Abstract
Formation of the 3′ ends of mature mRNAs requires recognition of the correct site within the last exon, cleavage of the nascent pre-mRNA, and, for most mRNAs, addition of a poly(A) tail. Several factors are involved in recognition of the correct 3′-end site. The cleavage stimulation factor (CstF) has three subunits, CstF-50 (gene symbol Cstf1), CstF-64 (Cstf2), and CstF-77 (Cstf3). Of these, CstF-64 is the RNA-binding subunit that interacts with the pre-mRNA downstream of the cleavage site. In male germ cells where CstF-64 is not expressed, a paralog, τCstF-64 (gene symbol Cstf2t) assumes its functions. Accordingly, Cstf2t knockout (Cstf2t−/−) mice exhibit male infertility due defective development of spermatocytes and spermatids. To discover differentially expressed genes responsive to τCstF-64, we performed RNA-seq in seminiferous tubules from wild type and Cstf2t−/− mice, and found that several histone and histone-like mRNAs were reduced in Cstf2t−/− mice. We further observed delayed accumulation of the testis-specific histone, H1fnt (formerly, H1t2 or Hanp1) in Cstf2t−/− mice. High-throughput sequence analysis of polyadenylation sites (A-seq) indicated reduced use of polyadenylation sites within a cluster downstream of H1fnt in knockout mice. However, high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) was not consistent with a direct role of τCstF-64 in polyadenylation of H1fnt. These findings together suggest that the τCstF-64 may control other reproductive functions that are not directly linked to the formation of 3′ ends of mature polyadenylated mRNAs during male germ cell formation.
Keywords: Testis-specific histone, protamine, H1fnt, polyadenylation, male germ cells
INTRODUCTION
Male gamete production entails a series of developmentally regulated changes to germ cells. Early stage male germ cells undergo self-renewal (Griswold, 2016; Nishimura and L’Hernault, 2017; Russell et al., 1990). A subset of those cells is induced to differentiate into spermatogonia. Both the primordial stage cells and spermatogonia divide mitotically. Subsequently, spermatogonia enter meiosis to become primary, then secondary spermatocytes. The secondary spermatocytes finish meiosis, differentiate further as haploid spermatids, and eventually become spermatozoa.
There are two major developmental goals for sperm production: restructuring of gene expression to enable post-natal germ cell differentiation, and compaction of the haploid genome into the sperm head. To accomplish the former, male germ cells express many specialized transcriptional, translational, and RNA-processing factors (Chalmel and Rolland, 2015; Eddy, 2002; Iguchi et al., 2006; Lee et al., 2009; Licatalosi, 2016; MacDonald and Grozdanov, 2017; MacDonald and McMahon, 2010; Zagore et al., 2015; Zhu et al., 2016). Notably, male germ cells express mRNAs that differ in their 3′ ends due to alternative polyadenylation (Huber et al., 2005; Li et al., 2016; Liu et al., 2007; McMahon et al., 2006; Wallace et al., 1999). An important gene contributing to alternative polyadenylation in male germ cells is Cstf2t, which encodes the testis- and brain-expressed polyadenylation protein, τCstF-64. Cstf2t is a paralog of the ubiquitously expressed 64,000 Mr subunit of the cleavage stimulation factor, CstF-64 (gene symbol Cstf2). τCstF-64 is expressed most abundantly in male germ cells and brain (Huber et al., 2005; McMahon et al., 2006; Monarez et al., 2007; Wallace et al., 1999; Wallace et al., 2004), although it also functions in other cell types (Ruepp et al., 2011; Yao et al., 2013; Youngblood et al., 2014). Targeted deletion of Cstf2t in mice leads to male infertility due to defects in post-meiotic germ cell development: smaller numbers of primary spermatocytes, round, and elongating spermatids, such that the resulting epididymal contents contained few motile sperm cells and were unable to fertilize mouse eggs in vitro (Dass et al., 2007; Hockert et al., 2011; Tardif et al., 2010). In part, these changes were due to altered polyadenylation and splicing of specific regulatory genes important for spermiogenesis (Grozdanov et al., 2016; Li et al., 2012).
To accomplish genome compaction, germ cells exchange the somatic cohort of histone proteins, first with testis-specific histones, second with transition proteins, and finally with protamines (Bao and Bedford, 2016; Barral et al., 2017; Bošković and Torres-Padilla, 2013; Hoghoughi et al., 2017; Pradeepa and Rao, 2007; Sassone-Corsi, 2002; Wang et al., 2017). These changes in chromatin packaging require precise deployment of the testis-specific histones and histone-like proteins such as protamines (collectively, “histone-like genes”) to make sperm cells that are competent for fertilization (Churikov et al., 2004; Govin et al., 2004; Meistrich et al., 2003; Pradeepa and Rao, 2007). In previous studies, we noted that expression of intronless small genes (ISGs) was reduced in testes from Cstf2t−/− mice (Dass et al., 2007; Li et al., 2012). ISGs consist of small (the lowest 20% in length) protein-coding genes, and include the histone-like genes in male germ cells. Further, histone mRNA processing involves Cstf2 (Kolev and Steitz, 2005; Ruepp et al., 2011; Yang et al., 2013; Youngblood et al., 2014), suggesting that Cstf2t might also contribute to production of histone-like genes in testis.
In somatic cells, histones are regulated at the transcriptional and post-transcriptional levels. Specifically, replication-dependent histone mRNA transcription, translation, and stability are governed by formation of histone mRNA 3′ ends that is recognized by specialized machinery, distinct from the cleavage and polyadenylation machinery that acts on 3′ ends of most other mRNAs (Dominski and Marzluff, 2007; Marzluff et al., 2008). Nonetheless, there is overlap in the components of the machinery that forms 3′ ends of most mRNAs and histone mRNAs. That overlap includes subunits of the cleavage and polyadenylation specificity factor (CPSF, (Dominski et al., 2005; Kolev and Steitz, 2005; Sabath et al., 2013; Sullivan et al., 2009; Yang et al., 2013)) and of the cleavage stimulation factor (CstF, (Romeo et al., 2014; Sabath et al., 2013; Yang et al., 2013)). Such components regulate, for example, 3′ end formation of histone mRNAs necessary for entry into S phase of the cell cycle (Youngblood et al., 2014). While much is known about histone gene regulation in somatic cells, less is known about the regulation of testis-specific histones and histone-like proteins (Bao and Bedford, 2016; Gou et al., 2017; Kutchy et al., 2017; Rathke et al., 2014).
Here we show that loss of Cstf2t results in altered histone-like gene expression contributing to the infertile phenotype of male Cstf2t−/− mice. We observed reduced mRNAs encoding members of every histone gene group, particularly the testis-specific histones and protamines in Cstf2t−/− testes. Concurrently, we observed delayed accumulation of testis-specific histone proteins, including H1fnt (formerly H1t2 or Hanp1 (Martianov et al., 2005; Tanaka et al., 2005)), and Hils1 (Mishra et al., 2015; Yan et al., 2003). For some testis-specific histones, we cannot account for the changes by a direct effect of Cstf2t on altered cleavage and polyadenylation, and must be due to indirect effects. These results point to an important role for Cstf2t in control of gene expression during spermiogenesis.
METHODS AND MATERIALS
Animal Use and Generation of Cstf2ttm1Ccma Mice
All animal treatments and tissues obtained in the study were performed according to protocols approved by the Institutional Animal Care and Use Committee at the Texas Tech University Health Sciences Center in accordance with the National Institutes of Health animal welfare guidelines. TTUHSC’s vivarium is AAALAC-certified and has a 14/10-hour light/dark cycle with temperature and relative humidity of 20–22°C and 40–70%, respectively.
Deletion of the entire Cstf2t coding region from chromosome 19, breeding, and genotyping were described previously (Dass et al., 2007). Briefly, the knockout targeting vector was created using the Cstf2t coding region from chromosome 19 with pGK-Neo, electroporated into 129SvEv ES cells, and G418-resistant colonies in which Neo had replaced Cstf2t were identified by PCR. These cells were microinjected into C57BL/6 embryos and reimplanted into pseudopregnant females. Germ-line transmission was confirmed by PCR analysis of F1 animals. Cstf2ttm1Ccma mice used in these studies were therefore of mixed C57BL/6-129SvEv background. Mutants were maintained as a congenic strain by repeated backcrossing (every 4–5 generations) to C57BL/6NCrl (Charles River) and otherwise breeding exclusively within the colony. At the time of this study, mice were bred to approximately 50 generations, with at least ten backcrosses to C57BL/6NCrl.
Genotyping of Cstf2ttm1Ccma Mice by PCR
Genomic DNA was extracted from tail snips of Cstf2ttm1Ccma mice by proteinase K digestion followed by isopropanol precipitation. PCRs were performed using wild type- and Cstf2ttm1Ccma-specific primers to determine the presence of the transgene (Dass et al., 2007). Where indicated, absence of τCstF-64 protein in Cstf2ttm1Ccma/tm1Ccma (herein, Cstf2t−/−) mice was confirmed by Western blot using the τCstF-64-specific antibody, 6A9 (Dass et al., 2007; Wallace et al., 1999).
Isolation of seminiferous tubules
Seminiferous tubules were isolated as follows: a small incision was made in the tunica albuginea of the testis and the contents (mainly seminiferous tubules) were gently collected in ~5 ml of ice-cold Dulbecco’s Phosphate-Buffered Saline (DPBS, Life Technologies) supplemented with phenylmethanesulfonyl fluoride (PMSF). The contents were vigorously shaken, breaking apart the tissue. Tissue parts were allowed to settle at for 5 min on ice, and the supernatant (containing mainly Leydig cells) was removed. The procedure was repeated two more times for a total of three times. After the final settlement, the tissue was spun briefly in a micro-centrifuge at 500×g. The obtained seminiferous tubules were used to isolate either total RNA or protein.
Protein isolation and immunoblots
Seminiferous tubules from one animal were lysed in either 250 μl extraction buffer [DPBS, 0.5 % Triton X-100 (v/v), 2 mM PMSF, 0.02% NaN3] or RIPA buffer [50 mM Tris-HCl pH:8.8, 150 mM NaCl, 0.1% sodium dodecyl sulphate (SDS), 0.5% deoxycholate, 0.5% NP-40], briefly sonicated, and incubated on ice for 10 min. Lysates were spun down for 10 min at 400×g at 4°C for 10 min. Equal amounts of protein from wild type and Cstf2t−/− animals were loaded on pre-cast NuPAGE Novex 4–12% Bis-Tris Gels (Life Technologies), followed by a semi-dry transfer to polyvinylidene fluoride membranes. Membranes were incubated with primary antibody, followed by the appropriate secondary antibody conjugated to horseradish peroxidase. Antibody-antigen interaction was revealed using SuperSignal West Pico Chemiluminescence kit (Thermo Scientific).
The H1fnt antibody (a gift from Igor Martianov) was used as described for Western blots (Martianov et al., 2005). For immunofluorescence, we used an antibody against H1fnt from Santa Cruz Biotechnology (L-14, sc-136700). The antibody against Hils1 was used as described (Yan et al., 2003). Goat anti-protamine 2 antibody was purchased from Santa Cruz Biotechnology (C-14, sc-23104) and used according manufacture’s recommendations. The antibodies against H2B (#8135) and H4 histones (#2592) were obtained from Cell Signaling Technology and used according to the recommendations. Mouse anti-CstF-64 (3A7), mouse anti-CstF-64 (6A9) and E7 anti-β-tubulin monoclonal antibody were used as described (Dass et al., 2007).
Immunofluorescence
Testes for wild type and Cstf2t−/− mice were collected from age-matched animals and incubated in 10% formalin solution overnight. Tissues were paraffin embedded and five μm sections were obtained. Tissue sections were then deparaffinized, and the antigen was retrieved using heat and citrate buffer. Immunostaining was performed as previously described (Grozdanov et al., 2006). Briefly, tissue sections were blocked in 5% donkey serum, 1% BSA, 0.1M Tris-HCl pH 7.4, 015 M NaCl, 0.1% Tween 20 for one hour at room temperature. Primary anti-H1fnt antibody (Santa Cruz Biotechnology) was pre-incubated in 2% donkey serum, 1% BSA, 0.1M Tris-HCl pH 7.4, 015 M NaCl, 0.1% Tween 20 for 30 min on ice and applied on the tissue for one hour at room temperature. Tissue sections were washed three times in 0.1M Tris-HCl pH 7.4, 015 M NaCl for 10 min each and secondary anti-rabbit Cy3-conjugated antibody was applied in 2% mouse serum, 1% BSA, 0.1M Tris-HCl pH 7.4, 015 M NaCl, 0.1% Tween 20 for one hour at room temperature. Sections were washed again in 0.1M Tris-HCl pH 7.4, 015 M NaCl for 10 min each and mounted in ProLong Diamond Antifade Mountant with DAPI (ThermoFischer Scientific). Microscopy was performed on inverted Nikon Ti microscope using a confocal A1 module. Images were adjusted for clarity in ImageJ software.
RNA isolation, RNA-seq, and A-seq
RNA-seq (Grozdanov et al., 2016; Youngblood et al., 2014) and A-seq were carried out essentially as described previously (Grozdanov et al., 2016; Martin et al., 2012; Youngblood et al., 2014). The resulting cDNA libraries were sequenced on Illumina platform with 50-nucleotide coverage (SE50). The obtained reads were strand-specific and coincided with the sense strand of the mRNAs.
HITS-CLIP library preparation
Cross-linking and immunoprecipitation combined with high-throughput sequencing (HITS-CLIP) was performed exactly as previously described (Grozdanov and Macdonald, 2014) using τCstF-64-specific 6A9 monoclonal antibody.
UCSC genome browser tracks for RNA-Seq data
To visualize the mapping of RNA-Seq data from testes for wild-type and Cstf2t−/− mice, the UCSC genome browser tracks were generated as follows. The raw RNA-Seq reads were aligned to the mouse (mm9) genome using STAR (version 2.3.1t) (Dobin et al., 2013) with default settings. Only uniquely mapped reads were retained to compute the coverage of the genes in the genome. The reads coverage was calculated using the tool genomeCoverageBed in BEDTools (Quinlan and Hall, 2010). The generated wiggle files were transformed to BigWig files using the wigToBigWig utility (Kent et al., 2010). The BigWig files of reads coverage for wild-type and Cstf2t−/− mice in mouse genome were visualized in UCSC Genome Browser (Kent et al., 2002).
UCSC genome browser tracks for HITS-CLIP-Seq data
To visualize the mapping of HITS-CLIP-Seq data from testes for wild-type and Cstf2t−/− mice, the UCSC genome browser tracks were generated as follows. The raw HITS-CLIP-Seq reads were aligned to the mouse (mm9) genome using GSNAP (Wu and Nacu, 2010) with default settings. Only uniquely mapped reads were retained to compute the coverage of the genes in the genome. The reads coverage was calculated using the tool genomeCoverageBed in BEDTools (Quinlan and Hall, 2010). The generated wiggle files were transformed to BigWig files using the wigToBigWig utility (Kent et al., 2010). The BigWig files of reads coverage for wild-type and Cstf2t−/− mice in mouse genome were visualized in UCSC Genome Browser (Kent et al., 2002).
Differential histone gene expression and A-seq data processing
RNA-seq reads obtained from each biological replicate were independently aligned on the mouse reference genome (Mouse Genome v37.2, MGSCv37, mm9) using the SeqMan NGen v.10 software (DNASTAR Inc.). The assembly files generated by SeqMan NGen were used to identify the differentially expressed histone genes using the QSeq part of ArrayStar package (DNASTAR Inc) as previously described (Youngblood et al., 2014).
A-seq data were aligned on the mouse reference genome (Mouse Genome v37.2, MGSCv37, mm9) using the SeqMan NGen v.10 software (DNASTAR Inc.). The A-seq reads aligning with the 3′ ends of H1fnt and Hils1 were manually counted and the bar graph was built using the Microsoft Excel software.
Reverse transcription and TaqMan PCR
Complementary DNA for TaqMan quantitative RT-PCR (qRT-PCR) was prepared from about 80 ng of total RNA using SuperScript VILO MasterMix (ThermoFisher Scientific) to verify gene expression levels. A pre-dispensed panel of 16 TaqMan probes and primers (see Fig. 2) was used to determine the expression of the genes in testis tissues at the indicated genotypes and ages. qRT-PCR was performed using TaqMan Fast Advanced Master Mix (ThermoFisher Scientific) on QuantStudio™ 12K Flex Real-Time PCR System. The relative expression in the total RNA was calculated using the comparative Ct method using 18S rRNA as a reference for the gene expression. Gene expression of RNA isolated from testis obtained from wild type animals at age of 25 dpp was normalized to one (Livak and Schmittgen, 2001). The average of three biological replicates was used at each age (25, 28, 31, 35, and 77 dpp) and wild type and Cstf2t−/− genotype to generate a heat-map of the resulting values using the R software package. The polyadenylation signature of the selected replication-dependent histone genes and the testis specific genes were determined by the following: Polyadenylated and non-polyadenylated RNA was isolated using Dynabeads mRNA DIRECT Purification Kit and 80 ng of the corresponding RNA was used to generate cDNA using SuperScript VILO MasterMix. Ct values of different ages and both genotypes for a particular gene were combined to obtain a box and whiskers plot. Smaller Ct values represent more abundant mRNA and a value close to 40 indicates mRNA that is expressed at very low levels.
Figure 2.
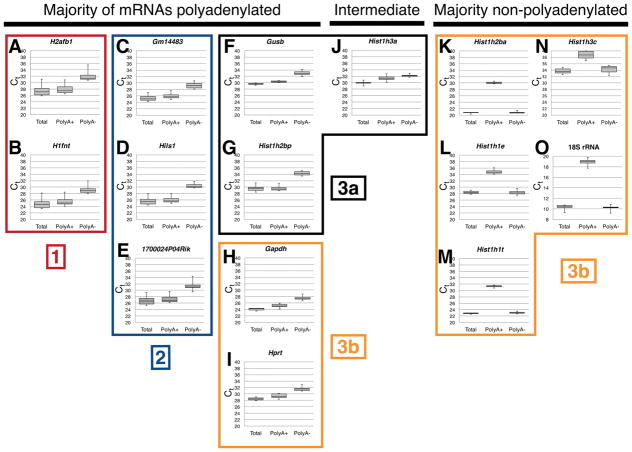
Testis-expressed histone-like gene mRNAs are generally polyadenylated. Complementary DNA was prepared from seminiferous tubules of wild type C57BL/6NCrl mice from either total RNA, poly(A)+, or poly(A)− RNA as indicated. Shown are threshold cycle (Ct) values for the indicated genes H2afb1 (A), H1fnt (B), Gm14483 (C), Hils1 (D), 1700024P04Rik (E), Gusb (F), Hist1h2bp (G), Gapdh (H), Hprt (I), Hist1h3a (J), Hist1h2ba (K), Hist1h1e (L), Hist1h1t (M), Hist1h3c (N), and 18S ribosomal RNA (O). Note that larger Ct values in qRT-PCR correspond with lower levels of cDNA. Boxed groups are numbered ([1], [2], [3a], [3b]) to correspond with the clustered groupings in Figure 1B.
RESULTS
Testis-specific histones are down-regulated in Cstf2t−/− mouse testes
Previously, we performed high-throughput sequencing of cDNAs (RNA-seq) and noted a preponderance of intronless small genes (ISGs) that were down-regulated in seminiferous tubules from 25-day postpartum (dpp) Cstf2t−/− mice (Li et al., 2012). Prominent among those ISGs were histone and histone-like genes. The composition of testicular cell types does not differ between wild type and Cstf2t−/− mice at 25-dpp (Dass et al., 2007; Hockert et al., 2011), so the RNA-seq differences reflected actual mRNA abundances in germ cells at these stages. Because we have also noted roles for CstF-64 in histone mRNA processing (Youngblood et al., 2014), we decided to examine the effects of τCstF-64 on histone and histone-like genes in male germ cells.
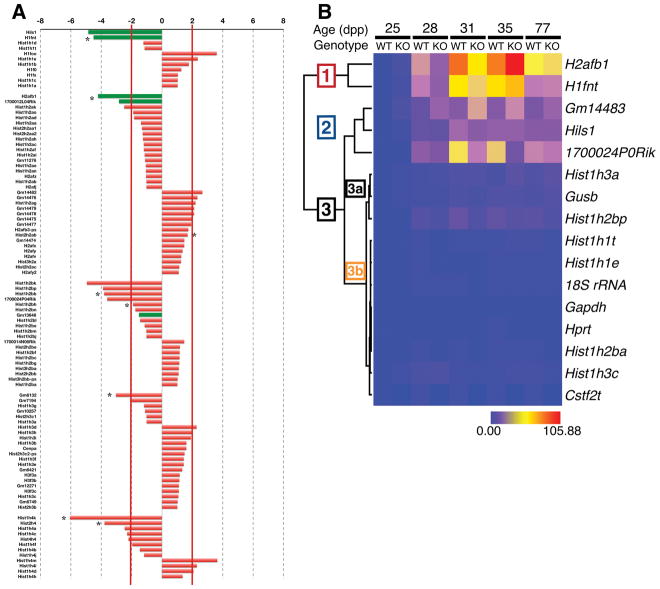
In further analyses of RNA-seq from seminiferous tubules of 25 dpp wild type or Cstf2t−/− mice (Grozdanov et al., 2016), we examined histone and histone-like gene mRNAs. We saw that almost half of these genes (47/99) were down-regulated in Cstf2t−/− tubules, representing all major histone gene families (Fig. 1A). Messenger RNAs for testis-specific histone-like genes (H1fnt, Hils1, H2afb1, 1700012L04Rik, Gm13646) were down-regulated, suggesting reproductive specificity of the effect even at 25 dpp, when the testis-specific histone-like proteins are not yet expressed detectably (see below).
Figure 1.
The majority of histone and testis-specific histone mRNAs are down-regulated in seminiferous tubules of Cstf2t−/− mice. (A) RNA-seq analyses of histone and testis-specific histone genes in seminiferous tubules isolated from wild type and Cstf2t−/− mice. The bars representing testis-specific histones are indicated in green. Asterisks indicate genes whose expression is significantly different between the groups (p < 0.01). (B) Heat map showing clustered differences in mRNA expression of histone-like genes in wild type and Cstf2t−/− mouse seminiferous tubules. Clustered expression is indicated as groups [1], [2], or [3], with subgroups [3a] and [3b].
To verify these results, we performed real-time RT-PCR quantification (qRT-PCR) of the total mRNAs for eleven testis specific histone-like genes, Cstf2t, and several control genes (Gapdh, Gusb, Hprt, and 18S rRNA) at 25, 28, 31, 35 and 77 dpp on wild type and Cstf2t−/− tubules. Analysis of these results indicated that expression of the majority of testis-specific histone-like genes clustered into two groups: H2afb1 with H1fnt (Fig. 1B, Cluster 1), and Gm14483, Hils1, and 1700024P0Rik (Cluster 2). The testis-specific Hist1h1t clustered with the remaining non-testis-specific histone genes and the reference genes (Gapdh, Hprt, Gusb, and 18s rRNA, Cluster 3). These results suggest that the testis-specific histone like genes have similar physiological roles during spermatogenesis.
Most testis-specific histone mRNAs are polyadenylated in seminiferous tubules
Replication-dependent histones in somatic cells and pre-meiotic germ cells are generally not polyadenylated, but instead have a unique 3′ end stem loop that governs their synthesis and metabolism during the cell cycle (Marzluff et al., 2008). However, some of those mRNAs receive poly(A) tails by the cleavage and polyadenylation mechanism (Kari et al., 2013; Shepard et al., 2011; Youngblood et al., 2014). Replication-independent and most testis-expressed histone-like genes are also not polyadenylated (Sun and Qi, 2014). We examined the polyadenylation states of histone and histone-like gene mRNAs in wild type and Cstf2t−/− seminiferous tubules using TaqMan quantitative RT-PCR, comparing total, polyadenylated, and non-polyadenylated transcripts (Fig. 2). Examination of threshold cycle (Ct) values for 18S ribosomal RNA (rRNA) revealed that the majority of 18S rRNA was non-polyadenylated in testes from mice of all ages and genotypes tested (poly(A)− Ct = 10.26, Fig. 2O). This result is consistent with the lack of polyadenylation of rRNAs. In contrast, the majority of Gapdh and Hprt1 mRNAs were polyadenylated (poly(A)+ Ct = 25.17 and 29.35, respectively, Fig. 2H, I). The replication-dependent histone mRNAs (Hist1h2ba, Hist1h1e, Hist1h3c, and Hist1h1t) were most abundant in the poly(A)− samples, showing the poly(A)− signature (Fig. 2K, L, N). However, testis-expressed histone mRNAs (H1fnt, Hils1, H2afb, Gm14483, and 1700024P0Rik) were most abundant in the poly(A)+ samples (Fig. 2A–E, G, J), indicating that they were polyadenylated. As an outlier, the testis-expressed histone Hist1h1t, is not polyadenylated (Fig. 2M). However, this gene, though testis-expressed (Nayernia et al., 2003), is part of the histone H1 cluster and is expressed in other cell types, as well (Sun and Qi, 2014). Overall, these observations support the conclusions that this family of testis-specific histone-like genes are polyadenylated in seminiferous tubules, and thus susceptible to regulation by τCstF-64. They also suggest that testis-specific histone-like genes have a function uncoupled from the cell cycle and in this way, differ from the cell-cycle dependent histone genes.
Accumulation of the testis-specific histone H1fnt is delayed in Cstf2t−/− mice
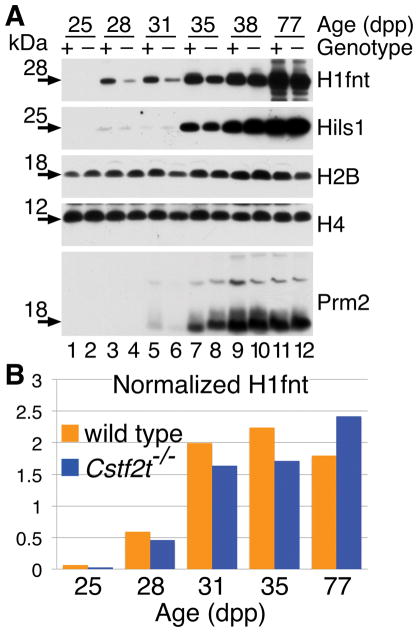
Antibodies are often unable to distinguish individual histone protein variants, so we could not recognize all histones or histone-like protein variants in these experiments. However, where we could, we examined expression of ubiquitous and testis-specific histones at 25, 28, 31, 35, 38, and 77 dpp (Fig. 3). Ubiquitously expressed histones H2B and H4 do not vary greatly between wild type and Cstf2t−/− tubules at any age, consistent with the results obtained by TaqMan qRT-PCR. However, we saw a reduction in the expression of the H1fnt protein in Cstf2t−/− tubules compared to wild type at 28–35 dpp (Fig. 3B). At 77 dpp, we saw an increase of H1fnt protein in Cstf2t−/− tubules, which might reflect the increased representation of earlier cell types expressing H1fnt in older mice (Hockert et al., 2011). We saw a similar, but less pronounced reduction in Hils1 protein at the same times. This developmental delay in testis-specific histone protein expression echoes the reduced mRNA amounts for H1fnt, suggesting that τCstF-64 is important for its correct mRNA production of the gene. The same might be true for Hils1, though reduced Hils1 protein amounts are not as reflective of the greatly reduced mRNA (Fig. 1A). This supports the conclusion that the changes in H1fnt and Hils1 protein levels are associated with τCstF-64 in germ cells.
Figure 3.
Accumulation of the testis-specific histone H1fnt is delayed in Cstf2t−/− mice. (A) Representative Western blot analysis of selected histone (H2B and H4), testis-specific histones (H1fnt and Hils1), and protamine 2 (Prm2) proteins in seminiferous tubules from wild type and Cstf2t knockout mice. The age of the animals is shown as days postpartum (dpp) and does not correspond to days of spermatogenesis. (B) Western blots as above were scanned and quantified for 25, 28, 35, and 77 dpp.
H1fnt does not localize differently in testes of Cstf2t−/− mice
To determine whether the decreased abundance of H1fnt was associated with altered localization within germ cells, we examined immunofluorescence using an anti-H1fnt antibody (see Materials and Methods) in 31 and 35 dpp (ages chosen because of the start of protein synthesis in testis of the data in Fig. 3) testis samples (Fig. 4). Despite the overall lower amounts of H1fnt protein in Cstf2t−/− testes at these developmental times, we did not observe an abnormal distribution of H1fnt in spermatogenic cells. This suggests that mechanisms driving H1fnt localization and DNA deposition are not altered in Cstf2t−/− mice, only its abundance.
Figure 4.
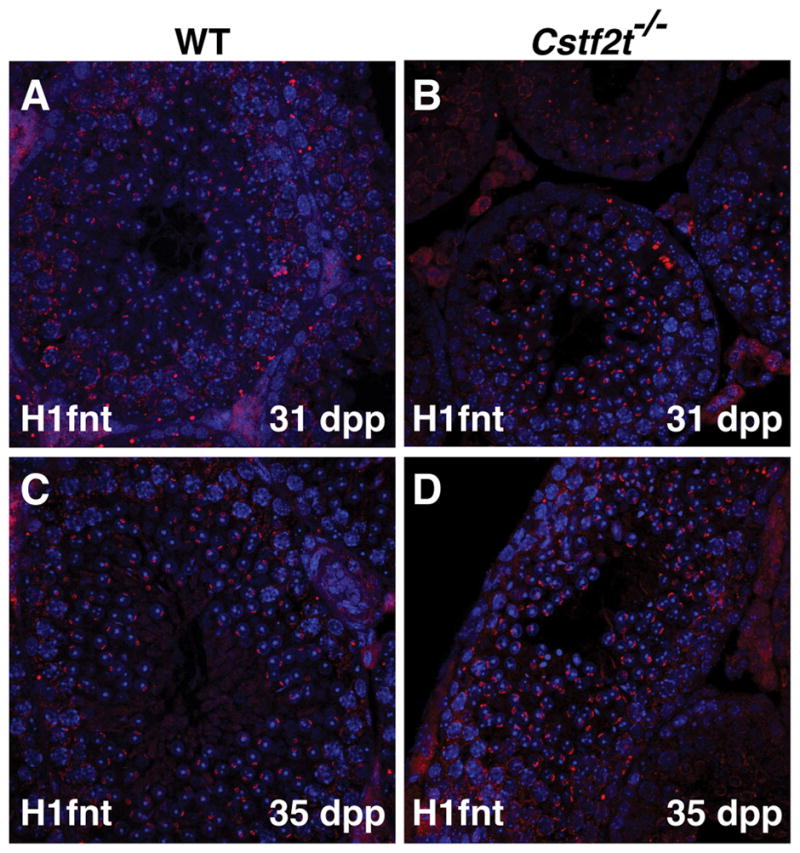
Localization of H1fnt does not vary within wild type and Cstf2t−/− mouse male germ cells. Immunofluorescence using the anti-H1fnt antibody of testis sections from wild type (A and C) and Cstf2t−/− (B and D) mice at 31 (A and B) and 35 (C and D) dpp.
Cleavage and polyadenylation sites in H1fnt and Hils1 genes are not affected in Cstf2t−/− mice, only the abundance of the mRNAs
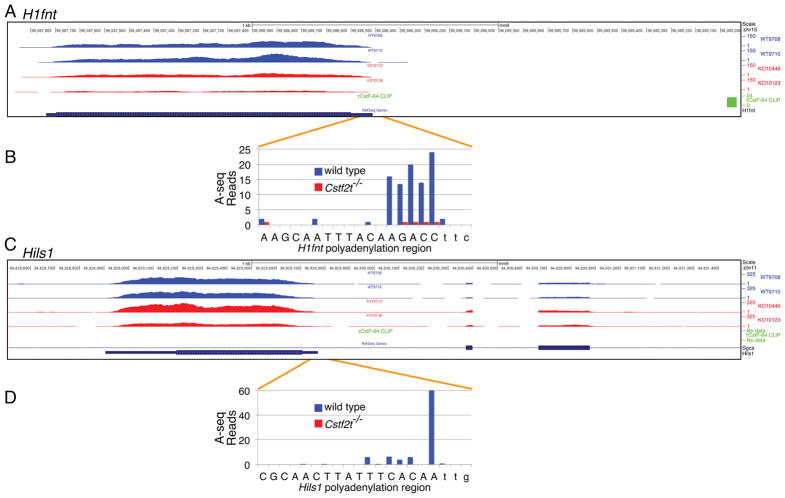
τCstF-64 regulates polyadenylation site choice (Grozdanov et al., 2016; Hwang et al., 2016; Kargapolova et al., 2017; Liu et al., 2007; McMahon et al., 2006), so we wanted to determine the extent to which loss of Cstf2t affected polyadenylation site choice in these genes. We examined sites of cleavage/polyadenylation in H1fnt and Hils1 using high-throughput sequencing (A-seq, (Grozdanov et al., 2016; Martin et al., 2012)). In examining closely spaced sites of poly(A) addition in H1fnt, we did not note changes in localization of any of the five major sites in tubules from Cstf2t−/− mice (Fig. 5A, B), only reduced expression of the mRNA. Hils1 mRNA showed a similar lack of change in the use of cleavage/polyadenylation sites (Fig. 5C, D) and reduction in number of reads in the 25-dpp Cstf2t−/− mice. We cannot differentiate whether the reductions reflect either direct effects of τCstF-64 on polyadenylation site choice for these genes or the overall reduction in RNA-seq reads at the loci.
Figure 5.
RNA-seq, polyadenylation sites and τCstF-64-binding sites in the mouse H1fnt and Hils1 genes. Shown are high-throughput RNA-seq tracks for two wild type and two Cstf2t−/− mouse seminiferous tubules for (A) H1fnt and (C) Hils1. HITS-CLIP sites were determined using an antibody against τCstF-64 to perform immunoprecipitation of the protein-RNA complex in seminiferous tubules from wild type mice (τCstF-64 CLIP). Cleavage and polyadenylation sites were determined using A-seq for (B) H1fnt and (D) Hils1. Capital letters represent the annotated 3′ ends of each gene; lower case letters represent downstream sequences.
τCstF-64 binds downstream of the mouse H1fnt gene, but is not likely to be directly involved in its polyadenylation
Although we did not observe alteration in the use of the cleavage and polyadenylation sites in either H1fnt or Hils1, we tested whether there might be direct effects of τCstF-64 on expression of either gene. Such effects might be direct (co-transcriptional binding of τCstF-64 to the H1fnt mRNA) or indirect (a consequence of effects of τCstF-64 on other genes that impact H1fnt expression). We performed high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP (Licatalosi et al., 2008)) using an antibody to τCstF-64 (Grozdanov et al., 2016; Grozdanov and Macdonald, 2014). Examination of HITS-CLIP for τCstF-64 in the region downstream of the H1fnt open reading frame showed one τCstF-64 CLIP site approximately ~1.5 kb downstream of the sites of poly(A) addition (Fig. 5A). Because this site is relatively distant from the body of the gene, we do not believe it is participating in H1fnt polyadenylation in a meaningful way. There were not any τCstF-64 CLIP sites associated with Hils1 mRNA within 2 kb downstream of the gene (Fig. 5C). Therefore, it is unlikely that τCstF-64 binding in either H1fnt or Hils1 has a direct effect on their polyadenylation. Reductions in both H1fnt and Hils1 mRNAs must therefore occur at another level, possibly τCstF-64-dependent global changes in gene expression (Li et al., 2012). Examination of the other testis-specific histone mRNAs did not reveal τCstF-64 CLIP sites within 1.5 kb of their 3′ ends, suggesting they follow the same patterns as H1fnt and Hils1.
DISCUSSION
One of the several mechanisms by which Cstf2t−/− male mice were known to be infertile was due to decreased expression of intronless small genes including the histone-like genes (Li et al., 2012). This is in contrast to other mechanisms by which alternatively spliced genes are preferentially expressed in male germ cells (Naro et al., 2017). Here we examined expression of these small intronless histone-like genes more closely and tested possible mechanisms for the role of τCstF-64 in their expression. We saw that one-half of the histone mRNAs were reduced in seminiferous tubules of Cstf2t−/− mice at early stages (25 dpp) before cell type differences were evident between the genotypes. We previously reported that Tnp1, Tnp2 and protamine mRNAs were reduced in Cstf2t−/− testes (Grozdanov et al., 2016). Similarly, all five testis-specific histone mRNAs surveyed were reduced (Fig. 1), suggesting roles for τCstF-64 in expression of this class of histone-like genes in male germ cells. The majority of testis-specific histone mRNAs are polyadenylated (Fig. 2), suggesting that polyadenylation misregulation could have a preferential effect on those. Corresponding to the reduced histone-like gene mRNAs in Cstf2t−/− mice, we found that expression of the H1fnt and Hils1 proteins was delayed in the Cstf2t−/− tubules (Fig. 3). We attempted to account for these changes in H1fnt protein levels, by noting reduced overall polyadenylation in the H1fnt gene. However, these changes in polyadenylation did not correlate strongly with sites of τCstF-64 binding in the nascent mRNA (Fig. 5A, B). Similarly, Hils1 did not show altered polyadenylation, only reduction (Fig. 5C). Thus, we cannot support a direct role for τCstF-64 in control of histone-like gene expression through promoting correct cleavage/polyadenylation site choice for these genes (McMahon et al., 2006).
H1fnt is a replication-independent variant of the linker histone H1 that is expressed in round and elongating spermatids in mice, where it is localized in the nucleus at the apical pole (Martianov et al., 2005), ultimately affecting sperm motility (Tanaka et al., 2005). Thus, H1fnt contributes to correct chromosomal condensation and maintenance of intact DNA in step 12 and 13 spermatids. H1fnt is also necessary for nuclear localization of other chromatin components (Catena et al., 2009; Catena et al., 2006; Lambrot et al., 2012). In Cstf2t−/− mice, we observed defects in chromosomal condensation, though such errors initiated earlier in secondary spermatocytes (Dass et al., 2007). This suggests that delayed expression of H1fnt probably contributes to the infertility in the Cstf2t−/− male mice, but that other errors must contribute as well. Notably, H1fnt contributes to protamine deposition (Tanaka et al., 2005), errors in expression of which we previously observed in Cstf2t−/− mice (Grozdanov et al., 2016).
We saw smaller magnitude changes in expression of Hils1 protein in tubules of Cstf2t−/− mice (Fig. 3). Hils1 is a testis-expressed histone H1 variant (Yan et al., 2003) that is heavily modified post-translationally (Mishra et al., 2015). Hils1 expression begins later in spermiogenesis than H1fnt, though both are expressed in elongating spermatids (Yan et al., 2003). Interestingly, Hils1 may act as both a linker and a DNA-compaction histone, contributing to the high condensation of DNA in sperm heads (Yan et al., 2003), suggesting that small changes in Hils1 expression might lead to cascading effects on male germ cell development in Cstf2t−/− mice.
Reduction of CSTF2 (Ruepp et al., 2011; Takagaki and Manley, 1998; Yao et al., 2013; Youngblood et al., 2014) or CSTF2T (Hwang et al., 2016; Kargapolova et al., 2017) in human, mouse, or avian cell lines did not overtly impair cell viability, although expression of individual mRNAs varied greatly. Similarly, Cstf2t−/− mice are viable and mostly normal in the absence of τCstF-64, despite infertility in males (Dass et al., 2007; Hockert et al., 2011; Tardif et al., 2010) and enhanced spatial memory in females (Harris et al., 2016). This suggests that τCstF-64 function is important in specific tissues, but not essential for general cellular processes. Similarly, mouse embryonic stem cells lacking Cstf2 were able to differentiate into neuronal but not endodermal cell lineages (Youngblood and MacDonald, 2014); in these cells, τCstF-64 subsumed many but not all functions of CstF-64 (Shankarling et al., 2009). The distribution of functions between CstF-64 and τCstF-64 seems reasonable, because τCstF-64 is present only in eutherian mammals (Dass et al., 2002; Dass et al., 2001), and thus would be expected to take on ancillary but not ancient functions.
CstF-64 is also involved in replication-dependent histone mRNA 3′ end processing (Kolev and Steitz, 2005). We noted in a previous study that CstF-64 supports stem cell pluripotency by licensing entry into cellular S-phase (Youngblood et al., 2014). In Cstf2-null embryonic stem cells, loss of CstF-64 is partially but not completely compensated by increased expression of τCstF-64. In particular, τCstF-64 participates in the histone mRNA 3′ end processing complex in the absence of CstF-64. This suggests a model in germ cells whereby τCstF-64 participates in 3′ end processing of both somatic histone genes and testis-specific ones in meiotic and postmeiotic germ cells where CstF-64 is absent. It also suggests that the cohort of testis-specific histone mRNAs are processed differently than in somatic cells as an adaptation to the absence of CstF-64.
In support of this hypothesis, Kargapolova et al. (Kargapolova et al., 2017) published a study in which they noted binding of τCstF-64 (via a modified CLIP protocol) to both replication-dependent and replication-independent histone mRNA precursors in a human neuroblastoma cell line. In agreement with our data on H1fnt (Fig. 5A), those authors noted clustering of τCstF-64 binding at locations 3′ of the polyadenylation sites in replication-independent histone genes, of which H1fnt is a member. Neither Kargapolova et al. nor we saw τCstF-64 clustering in Hils1 (Fig. 5C). Interestingly, Kargapolova et al. observed τCstF-64 binding within the 5′ ends of replication-dependent histone genes. Previously, we noted decreased participation of τCstF-64 in the histone 3′ end processing complex when CstF-64 was abundant; τCstF-64 was only recruited to that complex when CstF-64 was absent (Youngblood et al., 2014). Thus, we propose that CstF-64 probably plays a primary role in 3′ end processing of replication-dependent histone genes, acting as an important component of the histone mRNA 3′ processing complex (Kolev and Steitz, 2005; Ruepp et al., 2011; Yang et al., 2013; Youngblood et al., 2014). In contrast, τCstF-64 plays only a secondary or passive role in processing of replication-dependent histone genes. Instead, it plays a more important role in 3′ end cleavage and polyadenylation in the replication-independent histone genes. This is most important in spermiogenesis, where replication-independent histones and histone-like genes are essential to the correct developmental program.
Loss of Cstf2t in mice results in infertility for many reasons including large-scale changes in gene expression, genomic misregulation, altered splicing, and altered polyadenylation (Dass et al., 2007; Grozdanov et al., 2016; Hockert et al., 2011; Li et al., 2012; Tardif et al., 2010). Alterations in expression of histone-like genes in Cstf2t−/− mouse germ cells could account for altered chromosomal structures (Dass et al., 2007) and derepression of intergenic chromosomal expression (Li et al., 2012). Future work will examine consequences of altered histone-like proteins in structural remodeling of chromatin in wild-type and Cstf2t−/− mouse testes.
Acknowledgments
The authors want to thank Igor Martianov for the gift of the Hils1 antibody. Real-time PCR data were generated in the Molecular Biology Core Facility supported in part by TTUHSC. Research reported in this publication was supported in part by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under award number R01HD037109. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional support was from the Texas Tech University Health Sciences Center’s School of Medicine, and the Department of Cell Biology & Biochemistry.
References
- Bao J, Bedford MT. Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction. 2016;151:R55–70. doi: 10.1530/REP-15-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral S, Morozumi Y, Tanaka H, Montellier E, Govin J, de Dieuleveult M, Charbonnier G, Coute Y, Puthier D, Buchou T, et al. Histone Variant H2A.L.2 Guides Transition Protein-Dependent Protamine Assembly in Male Germ Cells. Mol Cell. 2017;66:89–101. e108. doi: 10.1016/j.molcel.2017.02.025. [DOI] [PubMed] [Google Scholar]
- Bošković A, Torres-Padilla ME. How mammals pack their sperm: a variant matter. Genes Dev. 2013;27:1635–1639. doi: 10.1101/gad.226167.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catena R, Escoffier E, Caron C, Khochbin S, Martianov I, Davidson I. HMGB4, a novel member of the HMGB family, is preferentially expressed in the mouse testis and localizes to the basal pole of elongating spermatids. Biol Reprod. 2009;80:358–366. doi: 10.1095/biolreprod.108.070243. [DOI] [PubMed] [Google Scholar]
- Catena R, Ronfani L, Sassone-Corsi P, Davidson I. Changes in intranuclear chromatin architecture induce bipolar nuclear localization of histone variant H1T2 in male haploid spermatids. Developmental biology. 2006;296:231–238. doi: 10.1016/j.ydbio.2006.04.458. [DOI] [PubMed] [Google Scholar]
- Chalmel F, Rolland AD. Linking transcriptomics and proteomics in spermatogenesis. Reproduction. 2015;150:R149–157. doi: 10.1530/REP-15-0073. [DOI] [PubMed] [Google Scholar]
- Churikov D, Zalenskaya IA, Zalensky AO. Male germline-specific histones in mouse and man. Cytogenet Genome Res. 2004;105:203–214. doi: 10.1159/000078190. [DOI] [PubMed] [Google Scholar]
- Dass B, McDaniel L, Schultz RA, Attaya E, MacDonald CC. The gene CSTF2T encoding the human variant CstF-64 polyadenylation protein τCstF-64 is intronless and may be associated with male sterility. Genomics. 2002;80:509–514. [PubMed] [Google Scholar]
- Dass B, McMahon KW, Jenkins NA, Gilbert DJ, Copeland NG, MacDonald CC. The gene for a variant form of the polyadenylation protein CstF-64 is on chromosome 19 and is expressed in pachytene spermatocytes in mice. Journal of Biological Chemistry. 2001;276:8044–8050. doi: 10.1074/jbc.M009091200. [DOI] [PubMed] [Google Scholar]
- Dass B, Tardif S, Park JY, Tian B, Weitlauf HM, Hess RA, Carnes K, Griswold MD, Small CL, MacDonald CC. Loss of polyadenylation protein τCstF-64 causes spermatogenic defects and male infertility. Proceedings of the National Academy of Science, USA. 2007;104:20374–20379. doi: 10.1073/pnas.0707589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Marzluff WF. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Eddy EM. Male germ cell gene expression. Recent Progress in Hormone Research. 2002;57:103–128. doi: 10.1210/rp.57.1.103. [DOI] [PubMed] [Google Scholar]
- Gou LT, Kang JY, Dai P, Wang X, Li F, Zhao S, Zhang M, Hua MM, Lu Y, Zhu Y, et al. Ubiquitination-Deficient Mutations in Human Piwi Cause Male Infertility by Impairing Histone-to-Protamine Exchange during Spermiogenesis. Cell. 2017;169:1090–1104. e1013. doi: 10.1016/j.cell.2017.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur J Biochem. 2004;271:3459–3469. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- Griswold MD. Spermatogenesis: The Commitment to Meiosis. Physiol Rev. 2016;96:1–17. doi: 10.1152/physrev.00013.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov PN, Amatullah A, Graber JH, MacDonald CC. TauCstF-64 Mediates Correct mRNA Polyadenylation and Splicing of Activator and Repressor Isoforms of the Cyclic AMP-Responsive Element Modulator (CREM) in Mouse Testis. Biol Reprod. 2016;94:34. doi: 10.1095/biolreprod.115.134684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov PN, Macdonald CC. High-throughput sequencing of RNA isolated by cross-linking and immunoprecipitation (HITS-CLIP) to determine sites of binding of CstF-64 on nascent RNAs. Methods in molecular biology. 2014;1125:187–208. doi: 10.1007/978-1-62703-971-0_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov PN, Yovchev MI, Dabeva MD. The oncofetal protein glypican-3 is a novel marker of hepatic progenitor/oval cells. Lab Invest. 2006;86:1272–1284. doi: 10.1038/labinvest.3700479. [DOI] [PubMed] [Google Scholar]
- Harris JC, Martinez JM, Grozdanov PN, Bergeson SE, Grammas P, MacDonald CC. The Cstf2t Polyadenylation Gene Plays a Sex-Specific Role in Learning Behaviors in Mice. PLoS One. 2016;11:e0165976. doi: 10.1371/journal.pone.0165976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockert KJ, Martincic K, Mendis-Handagama SMLC, Borghesi LA, Milcarek C, Dass B, MacDonald CC. Spermatogenetic but not immunological defects in mice lacking the τCstF-64 polyadenylation protein. Journal of Reproductive Immunology. 2011;89:26–37. doi: 10.1016/j.jri.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoghoughi N, Barral S, Vargas A, Rousseaux S, Khochbin S. Histone variants: essential actors in the male genome programing. J Biochem. 2017 doi: 10.1093/jb/mvx079. [DOI] [PubMed] [Google Scholar]
- Huber Z, Monarez RR, Dass B, MacDonald CC. The mRNA encoding τCstF-64 is expressed ubiquitously in mouse tissues. Annals of the New York Academy of Sciences. 2005;1061:163–172. doi: 10.1196/annals.1336.017. [DOI] [PubMed] [Google Scholar]
- Hwang HW, Park CY, Goodarzi H, Fak JJ, Mele A, Moore MJ, Saito Y, Darnell RB. PAPERCLIP Identifies MicroRNA Targets and a Role of CstF64/64tau in Promoting Non-canonical poly(A) Site Usage. Cell Rep. 2016;15:423–435. doi: 10.1016/j.celrep.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi N, Tobias JW, Hecht NB. Expression profiling reveals meiotic male germ cell mRNAs that are translationally up- and down-regulated. Proceedings of the National Academy of Science, USA. 2006;103:7712–7717. doi: 10.1073/pnas.0510999103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargapolova Y, Levin M, Lackner K, Danckwardt S. sCLIP-an integrated platform to study RNA-protein interactomes in biomedical research: identification of CSTF2tau in alternative processing of small nuclear RNAs. Nucleic Acids Res. 2017;45:6074–6086. doi: 10.1093/nar/gkx152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kari V, Karpiuk O, Tieg B, Kriegs M, Dikomey E, Krebber H, Begus-Nahrmann Y, Johnsen SA. A Subset of Histone H2B Genes Produces Polyadenylated mRNAs under a Variety of Cellular Conditions. PLoS One. 2013;8:e63745. doi: 10.1371/journal.pone.0063745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutchy NA, Velho A, Menezes ESB, Jacobsen M, Thibaudeau G, Wills RW, Moura A, Kaya A, Perkins A, Memili E. Testis specific histone 2B is associated with sperm chromatin dynamics and bull fertility—a pilot study. Reprod Biol Endocrinol. 2017;15:59. doi: 10.1186/s12958-017-0274-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrot R, Jones S, Saint-Phar S, Kimmins S. Specialized distribution of the histone methyltransferase Ezh2 in the nuclear apical region of round spermatids and its interaction with the histone variant H1t2. J Androl. 2012;33:1058–1066. doi: 10.2164/jandrol.111.013870. [DOI] [PubMed] [Google Scholar]
- Lee TL, Pang AL, Rennert OM, Chan WY. Genomic landscape of developing male germ cells. Birth Defects Res C Embryo Today. 2009;87:43–63. doi: 10.1002/bdrc.20147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Park JY, Zheng D, Hoque M, Yehia G, Tian B. Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. BMC Biol. 2016;14:6. doi: 10.1186/s12915-016-0229-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yeh HJ, Shankarling GS, Ji Z, Tian B, MacDonald CC. The tauCstF-64 polyadenylation protein controls genome expression in testis. PLoS One. 2012;7:e48373. doi: 10.1371/journal.pone.0048373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD. Roles of RNA-binding Proteins and Post-transcriptional Regulation in Driving Male Germ Cell Development in the Mouse. Adv Exp Med Biol. 2016;907:123–151. doi: 10.1007/978-3-319-29073-7_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, MacDonald CC, Graber JH. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Research. 2007;35:234–246. doi: 10.1093/nar/gkl919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- MacDonald CC, Grozdanov PN. Nonsense in the Testis: Multiple Roles for Nonsense-Mediated Decay Revealed in Male Reproduction. Biol Reprod. 2017;96:939–947. doi: 10.1093/biolre/iox033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald CC, McMahon KW. Tissue-Specific Mechanisms of Alternative Polyadenylation: Testis, Brain and Beyond. WIREs RNA. 2010;1:494–501. doi: 10.1002/wrna.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc Natl Acad Sci U S A. 2005;102:2808–2813. doi: 10.1073/pnas.0406060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Gruber AR, Keller W, Zavolan M. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Reports. 2012;1:753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KW, Hirsch BA, MacDonald CC. Differences in polyadenylation site choice between somatic and male germ cells. BMC Molecular Biology. 2006;7:35. doi: 10.1186/1471-2199-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich ML, Mohapatra B, Shirley CR, Zhao M. Roles of transition nuclear proteins in spermiogenesis. Chromosoma. 2003;111:483–488. doi: 10.1007/s00412-002-0227-z. [DOI] [PubMed] [Google Scholar]
- Mishra LN, Gupta N, Rao SM. Mapping of post-translational modifications of spermatid-specific linker histone H1-like protein, HILS1. J Proteomics. 2015;128:218–230. doi: 10.1016/j.jprot.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Monarez RR, MacDonald CC, Dass B. Polyadenylation proteins CstF-64 and τCstF-64 exhibit differential binding affinities for RNA polymers. Biochemical Journal. 2007;401:651–658. doi: 10.1042/BJ20061097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naro C, Jolly A, Di Persio S, Bielli P, Setterblad N, Alberdi AJ, Vicini E, Geremia R, De la Grange P, Sette C. An Orchestrated Intron Retention Program in Meiosis Controls Timely Usage of Transcripts during Germ Cell Differentiation. Dev Cell. 2017;41:82–93. e84. doi: 10.1016/j.devcel.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayernia K, Meinhardt A, Drabent B, Adham IM, Muller C, Steckel M, Sancken U, Engel W. Synergistic effects of germ cell expressed genes on male fertility in mice. Cytogenet Genome Res. 2003;103:314–320. doi: 10.1159/000076819. [DOI] [PubMed] [Google Scholar]
- Nishimura H, L’Hernault SW. Spermatogenesis. Curr Biol. 2017;27:R988–R994. doi: 10.1016/j.cub.2017.07.067. [DOI] [PubMed] [Google Scholar]
- Pradeepa MM, Rao MR. Chromatin remodeling during mammalian spermatogenesis: role of testis specific histone variants and transition proteins. Soc Reprod Fertil Suppl. 2007;63:1–10. [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R. Chromatin dynamics during spermiogenesis. Biochim Biophys Acta. 2014;1839:155–168. doi: 10.1016/j.bbagrm.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Romeo V, Griesbach E, Schumperli D. CstF64: cell cycle regulation and functional role in 3′ end processing of replication-dependent histone mRNAs. Mol Cell Biol. 2014;34:4272–4284. doi: 10.1128/MCB.00791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp MD, Schweingruber C, Kleinschmidt N, Schümperli D. Interactions of CstF-64, CstF-77, and symplekin: implications on localisation and function. Mol Biol Cell. 2011;22:91–104. doi: 10.1091/mbc.E10-06-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell LD, Ettlin RA, Sinha Hikim AP, Clegg ED. Histological and Histopathological Evaluation of the Testis. Clearwater, FL: Cache River Press; 1990. [Google Scholar]
- Sabath I, Skrajna A, Yang XC, Dadlez M, Marzluff WF, Dominski Z. 3′-End processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors. RNA. 2013;19:1726–1744. doi: 10.1261/rna.040360.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- Shankarling GS, Coates PW, Dass B, MacDonald CC. A family of splice variants of CstF-64 expressed in vertebrate nervous systems. BMC Mol Biol. 2009;10:22. doi: 10.1186/1471-2199-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Steiniger M, Marzluff WF. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell. 2009;34:322–332. doi: 10.1016/j.molcel.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Qi H. Dynamic expression of combinatorial replication-dependent histone variant genes during mouse spermatogenesis. Gene Expr Patterns. 2014;14:30–41. doi: 10.1016/j.gep.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Molecular Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Iguchi N, Isotani A, Kitamura K, Toyama Y, Matsuoka Y, Onishi M, Masai K, Maekawa M, Toshimori K, et al. HANP1/H1T2, a novel histone H1-like protein involved in nuclear formation and sperm fertility. Mol Cell Biol. 2005;25:7107–7119. doi: 10.1128/MCB.25.16.7107-7119.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif S, Akrofi A, Dass B, Hardy DM, MacDonald CC. Infertility with impaired zona pellucida adhesion of spermatozoa from mice lacking τCstF-64. Biol Reprod. 2010;83:464–472. doi: 10.1095/biolreprod.109.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AM, Dass B, Ravnik SE, Tonk V, Jenkins NA, Gilbert DJ, Copeland NG, MacDonald CC. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proceedings of the National Academy of Science, USA. 1999;96:6763–6768. doi: 10.1073/pnas.96.12.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AM, Denison T, Attaya EN, MacDonald CC. Developmental differences in expression of two forms of the CstF-64 polyadenylation protein in rat and mouse. Biology of Reproduction. 2004;70:1080–1087. doi: 10.1095/biolreprod.103.022947. [DOI] [PubMed] [Google Scholar]
- Wang L, Xu Z, Khawar MB, Liu C, Li W. The histone codes for meiosis. Reproduction. 2017;154:R65–R79. doi: 10.1530/REP-17-0153. [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Ma L, Burns KH, Matzuk MM. HILS1 is a spermatid-specific linker histone H1-like protein implicated in chromatin remodeling during mammalian spermiogenesis. Proc Natl Acad Sci U S A. 2003;100:10546–10551. doi: 10.1073/pnas.1837812100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Sabath I, Debski J, Kaus-Drobek M, Dadlez M, Marzluff WF, Dominski Z. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′-end processing. Molecular and Cellular Biology. 2013;33:28–37. doi: 10.1128/MCB.00653-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Choi EA, Weng L, Xie X, Wan J, Xing Y, Moresco JJ, Tu PG, Yates JR, 3rd, Shi Y. Overlapping and distinct functions of CstF64 and CstF64τ in mammalian mRNA 3′ processing. RNA. 2013;109:18773–18778. doi: 10.1261/rna.042317.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood BA, Grozdanov PN, MacDonald CC. CstF-64 supports pluripotency and regulates cell cycle progression in embryonic stem cells through histone 3′ end processing. Nucleic Acids Res. 2014;42:8330–8342. doi: 10.1093/nar/gku551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood BA, MacDonald CC. CstF-64 is necessary for endoderm differentiation resulting in cardiomyocyte defects. Stem cell research. 2014;13:413–421. doi: 10.1016/j.scr.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagore LL, Grabinski SE, Sweet TJ, Hannigan MM, Sramkoski RM, Li Q, Licatalosi DD. RNA Binding Protein Ptbp2 Is Essential for Male Germ Cell Development. Mol Cell Biol. 2015;35:4030–4042. doi: 10.1128/MCB.00676-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Li C, Yang S, Tian R, Wang J, Yuan Q, Dong H, He Z, Wang S, Li Z. Dynamics of the Transcriptome during Human Spermatogenesis: Predicting the Potential Key Genes Regulating Male Gametes Generation. Sci Rep. 2016;6:19069. doi: 10.1038/srep19069. [DOI] [PMC free article] [PubMed] [Google Scholar]