 minutes.
-
11
Incubate the plate for 60 minutes at 37 °C at 5% CO2.
-
12
Remove the media by aspiration and wash the cells 4 times with 2 mL sterile PBS warmed to 37 °C.
-
13
Add MEM containing 20 μg / mL gentamicin.
-
14
Incubate the plate for 120 minutes at 37 °C at 5% CO2.
-
15
Remove the media by aspiration and wash the cells 2 times with 2 mL sterile PBS warmed to 37 °C.
-
16
Add 500 μL Wright-Giemsa and let the plate stain for 2 minutes.
-
17
Add 2 mL distilled water to each well then remove all the liquid by aspiration.
-
18
Let the plates dry for at least 30 minutes. Examine the Henle-407 cells using brightfield microscopy using a 100x objective.
-
19
Assess 100 Henle-407 cells per well and score cells as positively spread if an infected Henle-407 cell (containing 3 or more Shigella) is adjacent to another infected cell.
BASIC PROTOCOL 2. SHIGELLA INTRACELLULAR GROWTH RATE
After Shigella invades a host cell, it replicates to high cell density within the host cell cytosol. Different Shigella strains can exhibit differences in intracellular doubling time, such as mutants defective in specific metabolic pathways (Kentner et al., 2014). The Shigella intracellular doubling time can be quantified by first infecting host cell monolayers, and then counting infected cells, lysing them, and enumerating intracellular bacteria by plating; this procedure is repeated over a timecourse to determine bacterial growth rate.
Materials
Shigella glycerol freezer stock
Tryptic soy agar plates with 0.01% w/v congo red (TSA + CR)
37 °C incubator
Luria-Bertani broth (LB)
Sodium deoxycholate
MEM (see recipe)
Henle-407 (ATCC CCL6) cultured in a T25 flask
37 °C humidified incubator with 5% CO2
6-well tissue-culture treated plate (Corning #3516)
-
Trypsin
0.05% final concentration
Spectrophotometer
Centrifuge capable of spinning plates
Phosphate buffered saline (PBS)
-
Gentamicin sulfate
20 μg / mL final concentration
Automated cell counter or hemocytometer
Stage 1: Preparing Shigella and host cells
Note: This protocol requires a 6-well plate of Henle-407 cells for each replicate of each strain or condition being tested.
-
1
Prepare Shigella cultures as detailed in Basic Protocol 1; Stage 1.
-
2
3–5 days prior to infection, trypsinize a confluent monolayer of Henle-407 cells.
-
3
Dilute Henle-407 cells 1:5 in 12 mL MEM.
-
4
Dispense 2 mL cells per well of a 6-well plate.
-
5
Incubate the plate at 37 °C at 5% CO2 until the cells are 70–90% confluent.
Stage 2: Infecting Henle-407 cells
-
6
Inoculate new tubes with 5 mL LB with 0.1% deoxycholate with 50 μL overnight Shigella culture.
-
7
Incubate the Shigella cultures at 37 °C with shaking until the cultures are mid-exponential phase, approximately 3–3.5 hours.
Cultures are at mid-exponential phase between an OD650 (10 mm lightpath) of 0.6–0.9
-
8
Read the OD650 of the Shigella cultures and dilute or concentrate them to 2 x 108 cfu / mL in sterile saline or PBS.
An OD650 of 1.0 (10 mm lightpath) is equivalent to 8 x 108 cfu / mL.
Shigella can be concentrated by centrifuging cultures in 1.5 mL Eppendorf tubes at 16,000 x g for 2 minutes, then aspirating away the media and resuspending the pellet in the appropriate volume of saline.
-
9
Add 100 μL Shigella to each well of the 6-well plate.
-
10
Centrifuge the plate at 200 x g for 10 minutes.
-
11
Incubate the plate for 30 minutes at 37 °C at 5% CO2.
-
12
Remove the media by aspiration and wash the cells 4 times with 2 mL sterile PBS warmed to 37 °C.
-
13
Add MEM containing 20 μg / mL gentamicin.
-
14
Incubate the plate for 60 minutes at 37 °C at 5% CO2.
-
15
For 1 well of the 6-well plate, remove the media by aspiration and wash once with 2 mL PBS warmed to 37 °C.
-
16
Trypsinize the monolayer and resuspend in 1 mL MEM.
-
17
Count the cells in suspension using an automated cell counter.
-
18
Centrifuge the cells at 1000 x g for 5 minutes.
-
19
Remove the MEM and add 1 mL sterile PBS + deoxycholate (1% w/v).
-
20
Pipet the liquid over the monolayer 5–10 times and transfer to a 1.5 mL eppendorf tube.
-
21
Vortex the liquid and serially dilute the enteroid cell lysate 1:10 in sterile saline or PBS for a total of 5 dilutions. Spot plate the dilutions on TSA + CR.
-
22
Repeat steps 10–13 five times every 30 minutes.
-
23
Incubate the plates overnight at 37 °C.
-
24
Count colonies to determine Shigella cfu / mL.
-
25
Calculate the number of Shigella cells per Henle-407 cell, then determine Shigella doubling time using equation 1 where t is time, y1 is Shigella / Henle-407 at 1 h.p.i., and y2 is Shigella / Henle-407 at 3.5 h.p.i.
- The doubling time of WT S. flexneri in Henle-407 cells is 31 minutes.
Equation 1
BASIC PROTOCOL 3. SHIGELLA PLAQUE FORMATION IN HOST CELL MONOLAYERS
When given sufficient time, Shigella will form plaques in tissue culture cell monolayers. The size of these plaques is a function of both the ability of Shigella to replicate within the host cell cytosol, and the ability of Shigella to spread to adjacent cells. Near confluent monolayers of Henle-407 cells are infected with Shigella at a low MOI, and gentamicin prevents extracellular replication. Shigella is allowed to replicate and spread for 48 hours, and then resulting plaques are visualized using Wright-Geimsa stain. The protocol detailed below describes plaque formation in Henle-407 cells but this protocol has been optimized for CoN-841 cells as well.
Materials
Shigella glycerol freezer stock
Tryptic soy agar plates with 0.01% w/v congo red (TSA + CR)
37 °C incubator
Luria-Bertani broth (LB)
MEM (see recipe)
Henle-407 (ATCC CCL6) cultured in a T25 flask
37 °C humidified incubator with 5% CO2
6-well tissue-culture treated plate (Corning #3516)
-
Trypsin
0.05% final concentration
Spectrophotometer
Centrifuge capable of spinning plates
Phosphate buffered saline (PBS)
-
Gentamicin sulfate
20 μg / mL final concentration
-
Glucose
0.2% w/v final concentration
Camco Quik Stain® II (Wright - Giemsa)
Camera
Stage 1: Preparing Shigella and host cells
Note: This protocol requires 3 wells of a 6-well plate of Henle-407 cells for each replicate of each strain or condition being tested.
-
1
Prepare Shigella cultures as detailed in Basic Protocol 1; Stage 1.
-
2
3–5 days prior to infection, trypsinize a confluent monolayer of Henle-407 cells.
-
3
Dilute Henle-407 cells 1:5 in 12 mL MEM.
-
4
Dispense 2 mL cells per well of a 6-well plate.
-
5
Incubate the plate at 37 °C at 5% CO2 until the cells are 70–90% confluent.
Stage 2: Infecting Henle-407 cells
-
6
Inoculate new tubes with 5 mL LB with 50 μL overnight Shigella culture.
-
7
Incubate the Shigella cultures at 37 °C with shaking until the cultures are mid-exponential phase, approximately 3–3.5 hours.
Cultures are at mid-exponential phase between an OD650 (10 mm lightpath) of 0.6–0.9
-
8
Read the OD650 of the Shigella cultures and dilute them to 5 x 104 cfu / mL in sterile saline or PBS.
An OD650 of 1.0 (10 mm lightpath) is equivalent to 8 x 108 cfu / mL.
Shigella can be concentrated by centrifuging cultures in 1.5 mL Eppendorf tubes at 16,000 x g for 2 minutes, then aspirating away the media and resuspending the pellet in the appropriate volume of saline.
-
9
Add 100 μL Shigella to each well of the 6-well plate.
-
10
Centrifuge the plate at 200 x g for 10 minutes.
-
11
Incubate the plate for 60 minutes at 37 °C at 5% CO2.
-
12
Remove the media by aspiration and wash the cells 4 times with 2 mL sterile PBS warmed to 37 °C.
-
13
Add MEM containing 20 μg / mL gentamicin and supplemented with additional 0.2% w/v glucose.
-
14
Incubate the plate for 48 hours at 37 °C at 5% CO2.
For Shigella strains with reduced plaque formation, the media can be replaced at 48 hours with new MEM + gentamicin and incubated an additional 24 hours.
-
15
Remove the media by aspiration and wash the cells 2 times with 2 mL sterile PBS warmed to 37 °C.
-
16
Add 500 μL Wright-Giemsa and let the plate stain for 2 minutes.
-
17
Add 2 mL distilled water to each well then remove all the liquid by aspiration.
-
18
Let the plates dry for at least 30 minutes.
Example of S. flexneri plaques can be seen in Figure 2.
-
19
Capture images of the 6-well plate using a camera.
-
20
Measure plaque size from captured plaque images using ImageJ software.
The diameter of the 6-well plate (34.8 mm) can be measured as a known size reference for scale.
Figure 2.
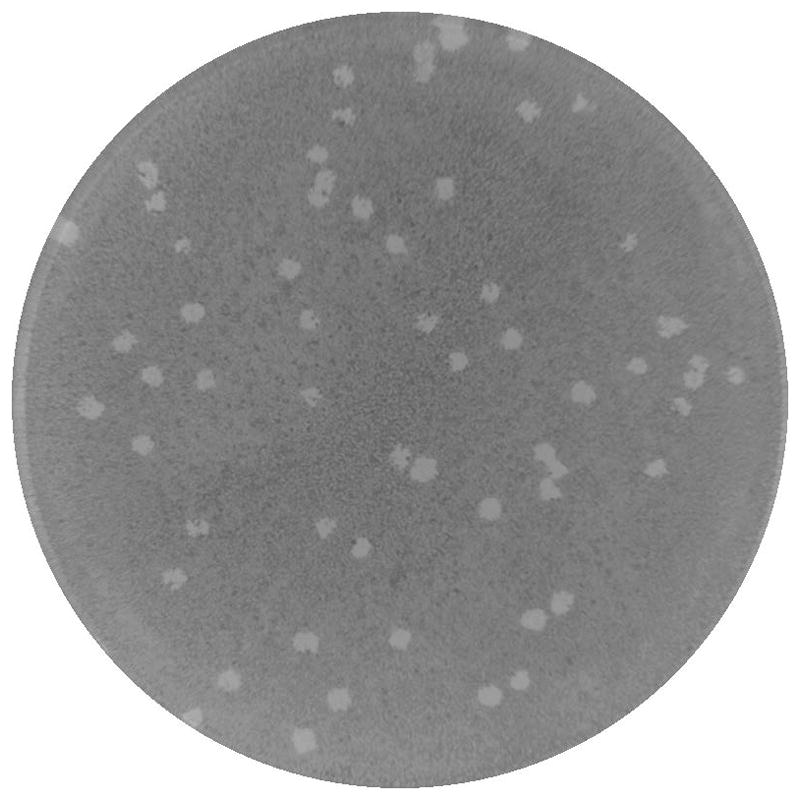
S. flexneri plaque formation in a Henle-407 monolayer. The monolayer was stained with Wright-Geimsa after 48 hours post infection. Light spots are lesions in the monolayer caused by S. flexneri.
BASIC PROTOCOL 4. SHIGELLA BASOLATERAL INVASION OF HUMAN INTESTINAL ENTEROIDS
Recent advances in tissue culture have led to the culture of intestinal enteroids. Stem cells are isolated from intestinal biopsies, expanded in vitro, and differentiated into monolayers. While difficult to manipulate, intestinal enteroids are currently the tissue culture model that most closely reflects the human host, as these cells are primary and untransformed. Intestinal enteroid monolayers are polarized, and Shigella only invades polarized cells from the basolateral surface. Therefore, intestinal enteroids must be grown on transwell inserts so that Shigella can be introduced to the basolateral surface of the monolayer.
Materials
See the chapter titled “Human Intestinal Enteroids for the Study of Bacterial Adherence, Invasion and Translocation” for materials to propagate intestinal enteroid monolayers.
Shigella glycerol freezer stock
Tryptic soy agar plates with 0.01% w/v congo red (TSA + CR)
37 °C incubator
Luria-Bertani broth (LB)
Sodium deoxycholate
37 °C humidified incubator with 5% CO2
Spectrophotometer
Phosphate buffered saline (PBS)
-
Gentamicin sulfate
20 μg / mL final concentration
CMGF-
Sterile forceps
-
Saponin, 5% w/v in PBS, filter sterilized
Saponin solution should be made fresh weekly.
Stage 1: Preparing Shigella and host cells
Note: This protocol has been optimized for differentiated intestinal enteroid monolayers derived from colon tissue, but we have demonstrated Shigella invades undifferentiated monolayers and monolayers derived from small intestine tissues as well.
-
1
Prepare Shigella cultures as detailed in Basic Protocol 1; Stage 1.
-
2
Prepare human intestinal enteroid monolayers on transwells as detailed in the separate chapter “Human Intestinal Enteroids for the Study of Bacterial Adherence, Invasion and Translocation.”
Stage 2: Infecting intestinal enteroid monolayers
-
3
Inoculate new tubes with 5 mL LB with 0.1% deoxycholate with 50 μL overnight Shigella culture.
-
4
Incubate the Shigella cultures at 37 °C with shaking until the cultures are mid-exponential phase, approximately 3–3.5 hours.
Cultures are at mid-exponential phase between an OD650 (10 mm lightpath) of 0.6–0.9
-
5
Read the OD650 of the Shigella cultures and dilute or concentrate them to 1 x 108 cfu / mL in sterile saline or PBS.
An OD650 of 1.0 (10 mm lightpath) is equivalent to 8 x 108 cfu / mL.
Shigella can be concentrated by centrifuging cultures in 1.5 mL Eppendorf tubes at 16,000 x g for 2 minutes, then aspirating away the media and resuspending the pellet in the appropriate volume of saline.
-
6
Remove the media from both the upper and lower chamber of the transwell monolayers and replace the media in the upper chamber of the transwell monolayers with CMGF-.
-
7
Using sterile forceps, flip and transfer each transwell to individual wells of a 12-well plate.
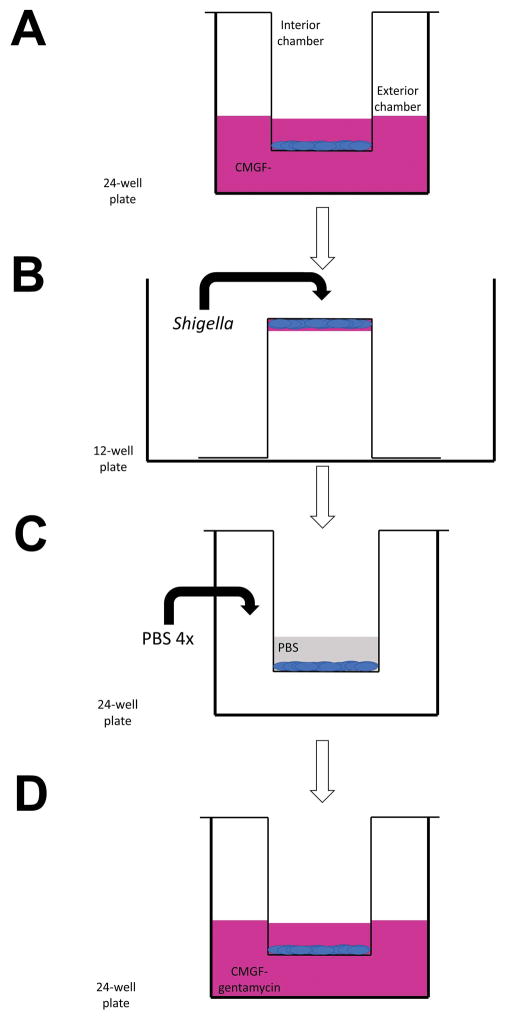
This process is diagrammed in Figure 3.
-
8
Add 10 μL Shigella to the base (now upward facing) of each transwell.
-
9
Incubate the plate for 60 minutes at 37 °C at 5% CO2.
-
10
Using sterile forceps, flip and transfer the transwell into a new well of a 24-well plate.
-
11
Add 100 μL sterile PBS to the upper chamber of the transwell.
-
12
Wash the lower chamber of the transwell 4 times with 600 μL sterile PBS warmed to 37 °C.
-
13
Remove the sterile PBS from the upper and lower chambers of the transwell and add CMGF- containing 20 μg / mL gentamicin to the transwell, 100 μL in the upper chamber and 600 μL in the lower chamber.
-
14
Incubate the plate for 120 minutes at 37 °C at 5% CO2.
-
15
Remove the media from the upper and lower chambers by aspiration and wash both chambers 2 times with sterile PBS warmed to 37 °C, 100 μL in the upper chamber and 600 μL in the lower chamber.
-
16
Remove the PBS from both chambers and add 100 μL PBS with 5% w/v saponin warmed to 37 °C to the upper chamber.
-
17
Pipet the liquid over the monolayer 5–10 times and transfer to a sterile 1.5 mL eppendorf tube.
-
18
Vortex the liquid and serially dilute the enteroid cell lysate 1:10 in sterile saline or PBS for a total of 5 dilutions. Spot plate the dilutions on TSA + CR.
-
19
Incubate the plates overnight at 37 °C.
-
20
Count cfu to determine Shigella infection.
Figure 3.
Schematic walkthrough of Shigella infection of enteroid monolayers grown on transwells. (A) The media of the interior and exterior chambers is replaced with CMGF-. (B) The transwell is inverted and aseptically transferred to a new well of a 12-well plate. The surface tension of the CMGF- will keep some of the media on the apical face of the enteroid monolayer. 10 μL Shigella is added to the upward facing side of the transwell (basolateral face of the monolayer). (C) After incubation, the transwell is transferred back to a 24-well plate. PBS is added to the interior chamber, and the exterior chamber is washed 4 times with PBS. (D) CMGF- supplemented with gentamicin is added to both the interior and exterior chambers.
REAGENTS AND SOLUTIONS
MEM
MEM powder (GIBCO 61100-087)
Tryptose phosphate broth (TPB, DIFCO 260300)
Sodium Bicarbonate (NaHCO3, Sigma S5761)
NEAA solution (Sigma M7145)
Glutamine (Sigma G7513)
Fetal bovine serum (FBS)
Add 23.8 g of MEM powder, 7.38 g of TPB powder, 5.5 g NaHCO3 in 2 L water. Dissolve and bring the pH to 7.3. Bring the total volume of 2.5 L and filter sterilize in 440 mL aliquots. Immediately before use, add 50 mL FBS, 5 mL NEAA solution, and 5 mL glutamine. Store cold for a maximum of 4 weeks.
CMGF
500 mL Advanced DMEM/F12 (Invitrogen 12634-010)
5 mL HEPES (invitrogen 15630080)
5 mL Glutamax (invitrogen 35050-079)
Store cold for a maximum of 4 weeks.
COMMENTARY
Background Information
As there is no small animal model of Shigellosis, tissue culture models have been used to study Shigella pathogenesis for more than thirty years (Oaks et al., 1985). Experiments performed with tissue culture models have revealed a great deal about Shigella virulence, including how Shigella invades host cells (Ménard et al., 1993), how Shigella spreads (Bernardini et al., 1989; Rossi et al., 2017), how host cells respond to cytosolic Shigella (Lucchini et al., 2005; Mantis et al., 1996), and how Shigella metabolizes host carbon (Kentner et al., 2014; Pieper et al., 2013; Waligora et al., 2014). The assays described here detail how to enumerate Shigella parameters of virulence including invasion, cell-cell spread, and plaque formation, but can and have been modified for more specialized applications, depending on the experimental question.
Critical Parameters and Troubleshooting
Much of the experimental variability of Shigella infections can usually be attributed to the state of the host cells. It is important to use host cells grown to similar confluence for repeated experiments, and using host cells at a later passage number can reduce Shigella infectivity. Additionally, different host cell types exhibit different permeability to gentamicin; therefore, it may be necessary to lower the gentamicin concentration in order to facilitate intracellular Shigella growth.
Depending on the experimental application, it is sometimes beneficial to increase Shigella invasion; in these cases, sodium deoxycholate should be supplemented to the Shigella LB culture prior to host cell invasion. After invading a host cell, Shigella can differentially impact host cell immune response depending on the intracellular bacterial density. Therefore, when using a new host cell line or Shigella strain, it may be necessary to adjust the MOI for each experiment.
Time Considerations
The time to execute the experiments described here from conception to results is largely dependent on preparing host cells in tissue culture. Tissue culture preparation of host cells is widely variable depending on the host cell line, the state of host cells preserved in liquid nitrogen, and the scale of the experiment. Once the appropriate host cells are prepared, the timeline of Shigella infection is consistent. Invasion and cell-cell spread assays can be performed over 2 days, growth rate assays can be performed over 3 days, plaque assays can be performed over 4 days, and enteroid infection experiments can be performed over 3 days. For invasion, spread, and plaque experiments, plates that have been stained with Giemsa will last indefinitely and can be analyzed at leisure.
Significance Statement.
There are no small animal models to study Shigellosis, thus tissue culture models are critical to understanding various aspects of Shigella host-pathogen interactions and developing novel antimicrobial therapies.
Acknowledgments
This work was supported by NIH grants AI16935 and AI131497 to S.M.P.
Footnotes
INTERNET RESOURCES
Site to download the image analysis software ImageJ, used for digitally measuring plaque size.
LITERATURE CITED
- Bernardini ML, Mounier J, d’Hauteville H, Coquis-Rondon M, Sansonetti PJ. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc Natl Acad Sci U S A. 1989;86:3867–3871. doi: 10.1073/pnas.86.10.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA. Bad Bug Book, Foodborne Pathogenic Microorganisms and Natural Toxins. 2012. pp. 22–25. [Google Scholar]
- Hsu PP, Sabatini DM. Cancer Cell Metabolism: Warburg and Beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Kentner D, Martano G, Callon M, Chiquet P, Brodmann M, Burton O, Wahlander A, Nanni P, Delmotte Nl, Grossmann J, Limenitakis J, Schlapbach R, Kiefer P, Vorholt JA, Hiller S, Bumann D. Shigella reroutes host cell central metabolism to obtain high-flux nutrient supply for vigorous intracellular growth. Proc Natl Acad Sci U S A. 2014;111:9929–9934. doi: 10.1073/pnas.1406694111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini S, Liu H, Jin Q, Hinton JCD, Yu J. Transcriptional Adaptation of Shigella flexneri during Infection of Macrophages and Epithelial Cells: Insights into the Strategies of a Cytosolic Bacterial Pathogen. Infect Immun. 2005;73:88–102. doi: 10.1128/IAI.73.1.88-102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantis N, Prévost MC, Sansonetti P. Analysis of epithelial cell stress response during infection by Shigella flexneri. Infect Immun. 1996;64:2474–2482. doi: 10.1128/iai.64.7.2474-2482.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson KP, Chanin RB, Sistrunk JR, Rasko DA, Fink PJ, Barry EM, Nataro JP, Faherty CS. Analysis of Shigella flexneri Resistance, Biofilm Formation, and Transcriptional Profile in Response to Bile Salts. Infect Immun. 2017:85. doi: 10.1128/IAI.01067-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oaks EV, Wingfield ME, Formal SB. Plaque formation by virulent Shigella flexneri. Infect Immun. 1985;48:124–129. doi: 10.1128/iai.48.1.124-129.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper R, Fisher CR, Suh MJ, Huang ST, Parmar P, Payne SM. Analysis of the Proteome of Intracellular Shigella flexneri Reveals Pathways Important for Intracellular Growth. Infect Immun. 2013;81:4635–4648. doi: 10.1128/IAI.00975-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope LM, Reed KE, Payne SM. Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect Immun. 1995;63:3642–3648. doi: 10.1128/iai.63.9.3642-3648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi RM, Yum L, Agaisse H, Payne SM. Cardiolipin Synthesis and Outer Membrane Localization Are Required for Shigella flexneri Virulence. MBio. 2017:8. doi: 10.1128/mBio.01199-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Waligora EA, Fisher CR, Hanovice NJ, Rodou A, Wyckoff EE, Payne SM. Role of Intracellular Carbon Metabolism Pathways in Shigella flexneri Virulence. Infect Immun. 2014;82:2746–2755. doi: 10.1128/IAI.01575-13. [DOI] [PMC free article] [PubMed] [Google Scholar]