Abstract
Objective
To determine whether systemic juvenile idiopathic arthritis (sJIA) susceptibility loci identified by candidate gene studies demonstrated association with sJIA in the largest study population assembled to date.
Methods
Single nucleotide polymorphisms (SNPs) from 11 previously reported sJIA risk loci were examined for association in 9 populations, including 770 sJIA cases and 6947 control subjects. The effect of sJIA-associated SNPs on gene expression was evaluated in silico in paired whole genome and RNA sequencing data from lymphoblastoid cell lines (LCL) of 373 European 1000 Genomes Project subjects. The relationship between sJIA-associated SNPs and response to anakinra treatment was evaluated in 38 US patients for whom treatment response data were available.
Results
We found no association of the 26 SNPs previously reported as sJIA-associated. Expanded analysis of the regions containing the 26 SNPs revealed only one significant association, the promoter region of IL1RN (p<1E-4). sJIA-associated SNPs correlated with IL1RN expression in LCLs, with an inverse correlation between sJIA risk and IL1RN expression. The presence of homozygous IL1RN high expression alleles correlated strongly with non-response to anakinra therapy (OR 28.7 [3.2, 255.8]).
Conclusion
IL1RN was the only candidate locus associated with sJIA in our study. The implicated SNPs are among the strongest known determinants of IL1RN and IL1RA levels, linking low expression with increased sJIA risk. Homozygous high expression alleles predicted non-response to anakinra therapy, nominating them as candidate biomarkers to guide sJIA treatment. This is an important first step towards the personalized treatment of sJIA.
Keywords: systemic juvenile idiopathic arthritis, genetics, IL1RN, anakinra, biomarker, personalized medicine
Systemic juvenile idiopathic arthritis (sJIA) is a rare, severe childhood inflammatory disease1, 2 that develops in the absence of an identifiable cause. sJIA is marked by the presence of chronic arthritis that occurs in the context of profound systemic inflammation, including quotidian fever, lymphadenopathy, hepatosplenomegaly, a salmon pink evanescent skin rash and serositis. It may also be accompanied by life threatening complications, including pericardial effusion, interstitial lung disease, amyloidosis, and macrophage activation syndrome, a highly lethal secondary form of hemophagocytic lymphohistiocytosis. Among children with sJIA, approximately half develop a destructive form of chronic arthritis that persists throughout their lives.
Despite its unifying inflammatory characteristics, sJIA is a heterogeneous condition with three distinct disease courses and variable expression of clinical manifestations and complications3. Regardless of the disease course and specific manifestations, the goal of sJIA treatment is to extinguish the systemic inflammation as rapidly as possible, taking advantage of the early therapeutic “window of opportunity” in an effort to avoid the development of persistent arthritis4. Achievement of this goal is often complicated by the fact that children with sJIA do not respond uniformly to the currently available therapies5, 6. A subset of sJIA responds to treatments targeting interleukin (IL)-1, a subset responds to IL-6 directed therapies, a subset responds to tumor necrosis factor (TNF)-α blockade, and a subset does not respond to any of these treatment strategies. Importantly, there is no objective determinant or biomarker that assists in predicting which therapeutic approach will be successful in individual patients, and thus there are often delays in ameliorating the systemic inflammation.
The pathophysiology of sJIA is poorly understood, as is the basis of its phenotypic heterogeneity. Due to its rare nature, most genetic studies of sJIA have utilized a candidate gene approach to examine small case-control collections. These studies produced a list of over two dozen single nucleotide polymorphisms (SNPs) at 11 distinct susceptibility loci that were reported as sJIA-associated loci. These include the IL1A/B7,8, GLI27, IL1RN/PSD47, IL1R27, IL10/209,10, IL611,12, MVK8, CCR513, MIF14, SLC26A215, and TAPBP16 loci (Table 1). Importantly, the original evidence supporting these associations was modest, and in many cases, the associations were not observed in studies of independent populations. Despite these facts, these associations are regularly included in discussions of sJIA pathophysiology.
Table 1.
Association results of 26 systemic juvenile idiopathic arthritis candidate SNPs in INCHARGE sJIA study collection
| Previous Study | INCHARGE Study | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Previous Study | SNP | Gene | P value | OR (95 C.I.) | P value | OR (95 C.I.) | i2 | N / n |
| Stock et. al | rs6712572 | IL1 Ligand (CKAP2L) | 0.0045 | 1.62 (1.16, 2.29) | 0.66 | 1.03 (0.91, 1.15) | 0.34 | 9/7708 |
| rs2071374 | IL1 Ligand (IL1A) | 0.0060 | 1.65 (1.15, 2.37) | 0.11 | 1.11 (0.98, 1.25) | 0 | 9/7711 | |
| rs3783516 | IL1 Ligand (IL1A/IL1B) | 0.0053 | 1.64 (1.15, 2.27) | 0.04 | 1.13(0.80, 1.26) | 0.30 | 9/7711 | |
| rs4848123 | IL1 Ligand (GLI2) | 0.0030 | 1.70 (1.19, 2.44) | 0.24 | 0.27 (0.12, 2.38) | 0.80 | 2/449 | |
| rs3917368 | IL1 Ligand (IL1B) | 0.0096 | 1.57 (1.11, 2.22) | 0.18 | 1.08 (0.96, 1.22) | 0.43 | 9/7715 | |
| rs1688075 | IL1 Ligand (IL1RN) | 0.0089 | 3.04 (1.58, 5.85) | 0.64 | 0.95 (0.77, 1.17) | 0.06 | 8/7603 | |
| rs4849159 | IL1 Ligand (PSD4) | 0.040 | 1.61 (1.02, 2.54) | 0.17 | 0.89 (0.75, 1.05) | 0 | 8/5755 | |
| rs6760120 | IL1 Ligand (PSD4) | 0.020 | 1.49 (1.06, 2.21) | 0.33 | 0.92 (0.77, 1.09) | 0.19 | 9/7713 | |
| rs12712122 | IL1 Receptor (IL1R2) | 0.0031 | 1.71 (1.21, 2.41) | 0.04 | 1.32 (1.03, 1.69) | 0 | 9/7710 | |
| rs4851531 | IL1 Receptor (IL1R2) | 0.0087 | 1.59 (1.11, 2.28) | 0.58 | 0.97 (0.86, 1.09) | 0.13 | 9/7708 | |
|
| ||||||||
| Omoyinmi et. al. | rs1400986 | IL-10 Family (IL20) | 0.0004 | 1.53 (1.21, 1.93) | 0.27 | 1.11 (0.93, 1.32) | 0.48 | 8/7519 |
| rs4129024 | IL-10 Family (MAPKAPK2) | 0.0027 | 0.68 (0.53, 0.88) | 0.05 | 0.87 (0.75, 1.00) | 0.08 | 9/7712 | |
|
| ||||||||
| Fife et. al. | rs1800896 | IL-10 Family (IL10) | 0.031 | 1.34 (n.p.) | 0.02 | 1.15 (1.02, 1.28) | 0 | 9/7716 |
| rs1400986 | IL-10 Family (IL20) | 0.028 | 1.51 (n.p.) | 0.27 | 1.11 (0.93, 1.32) | 0.48 | 8/7519 | |
|
| ||||||||
| Fishman et. al. | rs1800795 | IL6 | 0.03 | n.p. | 0.34 | 0.94 (0.84, 1.06) | 0.32 | 9/7710 |
|
| ||||||||
| Hinks et. al. | rs2071374 | IL1A | 0.001 | 1.50 (1.16, 1.92) | 0.11 | 1.11 (0.98, 1.25) | 0 | 9/7711 |
| rs11836136 | MVK | 0.03 | 1.34 (1.03, 1.74) | 0.58 | 1.05 (0.89, 1.23) | 0 | 9/7717 | |
|
| ||||||||
| Scheibel et. al. | rs333 | CCR5 | 0.004 | n.p. | 0.16 | 0.86 (0.69, 1.06) | 0.63 | 4/7009 |
|
| ||||||||
| De Benedetti et. al. | rs755622 | MIF | 0.017 | n.p. | 0.11 | 0.88 (0.76, 1.03) | 0 | 8/7513 |
|
| ||||||||
| Lamb et. al. | rs1541915 | SLC26A2 | 0.0003 | 2.3 (1.4, 3.7) | 0.76 | 0.98 (0.87, 1.11) | 0.19 | 8/7516 |
| rs245056 | SLC26A2 | 0.00002 | 2.8 (1.7, 4.6) | 0.72 | 1.03 (0.86, 1.23) | 0.26 | 8/7513 | |
| rs245055 | SLC26A2 | 0.004 | 2.5 (1.2, 5.0) | 0.56 | 0.95 (0.81, 1.12) | 0 | 9/7709 | |
| rs245051 | SLC26A2 | 0.0005 | 2.3 (1.4, 3.7) | 0.42 | 0.95 (0.85, 1.07) | 0.44 | 9/7708 | |
| rs245076 | SLC26A2 | 0.0015 | 2.7 (1.3, 5.6) | 0.46 | 0.94 (0.80, 1.11) | 0 | 9/7715 | |
| rs8073 | SLC26A2 | 0.04 | 2.3 (0.9, 5.6) | 0.25 | 0.91 (0.77, 1.07) | 0 | 9/7714 | |
|
| ||||||||
| Bukulmez et. al. | rs2071888 | TAPBP | 0.04 (TDT) | n.p. | 0.15 | 1.09 (0.97, 1.22) | 0 | 9/7715 |
SNP, single nucleotide polymorphism; INCHARGE, International Childhood Arthritis Genetic Consortium; OR, odds ratio, 95 CI, 95% confidence interval; i2, i2 test for heterogeneity; N, number of strata included in meta-analysis; n, number of samples included in meta-analysis; n.p., not provided; TDT, transmission disequilibrium testing
We have recently performed the largest genetic study of sJIA, a multi-national effort that included children with sJIA from 9 countries17,18. We identified two bona fide sJIA susceptibility loci and 24 additional loci suggestively associated with sJIA, however there was no overlap between the peak sJIA susceptibility loci in our studies and those reported in the earlier candidate gene studies. To evaluate the relationship between sJIA risk and the sJIA susceptibility loci identified by candidate gene studies, we have undertaken a regional association study of the 11 reported candidate susceptibility loci in the International Childhood Arthritis Genetics Consortium (INCHARGE) sJIA case-control collection.
Materials and Methods
Study design and participants
Directly observed and imputed SNP genotype data from the 9 case-control populations of the INCHARGE sJIA collection were evaluated for this study17,18. The INCHARGE sJIA collection includes children fulfilling International League of Associations for Rheumatology (ILAR) criteria for sJIA and control subjects from the United States, the United Kingdom, Germany, Turkey, Italy, Brazil, Argentina, Canada and Spain. SNP genotyping of genomic DNA from cases and controls was performed using Human Omni1M arrays and an iScan reader (Illumina). SNP genotypes were stratified by country of origin and, where available, combined with existing SNP genotype datasets, in silico, from geographically-matched healthy control individuals. Each geographically-defined stratum was subjected to rigorous quality control processes to remove samples and SNPs of poor quality using standard metrics. Ancestral outliers were removed from each geographically-defined stratum using a combination of principal components analysis and multidimensional scaling. The degree of matching was assessed using genomic control inflation factors (λGC), which were < 1.004 for each of the 9 strata. Detailed information about case and control populations included in the INCHARGE collection, along with technical descriptions and visualizations of the quality control processes and their results can be found in the supplementary material of our earlier papers17,18. For the present study, genotypes of SNPs residing in 11 candidate loci (Table S1) were examined in 770 children with sJIA and 6947 control samples from the United States, the United Kingdom, Germany, Turkey, Italy, Brazil, Argentina, Canada and Spain. For candidate loci defined by a single sJIA-associated SNP, the study interval was defined as ± 100 kilobases (Kb) from the position of that SNP. When more than one SNP association was present within a locus, the study interval was defined as ± 100 Kb from the mean of the positions of the reported sJIA-associated SNPs.
Statistical analysis
Association testing of candidate SNPs was performed under the additive model, adjusted for sex and ancestry-informative principal components, in each of the 9 sJIA case-control collections using SNPTESTv219. Association results were then combined across collections using fixed-effect meta-analysis with GWAMA software20. Heterogeneity was evaluated using the i2 statistic and SNPs exhibiting moderate evidence of heterogeneity (i2 > 0.5) were excluded from our analysis. Association data were visualized using SNP and Variation Suite 8 (SVS8, Golden Helix, Bozeman, Montana) and custom R scripts (R version 3.4.0). Haplotype analysis and examination of LD were performed using Haploview21. The SNP set was pruned for pairwise linkage disequilibrium (LD) of r2 < 0.5 by the Estimation-Maximization method with PLINK22 to determine the number of independent SNPs in the study. The threshold for study-wide significance was defined by a Bonferroni correction for the total number of independent SNPs across all candidate loci.
Gene expression analysis
The effect of sJIA-associated SNPs on gene and/or protein expression was examined using the Haploregv4.1 database23. The correlation of sJIA-associated SNPs with gene expression was investigated by an integrated examination of RNA-sequencing (RNA-seq) and whole genome sequencing (WGS) data from 1000 Genomes Project subjects24,25. RNA-seq data from the set of 373 lymphoblastoid cell lines (LCLs) of European 1000 Genomes Project subjects were downloaded from the Geuvadis website (http://www.geuvadis.org/web/geuvadis/RNAseq-project) and WGS data from the corresponding individuals was downloaded from the 1000 Genomes Project website (http://www.internationalgenome.org/data/). RNA-seq data (normalized reads per kilobase per million reads or RPKM) were stratified by sJIA risk allele genotype and the difference in relative expression between genotypes was evaluated using the non-parametric Kruskal-Wallis test. Box plots of relative expression were generated using R.
Therapeutic response analysis
The relationship between sJIA-associated IL1RN SNPs and therapeutic response to human recombinant IL-1RA (anakinra) or tocilizumab treatment was examined in sJIA patients from the U.S. stratum for whom therapeutic response data were available. This included 38 anakinra treated subjects and 14 subjects treated with tocilizumab. Treatment response data were extracted from medical records by the treating pediatric rheumatologist, who encoded either “no response” or “any response” for each subject. “No response” was defined as no improvement of either fever (if present) or arthritis. “Any response” was defined as any degree of improvement in either fever or arthritis. Treatment response was then tested for association with sJIA-associated SNPs by logistic regression under the dominant model using SVS8. The threshold of significance for the association test was defined by a Bonferroni correction for the number of independent SNPs tested, as defined by pairwise LD pruning (r2 < 0.5).
Results
Association testing of sJIA candidate SNPs and loci
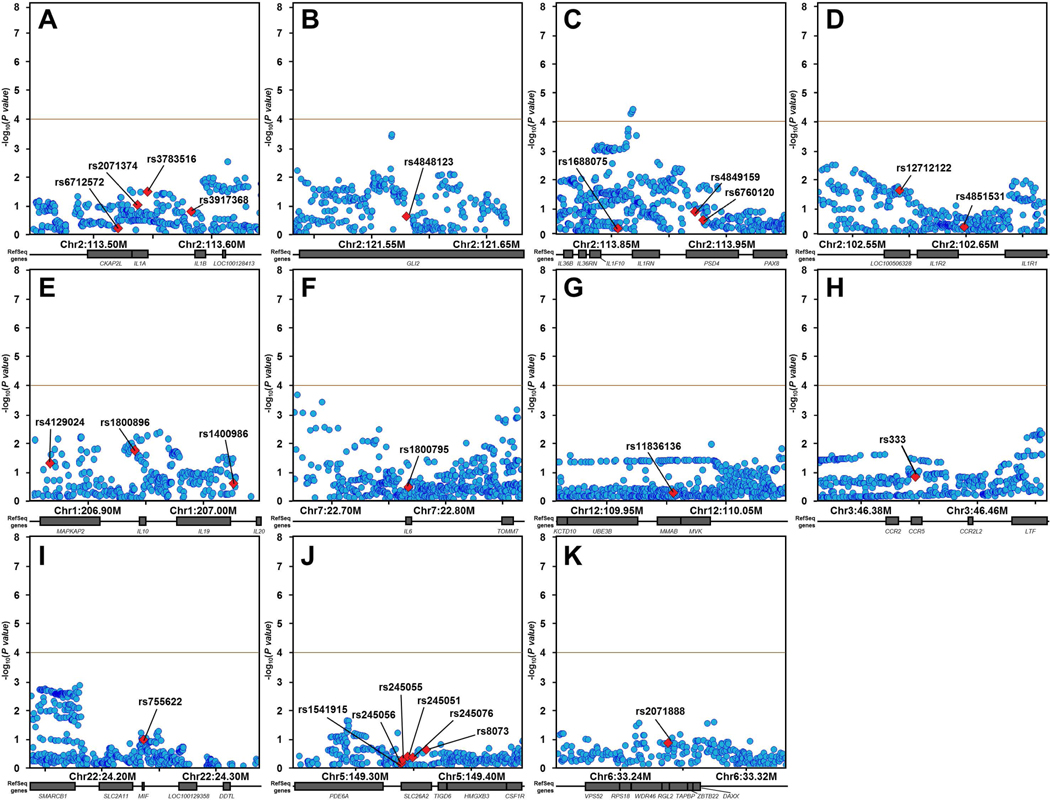
We first performed association testing of the 26 SNPs for which associations with sJIA had been previously reported (Table 1). After applying Bonferroni correction for 26 SNPs, association meta-analysis of the 9 INCHARGE sJIA study populations revealed no significant associations with sJIA (p < 0.05 ÷ 26 = 1.9 × 10−3, Table 1, Figure S1). To evaluate whether the 11 candidate loci containing these 26 SNPs harbored sJIA risk SNPs distinct from those previously described, we extended our analysis to test all SNPs within these candidate risk loci for association with sJIA. The candidate regions included a total of 5479 SNPs (Table S2), but LD pruning at a level of r2 < 0.5 determined that only 500 of them were independent. This defined the threshold of study-wide significance (p < 0.05 ÷ 500 = 1.0 × 10−4). By this standard, association meta-analyses of these 11 loci revealed a single significant association signal within the IL1RN locus (Figure 1). The association peak was located 4.3 Kb upstream from IL1RN, with 3 SNPs exceeding the significance threshold and the top 7 SNPs in strong LD with one another (Figure 2). In fact, LD mapping and haplotype analysis of the top 25 SNPs within this locus revealed that the top 7 sJIA-associated SNPs were inherited as a part of a common haplotype (Figure 2).
Figure 1. INCHARGE sJIA case-control regional association plots of loci previously implicated by candidate gene studies.
Regional association plots for previously reported sJIA candidate susceptibility loci near IL1A/B (A), GLI2 (B), IL1RN/PSD4 (C), IL1R2 (D), IL10/20 (E), IL6 (F), MVK (G), CCR5 (H), MIF (I), SLC26A2 (J), and TAPBP (K) show minimal significance in INCHARGE case-control dataset, except for a cluster of SNPs in the IL1RN/PSD4 region (C). None of the top SNPs from previous candidate studies (labeled and denoted by red diamonds) showed even nominal significance with sJIA. Other SNPs in the candidate loci are shown as blue circles. The brown horizontal line demonstrates the study-wide significance threshold.
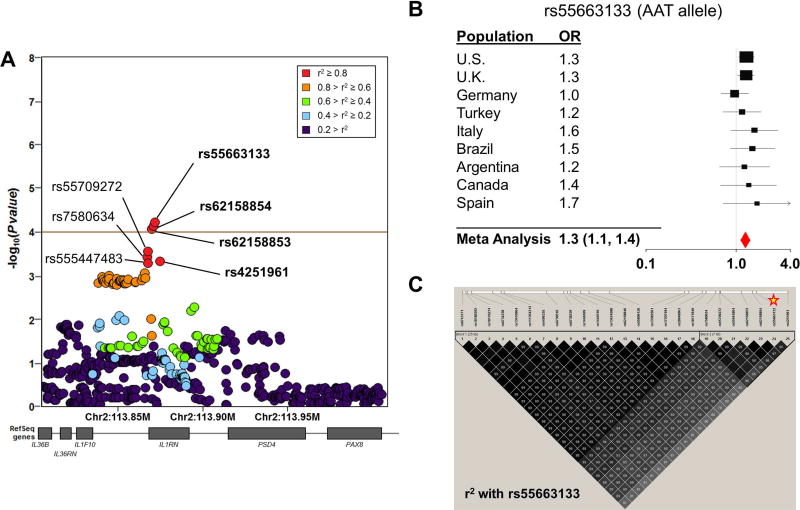
Figure 2. Variants of the IL1RN locus are associated with sJIA in the INCHARGE case-control collection.
SNP associations within the IL1RN locus are shown, colored by pairwise linkage disequilibrium (LD) with the most strongly associated SNP, rs55663133 (A). The brown horizontal line demonstrates the study-wide significance threshold. In Panel B, a forest plot demonstrates the effect size of rs55663133 by meta-analysis and in individual study populations. Panel C displays pairwise LD with the peak sJIA-associated SNP, rs55663133 (star) in the U.S. case-control population. The top 7 sJIA associated markers (19–25) form a strong LD block.
sJIA-associated IL1RN variants and gene expression
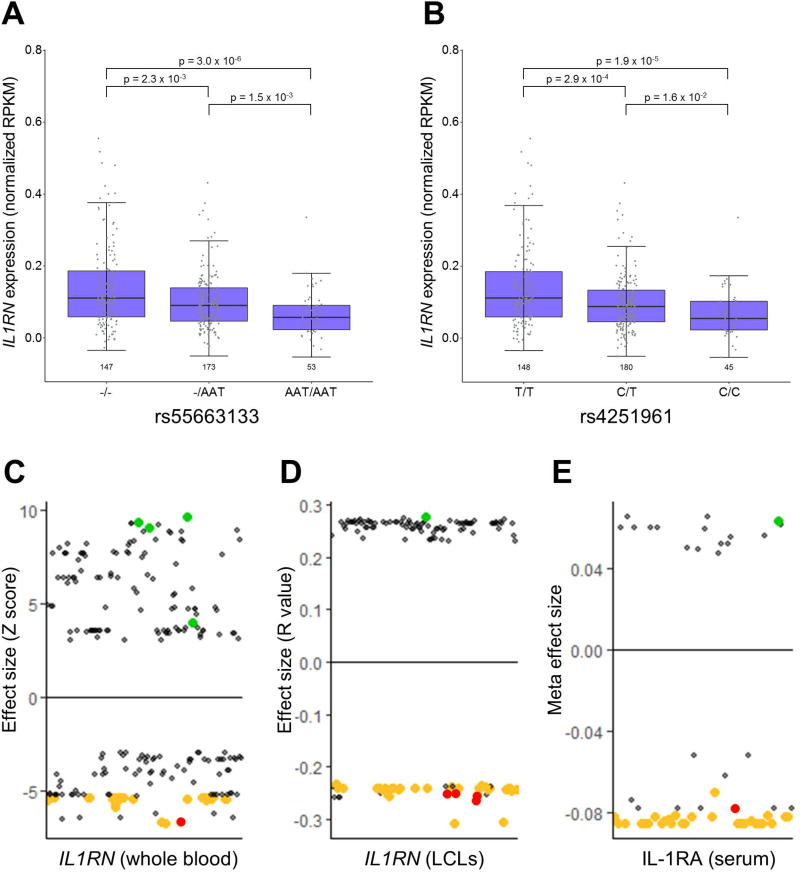
A query of the HaploRegv4.1 database revealed that many of the top sJIA-associated SNPs were known expression quantitative trait loci (eQTL) for IL1RN in whole blood26 and LCLs25 (Table 2). Moreover, a review of the literature found that sJIA-associated SNPs also correlated with IL-1RA protein levels in the largest study of genetic predictors of IL-1RA levels27. The SNP that most strongly correlated with IL-1RA in that study, rs4251961, was one of the top sJIA-associated SNPs and was a constituent of the 7 SNP haplotype (Figure 2, Table 2). These observations were corroborated by our direct analyses of LCL RNA-seq data from 1000 Genomes Project subjects25, which found that the sJIA-associated SNPs were strongly correlated with IL1RN expression (Figures 3 and S2). Specifically, alleles that were protective against sJIA correlated with high IL1RN expression and those that were risk factors for sJIA correlated with reduced IL1RN expression (Figure 3). Importantly, all three of the studies mentioned above parsimoniously demonstrated that sJIA risk alleles of the top 42 sJIA-associated SNPs were correlated with decreased levels of IL1RN expression or circulating IL-1RA protein (Figure 3).
Table 2.
Association of IL1RN SNPs with sJIA risk and their effect on IL1RN expression / IL-1RA concentration
|
IL1RN in
LCLs Lappalainen et al. 25 |
IL1RN in whole
blood Westra et al. 26 |
IL-1RA in serum Herder et al. 27 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| SNP | Risk Allele |
Meta P | Meta OR (95 CI) |
r2 | P-value | Effect Size |
P-value | Effect Size |
P-value | Effect Size |
| rs55663133 | AAT | 5.9 × 10−5 | 1.3 (1.1, 1.4) | 1 | 1.0 × 10−6 | −0.25 | ||||
| rs62158854 | G | 7.2 × 10−5 | 1.3 (1.1, 1.4) | 1 | 6.5 × 10−7 | −0.25 | ||||
| rs62158853 | T | 8.3 × 10−5 | 1.3 (1.1, 1.4) | 1 | 2.6 × 10−7 | −0.26 | ||||
| rs55709272 | C | 2.8 × 10−4 | 1.2 (1.1, 1.4) | 0.87 | ||||||
| rs7580634 | T | 3.7 × 10−4 | 1.2 (1.1, 1.4) | 0.91 | 2.8 × 10−6 | −0.24 | ||||
| rs4251961 | C | 4.6 × 10−4 | 1.2 (1.1, 1.4) | 0.86 | 1.0 × 10−6 | −0.25 | 1.6 × 10−11 | −6.74 | 2.2 × 10−34 | −0.08 |
| rs555447483 | A | 5.0 × 10−4 | 1.2 (1.1, 1.4) | 0.89 | ||||||
| rs28648961 | A | 8.5 × 10−4 | 1.2 (1.1, 1.4) | 0.75 | 2.8 × 10−6 | −0.24 | ||||
| rs111354213 | - | 9.8 × 10−4 | 1.2 (1.1, 1.4) | 0.74 | ||||||
| rs6743171 | C | 1.1 × 10−3 | 1.2 (1.1, 1.4) | 0.77 | 3.2 × 10−6 | −0.24 | 5.8 × 10−8 | −5.42 | 6.4 × 10−13 | −0.09 |
| rs17207494 | C | 1.1 × 10−3 | 1.2 (1.1, 1.4) | 0.75 | 2.9 × 10−6 | −0.24 | 3.8 × 10−8 | −5.50 | 1.3 × 10−11 | −0.08 |
| rs10171849 | C | 1.1 × 10−3 | 1.2 (1.1, 1.4) | 0.75 | 5.5 × 10−6 | −0.23 | 2.6 × 10−8 | −5.57 | 1.4 × 10−11 | −0.08 |
| rs4496335 | T | 1.2 × 10−3 | 1.2 (1.1, 1.4) | 0.77 | 3.0 × 10−6 | −0.24 | 5.2 × 10−8 | −5.44 | 6.4 × 10−13 | −0.09 |
| rs6730516 | T | 1.2 × 10−3 | 1.2 (1.1, 1.4) | 0.77 | 3.5 × 10−6 | −0.24 | 6.2 × 10−8 | −5.41 | 6.4 × 10−13 | −0.09 |
| rs55896126 | C | 1.2 × 10−3 | 1.2 (1.1, 1.4) | 0.75 | 2.7 × 10−6 | −0.24 | ||||
| rs6734238 | G | 1.2 × 10−3 | 1.2 (1.1, 1.4) | 0.73 | 2.4 × 10−8 | −5.58 | 1.1 × 10−12 | −0.08 | ||
| rs13410964 | A | 1.2 × 10−3 | 1.2 (1.1, 1.4) | 0.77 | 2.7 × 10−6 | −0.24 | 6.4 × 10−8 | −5.41 | 6.4 × 10−13 | −0.09 |
| rs13424580 | A | 1.2 × 10−3 | 1.2 (1.1, 1.4) | 0.75 | 2.4 × 10−6 | −0.24 | 5.3 × 10−8 | −5.44 | 1.4 × 10−11 | −0.08 |
| rs1446510 | T | 1.2 × 10−3 | 1.2 (1.1, 1.4) | 0.77 | 2.8 × 10−6 | −0.24 | 6.2 × 10−8 | −5.41 | 6.5 × 10−13 | −0.09 |
| rs10176274 | G | 1.3 × 10−3 | 1.2 (1.1, 1.4) | 0.77 | 2.7 × 10−6 | −0.24 | 5.8 × 10−8 | −5.42 | 6.4 × 10−13 | −0.09 |
| rs10188292 | T | 1.3 × 10−3 | 1.2 (1.1, 1.4) | 0.77 | 2.6 × 10−6 | −0.24 | 5.8 × 10−8 | −5.42 | 6.4 × 10−13 | −0.09 |
| rs1446509 | T | 1.3 × 10−3 | 1.2 (1.1, 1.4) | 0.77 | 2.0 × 10−6 | −0.24 | 6.2 × 10−8 | −5.41 | 6.5 × 10−13 | −0.09 |
| rs62158846 | T | 1.3 × 10−3 | 1.2 (1.1, 1.4) | 0.75 | ||||||
| rs6738239 | A | 1.3 × 10−3 | 1.2 (1.1, 1.4) | 0.77 | 4.7 × 10−6 | −0.23 | 6.1 × 10−8 | −5.42 | 6.5 × 10−13 | −0.09 |
| rs13382561 | G | 1.3 × 10−3 | 1.2 (1.1, 1.4) | 0.75 | 1.9 × 10−6 | −0.24 | 3.7 × 10−8 | −5.51 | 1.4 × 10−11 | −0.08 |
| rs7587033 | G | 0.001296 | 1.2 (1.1, 1.4) | 0.77 | 2.9 × 10−6 | −0.24 | ||||
| rs6750559 | A | 0.001325 | 1.2 (1.1, 1.4) | 0.77 | 2.5 × 10−6 | −0.24 | 6.1 × 10−8 | −5.42 | 6.4 × 10−13 | −0.09 |
| rs7574427 | A | 0.001344 | 1.2 (1.1, 1.4) | 0.77 | 2.1 × 10−6 | −0.24 | 4.3 × 10−8 | −5.48 | 1.4 × 10−11 | −0.08 |
| rs6722922 | T | 0.001374 | 1.2 (1.1, 1.4) | 0.77 | 2.7 × 10−6 | −0.24 | 6.1 × 10−8 | −5.42 | 6.4 × 10−13 | −0.09 |
| rs6741180 | A | 0.001376 | 1.2 (1.1, 1.4) | 0.77 | 3.1 × 10−6 | −0.24 | 6.0 × 10−8 | −5.42 | 6.4 × 10−13 | −0.09 |
| rs7574159 | A | 0.001393 | 1.2 (1.1, 1.4) | 0.75 | 2.1 × 10−6 | −0.24 | 5.3 × 10−8 | −5.44 | 1.2 × 10−11 | −0.08 |
| rs13398728 | C | 0.001434 | 1.2 (1.1, 1.4) | 0.77 | 2.7 × 10−6 | −0.24 | 6.0 × 10−8 | −5.42 | 6.4 × 10−13 | −0.09 |
| rs13409371 | A | 0.001445 | 1.2 (1.1, 1.4) | 0.79 | 6.3 × 10−7 | −0.25 | 5.2 × 10−9 | −5.84 | 3.8 × 10−12 | −0.08 |
| rs13409360 | A | 0.001467 | 1.2 (1.1, 1.4) | 0.79 | 6.5 × 10−7 | −0.25 | 3.8 × 10−9 | −5.89 | 7.8 × 10−13 | −0.08 |
| rs12329129 | A | 0.001468 | 1.2 (1.1, 1.4) | 0.77 | 2.8 × 10−6 | −0.24 | 6.1 × 10−8 | −5.42 | 6.4 × 10−13 | −0.09 |
| rs12328368 | G | 0.001473 | 1.2 (1.1, 1.4) | 0.77 | 2.7 × 10−6 | −0.24 | 6.1 × 10−8 | −5.42 | 6.4 × 10−13 | −0.09 |
| rs7596350 | G | 0.001475 | 1.2 (1.1, 1.4) | 0.75 | 1.7 × 10−6 | −0.24 | ||||
| rs6746979 | A | 0.001485 | 1.2 (1.1, 1.4) | 0.75 | 2.1 × 10−6 | −0.24 | 5.1 × 10−8 | −5.45 | 1.4 × 10−11 | −0.08 |
| rs58865280 | A | 0.001494 | 1.2 (1.1, 1.4) | 0.75 | ||||||
| rs9973741 | G | 0.001514 | 1.2 (1.1, 1.4) | 0.75 | 2.1 × 10−6 | −0.24 | ||||
| rs12328766 | G | 0.001562 | 1.2 (1.1, 1.4) | 0.77 | 3.0 × 10−6 | −0.24 | 6.2 × 10−8 | −5.41 | 6.4 × 10−13 | −0.09 |
| rs550593914 | T | 0.001593 | 1.2 (1.1, 1.4) | 0.77 | ||||||
SNP, single nucleotide polymorphism; Meta P, fixed effect meta-analysis P value; Meta OR, fixed effect meta-analysis odds ratio; 95 CI, 95% confidence interval; r2, pairwise r2 with rs55663133 using the Estimation-Maximization method in the U.S. case-control population; LCL, lymphoblastoid cell line. The top 7 sJIA-associated SNPs, which are inherited as an LD block, are shown in bold italics.
Figure 3. Relationship of IL1RN expression and IL-1RA protein levels with sJIA-associated SNPs.
IL1RN expression by RNA sequencing from the study of Lappalainen et al. 25 is shown, stratified by genotype, for representative sJIA-associated SNPs (A and B). Dot plots depict all SNPs with reported correlations with IL1RN expression (C and D) or IL-1RA protein levels (E) in the studies by Westra et al. 26, Lappalainen et al. 25, and Herder et al. 27, respectively. SNPs among the top 42 sJIA-associated SNPs are highlighted in green (sJIA protective alleles) and gold (sJIA risk alleles), and the top 7 sJIA-associated SNPs are highlighted in red.
sJIA-associated IL1RN variants and response to anakinra therapy in sJIA
Given that the response of sJIA to treatment with recombinant human IL-1RA (anakinra) is variable, we hypothesized that individuals with the highest genetically-encoded levels of IL-1RA may fail to respond to anakinra treatment more often than those with lower genetically-encoded levels. To evaluate this possibility, we examined clinical and SNP genotype data in 38 sJIA patients from the U.S. collection that had received anakinra and for whom clinical data were available. Within this group of anakinra treated subjects, there were 9 non-responders and 29 “any responders”. An examination of the top 7 sJIA-associated IL1RN SNPs found that for each SNP, homozygosity for the IL1RN high expression alleles was associated with non-response to anakinra treatment (p < 0.05, Table 3). rs555447483 showed the strongest association with anakinra non-response (p = 7.7 × 10−4; OR 28.7 [3.2, 255.8], with homozygous high expression alleles predicting non-response with a sensitivity of 92% and a specificity of 71%.
Table 3.
Association between sJIA-associated quantitative trait loci for IL1RN expression (and serum levels of IL-1RA protein) and response to anakinra therapy in 38 patients from the INCHARGE U.S. population.
| Homozygote frequency
|
|||||
|---|---|---|---|---|---|
| SNP | Effect allele (high expression) |
Non-responder (n=9) |
Any responder (n=29) |
P value | OR (95CI) |
| rs55663133 | - | 0.67 | 0.22 | 1.6 × 10−2 | 7.0 (1.3, 36.7) |
| rs62158854 | T | 0.67 | 0.22 | 1.6 × 10−2 | 7.0 (1.3, 36.7) |
| rs62158853 | C | 0.67 | 0.24 | 2.1 × 10−2 | 6.3 (1.2, 32) |
| rs55709272 | T | 0.67 | 0.1 | 9.8 × 10−4 | 17.3 (2.8, 108.1) |
| rs7580634 | G | 0.67 | 0.1 | 9.8 × 10−4 | 17.3 (2.8, 108.1) |
| rs4251961 | T | 0.78 | 0.21 | 1.8 × 10−3 | 13.4 (2.2, 82) |
| rs555447483 | - | 0.71 | 0.08 | 7.7 × 10−4 | 28.7 (3.2, 255.8) |
SNP, single nucleotide polymorphism; OR, odds ratio; 95CI, 95% confidence interval.
To determine whether the relationship between these SNPs and sJIA treatment failure were specific to anakinra, we performed an identical examination of the 14 sJIA patients from the U.S. collection who were treated tocilizumab, an anti-IL-6 monoclonal antibody. Within this group, which included 3 tocilizumab non-responders and 11 tocilizumab “any responders,” we found no association between sJIA-associated IL1RN SNPs and response to tocilizumab treatment (Table S3). Moreover, 8 of the 14 patients treated with tocilizumab were anakinra non-responders who received tocilizumab as second line treatment. Among the 8 anakinra non-responders, 6 were tocilizumab “any responders.” Taken together, these facts support the hypothesis that sJIA-associated IL1RN SNPs specifically predict non-response of sJIA to anakinra treatment, as opposed to identifying individuals whose sJIA is more broadly refractory to treatment.
Discussion
Through an examination of common genetic variants at 11 previously reported sJIA susceptibility loci in the INCHARGE sJIA collection, this study has yielded three important observations. First, this study has demonstrated that the IL1RN locus is a bona fide sJIA susceptibility locus. Second, it has revealed that genetically-encoded high expression of IL1RN and production of IL-1RA are protective against sJIA (and conversely that genetically-encoded low expression/production are risk factors for developing sJIA.) Most importantly, it has shown that homozygosity for the high expression alleles of sJIA-associated IL1RN SNPs is strongly associated with unresponsiveness to anakinra treatment in sJIA patients.
The original studies describing these 11 candidate loci reported modest associations that were identified in small case-control collections7–16. At most of these loci, the associations with sJIA were not observed in subsequent studies of other populations, calling their proposed relationships with sJIA into question. We sought to evaluate these associations more rigorously by using the INCHARGE sJIA collection, which provided greater statistical power than any previous study of these loci while also allowing for internal validation through the examination of 9 independent populations. Using this approach, we found that only one of these candidate loci, IL1RN, was associated with sJIA. At this locus, we observed 3 sJIA-associated SNPs that tagged a 7 SNP haplotype in the promoter region of IL1RN, as well as a cluster of 39 other SNPs with intermediate evidence of association with sJIA. Importantly, the IL1RN association signal identified in the present study did not include any of the SNPs that were previously reported as sJIA-associated (Table 1) or any SNPs that were in strong LD with those SNPs (Figures 1 and S1). This observation suggests that the historical candidate gene studies of sJIA could have been negatively impacted by poor statistical power, as has been the case in other genetically complex diseases, such as schizophrenia28.
Given that the association signal of the IL1RN locus was within the promoter region, we hypothesized that these SNPs may influence sJIA risk by altering gene expression. By examining previously published gene expression studies and integrating our association data with publicly available gene expression datasets, we found that the risk alleles of the top 42 sJIA-associated IL1RN SNPs correlated with reduced IL1RN expression and circulating IL-1RA levels (Figure 2, Table 2). Furthermore, we observed that the top 7 sJIA-associated SNPs were among the SNPs most strongly associated with IL1RN expression levels in whole blood and LCLs, and with circulating levels of IL-1RA protein, in published studies (Table 2)25–27. Taken together these observations suggest that the sJIA-associated IL1RN SNPs influence sJIA risk through their effect on IL1RN expression and production of IL-1RA.
IL-1RA is a well-documented positive acute phase protein29 and it has been shown to be highly expressed in the blood of children with active sJIA30–31. Therefore, one would expect that gene expression studies should find increased expression of IL1RN in children with active sJIA compared to healthy subjects or children with quiescent sJIA. There have been several studies that have examined gene expression in sJIA peripheral blood mononuclear cells. In one of these studies, the expression of positive acute phase genes was upregulated in children with sJIA and the authors noted that IL1RN was among this cluster32. However, three other studies found no relationship between sJIA and IL1RN expression33–35. There are a couple of potential reasons for these conflicting results. These studies were undertaken in relatively small numbers of sJIA cases, so it is possible that they lacked the statistical power to identify a relationship between sJIA and IL1RN expression. It is also possible that these studies were affected by confounding variables that altered IL1RN expression in the sJIA patients, such as the duration of sJIA, the level of sJIA disease activity or the treatment(s) administered for sJIA. By examining the correlation between sJIA-associated IL1RN variants and gene expression in healthy individuals, the present study could identify the relationship between IL1RN expression and sJIA without the interference of these potential confounders.
Looking beyond disease risk, we also observed that high expression alleles of sJIA-associated IL1RN SNPs were strongly associated with non-response to anakinra therapy. The lack of association between these SNPs and non-response to tocilizumab treatment suggests that these SNPs are specifically associated with anakinra non-responsiveness, as opposed to being associated with more global therapeutic recalcitrance. In the context of the bi-phasic hypothesis of sJIA pathophysiology, new onset sJIA is treated with the goal of rapidly inducing remission within the therapeutic window of opportunity4. Anakinra is commonly chosen as the first line treatment because its effects can be observed within days of initiation and because its dosing can be rapidly escalated, but it is not effective in all patients36. In the subset of sJIA cases that ultimately don’t respond to anakinra, their time to remission is extended by the failed therapeutic course of anakinra. The findings of this study can be used to identify the subset of children with sJIA that are unlikely to respond to anakinra and facilitate the selection of an alternative treatment. In doing so, one can avoid the delay associated with a first-line therapeutic failure and reduce the time to remission, as well as prevent unnecessary exposure to the risks of anakinra treatment. This is the first candidate biomarker that can prospectively guide therapeutic decision making in sJIA.
Despite the strength of our findings, it is important to consider potential limitations of our study. This study evaluated genetic associations in 9 independent sJIA case-control collections. It will be important to examine the IL1RN region in larger, independent groups of patients. There were several limitations to the evaluation of genetic predictors of anakinra response. Therapeutic response to anakinra was examined in 38 sJIA patients, which is a relatively small group. The anakinra treated patients were not treated in a standardized fashion, with potential variation in the drug dosing and duration, timing of dose escalation and co-administration of other agents (ie. glucocorticoids). The clinical response data were extracted from medical records in a post hoc analysis and clinical response metrics were not standardized. We anticipated that these factors would complicate differentiating incomplete and complete response, but should not influence the identification of non-response. Therefore, we chose to compare non-response to “any response.” Nonetheless, it will be important to evaluate the correlation between IL1RN SNPs and response to anakinra in prospective studies of larger numbers of patients treated and monitored in a standardized manner.
By identifying a prospective biomarker capable of guiding the treatment of sJIA, this study brings precision medicine to the rheumatology clinic. Looking forward, it will be important to determine whether these findings are generalizable beyond anakinra and sJIA. For example, can the IL1RN SNPs predict therapeutic response with other IL-1 directed therapies, such as monoclonal anti-IL-1β antibodies (canakinumab) or the IL-1 trap (rilonacept), in sJIA. Similarly, these SNPs may predict therapeutic response to anakinra (or other IL-1 directed therapies) in conditions other than sJIA, such as adult-onset Still’s disease or monogenic autoinflammatory diseases. Given that recently published trials have found that canakinumab treatment significantly reduces the risk of recurrent cardiovascular events37, as well as the incidence of and mortality from lung cancer38, it is even possible that the utility of this prospective biomarker may extend beyond the field of rheumatology.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Programs of the National Institute of Arthritis and Musculoskeletal and Skin Diseases and the National Human Genome Research Institutes of the National Institutes of Health (NIH). The collection of UK sJIA samples was supported by Arthritis Research UK (20385) and by the National Institute for Health Research (NIHR) Biomedical Research Centre. The CAPS study was funded by Arthritis Research UK (20542). SPARKS-CHARMS was funded by grants from SPARKS UK (08ICH09 and 12ICH08) and the Medical Research Council (MR/M004600/1), and supported by the NIHR Biomedical Research Centres at Great Ormond Street Hospital for Children National Health Service (NHS) Foundation Trust and University College London Hospitals Trust, and the NIHR-Clinical Research Network. The BBOP study was supported by the Canadian Institutes of Health Research and the Arthritis Society (CIHR funding reference number 82517) and the Canadian Arthritis Network (funding reference SRI-IJD-01). This research was supported in part by the Cincinnati Children’s Research Foundation and its Cincinnati Genomic Control Cohort. The authors acknowledge the use of DNA from the UK Blood Services collection of Common Controls (UKBS-CC collection), which is funded by Wellcome Trust grant 076113/C/04/Z and by the USA NIH research program grant to the National Health Service Blood and Transplant (RP-PG-0310-1002). The authors acknowledge the use of DNA from the British 1958 Birth Cohort collection, which is funded by the UK Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. This work utilized the computational resources of the NIH High Performance Computing (HPC) Biowulf cluster (http://hpc.nih.gov). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the UK Department of Health.
Financial Support and Disclosure of Conflict of Interest: This work was supported by the Intramural Research Programs of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (Z01-AR041198 to MJO) and the National Human Genome Research Institute (Z01-HG200370 to DLK) of the National Institutes of Health (NIH). Additional support was provided by NIH grants R01-AR059049 (AAG), R01-AR061297 (EDM), and R01-AR060893 (SP); Arthritis Research UK Grant 20385 (WT); the German Federal Ministry of Education and Research (BMBF projects 01ER0813 and 01ER0812 to KM and DF, BMBF project 01ER0828 to KM); the Val A. Browning Charitable Foundation (JFB); and the Marcus Foundation (SP). WT and AH are supported by the Manchester Academic Health Sciences Centre (MAHSC) and are funded by the National Institute for Health Research (NIHR) Biomedical Research Unit Funding Scheme. LRW is supported by the NIHR Great Ormond Street Hospital Biomedical Research Centre.
This work did not receive support from any commercial source. Dr. Grom has received consulting fees from Novartis and Novimmune. Dr. Martini is on the speakers bureau for Abbott, Abbvie, Amgen, Baxalta Biosimilars, Biogenidec, Bristol Meyers Squibb, Astellas, Boehringer, Italfarmaco, Janssen, MedImmune, Novartis, NovoNordisk, Pfizer, Sanofi, Roche, Servier, Takeda, UCB Biosciences and GmbH. He has been the recipient of research grants from Abbott, Bristol Meyers Squibb, Francesco Angelini S.P.A., Glaxo Smith Kline, Janssen Biotech Inc., Novartis, Pfizer, Roche, Sanofi, Aventis, Schwarz Biosciences and GmbH. Dr. Gattorno has received consulting fees and research grants Novartis and SOBI. Dr. Prahalad has received consulting fees from Novartis and Medac Pharma. Dr. Zeft owns stock in Merck Pharmaceuticals, OPKO and ARNI. Dr. Ilowite has received consulting fees from SOBI and Novartis. Dr. Mellins has received consulting fees and research grants from Novartis. Dr. Minden has received consulting fees from Chugai and research grants from Pfizer, Abbvie and Roche.
References
- 1.Woo P. Systemic juvenile idiopathic arthritis: diagnosis, management, and outcome. Nat Clin Pract Rheumatol. 2006;2(1):28–34. doi: 10.1038/ncprheum0084. [DOI] [PubMed] [Google Scholar]
- 2.Cimaz R. Systemic-onset juvenile idiopathic arthritis. Autoimmun Rev. 2016 doi: 10.1016/j.autrev.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Mellins ED, Macaubas C, Grom AA. Pathogenesis of systemic juvenile idiopathic arthritis: some answers, more questions. Nat Rev Rheumatol. 2011;7(7):416–26. doi: 10.1038/nrrheum.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nigrovic PA. Review: is there a window of opportunity for treatment of systemic juvenile idiopathic arthritis? Arthritis Rheumatol. 2014;66(6):1405–13. doi: 10.1002/art.38615. [DOI] [PubMed] [Google Scholar]
- 5.Beukelman T. Treatment advances in systemic juvenile idiopathic arthritis. F1000Prime Rep. 2014;6:21. doi: 10.12703/P6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janow G, Schanberg LE, Setoguchi S, Hasselblad V, Mellins ED, Schneider R, et al. The Systemic Juvenile Idiopathic Arthritis Cohort of the Childhood Arthritis and Rheumatology Research Alliance Registry: 2010–2013. J Rheumatol. 2016 doi: 10.3899/jrheum.150997. [DOI] [PubMed] [Google Scholar]
- 7.Stock CJ, Ogilvie EM, Samuel JM, Fife M, Lewis CM, Woo P. Comprehensive association study of genetic variants in the IL-1 gene family in systemic juvenile idiopathic arthritis. Genes Immun. 2008;9(4):349–57. doi: 10.1038/gene.2008.24. [DOI] [PubMed] [Google Scholar]
- 8.Hinks A, Martin P, Thompson SD, Sudman M, Stock CJ, Thomson W, et al. Autoinflammatory gene polymorphisms and susceptibility to UK juvenile idiopathic arthritis. Pediatr Rheumatol Online J. 2013;11(1):14. doi: 10.1186/1546-0096-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fife MS, Gutierrez A, Ogilvie EM, Stock CJ, Samuel JM, Thomson W, et al. Novel IL10 gene family associations with systemic juvenile idiopathic arthritis. Arthritis Res Ther. 2006;8(5):R148. doi: 10.1186/ar2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omoyinmi E, Forabosco P, Hamaoui R, Bryant A, Hinks A, Ursu S, et al. Association of the IL-10 gene family locus on chromosome 1 with juvenile idiopathic arthritis (JIA) PLoS One. 2012;7(10):e47673. doi: 10.1371/journal.pone.0047673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102(7):1369–76. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogilvie EM, Fife MS, Thompson SD, Twine N, Tsoras M, Moroldo M, et al. The −174G allele of the interleukin-6 gene confers susceptibility to systemic arthritis in children: a multicenter study using simplex and multiplex juvenile idiopathic arthritis families. Arthritis Rheum. 2003;48(11):3202–6. doi: 10.1002/art.11300. [DOI] [PubMed] [Google Scholar]
- 13.Scheibel I, Veit T, Neves AG, Souza L, Prezzi S, Machado S, et al. Differential CCR5Delta32 allelic frequencies in juvenile idiopathic arthritis subtypes: evidence for different regulatory roles of CCR5 in rheumatological diseases. Scand J Rheumatol. 2008;37(1):13–7. doi: 10.1080/03009740701631935. [DOI] [PubMed] [Google Scholar]
- 14.De Benedetti F, Meazza C, Vivarelli M, Rossi F, Pistorio A, Lamb R, et al. Functional and prognostic relevance of the −173 polymorphism of the macrophage migration inhibitory factor gene in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2003;48(5):1398–407. doi: 10.1002/art.10882. [DOI] [PubMed] [Google Scholar]
- 15.Lamb R, Thomson W, Ogilvie EM, Donn R. Positive association of SLC26A2 gene polymorphisms with susceptibility to systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2007;56(4):1286–91. doi: 10.1002/art.22444. [DOI] [PubMed] [Google Scholar]
- 16.Bukulmez H, Fife M, Tsoras M, Thompson SD, Twine NA, Woo P, et al. Tapasin gene polymorphism in systemic onset juvenile rheumatoid arthritis: a family-based case-control study. Arthritis Res Ther. 2005;7(2):R285–90. doi: 10.1186/ar1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ombrello MJ, Remmers EF, Tachmazidou I, Grom A, Foell D, Haas JP, et al. HLA-DRB1*11 and variants of the MHC class II locus are strong risk factors for systemic juvenile idiopathic arthritis. Proc Natl Acad Sci U S A. 2015;112(52):15970–5. doi: 10.1073/pnas.1520779112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ombrello MJ, Arthur VL, Remmers EF, Hinks A, Tachmazidou I, Grom AA, et al. Genetic architecture distinguishes systemic juvenile idiopathic arthritis from other forms of juvenile idiopathic arthritis: clinical and therapeutic implications. Ann Rheum Dis. 2016 doi: 10.1136/annrheumdis-2016-210324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 20.Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–81. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501(7468):506–11. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45(10):1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herder C, Nuotio ML, Shah S, Blankenberg S, Brunner EJ, Carstensen M, et al. Genetic determinants of circulating interleukin-1 receptor antagonist levels and their association with glycemic traits. Diabetes. 2014;63(12):4343–59. doi: 10.2337/db14-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farrell MS, Werge T, Sklar P, Owen MJ, Ophoff RA, O’Donovan MC, et al. Evaluating historical candidate genes for schizophrenia. Mol Psychiatry. 2015;20(5):555–62. doi: 10.1038/mp.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gabay C, Gigley J, Sipe J, Arend WP, Fantuzzi G. Production of IL-1 receptor antagonist by hepatocytes is regulated as an acute-phase protein in vivo. Eur J Immunol. 2001;31(2):490–9. doi: 10.1002/1521-4141(200102)31:2<490::aid-immu490>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 30.Prieur AM, Kaufmann MT, Griscelli C, Dayer JM. Specific interleukin-1 inhibitor in serum and urine of children with systemic juvenile chronic arthritis. Lancet. 1987;2(8570):1240–2. doi: 10.1016/s0140-6736(87)91854-x. [DOI] [PubMed] [Google Scholar]
- 31.De Benedetti F, Pignatti P, Massa M, Sartirana P, Ravelli A, Martini A. Circulating levels of interleukin 1 beta and of interleukin 1 receptor antagonist in systemic juvenile chronic arthritis. Clin Exp Rheumatol. 1995;13(6):779–84. [PubMed] [Google Scholar]
- 32.Fall N, Barnes M, Thornton S, Luyrink L, Olson J, Ilowite NT, et al. Gene expression profiling of peripheral blood from patients with untreated new-onset systemic juvenile idiopathic arthritis reveals molecular heterogeneity that may predict macrophage activation syndrome. Arthritis Rheum. 2007;56(11):3793–804. doi: 10.1002/art.22981. [DOI] [PubMed] [Google Scholar]
- 33.Ogilvie EM, Khan A, Hubank M, Kellam P, Woo P. Specific gene expression profiles in systemic juvenile idiopathic arthritis. Arthritis Rheum. 2007;56(6):1954–65. doi: 10.1002/art.22644. [DOI] [PubMed] [Google Scholar]
- 34.Barnes MG, Grom AA, Thompson SD, Griffin TA, Pavlidis P, Itert L, et al. Subtype-specific peripheral blood gene expression profiles in recent-onset juvenile idiopathic arthritis. Arthritis Rheum. 2009;60(7):2102–12. doi: 10.1002/art.24601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macaubas C, Nguyen KD, Peck A, Buckingham J, Deshpande C, Wong E, et al. Alternative activation in systemic juvenile idiopathic arthritis monocytes. Clin Immunol. 2012;142(3):362–72. doi: 10.1016/j.clim.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gattorno M, Piccini A, Lasiglie D, Tassi S, Brisca G, Carta S, et al. The pattern of response to anti-interleukin-1 treatment distinguishes two subsets of patients with systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2008;58(5):1505–15. doi: 10.1002/art.23437. [DOI] [PubMed] [Google Scholar]
- 37.Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, et al. Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N Engl J Med. 2017;377(12):1119–31. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 38.Ridker PM, MacFadyen JG, Thuren T, Everett BM, Libby P, Glynn RJ, et al. Effect of interleukin-1beta inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet. 2017 doi: 10.1016/S0140-6736(17)32247-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.