Abstract
Background
T-box family proteins are DNA-binding transcriptional regulators that play crucial roles during germ layer formation in the early vertebrate embryo. Well-characterized members of this family, including the transcriptional activators Brachyury and VegT, are essential for the proper formation of mesoderm and endoderm, respectively. To date, T-box proteins have not been shown to play a role in the promotion of the third primary germ layer, ectoderm.
Results
Here, we report that the T-box factor Tbx2 is both sufficient and necessary for ectodermal differentiation in the frog Xenopus laevis. Tbx2 is expressed zygotically in the presumptive ectoderm, during blastula and gastrula stages. Ectopic expression of Tbx2 represses mesoderm and endoderm, while loss of Tbx2 leads to inappropriate expression of mesoderm- and endoderm-specific genes in the region fated to give rise to ectoderm. Misexpression of Tbx2 also promotes neural tissue in animal cap explants, suggesting that Tbx2 plays a role in both the establishment of ectodermal fate and its dorsoventral patterning.
Conclusions
Our studies demonstrate that Tbx2 functions as a transcriptional repressor during germ layer formation, and suggest that this activity is mediated in part through repression of target genes that are stimulated, in the mesendoderm, by transactivting T-box proteins. Taken together, our results point to a critical role for Tbx2 in limiting the potency of blastula-stage progenitor cells during vertebrate germ layer differentiation.
Keywords: Tbx2, T-box, germ layer suppression, ectoderm, mesoderm, endoderm
Introduction
In triploblastic organisms, all tissues, with the exception of the germ cells, derive from the three primary germ layers: ectoderm, mesoderm, and endoderm. Studies in embryos of the frog Xenopus laevis have been instrumental in our understanding of vertebrate germ layer formation. Prior to gastrulation, animal pole cells in the Xenopus blastula embryo are specified to become ectoderm, while vegetal pole cells are specified to become endoderm. The mesodermal layer arises from the so-called “marginal zone,” the equatorial cells at the border of the ectoderm- and endoderm-forming regions.
Several members of the T-box family of DNA-binding proteins, including Brachyury and VegT, have been shown to play a prominent role in the appropriate partitioning of the primary germ layers in the Xenopus embryo; notably, these proteins function in this context as transcriptional activators (Naiche et al., 2005). For example, VegT directly stimulates the transcription of endodermal genes, as well as genes encoding Nodal-related factors that are secreted and act on the overlying cells of the equatorial region (the marginal zone) to induce mesoderm (Lustig et al., 1996; Stennard et al., 1996; Horb and Thomsen, 1997; Zhang et al., 1998). Nodal-related mesoderm induction, in turn, stimulates expression of another T-box transcription factor, brachyury, in the marginal zone. In mice, homozygous mutants for brachyury display a loss of posterior mesoderm due to primitive streak defects, lack a notochord, and die in utero (Showell et al., 2004). In Xenopus, injection of brachyury mRNA results in expanded ventral mesoderm formation (Cunliffe and Smith, 1992). Injection of RNA encoding either an Engrailed repressor domain-brachyury fusion construct, or a brachyury mutant truncated at the C-terminus and incapable of transactivation, inhibit mesoderm formation, suggesting that Brachyury plays an essential role in mesoderm formation (Rao, 1994; Conlon et al., 1996); surprisingly, expression of the C-terminal truncation mutant, B304, encoding only the 304 N-terminal amino acid residues exhibits neuralizing activity (Rao, 1994). Another T-box protein, Eomesodermin (Eomes) is also important for mesoendoderm development: ectopic expression of Eomes in blastula stage Xenopus explants or zebrafish embryos activates transcription of mesodermal and endodermal genes, while inhibition of Eomesodermin function leads to defective mesoderm gene activation (Ryan et al., 1996; Conlon et al., 2001; Bjornson et al., 2005). Notably, chromatin profiling studies in Xenopus have demonstrated significant overlap in the genomic binding sites of VegT, Brachyury and Eomes, suggesting that these T-box proteins, and perhaps others, activate common target genes during early embryogenesis (Gentsch et al., 2013).
In the early Xenopus embryo, active restriction of inappropriate germ layer formation plays a critical role in the differentiation of the ectoderm. Maternal factors including Coco, Ectodermin, SRF, and the zygotic factor XFDL56 repress mesoderm in the early embryo via distinct mechanisms, thus allowing for proper ectodermal development (Bell et al., 2003; Dupont et al., 2005; Yun et al., 2007; Sasai et al., 2008). Studies in our lab and by others have demonstrated that misexpression of the Fox family DNA-binding protein Xema/Foxi1e stimulates ectodermal differentiation in cells fated to give rise to mesoderm, while Foxi1e knockdown leads to the ectopic formation of mesoderm and endoderm in the embryonic ectodermal field (Suri et al., 2005; Mir et al., 2007). These studies again point to a requirement for suppression of mesendodermal fate during ectodermal differentiation; as Foxi1e functions as a transcriptional activator, it likely mediates mesendoderm suppression indirectly, via activation of one or more transcriptional targets (Suri et al., 2005). A prominent role for transcriptional repression during ectodermal differentiation has not previously been demonstrated.
We report here the function of the T-box family factor Tbx2 during ectodermal development. tbx2 is expressed in the presumptive ectoderm at blastula and gastrula stages. Misexpression of Tbx2 inhibits differentiation of both ventral ectoderm and growth factor-induced mesendoderm, and promotes neuralization, the latter via repression of Bone Morphogenetic Protein (BMP) pathway activity. Tbx2 functions as a transcriptional repressor, and appears to regulate a set of target genes that overlap with those regulated by transactivating T-box proteins, including Brachyury and VegT. Finally, Tbx2 knockdown promotes ectopic mesendoderm differentiation; taken together, these studies demonstrate that Tbx2 is sufficient and necessary for the transcriptional suppression of inappropriate germ layer formation in the presumptive ectoderm.
Results
Expression of Xenopus tbx2
Previous studies demonstrated a requirement for Foxi1e, functioning as a transcriptional activator, in the suppression of ectopic mesendoderm in Xenopus laevis (Suri et al., 2005). We previously performed gene chip-based screens to isolate transcripts that are downregulated following Foxi1e knockdown; isolates from these screens are expected to include potential mediators of Foxi1e function that are expressed in the presumptive gastrula ectoderm (Sridharan et al., 2012). One transcript that was strongly downregulated following Foxi1e knockdown encodes the previously identified T-box protein, Tbx2 (Bollag et al., 1994). While important roles for Tbx2 have been suggested later in development, the function of Tbx2 during early Xenopus embryogenesis remains elusive (Schlosser and Ahrens, 2004; Cho et al., 2011; Oropeza and Horb, 2012). We performed animal cap assays on embryos injected with translation-blocking Xema morpholino oligonucleotides and confirmed that tbx2 is strongly downregulated following Foxi1e knockdown, confirming the results of our microarray analysis (Fig. 1A). We note that foxi1e overexpression in animal cap explants does not lead to an enrichment of tbx2 expression in gastrula stage explants (data not shown). These data indicate that Foxi1e is necessary but not sufficient for tbx2 expression.
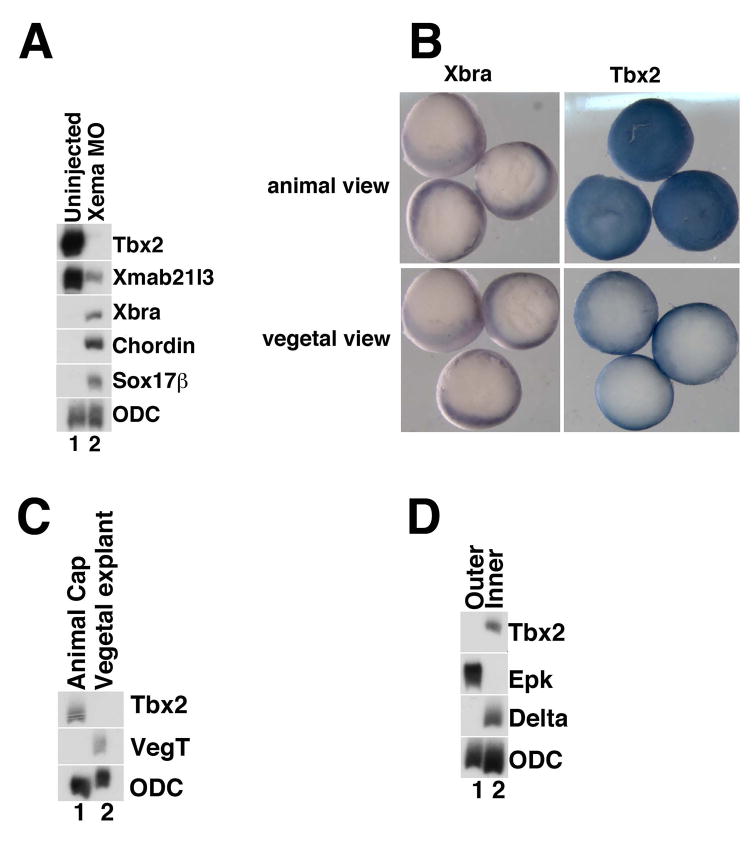
Fig 1. tbx2 expression in the blastula and gastrula stage ectoderm is dependent on Xema/Foxi1e activity.
(A) Xema morpholino-mediated knockdown (Xema MO) inhibits expression of tbx2. RT-PCR analysis of early gastrula stage explants. Ornithine decarboxylase (ODC) was used as a loading control (Bassez et al., 1990). (B) Whole-mount in situ hybridization of early gastrula stage embryos (stage 10.5). Tbx2 expression is seen as a blue stain throughout the animal pole of albino embryos; tbx2 expression is excluded from the vegetal pole. Expression of the panmesodermal marker Xbra is only detected in the marginal zone of gastrula stage embryos, as expected (Smith et al., 1991). (C) RT-PCR analysis of tbx2 in late blastula stage explants. tbx2 is expressed in the animal cap and excluded from vegetal pole explants; vegt is only expressed in vegetal explants, as expected (Lustig et al., 1996; Stennard et al., 1996; Zhang and King, 1996; Horb and Thomsen, 1997). (D) Inner and outer layers of early gastrula stage ectoderm was separated and analyzed by RT-PCR for expression of tbx2. tbx2 is expressed in the inner layer of the ectoderm; Delta is only expressed in the inner layer and serves as a control and epidermal keratin is only expressed in the outer layer (Jonas et al., 1985; Chalmers et al., 2002). All experiments shown or described in this figure, and throughout the manuscript, were performed at least three times.
Earlier studies have reported that Xenopus tbx2 expression initiates zygotically, and have detailed the spatial expression of this transcript from neurula through tadpole stage (Takabatake et al., 2000); our studies support these findings (data not shown). While preparing this manuscript, a study was published by Cho and colleagues, which reported the expression of tbx2 in the ventral ectoderm (Cho et al., 2017). Our whole mount in situ hybridization studies demonstrate that tbx2 is highly expressed throughout the animal pole ectoderm during early gastrula stages, in agreement with expression patterns reported in X. tropicalis (Fig. 1B) (Showell et al., 2006); Xbra expression is restricted to the marginal zone at similar stages, as expected (Fig. 1B) (Smith et al., 1991). To confirm the exclusion of tbx2 transcripts from vegetal pole cells, RT-PCR was performed on gastrula-stage animal and vegetal pole explants. tbx2 expression is observed in animal pole (“cap”) explants and is excluded from the vegetal pole, the latter the site of VegT expression (Fig. 1C) (Lustig et al., 1996; Stennard et al., 1996; Zhang and King, 1996; Horb and Thomsen, 1997; Clements et al., 1999). In contrast to X. tropicalis, where tbx2 expression is observed in the outer (epithelial) layer of the animal cap, X. laevis tbx2 expression is limited to cells in the deep (sensorial) layer (Fig. 1D) (Showell et al., 2006). In sum, tbx2 is expressed zygotically in the embryonic region fated to become ectoderm during Xenopus development.
Tbx2 inhibits mesoendoderm gene expression
Given that other T-box proteins play a critical role in the patterning of the mesodermal and endodermal germ layers, and that Tbx2, to our knowledge, is the first T-box factor with expression throughout the gastrula-stage ectoderm, we endeavored to establish the function of Tbx2 during germ layer development. Based on the spatial distribution of tbx2 and microarray analysis demonstrating that Foxi1e is required for early expression of tbx2, we hypothesized that Tbx2 might, like Foxi1e, suppress mesendoderm. Incubation of blastula stage animal caps with the TGFβ ligand Activin results in a dose-dependent induction of mesendodermal fate (Ariizumi et al., 1991a; Ariizumi et al., 1991b). Animal caps derived from embryos injected with tbx2 RNA at early cleavage stages show a marked reduction in levels of the Activin-induced ventral mesodermal markers Xbra and wnt-8, the dorsal mesodermal markers chordin and goosecoid and the endodermal marker sox17β (Fig. 2A) (Cho et al., 1991; Christian et al., 1991; Smith et al., 1991; Smith and Harland, 1991; Sasai et al., 1994; Hudson et al., 1997). These data demonstrate that tbx2, expressed in the animal pole ectoderm, can suppress mesodermal and endodermal fate.
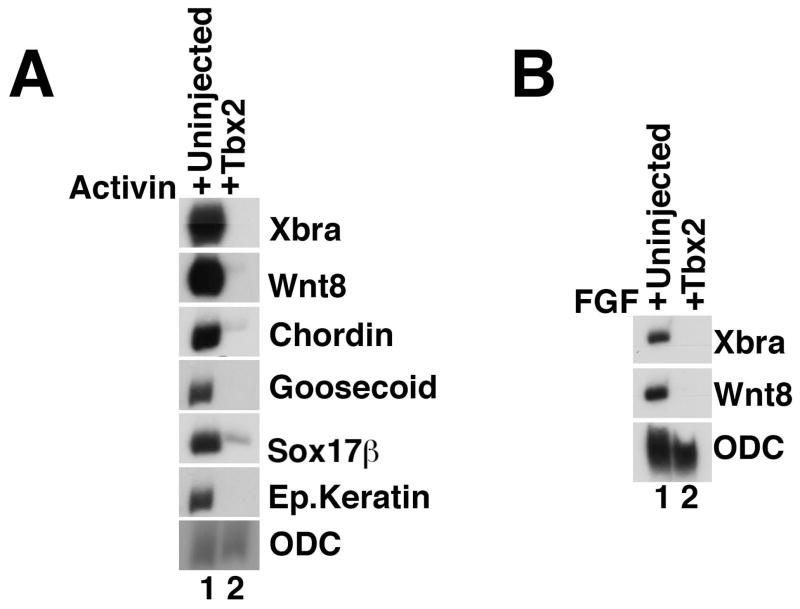
Fig 2. Ectopic Tbx2 suppresses mesendoderm induction.
(A) Tbx2 inhibits Activin-induced mesendoderm. RT-PCR analysis of animal cap explants dissected at late blastula stages and cultured until midgastrula stages. Activin (0.25ng/ml) was added to stage 8 animal caps excised from uninjected embryos or from embryos injected with tbx2 RNA into the animal pole of both blastomeres at the two-cell stage. (B) Inhibition of FGF-mediated mesoderm induction by Tbx2. RT-PCR analysis of animal cap explants dissected at late blastula stages and cultured until midgastrula stages. bFGF (10ng/ml) was added to stage 8 animal caps excised from uninjected embryos or from embryos injected with tbx2 RNA, as listed. Unless otherwise noted, 1ng of tbx2 RNA was used in this and in subsequent experiments.
The primary secreted mesendoderm-inducing signal in Xenopus is believed to be a member of the Nodal-related branch of the TGFβ superfamily (Whitman, 2001); FGF receptor activation can also induce mesoderm in animal cap explants, and FGF is required in the marginal zone for the differentiation of this germ layer (Harland and Gerhart, 1997). Misexpression of tbx2 effectively inhibits FGF-mediated mesoderm induction, as well, suggesting that inhibition of mesoderm by Tbx2 is downstream or independent of the Nodal-related and FGF signaling networks (Fig. 2B).
Tbx2 promotes neural fate
Expression of the epidermal-specific marker, epidermal keratin, is reduced in animal pole explants derived from tbx2 RNA-injected embryos in the presence or absence of Activin (Fig. 2A and data not shown) (Jonas et al., 1985). We therefore examined the role of Tbx2 in the patterning of the ectoderm itself. It is well established that the ectoderm gives rise to two major, distinct fates: epidermis, ventrally, and neural ectoderm, dorsally (Heasman, 2006). Since Tbx2 suppresses mesoderm, endoderm and ventral ectoderm, we asked if Tbx2 might play a role in promoting the development of neural tissue. To test this possibility, animal caps excised from embryos injected with tbx2 RNA were analyzed for neural markers. We find that Tbx2 induces the pre-neural markers sox2 and sox3, indicating that Tbx2 promotes dorsal ectodermal fate (Fig. 3A) (Uwanogho et al., 1995; Wills et al., 2010).
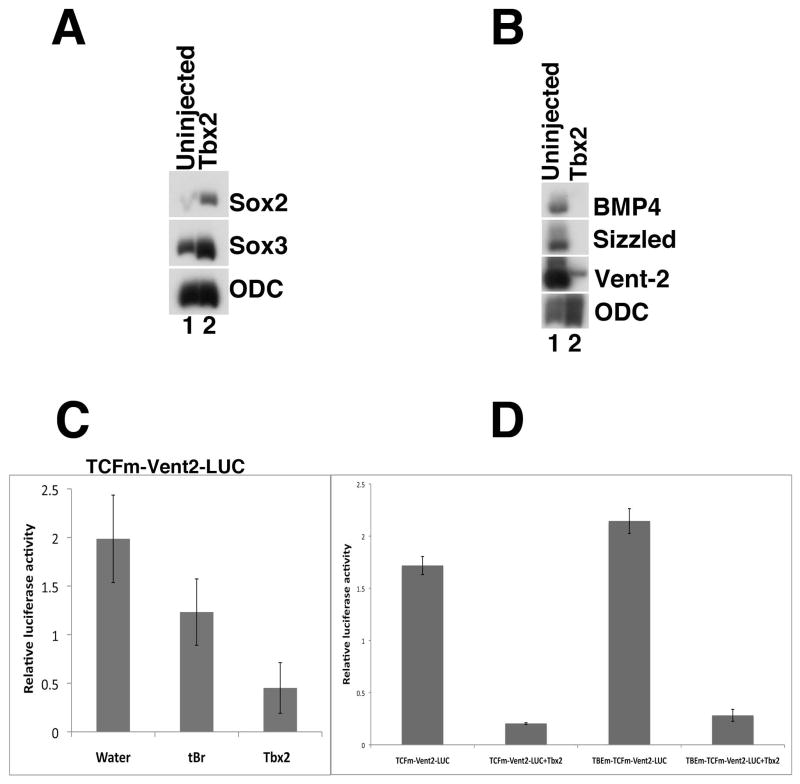
Fig 3. Tbx2 promotes neural fate.
(A) Ectopic tbx2 induces the pre-neural markers sox2 and sox3. RT-PCR analysis was performed on animal cap explants from uninjected embryos and from embryos injected with tbx2 RNA; explants were harvested at gastrula stages. (B) Tbx2 represses the BMP targets bmp4, sizzled and vent-2 (Lee et al., 2002; Collavin and Kirschner, 2003). RT-PCR analysis of mid-gastrula stage animal caps. (C). Tbx2 misexpression inhibits expression of a Vent-2 luciferase reporter fusion protein (TCFm-Vent2-LUC); this construct includes a mutation in a TCF binding site that renders it insensitive to Wnt activation (Hikasa et al., 2010). Truncated BMP receptor (tBR) was used a positive control (Graff et al., 1994). Embryos at the 2-cell stage were injected with 5pg of pRLTK, 50 pg of TCF and 1ng tbx2 RNA or 1ng tBR RNA in the animal pole. (D) Tbx2 misexpression inhibits expression of a Vent-2 luciferase reporter fusion protein with a mutation in the putative T-box binding element (TBEm-TCFm-Vent2-LUC). TCFm-Vent2-LUC and TBEm-TCFm-Vent2-LUC were injected in the absence or presence of tbx2 RNA at the 2-cell stage and collected for analysis of Luciferase expression. Samples of five whole embryo lysates were assayed in triplicate for Firefly and Renilla luciferase activity at mid-gastrula stages. 15 embryos were used for each trial, and each experiment was repeated at least 3 times to confirm the observed trends. Error bars indicate standard error.
Previous studies have shown that a critical, initial step in the specification of neural fate is the inhibition of BMP signaling (Weinstein and Hemmati-Brivanlou, 1999). To better understand how Tbx2 promotes neural fate, we first analyzed the effects of Tbx2 on targets of BMP signaling. We find that Tbx2 suppresses markers of BMP4 activity, including sizzled, Xvent-2 and bmp-4 (Fig. 3B) (Marom et al., 1999; Rastegar et al., 1999; Reversade et al., 2005; Rogers et al., 2009). To further explore this repression of BMP signaling, we examined the effects of Tbx2 on the expression of a modified Xvent-2-Luciferase promoter lacking Wnt-responsive TCF/LEF binding sites (TCFm -vent2-LUC), but responsive to BMP pathway activation (Hikasa et al., 2010). Analysis of whole embryo lysates co-injected with the TCFm -vent2-LUC construct and tbx2 RNA demonstrate that Tbx2 strongly represses TCFm -vent2-LUC expression (Fig. 3C).
Individual T-box proteins bind to distinct T-box binding elements (TBEs) in the promoters of target genes and regulate their expression (Abrahams et al., 2010). Since Tbx2 strongly represses TCFm -vent2-LUC, we examined the promoter sequence for candidate T-box binding elements (TBEs). Our analysis revealed a predicted TBE site (GGGTGA) that, in another context, is recognized by the Tbx2 protein (Carreira et al., 1998). To test whether Tbx2 can directly repress transcription of Xvent2 through the predicted TBE, this site was mutated (GGGTGA to GGGGTC) in TCFm -vent2-LUC using a PCR-based mutagenesis strategy (TBEm-TCFm-vent2-LUC), thereby likely eliminating the affinity of this site for Tbx2. Embryos were injected with either TCFm -vent2-LUC or TBEm-TCFm-vent2-LUC, both in the presence and absence of tbx2 RNA (Fig. 3D). We found that the presence or absence of the putative Tbx2-binding site has no significant effect on the ability of Tbx2 to repress Luciferase expression, suggesting that Tbx2 regulates Xvent-2 expression indirectly, via intermediary factors.
Tbx2 is required for the suppression of mesendoderm in the animal pole
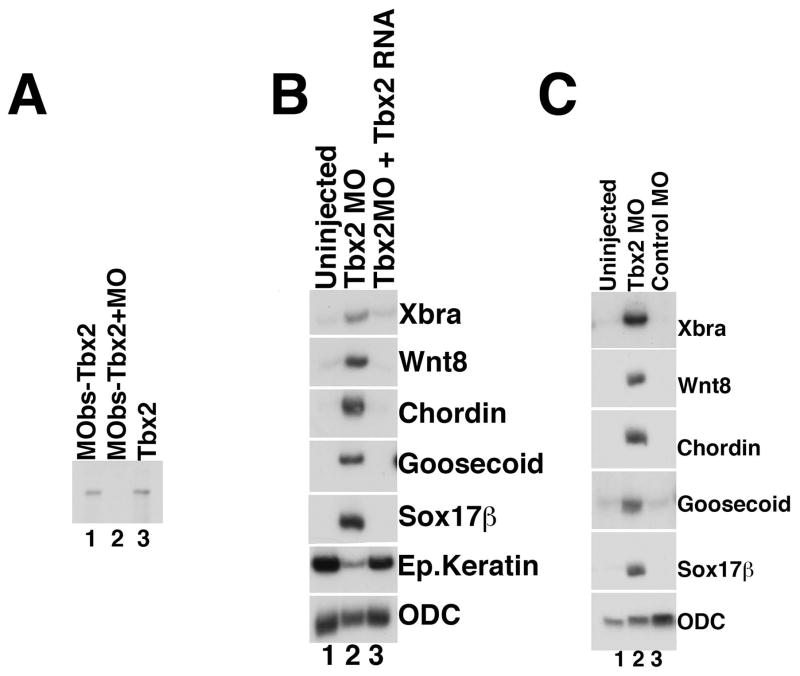
To determine whether Tbx2 is necessary for ectodermal development, we utilized a morpholino oligonucleotide designed to block translation of tbx2 RNA (Tbx2MO). We first confirmed that Tbx2 MO binds and inhibits translation of tbx2 RNA; as expected, Tbx2MO blocks translation of a tbx2 mRNA that contains the Tbx2MO binding site (MObs-Tbx2) in vitro (Fig. 4A, lane 2). Embryos injected with the Tbx2 morpholino do not develop normally beyond gastrulation, and form none of the hallmark embryonic structures seen at subsequent stages of development (data not shown); we were not, however, able to rescue this phenotype and are thus not able to definitively attribute these defects to the loss or reduction of Tbx2. We next attempted to determine the effects of Tbx2 knockdown in explant cultures. We reasoned that if Tbx2 suppresses mesendoderm, loss of Tbx2 might promote the differentiation of ectopic mesendoderm. Consistent with this hypothesis, we find that morpholino-mediated knockdown of Tbx2 leads to elevated expression of mesodermal markers, including Xbra, wnt-8, chordin, and goosecoid, and induction of the endodermal marker sox17β, in animal cap explants (Fig. 4B, lane 2); injection of a control, “scrambled” morpholino does not lead to elevated expression of mesodermal or endodermal markers (Fig. 4C, lane 3). Dose response studies performed at different concentrations of Tbx2 morpholino indicate that loss of Tbx2 leads to elevated ventral mesodermal and dorsal mesodermal markers at higher doses, and only ventral mesodermal markers at low doses (data not shown). To determine whether the changes in gene expression observed in these assays are a consequence of Tbx2 knockdown, we performed rescue experiments and found that tbx2 RNA indeed inhibits Tbx2MO-mediated mesendoderm formation (Fig. 4B, compare lanes 2 and 3). Overall, these results demonstrate that Tbx2 is required for the suppression of inappropriate mesoderm and endoderm in the presumptive ectoderm.
Fig 4. Tbx2 knockdown leads to ectopic expression of mesendodermal marker genes.
(A) Tbx2 morpholino (Tbx2MO) blocks translation of tbx2 RNA in vitro. MObs-Tbx2 is a tbx2 construct that contains the Tbx2MO-binding site. (B) Mesendodermal markers are upregulated in ectodermal explants derived from Tbx2 morpholino-injected embryos. RT-PCR analysis of animal cap explants dissected at late blastula stages and harvested at mid-gastrula stages; embryos were injected with Tbx2 morpholino (80ng) at early cleavage stages. For the rescue experiment, 80ng of Tbx2 morpholino and 1ng of tbx2 RNA were co-injected at early cleavage stages. (C) Injection of a control (80ng), “scrambled” morpholino does not lead to extra-ectodermal gene expression, while injection of Tbx2 morpholino (80ng) results in ectopic expression of both mesodermal and endodermal markers.
Tbx2 functions as a repressor
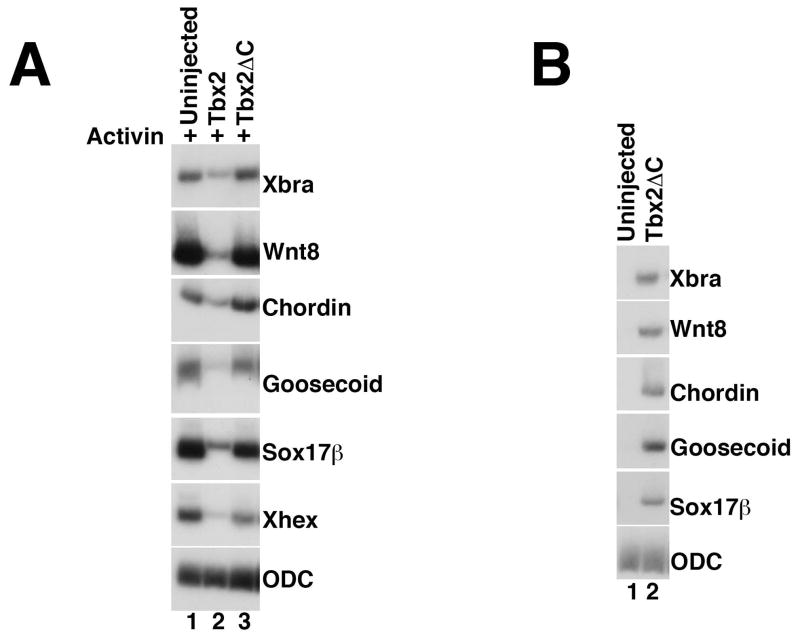
We next sought to identify the mechanisms by which Tbx2 regulates gene expression during early development. Tbx2 has been shown to function as a transcriptional repressor in several distinct biological contexts (Carreira et al., 1998; Jacobs et al., 2000; Sinha et al., 2000); however, studies by Paxton and colleagues have identified both activator and repressor activity for Tbx2 (Paxton et al., 2002). A repressor domain has been identified in the C-terminal domain of Tbx2 (Paxton et al., 2002; Cho et al., 2011); in order to determine whether this region mediates Tbx2 function during mesendoderm suppression, we designed a Tbx2 construct in which the C-terminal amino acids (519 -688) were deleted (Tbx2ΔC). RNA synthesized from this construct did not inhibit mesendoderm induction by Activin; similar concentrations of tbx2 RNA strongly inhibited Activin-mediated induction, suggesting that the Tbx2 C-terminus is necessary for mesendoderm suppression by Tbx2 (Fig. 5A). Notably, injection of RNA encoding tbx2ΔC resulted in ectopic expression of mesodermal markers, in the absence of exogenous growth factors (Fig. 5B); this result suggests that Tbx2ΔC functions as a dominant negative reagent that can inhibit the activity of native Tbx2 in the animal pole ectoderm.
Fig 5. The Tbx2 C-terminus is required for mesendoderm suppression.
(A) Expression of Tbx2ΔC does not suppress mesoderm induction by Activin. RT-PCR analysis of animal caps dissected at late blastula stages and cultured until mid-gastrula stages. tbx2ΔC RNA (1ng) was injected at early cleavage stages, as indicated. Activin (0.5ng/ml) was added to stage 8 animal caps, as indicated (B) Expression of Tbx2ΔC stimulates expression of mesodermal marker genes in the absence of exogenous growth factors. RT-PCR analysis of animal caps dissected at late blastula stages and cultured until mid-gastrula stages. tbx2ΔC (1ng) was injected at early cleavage stages, as indicated.
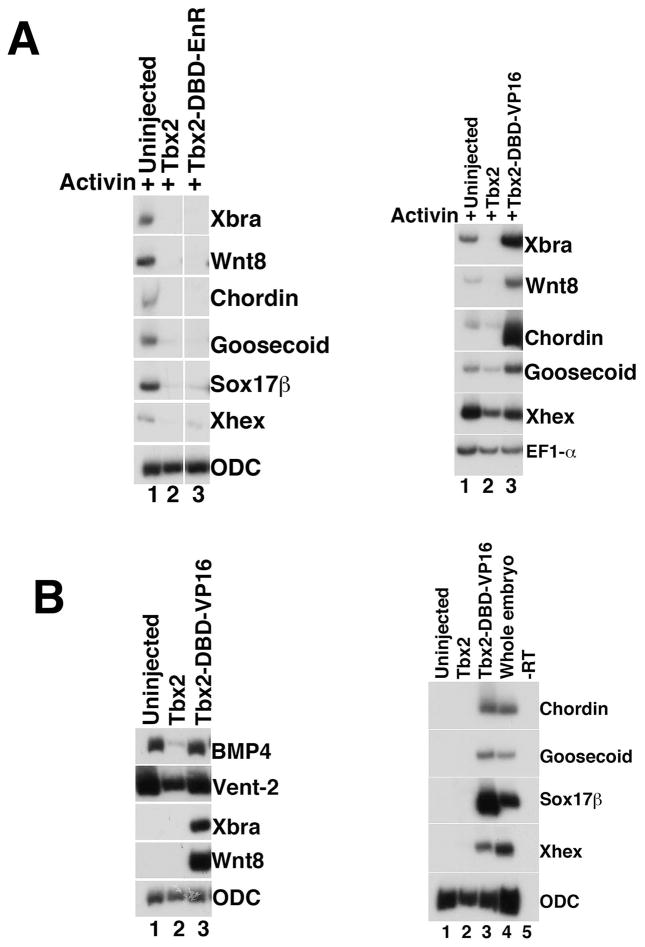
Our demonstration of Tbx2ΔC activity, along with previous studies in other systems demonstrating repressor activity in the Tbx2 C-terminus, suggest that Tbx2 functions as a repressor in the context of germ layer development. To test this hypothesis, we generated constructs in which the DNA-binding domain (DBD) of Tbx2 was fused to either the Engrailed (EnR) repressor domain or the VP16 activator domain (Kessler, 1997); RNA transcribed from these constructs was injected into embryos for analysis in explant assays. In Activin-treated animal caps, tbx2-DBD-EnR expression mimics the repression of mesendodermal genes observed following misexpression of wild-type tbx2, while misexpression of tbx2-DBD-VP16 does not inhibit Activin-mediated induction (Fig. 6A); these results suggest that Tbx2 functions as a repressor during early embryonic development. Animal cap explants from embryos injected with tbx2-DBD-VP16 show no reduction in BMP targets bmp4 and vent-2, as well as markedly increased expression of mesodermal and endodermal genes, in the absence of exogenous growth factors (Fig. 6B); these latter results resemble the effects seen following morpholino-mediated knockdown of Tbx2 (Fig. 4B). Taken together, these data suggest that Tbx2 functions as a repressor during early development.
Fig 6. Tbx2 functions as a repressor during germ layer differentiation.
(A) Expression of Tbx2-DBD-EnR inhibits mesendoderm induction by Activin (left panel); expression of Tbx2-DBD-Vp16 does not inhibit Activin-mediated mesendoderm induction (right panel). RT-PCR analysis of animal caps dissected at late blastula stages and cultured until mid-gastrula stages. Tbx2-DBD-EnR or Tbx2 DBD-VP16 RNA (1ng) was injected at early cleavage stages, as indicated. Activin (0.5ng/ml) was added to stage 8 animal caps, as indicated (B) Expression of Tbx2 DBD-VP16 inhibits BMP target gene expression (left panel), and stimulates expression of mesodermal and endodermal marker genes in the absence of exogenous growth factors (left and right panels). RT-PCR analysis of animal caps dissected at late blastula stages and cultured until mid-gastrula stages. Tbx2-DBD-VP16 RNA (1ng) was injected at early cleavage stages, as indicated. The “-RT” lane contains all reagents except reverse transcriptase and was used as a negative control.
Tbx2 represses direct targets of transactivating T-box proteins
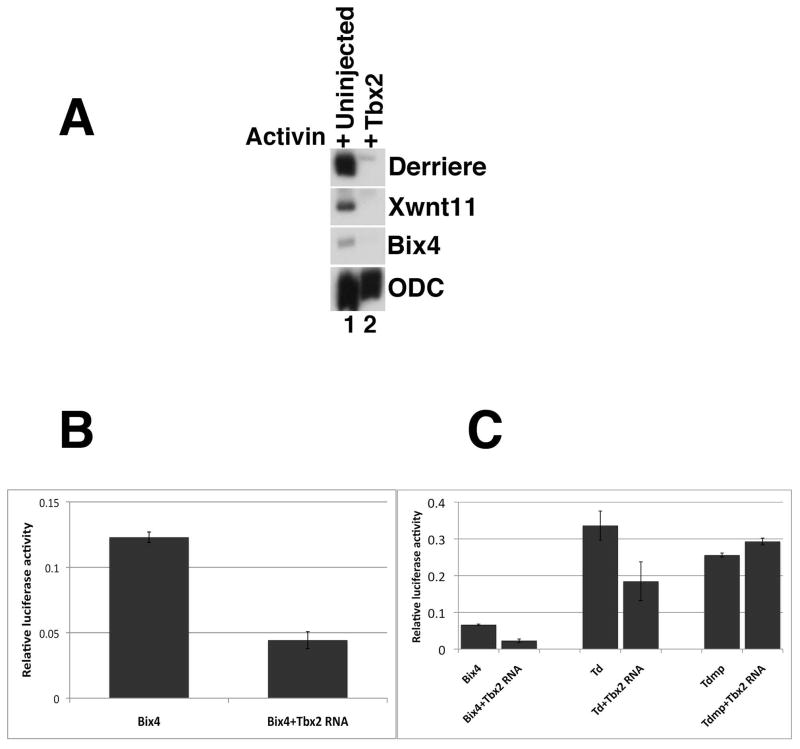
Identification of Tbx2 targets constitutes an important step in understanding how Tbx2 exerts its function. The DNA binding domains of T-box proteins are comprised of approximately 180 amino acid residues, and are highly conserved among closely-related T-box family members (Bollag et al., 1994); for example, the DNA binding domains of Tbx4 and Tbx5 share 94% identity (Papaioannou and Goldin, 2008). Consistently, target DNA sequences for closely related T-box proteins are also highly conserved (Conlon et al., 2001; Abrahams et al., 2010); for example, Brachyury, VegT and Eomesodermin preferentially bind to the same core motif TCACACCT (Tada et al., 1998; Casey et al., 1999; Conlon and Smith, 1999; Tada and Smith, 2001). Since Brachyury, VegT, and Eomesodermin share similarity in their DNA binding domains and bind to the same core sequence, we hypothesized that Tbx2, with a DNA binding domain that is very similar to that of the T-box transactivators listed above, may bind to an overlapping set of genomic targets (Tada et al., 1998; Tada and Smith, 2001; Showell et al., 2004; Abrahams et al., 2010). In support of this model, we find that ectopic Tbx2 represses Activin-induced induction of wnt11, a direct target of Brachyury, derriere, a direct target of VegT and bix4 (Brachyury inducible homeobox containing gene), a target of both Brachyury and VegT (Fig. 7A) (Zhang and King, 1996; Casey et al., 1999; Tada and Smith, 2000; White et al., 2002). These results suggest that Tbx2, VegT, and Brachyury share at least a subset of target genes, and raise the possibility that Tbx2 represses the expression of these genes in the presumptive ectoderm.
Fig 7. Tbx2 represses downstream targets of VegT and Brachyury.
(A) Tbx2 represses expression of derriere, xwnt11, and bix4 (Zhang and King, 1996; Casey et al., 1999). RT-PCR analysis of animal caps dissected at late blastula stages and cultured until mid-gastrula stages, as indicated. Activin (0.5ng/ml) was added to stage 8 animal caps, as indicated. (B) tbx2 represses expression of a Bix4 reporter construct. Embryos at the 2-cell stage were injected with bix4-LUC (20pg) and pRLTK (7pg), in the absence and presence of tbx2 RNA (250 pg). Whole embryo lysates from gastrula stage embryos were assayed in triplicate for Firefly and Renilla luciferase activity. (C) Mutation of three T-box binding sites renders bix4 insensitive to repression by Tbx2. Embryos were injected at the 2-cell stage with pRLTK (7pg) and bix4-LUC (20pg), Td-LUC (20pg), or Tdmp-LUC (20pg) in the absence and presence of tbx2 RNA. Tbx2 blocks the expression of Td-LUC less efficiently than it does bix4-LUC, while mutation of the three T-box sites renders Tdmp-LUC insensitive to repression by Tbx2. Samples of five whole embryo lysates from gastrula stage embryos were assayed in triplicate for Firefly and Renilla luciferase activity. Error bars indicate standard error.
Repression of bix4 by Tbx2 is mediated by T-box binding sites on the bix4 promoter
As described above, both Brachyury and VegT bind to regulatory regions of the bix4 gene and induce its expression in mesoderm and endoderm (Tada et al., 1998; Casey et al., 1999). Based on our earlier results, we reasoned that Tbx2 might directly regulate bix4 expression in the cells of the presumptive ectoderm. To address this possibility, we performed Luciferase assays on lysates derived from gastrula stage embryos that had been injected at early cleavage stages with a bix4-Luciferease reporter fusion construct (bix4-LUC) in the presence or absence of tbx2 RNA (Tada et al., 1998; Casey et al., 1999). As expected, ectopic Tbx2 represses expression of bix4-luciferase (Fig. 7B).
The Bix4 upstream regulatory region contains three T-box binding elements located within 200 base pairs of the transcription start site, labeled previously as Td, Tm and Tp (T-box distal, medial, and proximal, respectively) (Casey et al., 1999). Mutational studies of the three sites suggested that the most distal site (Td) likely plays a role in restricting expression of Bix4 (Casey et al., 1999). We speculated that Tbx2 might bind to the distal site (Td) of bix4 and repress its expression. To test this hypothesis, we injected embryos with the Td-mutant construct in the presence or absence of tbx2 RNA, and performed Luciferase assays on lysates derived at gastrula stages. In agreement with Casey and colleagues (1999), we find that the Td construct alone has higher basal levels of activity than does the wild-type construct (Fig. 7C). Expression of Td is reduced, but still prominent, upon co-injection of tbx2 RNA, suggesting that Tbx2 represses bix4 expression partially, but not completely, via the Td T-box binding site bix4 (Fig. 7C). We next generated a construct in which all three known T-Box binding sites, distal, middle and proximal, were mutated (Tdmp). Injected alone, this construct, like Td, has higher basal levels of activity than does wild-type bix4; Tdmp Luciferase activity, however, is insensitive to Tbx2-mediated repression (Fig. 7C). These results suggest that Tbx2 inhibits bix4 expression, at least in part, via multiple T-box binding sites on the bix4 promoter.
Discussion
We report here the localization and activity of the T-box DNA binding factor Tbx2 during germ layer differentiation. tbx2 is expressed in the deep layer of the presumptive ectoderm in Xenopus laevis embryos. Tbx2 knockdown results in the ectopic expression of mesodermal and endodermal marker genes, while Tbx2 misexpression inhibits epidermal fate, promotes neuralization, and suppresses both Activin-induced mesendoderm induction and FGF-induced mesoderm induction; Tbx2 functions as a transcriptional repressor during mesendoderm suppression. Taken together, our studies demonstrate that appropriate germ layer differentiation in the Xenopus embryo is dependent upon Tbx2-mediated repression of extra-ectodermal fate.
Homozygous inactivation of tbx2 in the mouse results in embryonic lethality (Harrelson et al., 2004). As the earliest reported expression of mouse tbx2 is in the allantois at post-gastrula stage E8.5, it is not surprising that disruption of germ layer patterning has not been observed in these animals (Mahlapuu et al., 2001). Other T-box factors including the closely-related Tbx3, expressed in the pre-gastrula mouse epiblast, may contribute to suppression of ectopic mesendoderm during mammalian embryogenesis (Weidgang et al., 2013). We note that homozygous inactivation of tbx3 does not disrupt germ layer patterning, suggesting the involvement of additional or alternative T-box factors in mammalian germ layer suppression (Davenport et al., 2003).
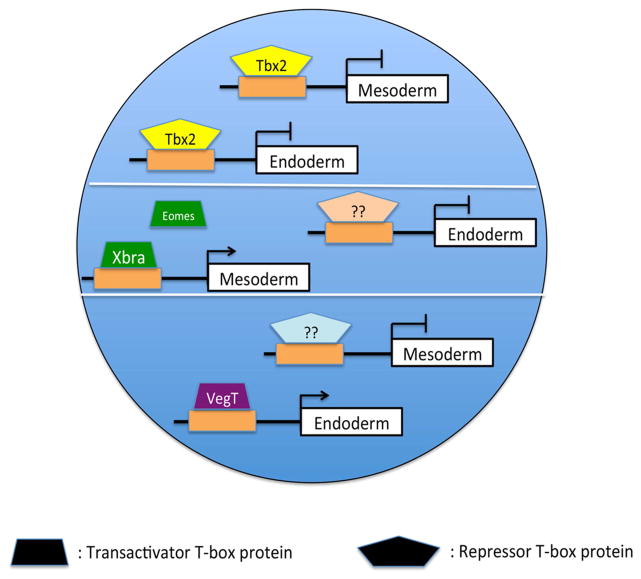
In Xenopus, endoderm and mesoderm differentiation are regulated by multiple transactivating T-box proteins, including VegT, Brachyury, and Eomesodermin, all of which share DNA target sequences similar to that of Tbx2 (Ryan et al., 1996; Gentsch et al., 2013). These T-box factors show restricted expression, with tbx2 expressed in the presumptive ectoderm, and brachyury, vegT and eomesodermin expressed in the presumptive endoderm and/or mesoderm (Showell et al., 2004). We report here that repression of bix4, a previously described target of both Brachyury and VegT, by Tbx2 is mediated via multiple T-box binding sites in the bix4 promoter (Tada et al., 1998; Casey et al., 1999). It is well established that transactivating T-box proteins bind to and induce expression of mesendodermal target genes, including bix4 (Tada and Smith, 2001); our studies build on and refine this model of T-box function, and suggest that repressor T-box proteins including Tbx2 suppress expression of these same genes, and thus inhibit mesendodermal differentiation, in the cells of the presumptive ectoderm in Xenopus (Fig. 8).
Fig 8. A model of Tbx2-mediated ectodermal specification.
Tbx2 binds to and represses target genes in animal pole progenitor cells, thereby inhibiting mesoderm and endoderm formation. In the marginal zone and vegetal pole, other T-box proteins bind to and activate an overlapping set of target genes, stimulating mesodermal and endodermal differentiation.
Direct repression of mesendodermal target genes may not be the only mechanism by which Tbx2 inhibits inappropriate germ layer differentiation. During preparation of this manuscript, Cho and colleagues reported that Tbx2 limits FGF-mediated neural caudalization via transcriptional suppression of the flrt3 gene (Cho et al., 2017). flrt3 does not appear to be expressed in the presumptive ectoderm at gastrula stages (Bottcher et al., 2004); nevertheless, this finding raises the possibility that Tbx2 inhibits mesendoderm both through direct binding to mesendoderm-specific target genes and through indirect regulation of FGF signaling, the latter shown previously to be required for mesodermal differentiation (Heasman, 2006).
Suppression of ectodermal BMP activity by Tbx2 may be mediated through direct repression of BMP target genes. The absence of de-repression following mutation of the sole candidate TBE site within the region of the vent-2 promoter shown to be sufficient for regulation by BMP, however, suggests that an alternate mechanism may underlie BMP signal inhibition by Tbx2 (Henningfeld et al., 2000; Lee et al., 2002). Studies have demonstrated that the T-box proteins Brachyury and Eomesodermin interact with the BMP signal transducer Smad1 during mesodermal and endodermal differentiation, respectively (Messenger et al., 2005; Faial et al., 2015). Another T-box protein, Tbx20, has been shown to inhibit BMP signaling by directly binding to Smad1 and Smad5, sequestering them from association with the co-factor Smad4; this association is mediated through the Tbx20 T-box domain (Henningfeld et al., 2000; Singh et al., 2009). These studies raise the possibility that Tbx2 may also physically associate with Smad1 or other intracellular mediators of BMP receptor activity to abrogate BMP signaling.
Although we have established a requirement for Tbx2 in the suppression of ectopic mesendoderm, the physiological significance of BMP target gene repression in this process is not clear. Tbx2-mediated BMP inhibition may contribute primarily to one of several previously described mechanisms that limit BMP activity in, and thus limit ventralization of, the early embryo prior to the secretion of extracellular antagonists from the Organizer (Onichtchouk et al., 1999; Zhu et al., 1999; Bell et al., 2003; Dupont et al., 2005; Heasman, 2006; Sridharan et al., 2012). As epidermal induction occurs in regions of the embryo that express native tbx2, however, it is at present difficult to conclude that Tbx2-mediated BMP suppression plays a central role in neuralization of the dorsal ectoderm.
Experimental Procedures
Gene chip analysis and isolation of Tbx2
Gene chip analysis was performed as described in Sridharan et al., 2012. Briefly, RNA from 80 animal cap explants, cultured to stage 11, derived from embryos injected with 1ng xema/foxi1e or β-galactosidase RNA, 63ng 1:2 Xema MO1: Xema MO2, or 62.5ng scrambled morpholino (CMO) were used to generate hybridization probes for use on Affymetrix GeneChip Xenopus laevis Genome Arrays (Suri et al., 2005; Sridharan et al., 2012). Microarray data were normalized by RMA (Irizarry et al., 2003) and analyzed using the affylmGUI Bioconductor package (Wettenhall et al., 2006).
Xenopus tbx2 was isolated in a microarray screen to identify transcriptional targets of Foxi1e (Suri et al., 2005; Sridharan et al., 2012). The probe set (XI.931.1) corresponding to tbx2 was down-regulated in Foxi1e morpholino-injected animal pole ectodermal explants at stage 11, when compared to uninjected or control scrambled morpholino-injected samples, respectively. Full-length tbx2 cDNA was a gift from the lab of Jin Kwan Han (Cho et al., 2011).
Preparation of Tbx2ΔC, repressor and activator fusion constructs
The Tbx2ΔC construct was designed to contain amino acids 1-518, following deletion of sequence encoding the putative repressor domain (aa 519 to 688) (Cho et al., 2011). For the Tbx2-DBD-VP16 and Tbx2-DBD-EnR constructs, sequence encoding residues 410-490 of the VP16 activator (Kessler, 1997) and residues 1-298 of the Drosophila Engrailed repressor (Kessler, 1997), respectively, were cloned downstream of the Tbx2 DNA-binding domain (sequence encoding residues 91-279).
RNA preparation, explant dissection, and cell culture
RNA was synthesized in vitro in the presence of cap analog using the mMessage mMachine kit (Ambion). Microinjection, explant dissection, and cell culture were performed as described (Hemmati-Brivanlou and Melton, 1994; Wilson and Hemmati-Brivanlou, 1995).
Luciferase assays
The BMP-responsive, Wnt-unresponsive Vent-2 promoter (TCFm-Luc) was a gift from the S. Sokol laboratory (Hikasa et al., 2010). Mutation in the putative T-box Binding Element (TBE) (GGGTGA to GGGGTC) was generated by GENEWIZ and independently sequenced. Embryos were injected at the 2-cell stage with 50 pg of TCFm-Luc reporter plasmid, with or without tbx2 RNA, (Hikasa et al., 2010); 5 pg of Renilla luciferase reporter was co-injected with all samples as an internal control.
The Bix4-Luc promoter was a gift from Elena M Silva (Tada et al., 1998; Casey et al., 1999). The Td construct was generated by GENEWIZ and independently sequenced. The mutations in Td-LUC were introduced as described in (Casey et al., 1999). The triple mutant, Tdmp-LUC, in which all three characterized T-elements in bix4 were mutated, was generated by GENEWIZ and independently sequenced; Tm and Tp mutations were introduced as described (Casey et al., 1999). For all luciferase assays, samples of five embryos each were collected in triplicate at stage 11 for analysis. Embryos were lysed in 200ul passive lysis buffer (Promega); 20 μl was assayed for luminescence.
Whole mount in situ hybridization
Whole mount in situ hybridization was carried out using standard protocols (Harland, 1991). BM Purple (Roche) was used for chromogenic reactions.
Morpholinos
Morpholino antisense oligonucleotides (GeneTools LLC) were designed to hybridize to the 5′ region of tbx2 mRNA to block translation. Morpholinos were heated at 65°C for 5 minutes, and then cooled on ice prior to microinjection. The Tbx2MO used in this study was designed as follows: 5′-TTCCAGAGCGAGATAGAGCCCTGTC. Since there is one nucleotide difference between the tbx2.L and tbx2.S alloalleles at the tbx2 morpholino target sequence, it is possible that the Tbx2MO preferentially inhibits translation of tbx2.L.
A morpholino-sensitive Tbx2 construct, MObs-Tbx2, was generated by PCR using KOD Hot Start Polymerase (EMD, Rockland, MA), with the following primers: MO-Tbx2-FP (Phosphorylated) TTTGTGTATGCACCGATGAGAGATCCAGCTTTCCCGGGGG and MO-Tbx2a-RP: TCCAGGAGGGAATGCAAAAAGAACAAGTAGCTTGTATTC.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Xenopus laevis embryos were staged according to Nieuwkoop and Faber, 1967 and harvested at appropriate stages according to morphological criteria. RNA was prepared using RNA Bee RNA isolation reagent (Tel-Test Inc.). RT-PCR was performed as described (Wilson and Hemmati-Brivanlou, 1995). Primer Sequences designed for this study are as follows: Xmab21l3-F: GGAGGATGAGTAGGATAAAGTGGTG, Xmab21l3-R: TATGCCGCTCTTCTGATGCCCAG; Xtbx2-F: CCTGGACAGCTGCCTTATTC, Xtbx2-R: CGGCTTCAACTAAGGATGGA; Bix4-F: AGGACCTCCTGTCTTGCCC, Bix4-R:AG ATGCTACAGGCTGGAGCAA.
The Xenopus laevis genome contains two homeologs for the tbx2 gene, designated as tbx2.L and tbx2.S. While the 3′UTR of tbx2.S is not annotated, there is a high degree of homology between the two homeologs; although the primers used in this study were designed to detect tbx2.L transcripts, these primers may also amplify sequences derived from tbx2.S transcripts.
All other primer sequences are as described: ODC, Xbra, Wnt8, chordin, goosecoid and Sox17β (Suri et al., 2004), sox2, sox3 and Sizzled (Sridharan et al., 2012). VegT (Horb and Thomsen, 1997; Zhang et al., 1998); Delta (Tao et al., 2005); Epidermal Keratin (Takahashi et al., 2015); Bmp4 (Fainsod et al., 1997); Xvent2 (Miyazaki et al., 2012); XWnt11 (Afouda and Hoppler, 2011); Derriere (Sun et al., 1999).
Acknowledgments
This work is supported by PHS Grants R03-HD077015 (DCW) and R15GM124577 (DCW) and with funds from Queens College of the City University of New York and the Professional Staff Congress-City University of New York.
We thank Dr. E.S. Casey and Dr. J.C. Smith for the Bix4-Luc construct. We thank Dr. S. Sokol for the Vent2-Luc construct. We thank Dr. J.K. Han for providing us with Xenopus Tbx2. We thank Dr. D.A. Melton for the truncated BMP receptor (tBr). In addition, we thank current and former members of the lab, particularly Tomomi Haremaki and Ye Jin, for thoughtful discussion, and Tomomi Haremaki for his contributions to early stages of this project. This work is supported by PHS Grants R03-HD077015 (DCW) and R15GM124577 (DCW) and with funds from Queens College of the City University of New York and the Professional Staff Congress-City University of New York.
References
- Abrahams A, Parker MI, Prince S. The T-box transcription factor Tbx2: its role in development and possible implication in cancer. IUBMB Life. 2010;62:92–102. doi: 10.1002/iub.275. [DOI] [PubMed] [Google Scholar]
- Afouda BA, Hoppler S. Different requirements for GATA factors in cardiogenesis are mediated by non-canonical Wnt signaling. Dev Dyn. 2011;240:649–662. doi: 10.1002/dvdy.22570. [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Moriya N, Uchiyama H, Asashima M. Concentration-dependent inducing activity of activin A. Roux Arch Dev Biol. 1991a;200:230–233. doi: 10.1007/BF00361342. [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Sawamura K, Uchiyama H, Asashima M. Dose and time-dependent mesoderm induction and outgrowth formation by activin A in Xenopus laevis. Int J Dev Biol. 1991b;35:407–414. [PubMed] [Google Scholar]
- Bassez T, Paris J, Omilli F, Dorel C, Osborne HB. Post-transcriptional regulation of ornithine decarboxylase in Xenopus laevis oocytes. Development. 1990;110:955–962. doi: 10.1242/dev.110.3.955. [DOI] [PubMed] [Google Scholar]
- Bell E, Muñoz-Sanjuán I, Altmann CR, Vonica A, Brivanlou AH. Cell fate specification and competence by Coco, a maternal BMP, TGFβ and Wnt inhibitor. Development. 2003;130:1381. doi: 10.1242/dev.00344. [DOI] [PubMed] [Google Scholar]
- Bjornson CRR, Griffin KJP, Farr GH, Terashima A, Himeda C, Kikuchi Y, Kimelman D. Eomesodermin Is a Localized Maternal Determinant Required for Endoderm Induction in Zebrafish. Developmental Cell. 2005;9:523–533. doi: 10.1016/j.devcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Bollag RJ, Siegfried Z, Cebra-Thomas JA, Garvey N, Davison EM, Silver LM. An ancient family of embryonically expressed mouse genes sharing a conserved protein motif with the T locus. Nat Genet. 1994;7:383–389. doi: 10.1038/ng0794-383. [DOI] [PubMed] [Google Scholar]
- Bottcher RT, Pollet N, Delius H, Niehrs C. The transmembrane protein XFLRT3 forms a complex with FGF receptors and promotes FGF signalling. Nat Cell Biol. 2004;6:38–44. doi: 10.1038/ncb1082. [DOI] [PubMed] [Google Scholar]
- Carreira S, Dexter TJ, Yavuzer U, Easty DJ, Goding CR. Brachyury-related transcription factor Tbx2 and repression of the melanocyte-specific TRP-1 promoter. Mol Cell Biol. 1998;18:5099–5108. doi: 10.1128/mcb.18.9.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey ES, Tada M, Fairclough L, Wylie CC, Heasman J, Smith JC. Bix4 is activated directly by VegT and mediates endoderm formation in Xenopus development. Development. 1999;126:4193–4200. doi: 10.1242/dev.126.19.4193. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Welchman D, Papalopulu N. Intrinsic differences between the superficial and deep layers of the Xenopus ectoderm control primary neuronal differentiation. Dev Cell. 2002;2:171–182. doi: 10.1016/s1534-5807(02)00113-2. [DOI] [PubMed] [Google Scholar]
- Cho GS, Choi SC, Park EC, Han JK. Role of Tbx2 in defining the territory of the pronephric nephron. Development. 2011;138:465–474. doi: 10.1242/dev.061234. [DOI] [PubMed] [Google Scholar]
- Cho GS, Park DS, Choi SC, Han JK. Tbx2 regulates anterior neural specification by repressing FGF signaling pathway. Dev Biol. 2017;421:183–193. doi: 10.1016/j.ydbio.2016.11.020. [DOI] [PubMed] [Google Scholar]
- Cho KWY, Blumberg B, Steinbeisser H, De Robertis EM. Molecular Nature of Spemann’s Organizer: the Role of the Xenopus Homeobox Gene goosecoid. Cell. 1991;67:1111–1120. doi: 10.1016/0092-8674(91)90288-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian JL, McMahon JA, McMahon AP, Moon RT. Xwnt-8, a Xenopus Wnt-1/int-1-related gene responsive to mesoderm-inducing growth factors, may play a role in ventral mesodermal patterning during embryogenesis. Development. 1991;111:1045–1055. doi: 10.1242/dev.111.4.1045. [DOI] [PubMed] [Google Scholar]
- Clements D, Friday RV, Woodland HR. Mode of action of VegT in mesoderm and endoderm formation. Development. 1999;126:4903–4911. doi: 10.1242/dev.126.21.4903. [DOI] [PubMed] [Google Scholar]
- Collavin L, Kirschner MW. The secreted Frizzled-related protein Sizzled functions as a negative feedback regulator of extreme ventral mesoderm. Development. 2003;130:805–816. doi: 10.1242/dev.00306. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Fairclough L, Price BM, Casey ES, Smith JC. Determinants of T box protein specificity. Development. 2001;128:3749–3758. doi: 10.1242/dev.128.19.3749. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Sedgwick SG, Weston KM, Smith JC. Inhibition of Xbra transcription activation causes defects in mesodermal patterning and reveals autoregulation of Xbra in dorsal mesoderm. Development. 1996;122:2427. doi: 10.1242/dev.122.8.2427. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Smith JC. Interference with brachyury function inhibits convergent extension, causes apoptosis, and reveals separate requirements in the FGF and activin signalling pathways. Dev Biol. 1999;213:85–100. doi: 10.1006/dbio.1999.9330. [DOI] [PubMed] [Google Scholar]
- Cunliffe V, Smith JC. Ectopic mesoderm formation in Xenopus embryos caused by widespread expression of a Brachyury homologue. Nature. 1992;358:427–430. doi: 10.1038/358427a0. [DOI] [PubMed] [Google Scholar]
- Davenport TG, Jerome-Majewska LA, Papaioannou VE. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development. 2003;130:2263–2273. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S. Germ-Layer Specification and Control of Cell Growth by Ectodermin, a Smad4 Ubiquitin Ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Faial T, Bernardo AS, Mendjan S, Diamanti E, Ortmann D, Gentsch GE, Mascetti VL, Trotter MW, Smith JC, Pedersen RA. Brachyury and SMAD signalling collaboratively orchestrate distinct mesoderm and endoderm gene regulatory networks in differentiating human embryonic stem cells. Development. 2015;142:2121–2135. doi: 10.1242/dev.117838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fainsod A, Deissler K, Yelin R, Marom K, Epstein M, Pillemer G, Steinbeisser H, Blum M. The dorsalizing and neural inducing gene follistatin is an antagonist of BMP-4. Mech Dev. 1997;63:39–50. doi: 10.1016/s0925-4773(97)00673-4. [DOI] [PubMed] [Google Scholar]
- Gentsch George E, Owens Nick D, Martin Stephen R, Piccinelli P, Faial T, Trotter Matthew W, Gilchrist Michael J, Smith James C. In Vivo T-Box Transcription Factor Profiling Reveals Joint Regulation of Embryonic Neuromesodermal Bipotency. Cell Reports. 2013;4:1185–1196. doi: 10.1016/j.celrep.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JM, Thies RS, Song JJ, Celeste AJ, Melton DA. Studies with a Xenopus BMP receptor suggest that ventral mesoderm-inducing signals override dorsal signals in vivo. Cell. 1994;79:169–179. doi: 10.1016/0092-8674(94)90409-x. [DOI] [PubMed] [Google Scholar]
- Harland R, Gerhart J. Formation and function of Spemann’s organizer. Annu Rev Cell Dev Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Harrelson Z, Kelly RG, Goldin SN, Gibson-Brown JJ, Bollag RJ, Silver LM, Papaioannou VE. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development. 2004:131. doi: 10.1242/dev.01378. [DOI] [PubMed] [Google Scholar]
- Heasman J. Maternal determinants of embryonic cell fate. Semin Cell Dev Biol. 2006;17:93–98. doi: 10.1016/j.semcdb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Melton DA. Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell. 1994;77:273–281. doi: 10.1016/0092-8674(94)90319-0. [DOI] [PubMed] [Google Scholar]
- Henningfeld KA, Rastegar S, Adler G, Knochel W. Smad1 and Smad4 are components of the bone morphogenetic protein-4 (BMP-4)-induced transcription complex of the Xvent-2B promoter. J Biol Chem. 2000;275:21827–21835. doi: 10.1074/jbc.M000978200. [DOI] [PubMed] [Google Scholar]
- Hikasa H, Ezan J, Itoh K, Li X, Klymkowsky MW, Sokol SY. Regulation of TCF3 by Wnt-dependent phosphorylation during vertebrate axis specification. Dev Cell. 2010;19:521–532. doi: 10.1016/j.devcel.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horb ME, Thomsen GH. A vegetally localized T-box transcription factor in Xenopus eggs specifies mesoderm and endoderm and is essential for embryonic mesoderm formation. Development. 1997;124:1689–1698. doi: 10.1242/dev.124.9.1689. [DOI] [PubMed] [Google Scholar]
- Hudson C, Clements D, Friday RV, Stott D, Woodland HR. Xsox17alpha and -beta mediate endoderm formation in Xenopus. Cell. 1997;91:397–405. doi: 10.1016/s0092-8674(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Keblusek P, Robanus-Maandag E, Kristel P, Lingbeek M, Nederlof PM, van Welsem T, van de Vijver MJ, Koh EY, Daley GQ, van Lohuizen M. Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nat Genet. 2000:26. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- Jonas E, Sargent TD, Dawid IB. Epidermal keratin gene expressed in embryos of Xenopus laevis. Proc Natl Acad Sci U S A. 1985;82:5413–5417. doi: 10.1073/pnas.82.16.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler DS. Siamois is required for formation of Spemann’s organizer. Proc Natl Acad Sci U S A. 1997;94:13017–13022. doi: 10.1073/pnas.94.24.13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Park MJ, Lee SY, Hwang YS, Lee H, Roh DH, Kim JI, Park JB, Lee JY, Kung HF, Kim J. Transcriptional regulation of Xbr-1a/Xvent-2 homeobox gene: analysis of its promoter region. Biochem Biophys Res Commun. 2002;298:815–823. doi: 10.1016/s0006-291x(02)02570-6. [DOI] [PubMed] [Google Scholar]
- Lustig KD, Kroll KL, Sun EE, Kirschner MW. Expression cloning of a Xenopus T-related gene (Xombi) involved in mesodermal patterning and blastopore lip formation. Development. 1996;122:4001. doi: 10.1242/dev.122.12.4001. [DOI] [PubMed] [Google Scholar]
- Mahlapuu M, Ormestad M, Enerback S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128:155. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- Marom K, Fainsod A, Steinbeisser H. Patterning of the mesoderm involves several threshold responses to BMP-4 and Xwnt-8. Mech Dev. 1999;87:33–44. doi: 10.1016/s0925-4773(99)00137-9. [DOI] [PubMed] [Google Scholar]
- Messenger NJ, Kabitschke C, Andrews R, Grimmer D, Nunez Miguel R, Blundell TL, Smith JC, Wardle FC. Functional specificity of the Xenopus T-domain protein Brachyury is conferred by its ability to interact with Smad1. Dev Cell. 2005;8:599–610. doi: 10.1016/j.devcel.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Mir A, Kofron M, Zorn AM, Bajzer M, Haque M, Heasman J, Wylie CC. FoxI1e activates ectoderm formation and controls cell position in the Xenopus blastula. Development. 2007;134:779–788. doi: 10.1242/dev.02768. [DOI] [PubMed] [Google Scholar]
- Miyazaki A, Ishii K, Yamashita S, Nejigane S, Matsukawa S, Ito Y, Onuma Y, Asashima M, Michiue T. mNanog possesses dorsal mesoderm-inducing ability by modulating both BMP and Activin/nodal signaling in Xenopus ectodermal cells. PLoS One. 2012;7:e46630. doi: 10.1371/journal.pone.0046630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-Box Genes in Vertebrate Development. Annual Review of Genetics. 2005;39:219–239. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- Onichtchouk D, Chen YG, Dosch R, Gawantka V, Delius H, Massague J, Niehrs C. Silencing of TGF-beta signalling by the pseudoreceptor BAMBI. Nature. 1999;401:480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]
- Oropeza D, Horb M. Transient expression of Ngn3 in Xenopus endoderm promotes early and ectopic development of pancreatic beta and delta cells. Genesis (New York, Ny: 2000) 2012;50:271–285. doi: 10.1002/dvg.20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxton C, Zhao H, Chin Y, Langner K, Reecy J. Murine Tbx2 contains domains that activate and repress gene transcription. Gene. 2002;283:117–124. doi: 10.1016/s0378-1119(01)00878-2. [DOI] [PubMed] [Google Scholar]
- Rao Y. Conversion of a mesodermalizing molecule, the Xenopus Brachyury gene, into a neuralizing factor. Genes & Development. 1994;8:939–947. doi: 10.1101/gad.8.8.939. [DOI] [PubMed] [Google Scholar]
- Rastegar S, Friedle H, Frommer G, Knochel W. Transcriptional regulation of Xvent homeobox genes. Mech Dev. 1999;81:139–149. doi: 10.1016/s0925-4773(98)00239-1. [DOI] [PubMed] [Google Scholar]
- Reversade B, Kuroda H, Lee H, Mays A, De Robertis EM. Depletion of Bmp2, Bmp4, Bmp7 and Spemann organizer signals induces massive brain formation in Xenopus embryos. Development. 2005;132:3381–3392. doi: 10.1242/dev.01901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Moody SA, Casey ES. Neural induction and factors that stabilize a neural fate. Birth Defects Res C Embryo Today. 2009;87:249–262. doi: 10.1002/bdrc.20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Garrett N, Mitchell A, Gurdon JB. Eomesodermin, a Key Early Gene in Xenopus Mesoderm Differentiation. Cell. 1996;87:989–1000. doi: 10.1016/s0092-8674(00)81794-8. [DOI] [PubMed] [Google Scholar]
- Sasai N, Yakura R, Kamiya D, Nakazawa Y, Sasai Y. Ectodermal factor restricts mesoderm differentiation by inhibiting p53. Cell. 2008;133:878–890. doi: 10.1016/j.cell.2008.03.035. [DOI] [PubMed] [Google Scholar]
- Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont LK, De Robertis EM. Xenopus chordin: a novel dorsalizing factor activated by organizer-specific homeobox genes. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser G, Ahrens K. Molecular anatomy of placode development in Xenopus laevis. Developmental Biology. 2004;271:439–466. doi: 10.1016/j.ydbio.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Showell C, Binder O, Conlon FL. T-box Genes in Early Embryogenesis. Developmental dynamics : an official publication of the American Association of Anatomists. 2004;229:201–218. doi: 10.1002/dvdy.10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showell C, Christine KS, Mandel EM, Conlon FL. Developmental expression patterns of Tbx1, Tbx2, Tbx5, and Tbx20 in Xenopus tropicalis. Dev Dyn. 2006;235:1623–1630. doi: 10.1002/dvdy.20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Horsthuis T, Farin HF, Grieskamp T, Norden J, Petry M, Wakker V, Moorman AF, Christoffels VM, Kispert A. Tbx20 interacts with smads to confine tbx2 expression to the atrioventricular canal. Circ Res. 2009;105:442–452. doi: 10.1161/CIRCRESAHA.109.196063. [DOI] [PubMed] [Google Scholar]
- Sinha S, Abraham S, Gronostajski RM, Campbell CE. Differential DNA binding and transcription modulation by three T-box proteins, T, TBX1 and TBX2. Gene. 2000;258:15–29. doi: 10.1016/s0378-1119(00)00417-0. [DOI] [PubMed] [Google Scholar]
- Smith JC, Price BM, Green JB, Weigel D, Herrmann BG. Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell. 1991;67:79–87. doi: 10.1016/0092-8674(91)90573-h. [DOI] [PubMed] [Google Scholar]
- Smith WC, Harland RM. Injected Xwnt-8 RNA acts early in Xenopus embryos to promote formation of a vegetal dorsalizing center. Cell. 1991;67:753–765. doi: 10.1016/0092-8674(91)90070-f. [DOI] [PubMed] [Google Scholar]
- Sridharan J, Haremaki T, Jin Y, Teegala S, Weinstein DC. Xmab21l3 mediates dorsoventral patterning in Xenopus laevis. Mech Dev. 2012;129:136–146. doi: 10.1016/j.mod.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennard F, Carnac G, Gurdon JB. The Xenopus T-box gene, Antipodean, encodes a vegetally localised maternal mRNA and can trigger mesoderm formation. Development. 1996;122:4179. doi: 10.1242/dev.122.12.4179. [DOI] [PubMed] [Google Scholar]
- Sun BI, Bush SM, Collins-Racie LA, LaVallie ER, DiBlasio-Smith EA, Wolfman NM, McCoy JM, Sive HL. derriere: a TGF-beta family member required for posterior development in Xenopus. Development. 1999;126:1467–1482. doi: 10.1242/dev.126.7.1467. [DOI] [PubMed] [Google Scholar]
- Suri C, Haremaki T, Weinstein DC. Inhibition of mesodermal fate by Xenopus HNF3β/FoxA2. Developmental biology. 2004;265:90–104. doi: 10.1016/j.ydbio.2003.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri C, Haremaki T, Weinstein DC. Xema, a foxi-class gene expressed in the gastrula stage Xenopus ectoderm, is required for the suppression of mesendoderm. Development. 2005;132:2733–2742. doi: 10.1242/dev.01865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M, Casey ES, Fairclough L, Smith JC. Bix1, a direct target of Xenopus T-box genes, causes formation of ventral mesoderm and endoderm. Development. 1998;125:3997–4006. doi: 10.1242/dev.125.20.3997. [DOI] [PubMed] [Google Scholar]
- Tada M, Smith JC. Xwnt11 is a target of Xenopus Brachyury: regulation of gastrulation movements via Dishevelled, but not through the canonical Wnt pathway. Development. 2000;127:2227–2238. doi: 10.1242/dev.127.10.2227. [DOI] [PubMed] [Google Scholar]
- Tada M, Smith JC. T-targets: clues to understanding the functions of T-box proteins. Dev Growth Differ. 2001;43:1–11. doi: 10.1046/j.1440-169x.2001.00556.x. [DOI] [PubMed] [Google Scholar]
- Takabatake Y, Takabatake T, Takeshima K. Conserved and divergent expression of T-box genes Tbx2-Tbx5 in Xenopus. Mechanisms of Development. 2000;91:433–437. doi: 10.1016/s0925-4773(99)00329-9. [DOI] [PubMed] [Google Scholar]
- Takahashi C, Kusakabe M, Suzuki T, Miyatake K, Nishida E. mab21-l3 regulates cell fate specification of multiciliate cells and ionocytes. Nat Commun. 2015;6:6017. doi: 10.1038/ncomms7017. [DOI] [PubMed] [Google Scholar]
- Tao J, Kuliyev E, Wang X, Li X, Wilanowski T, Jane SM, Mead PE, Cunningham JM. BMP4-dependent expression of Xenopus Grainyhead-like 1 is essential for epidermal differentiation. Development. 2005;132:1021–1034. doi: 10.1242/dev.01641. [DOI] [PubMed] [Google Scholar]
- Uwanogho D, Rex M, Cartwright EJ, Pearl G, Healy C, Scotting PJ, Sharpe PT. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech Dev. 1995;49:23–36. doi: 10.1016/0925-4773(94)00299-3. [DOI] [PubMed] [Google Scholar]
- Weidgang CE, Russell R, Tata PR, Kuhl SJ, Illing A, Muller M, Lin Q, Brunner C, Boeckers TM, Bauer K, Kartikasari AE, Guo Y, Radenz M, Bernemann C, Weiss M, Seufferlein T, Zenke M, Iacovino M, Kyba M, Scholer HR, Kuhl M, Liebau S, Kleger A. TBX3 Directs Cell-Fate Decision toward Mesendoderm. Stem Cell Reports. 2013;1:248–265. doi: 10.1016/j.stemcr.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein DC, Hemmati-Brivanlou A. Neural induction. Annu Rev Cell Dev Biol. 1999;15:411–433. doi: 10.1146/annurev.cellbio.15.1.411. [DOI] [PubMed] [Google Scholar]
- Wettenhall JM, Simpson KM, Satterley K, Smyth GK. affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics. 2006;22:897–899. doi: 10.1093/bioinformatics/btl025. [DOI] [PubMed] [Google Scholar]
- White RJ, Sun BI, Sive HL, Smith JC. Direct and indirect regulation of derriere, a Xenopus mesoderm-inducing factor, by VegT. Development. 2002;129:4867–4876. doi: 10.1242/dev.129.20.4867. [DOI] [PubMed] [Google Scholar]
- Whitman M. Nodal signaling in early vertebrate embryos: themes and variations. Dev Cell. 2001;1:605–617. doi: 10.1016/s1534-5807(01)00076-4. [DOI] [PubMed] [Google Scholar]
- Wills AE, Choi VM, Bennett MJ, Khokha MK, Harland RM. BMP antagonists and FGF signaling contribute to different domains of the neural plate in Xenopus. Dev Biol. 2010;337:335–350. doi: 10.1016/j.ydbio.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PA, Hemmati-Brivanlou A. Induction of epidermis and inhibition of neural fate by Bmp-4. Nature. 1995;376:331–333. doi: 10.1038/376331a0. [DOI] [PubMed] [Google Scholar]
- Yun CH, Choi SC, Park E, Kim SJ, Chung AS, Lee HK, Lee HJ, Han JK. Negative regulation of Activin/Nodal signaling by SRF during Xenopus gastrulation. Development. 2007;134:769–777. doi: 10.1242/dev.02778. [DOI] [PubMed] [Google Scholar]
- Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The Role of Maternal VegT in Establishing the Primary Germ Layers in Xenopus Embryos. Cell. 1998;94:515–524. doi: 10.1016/s0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]
- Zhang J, King ML. Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development. 1996;122:4119–4129. doi: 10.1242/dev.122.12.4119. [DOI] [PubMed] [Google Scholar]
- Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]