Abstract
Background
Siglec-8 is present at a high level on human blood eosinophils and low level on blood basophils. Engagement of Siglec-8 on blood eosinophils causes its internalization and results in apoptosis. Siglec-8 is a potential therapeutic target in eosinophilic asthma.
Objectives
To determine Siglec-8 levels on eosinophils and basophils recruited during lung inflammation.
Method
We analyzed surface Siglec-8 by flow cytometry on cells obtained by bronchoalveolar lavage (BAL) 48 h after segmental lung allergen challenge of human subjects with mild allergic asthma and used confocal microscopy to compare Siglec-8 distribution on BAL and blood eosinophils.
Results
Like their blood counterparts, BAL eosinophils had high unimodal surface Siglec-8, while BAL basophils had lower but detectable surface Siglec-8. BAL macrophages, monocytes, neutrophils, and plasmacytoid dendritic cells did not express surface Siglec-8. Microscopy of freshly isolated blood eosinophils demonstrated homogeneous Siglec-8 distribution over the cell surface. Upon incubation with IL-5, Siglec-8 on the surface of eosinophils became localized in patches both at the nucleopod tip and at the opposite cell pole. BAL eosinophils also had a patchy Siglec-8 distribution.
Conclusions
We conclude that 48 h after segmental allergen challenge, overall levels of Siglec-8 expression on airway eosinophils resemble those on blood eosinophils, but with a patchier distribution, a pattern consistent with activation. Thus, therapeutic targeting of Siglec-8 has the potential to impact blood as well as lung eosinophils, which may be associated with improved outcome in eosinophilic lung diseases.
Keywords: airway, blood, flow cytometry, eosinophil, basophil
2. Introduction
Asthma is frequently characterized by eosinophil-rich airway inflammation [1]. Airway eosinophilia is associated with asthma exacerbations [2,3] and appears to play a role in airway remodeling [4,5]. Eosinophils become activated, arrest, extravasate, and move through the bronchial tissue and lumen, and display increased survival in asthma [6–8]. Cell-surface glycan-binding proteins may modulate eosinophil function, particularly recruitment and survival, and such interactions may be exploited therapeutically in asthma and other eosinophilic diseases [9,10]. Siglec-8 is one such glycan-binding protein, highly expressed on the surface of human blood eosinophils, expressed at a low level on blood basophils, and also expressed on mast cells, e.g., cultured and derived from cord blood [9,11]. Siglec-8 levels on airway cells have not been characterized. As Siglec-8 is a potential therapeutic target and as airway cells, particularly eosinophils, are relevant effector cells for the clinical expression of asthma [1], it is necessary to determine whether human airway eosinophils and basophils, which likely are activated in vivo, retain surface Siglec-8, and whether other airway cells express surface Siglec-8.
Engagement of Siglec-8 on blood eosinophils causes its internalization and results in apoptosis [9,12,13]. Paradoxically, Siglec-8-dependent apoptosis is amplified when eosinophils have been primed or activated with IL-5, GM-CSF, or IL-33 [9,12,14,15], or primed in vivo during allergic inflammation [14]. The presence of an inhibitory motif in the cytoplasmic domain indicates that Siglec-8 should be involved in negative cell signaling [9]. Recent observations indicate that Siglec-8 can, after IL-5 priming, initiate cell signaling leading to eosinophil apoptosis via phosphorylation of intracellular signaling proteins and β2 integrin-dependent adhesion [9,16,17]. Siglec-8-directed therapeutics are currently being developed. A preliminary report involving a phase 1 study in healthy adult volunteers showed that a single intravenous infusion of a humanized afucosylated IgG1 monoclonal antibody (mAb) against Siglec-8 (called AK002) rapidly and selectively depletes blood eosinophils in a sustained manner for at least 84 days [18]. Given the selective expression of Siglec-8 on both eosinophils and mast cells, AK002 is currently undergoing clinical trials in eosinophil- and mast cell associated diseases including refractory chronic urticaria (NCT [national clinical trial] 03436797), severe forms of allergic conjunctivitis (NCT03379311), and indolent systemic mastocytosis (NCT02808793).
IL-5 causes blood eosinophils in suspension to undergo acute shape change and polarize, with granules moving to one pole and the nucleus to the opposite pole into a specialized uropod, the “nucleopod” [19]. The nucleopod is enriched in cell surface receptors including P-selectin glycoprotein ligand-1 (PSGL1, CD162); PSGL1 localization on the surface of IL-5-activated eosinophils can be regarded as a reporter of cell activation [19]. However, cell shape and polarization, and localization of cell surface receptors have not been studied on airway eosinophils.
In allergic human subjects, segmental bronchoprovocation with allergen is a model of allergic airway inflammation that induces intense local recruitment of inflammatory cells, including eosinophils and basophils [20]. We utilized this model to characterize Siglec-8 expression and localization on cells in bronchoalveolar lavage (BAL).
3. Materials and Methods
3.1. Subjects for segmental lung allergen challenge
Eight subjects with mild allergic asthma who were allergic to ragweed, house dust mite, or cat dander were screened and enrolled as described [21,22]. At least four weeks before bronchoscopy, subjects underwent a whole-lung inhaled allergen (ragweed, house dust mite, or cat dander) challenge to determine the provocative dose of antigen producing a 20% fall in forced expiratory volume in 1 s (AgPD20) [21,23,24]. These studies were approved by the University of Wisconsin-Madison Health Sciences Institutional Review Board. Informed written consent was obtained from each subject before participation (protocol No. 2016-0021 for bronchoalveolar lavage study and No. 2013-1570 for eosinophil purification).
3.2. Segmental bronchoprovocation with allergen, bronchoalveolar lavage, and blood draws
Allergen was administered by bronchoscopy as previously described [21]. Briefly, two bronchopulmonary segments were lavaged (160 ml sterile 0.9% NaCl) and then a dose of 10% of the subject’s AgPD20 was administered into one segment and, when this was well tolerated, a dose of 20% of the AgPD20 was instilled in the second segment [21]. Bronchoscopy with BAL was repeated 48 h later and BAL recovered from the two segments was pooled [21]. Blood was obtained from each subject on the same day as the second bronchoscopy, when BAL was performed.
3.3. Three-color flow cytometry detection of Siglec-8 on airway and blood cell populations
Antibodies and reagents used for three-color analysis were the following. Fluorescein isothiocyanate (FITC)-conjugated anti-CD14 mAb clone M5E2 and anti-CD16 clone 3G8 [23]; phycoerythrin (PE)-conjugated anti-CD11b (αM integrin) clone D12, anti-CD14 clone MΦP9, anti-CCR3 (CD193) clone 5E8, anti-CD206 clone 19.2, and mouse IgG1 clone MOPC-21; human Fc block; and Calibrite two-color kit were from BD Biosciences (San Jose, CA, USA). FITC-conjugated anti-CD45 clone 2D1 and RBC lysis buffer were from eBioscience/ThermoFisher Scientific (Waltham, MA, USA). Alexa Fluor (AF) 647-conjugated mouse IgG1 isotype control clone MOPC-21 was from BioLegend (San Diego, CA, USA). PE-conjugated anti-Siglec-8 clone 837535 was from R&D Systems (Minneapolis, MN, USA). AF647-conjugated F(ab’)2 of clone 2E2 [12] was a gift from Drs. Rustom Falahati, Nenad Tomasevic, and Christopher Bebbington (Allakos Inc., San Carlos, CA) to the University of Wisconsin-Madison. Paraformaldehyde 16% solution, EM grade, methanol-free, was from Electron Microscopy Services (Hatfield, PA, USA). Mid-range one-peak rainbow fluorescent beads [25] were from Spherotech (Lake Forest, IL, USA). AF647 fluorescence reference standard microbeads were from Bangs Laboratories (Fishers, IN, USA).
Whole unfractionated BAL cells (4 × 105) or blood (100 µl) from three subjects (BAL cells and blood from the same subjects) obtained 48 h after segmental lung allergen challenge were processed for flow cytometry as follows. In some tubes, BAL cells (in 50 µl) were first incubated with 5 µl FITC-conjugated anti-CD16 and 50 µl FACS buffer (2% BSA in PBS) in the dark for 30 minutes at 4°C. One ml of phosphate-buffered saline (PBS), pH 7.4, was then added, tubes were centrifuged for 10 minutes at 260 g (1200 rpm in a Sorvall Technospin R centrifuge, Du Pont, Wilmington, DE, USA), and the cell pellet was resuspended in 50 µl FACS buffer. Then or in other tubes that were not incubated initially with anti-CD16, the cells in 50 µl were incubated with 5 µl of BD Fc block for 10 minutes at room temperature. Another 50 µl FACS buffer was added plus a mixture of directly labeled antibodies (0.1 µg of AF647-anti-Siglec-8 or 5 µl isotype control and 5 µl of FITC-conjugated anti-CD14 [to anti-CD16-treated samples] or FITC-conjugated anti-CD45 [to samples not treated with anti-CD16] and 5 µl PE-conjugated anti-CCR3). In addition, in each experiment, one tube contained “leukogate”, i.e., FITC-anti-CD45 and 20 µl of PE-conjugated anti-CD14. The mixtures were incubated for 30 minutes at 4°C. Samples were then washed with 1 ml PBS. After centrifugation, cells stained with directly labeled primary antibodies were resuspended in 250 µl 1% paraformaldehyde in PBS, stored at 4°C, and then washed with 1 ml PBS and resuspended in 250 µl FACS buffer just before data collection.
Blood drawn into vacuum tubes containing CTAD (citrate, theophylline, adenosine, and dipyridamole) anticoagulant solution (BD Vacutainer Systems, Franklin Lakes, NJ, USA) as before [23] was incubated with 1 ml of the eBioscience/ThermoFisher RBC lysis buffer for 15 minutes at room temperature. Tubes were centrifuged at room temperature. The leukocyte pellet was resuspended in 500 µl PBS and centrifuged again. The blood leukocytes were then resuspended in 50 µl FACS buffer and processed as described above for BAL cells.
Data were collected (from 3 × 104 – 2 × 105 events), using a FACSCalibur (BD Biosciences) at the University of Wisconsin Carbone Cancer Center Flow Laboratory, analyzed with FlowJo 7.6.5, and are described as specific geometric mean channel fluorescence (gMCF) as before [21,23,25,26]. In one experiment, isotype controls were not included for all conditions. For these conditions in this experiment, the value of the mean of the isotype control from the relevant conditions in the other experiments was used. Rainbow beads were run at setup to set the sensitivity of the detectors at a fluorescence intensity standardized for all subjects [23,25,26]. Compensation was performed with unlabeled, FITC-labeled, and PE-labeled Calibrite (BD), and AF647 (Bangs) beads.
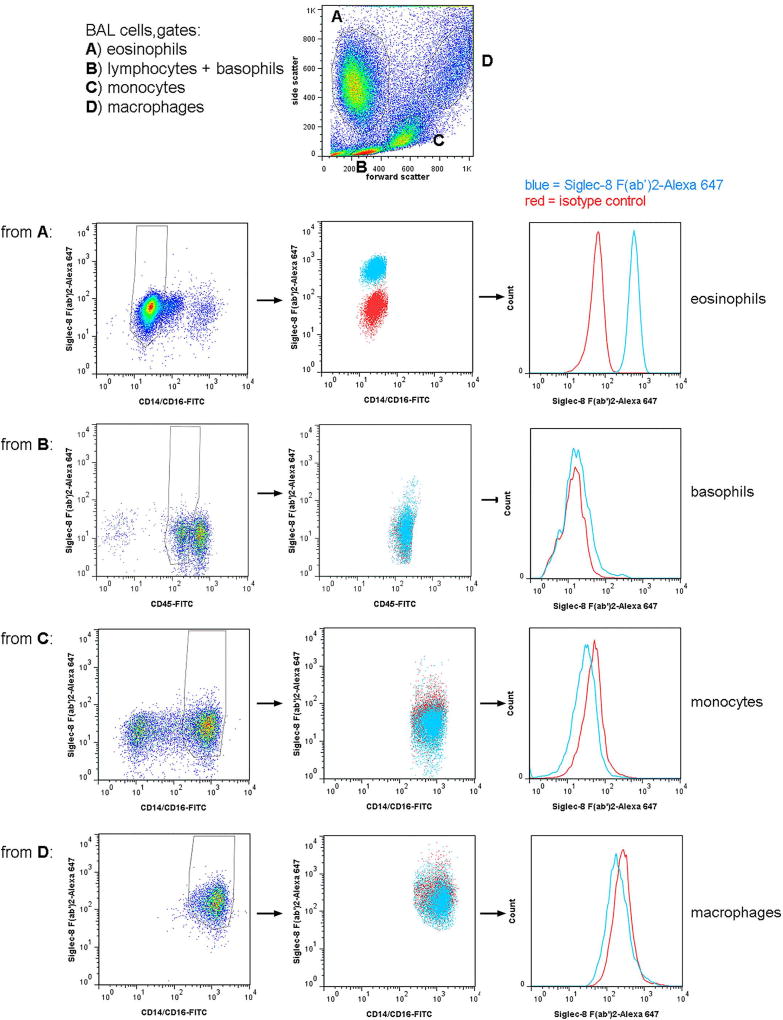
BAL cells were gated as shown in Figure 1 and described in the legend. Specifically, eosinophils were identified as having high side scatter (SSC) and were negative for CD14 and CD16, although had some autofluorescence in the FITC channel [21,24,27]. Regions within the SSC versus forward scatter (FSC) plot were compared to SSC versus FITC-anti-CD14/CD16 [21,23] and backgated on a “leukogate” plot of PE-anti-CD14 versus FITC-anti-CD45 as before [27], confirming that eosinophils are CD45-bright and CD14-negative but with some autofluorescence also in the PE channel [27], whereas monocytes are CD45-bright and CD14-very bright, macrophages are CD45-very bright and CD14-bright, and basophils CD45-intermediate (dimmer than lymphocytes [28]) and CD14-negative. In addition, in some experiments cells were also stained for CD11b and CD206, confirming that eosinophils are CCR3-positive, CD11b-positive, and CD206-negative; monocytes and macrophages CCR3-negative, CD11b-positive, and CD206-positive; and basophils CCR3-positive and CD11b-positive. Blood leukocytes were gated according to the same principles (not shown), also first by scatter, then compared to SSC versus CD14/CD16 [23,25,26,29] and backgated on CD14 versus CD45 [27], and resulted in the same populations, except that macrophages were absent and neutrophils were confirmed as having intermediate SSC and being CD16-positive [21] as well as CD45-intermediate and CD14-negative (not shown).
Fig 1.
Gating strategy for 3-color flow cytometry and Siglec-8 expression on BAL eosinophils, basophils, monocytes, and macrophages. Populations in a whole BAL cell preparation obtained 48 h after segmental lung allergen challenge were first gated based on side versus forward scatter (top dot plot) with gate A (high side scatter, low forward scatter) encompassing eosinophils, gate B (low side and forward scatter) including lymphocytes and basophils, gate C (intermediate side and forward scatter) encompassing monocytes, and gate D (high side and forward scatter) encompassing alveolar macrophages. Comparison to plots with side scatter versus CD14/CD16 and CD14 versus CD45, as well as expression or not of CCR3 (not shown), confirmed the locations of these populations. Row 1: Cells from gate A were gated further based on CD14/16 to include eosinophils (CD14/CD16-negative but autofluorescent) and exclude monocytes, macrophages, and possibly neutrophils (CD14/CD16-positive). Row 2: Cells from gate B were gated further based on CD45 to include basophils (CD45-intermediate) and exclude lymphocytes (CD45-bright) and other events (CD45-negative). Row 3: Cells from gate C were gated further based on CD14/CD16 to include monocytes (CD14-high) and exclude lymphocytes and eosinophils (CD14/CD16-negative). Row 4: Cells from gate D were gated further based on CD14/CD16 to include macrophages (CD14-bright). For each row: Left plots, AF647-conjugated isotype control versus CD14/CD16 or CD45; middle plot, Siglec-8 (AF647-conjugated) versus CD14/CD16 or CD45, blue = Siglec-8, red = isotype control; histograms (to the right) of the gated populations in the middle plots, blue = Siglec-8, red = isotype control. Siglec-8 expression shown in this figure is representative of data from 3 subjects.
3.4. Seven-color flow cytometry for confirmation of Siglec 8 expression on airway and blood cell populations
The following directly conjugated antibodies were used for the 7-color flow cytometry panel. FITC-conjugated antibodies to lineage markers (CD3 clone SK7, CD16 clone 3G3, CD19 clone 4G7, and CD56 clone NCAM-16.2; all from BD Biosciences); brilliant ultraviolet (BUV) 395-conjugated anti-CD14 (clone MφP9, BD Biosciences); brilliant violet (BV) 510-conjugated anti-CD45 (clone H130, BioLegend); AF647-conjugated anti-Siglec 8 F(ab’)2 fragment of clone 2E2 (gift from Allakos as above); PE-conjugated anti-CD203c (clone Np4D6, BD Biosciences), PE-anti-FceRI (high-affinity receptor for the Fc region of IgE, clone AER-37, eBioscience), or PE-cyanin (Cy) 7-conjugated anti-CD123 (clone 6H6, eBioscience/ThermoFisher); and BV421-conjugated anti-HLA-DR (HLA – antigen D related, clone G46-6, BD Biosciences).
The seven-color flow cytometry method was performed on blood and BAL obtained 48 h after segmental lung allergen challenge from three subjects who were different from the subjects used for the 3-color method described above in section 3.3. Blood was collected into EDTA and treated for 5 minutes at room temperature with a lysis buffer (0.8% NH4Cl, 10 mM KHCO3, 0.1 mM EDTA in double distilled H2O) to remove RBCs. Lysed whole blood or unseparated BAL cells (1–2 × 106) obtained 48 h after segmental lung allergen challenge were washed and resuspended at 2.5 million per tube in 80 µl of FACS buffer. Cells were stained for 15 minutes with a cocktail containing 5 µl each of FITC-conjugated mAbs to CD3, CD16, CD19, and CD56. Human BD Fc block (5 µl) was added for an additional 10 minutes. Cells were washed in FACS buffer and resuspended in 50 µl of BD brilliant staining buffer followed by the addition of 50 µl of a cocktail containing predetermined optimized concentrations of the above antibodies (generally 1–2.5 µl per tube) in brilliant staining buffer. Fluorescence minus one (FMO) controls were also prepared to allow accurate delineation of positively stained cells. Cells were incubated in the dark at room temperature for 30 min, washed twice with FACS buffer, and then fixed overnight in 1% formaldehyde. Data acquisition (of 4 × 105 – 1 × 106 events) was performed on a five-laser 18-detector BD LSRII flow cytometer at the University of Wisconsin Carbone Cancer Center Flow Laboratory. Automated compensation was performed with FlowJo 10.4 (TreeStar, Ashland, OR, USA), using data obtained from antibody-capture compensation beads (UltraComp eBeads, eBioscience) stained with the same antibody-fluorophore reagents used for the cells. Because there was poor binding of the AF647-conjugated anti-Siglec8 F(ab’)2 fragment to the compensation beads, an irrelevant AF647 antibody was used for compensation.
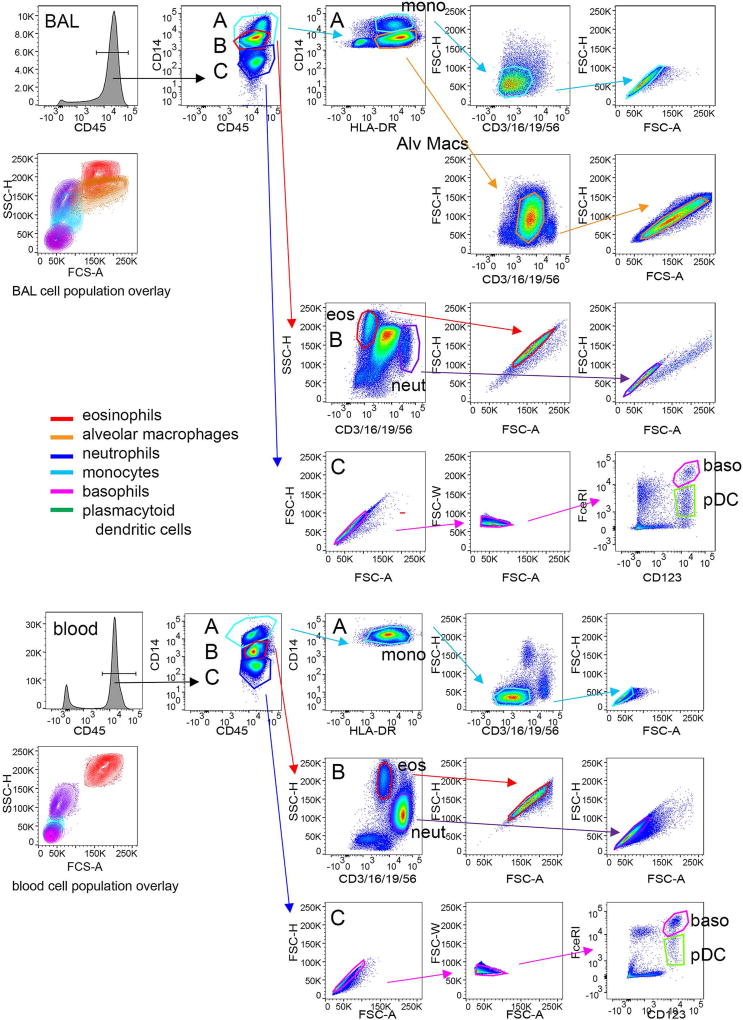
Cell populations were defined as shown in Figure 2. CD45-bright leukocytes were distinguished from non-cellular debris. Within the CD45-bright region, three populations were identified as CD14-bright (monocytes and alveolar macrophages), autofluorescent (eosinophils and neutrophils), and CD14-negative (basophils, dendritic cells, lymphocytes). The CD14-positive cells were further defined as CD14-very bright and HLA-DR-positive monocytes and CD14-moderate and HLA-DR-positive alveolar macrophages. Contaminating T cells, neutrophils, B cells, natural killer cells were eliminated using CD3/CD16/CD19/CD56 staining with FSC. Doublets were rejected using FSC height versus area. Autofluorescent cells within the BUV395 channel were distinguished using CD3/CD16/CD19/CD56 and SSC. Eosinophils were autofluorescent in the FITC channel with high SSC and neutrophils, which express CD16, were identified within the CD3/CD16/CD19/CD56-bright population as having moderate SSC. Doublets were eliminated using FSC height versus area. To identify basophils and plasmacytoid dendritic cells (pDCs) within the CD14-negative gate, doublets were removed using FSC height versus area and FSC width versus area. Basophils were defined as CD123-bright and FcεRI-bright, while pDCs were defined as CD123-bright and FcεRI-moderate. pDCs also expressed HLA-DR (not shown). Siglec-8 expression on each gated population within the fully stained sample was compared to comparable populations within the FMO sample containing all antibodies minus the one AF-anti-Siglec-8 antibody.
Fig. 2.
Gating strategy for 7-color flow cytometry. Populations in a whole BAL cell preparation obtained 48 h after segmental lung allergen challenge and lysed whole blood were stained with a panel of antibodies including a CD3/CD16/CD19/CD56-FITC cocktail, CD45-BV510, HLA-DR-BV421, CD14-BUV395, CD123-PE-Cy7, FcεRI-PE, and Siglec-8-AF647. CD45-bright leukocytes were distinguished from non-cellular debris. Within the CD45-bright region, three populations were identified as CD14-bright (A, light blue region encompassing monocytes and alveolar macrophages), autofluorescent (B, red region encompassing eosinophils and neutrophils), and CD14-negative (C, dark blue region encompassing basophils, dendritic cells, lymphocytes). The CD14-positive cells in region A were further defined as CD14-very bright and HLA-DR-positive monocytes (light blue region) and CD14-moderate and HLA-DR-positive alveolar macrophages (orange region). Contaminating T cells, neutrophils, B cells, and natural killer cells were eliminated using CD3/CD16/CD19/CD56-staining with forward side scatter (FSC). Doublets were removed based on forward scatter height (FSC-H) versus area (FSC-A). The autofluorescent cells in the BUV395 channel within region B were distinguished using CD3/CD16/CD19/CD56 and side scatter (SSC). Eosinophils (red region) were autofluorescent in the FITC channel with high SSC and neutrophils (purple region), which express CD16, were identified within the CD3/CD16/CD19/CD56-bright population as having moderate SSC. Doublets were eliminated using FSC-H versus FSC-A. Single cells within the CD14-negative cells in region C were identified using FSC-H versus FSC-A and then forward scatter width (FSC-W) versus area (FSC-A). Basophils were defined as CD123-bright and FcεRI-bright, while plasmacytoid dendritic cells (pDCs) were identified as CD123-bright and FcεRI-moderate. pDCs also expressed HLA-DR (not shown). A color overlay displaying contour plots of each cell population confirms appropriate FSC and SSC properties. This 7-color flow cytometry method was used for samples from 3 subjects and those subjects were different from those used in Figures 1 and 3.
3.5. Eosinophil purification and immunofluorescence confocal microscopy
For localization of Siglec-8 by confocal microscopy, BAL eosinophils obtained 48 h after segmental lung allergen challenge were purified, from two subjects who were different from the 6 subjects used for flow cytometry described above in sections 3.3 and 3.4, using a two-step Percoll gradient as described [22,24,30] and blood eosinophils were purified from heparinized blood from two control donors (not exposed to allergen challenge) by negative selection using a cocktail of anti-CD16, anti-CD14, anti-CD3, and anti-glycophorin beads as before [31]. After purification, eosinophils were resuspended at 2.5 × 106/ml in Roswell Park Memorial Institute (RPMI) medium with 0.1% BSA, and equilibrated for 1 h at 37°C [19]. Blood eosinophils were then incubated with or without recombinant IL-5 (R&D Systems) (50 ng/ml) for 10 minutes at 37°C. Cells were fixed by adding an equal volume of 7.4% paraformaldehyde in PBS (giving a final concentration of 3.7% paraformaldehyde) for 10 minutes at room temperature and quenched by centrifugation and resuspension in the same total volume of 0.1 M glycine in PBS [19] and incubation for 10 minutes at room temperature. One hundred µl (250,000) cells were cytospun as described [19] onto 12 mm coverslips, which previously had been coated with 0.1% poly-L-lysine solution (0.1%) (Sigma-Aldrich, St. Louis, MO, USA) for 1 h at room temperature, washed four times with H2O, washed with 95% ethanol, and dried. Coverslips were placed in wells in 24-well plates, blocked with 3% BSA in PBS containing 0.02% NaN3, and then stained, mounted, and viewed by immunofluorescence microscopy and photographed as described [31], using unlabeled anti-Siglec-8 (clone 2E2, gift from Allakos as above) or anti-PSGL1 (clone KPL-1 [31], BioLegend) and AF488-conjugated anti-mouse F(ab’)2 secondary antibody (Cell Signaling Technology, Danvers, MA, USA), and visualizing nuclei with 4’,6-diamidino-2-phenylindole (DAPI) (Life Technologies, Eugene, OR, USA). A 60× oil immersion objective was used [31]. Cell circumference and peripheral staining were quantified using the Fiji version of ImageJ (http://fiji.sc/Fiji) as before [31].
3.6. Statistical analysis
Paired t test was used to compare flow cytometry data between two groups of cells from the same subjects. Unpaired t test was used to compare quantified image data between two groups of cells. P ≤ 0.05 was considered significant. Analyses were performed using Prism (GraphPad, San Diego, CA, USA).
4. Results
4.1. Siglec-8 is expressed on BAL eosinophils and basophils
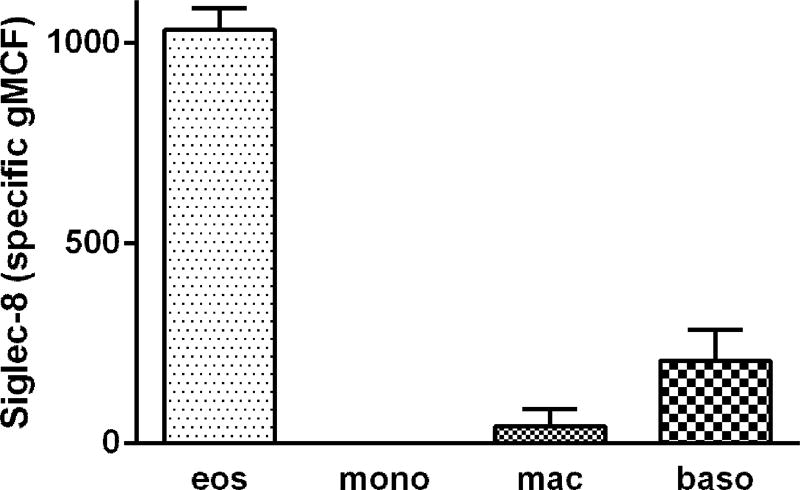
To determine Siglec-8 expression on human airway cells, we first used 3-color flow cytometry similar to protocols we have used before and analyzed cells recovered by BAL 48 h after segmental lung allergen challenge of subjects with allergic asthma [20]. BAL cells were gated as shown in Figure 1. BAL eosinophils had high unimodal surface Siglec-8 expression (Figures 1 and 3). Basophils, which like eosinophils are found in BAL after segmental antigen challenge [32], had detectable Siglec-8 expression, which was lower than that of eosinophils (Figures 1 and 3). BAL monocytes and macrophages had no Siglec-8 (Figures 1 and 3). BAL eosinophil Siglec-8 expression at 48 h was similar to that on blood eosinophils, i.e., there was no significant difference among eosinophil Siglec-8 levels in BAL and blood at 48 h (specific gMCF for BAL 1030 ± 50, blood 890 ± 100, means ± standard deviation [SD]). In addition to expression on blood eosinophils, Siglec-8 was detected at low levels on blood basophils as demonstrated before [9,11], but not on blood monocytes or neutrophils (not shown for the 3-color protocol, but see below), as expected [9].
Fig. 3.
Quantitation of Siglec-8 expression on BAL eosinophils, monocytes, macrophages, and basophils. Populations of BAL cells obtained 48 h after segmental lung allergen challenges were gated as in Figure 1. Siglec-8 expression presented as specific geometric channel fluorescence (gMCF), mean ± standard deviation (SD), n = 3 subjects.
4.2. Confirmation of Siglec-8 expression on cell subsets by 7-color flow cytometry
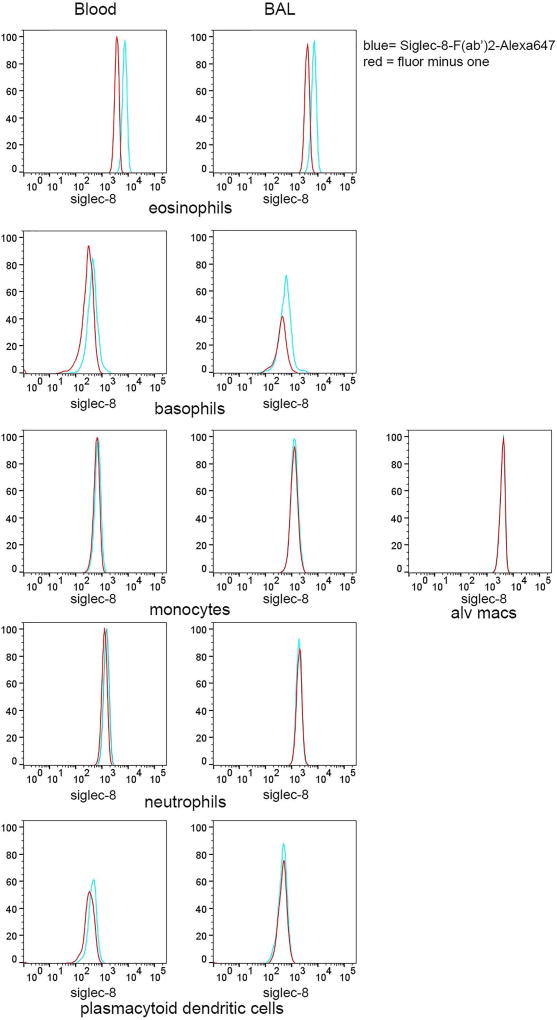
A 7-color flow cytometry panel was developed to more definitively identify individual BAL cell populations, particularly basophils, which have similar FSC and SSC as dendritic cells and lymphocytes in a different set of subjects. Utilization of the 7-color panel confirmed results from the 3-color flow cytometric analysis in the first set of subjects showing Siglec-8 expression on eosinophils and basophils in BAL and blood, with no detectable expression on monocytes, macrophages, neutrophils, and pDCs (Figure 4). There was no detectable Siglec-8 on cells within the CD14-negative gate that were positive for CD3/CD19/CD56 (not shown), indicating that T, B, and NK cells are also negative for Siglec-8. Cells fitting criteria of mast cells, which would be expected to have intermediate-high SSC and be FcεRI-positive [33] and which have been reported to express Siglec-8 [9,11], were not detected in the BAL 48 h after segmental allergen challenge.
Fig. 4.
Confirmation of Siglec-8 expression on cell subsets using 7-color flow cytometry. Populations of BAL and blood cells obtained 48 h after segmental lung allergen challenge were gated as in Figure 2. Histograms show Siglec-8 expression on each gated population within the fully-stained sample (blue) compared to the comparable population within the fluorescence minus one (FMO) control sample containing all antibodies minus the Siglec-8-AF647 antibody (red). Data are representative of experiments with 3 subjects who were different from the subjects used in Figures 1 and 3.
4.3. Siglec-8 localization on the surface of BAL eosinophils
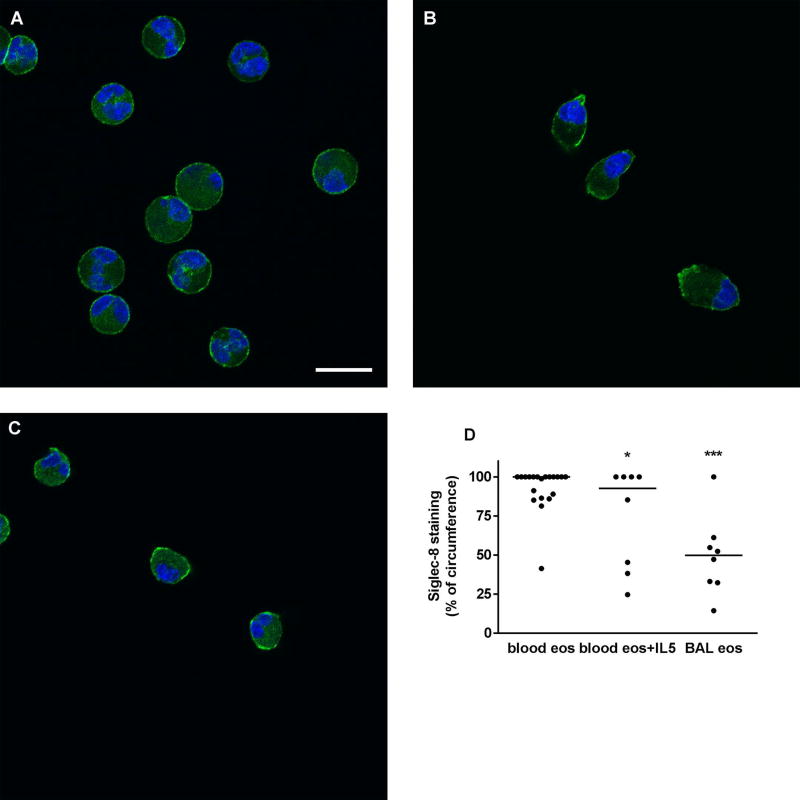
Immunofluorescence confocal microscopy was used to determine the localization of Siglec-8 on airway eosinophils compared to resting and IL-5-activated blood eosinophils. On unactivated blood eosinophils, Siglec-8 was homogeneously distributed around the cell periphery (Figure 5A). Upon acute IL-5 activation, blood eosinophils underwent characteristic shape change [19] and Siglec-8 became patchier and localized at the tip of the nucleopod and at the opposite pole of the cell (Figure 5B). On BAL eosinophils, Siglec-8 was also localized in patches at different sites around the cell periphery (Figure 5C). Quantitation of Siglec-8 localization, as percentage of cell circumference, confirmed the visual impression that on BAL eosinophils and IL-5-activated blood eosinophils Siglec-8 was more focused in patches and covered a significantly smaller proportion of the cell periphery than on unactivated blood eosinophils (Figure 5D). In addition, there was a trend to greater variation among BAL eosinophils (CV = 51%) than on unactivated or activated blood eosinophils (CV = 15% and 44%, respectively).
Fig. 5.
Localization of Siglec-8 on blood and BAL eosinophils. Localization of Siglec-8 (green) and nucleus (blue) in cytospun unactivated blood eosinophils (A), IL-5-activated blood eosinophils (IL-5, 50 ng/ml for 10 minutes as described in Materials and Methods) (B), and BAL eosinophils (C). Cells were analyzed by immunofluorescent staining using mAb to Siglec-8 and AF488-conjugated secondary antibody. Nuclei were stained with DAPI. Bar, 10 µm. Representative of two experiments (i.e., 2 subjects, these 2 subjects were different from the 6 subjects used in Figures 1–4). (D) Quantitation of Siglec-8 localization using Fiji software, peripheral Siglec-8 staining as percentage of circumference in unactivated blood eosinophils (blood eos), activated blood eosinophils (blood eos+IL5), and BAL eosinophils (BAL eos), *p (probability) < 0.05, ***p < 0.001 versus unactivated blood eosinophils.
4.4. PSGL1 localization on the surface of BAL eosinophils
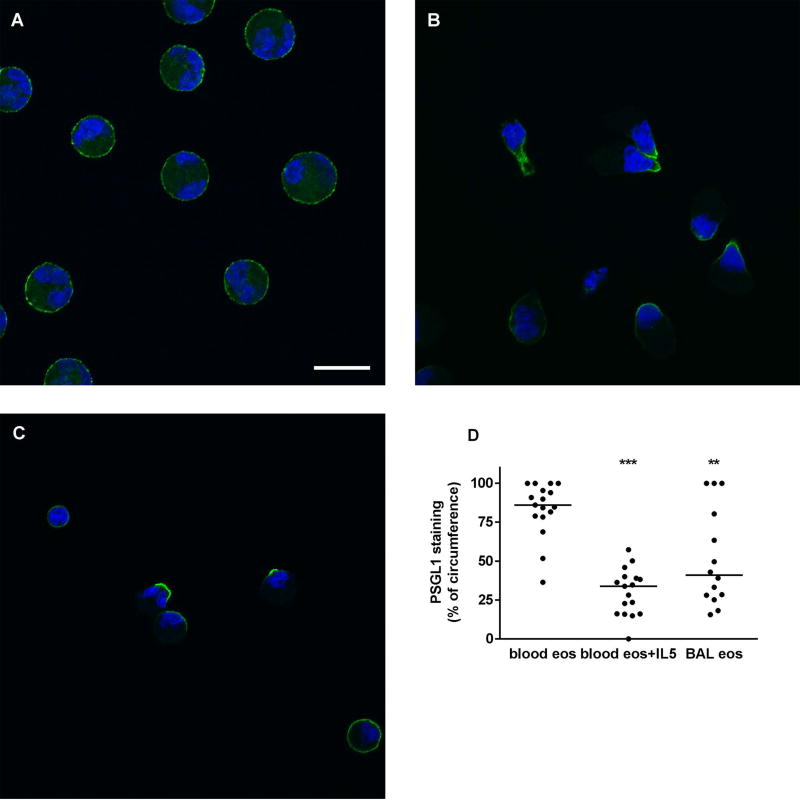
In order to compare the Siglec-8 cellular distribution pattern to a reference protein that we have imaged before in cytospun unactivated and IL-5-activated blood eosinophils [19] but not in BAL eosinophils, we also stained eosinophils for PSGL1, whose localization can be regarded as a reporter of cell activation and a positive control that activation has occurred. On unactivated blood eosinophils, PSGL1, like Siglec-8 and as seen previously [19], was distributed evenly around the cell circumference (Figure 6A). On IL-5-activated eosinophils, it was, also as before [19], concentrated at the nucleopod tip (Figure 6B). On BAL eosinophils, PSGL1 localization was found to be heterogeneous with some cells having PSGL1 localized at the tip, some cells having PSGL1 in a crescent, and some cells appearing similar to unactivated ones (Figure 6C). PSGL1 and Siglec-8 distribution on IL-5-activated blood eosinophils and BAL eosinophils were different in that Siglec-8 often had a distribution in several patches per cell while PSGL1 localization was focused at one site per cell (compare Figure 5B and C with Figure 6B and C). Quantitation confirmed that on BAL eosinophils and IL-5-activated blood eosinophils PSGL1 covered a significantly smaller part of the cell circumference than on unactivated blood eosinophils (Figure 6D). Further, as with Siglec-8, there was a trend to greater variation in PSGL1 staining on BAL eosinophils (CV = 61%) than on unactivated or activated blood eosinophils (CV = 21% and 49%, respectively). Finally, quantitative comparison between Siglec-8 and PSGL1 demonstrated that on IL-5-activated blood eosinophils, PSGL-1 covered a significantly smaller proportion than did Siglec-8 (p < 0.001), confirming the visual impression that PSGL1 localization is more focused and Siglec-8 is localized in several patches.
Fig. 6.
Localization of PSGL1 on blood and BAL eosinophils. Localization of P-selectin glycoprotein ligand-1 (PSGL1) (green) and nucleus (blue) in cytospun unactivated blood eosinophils (A), IL-5-activated blood eosinophils (IL-5, 50 ng/ml for 10 minutes as described in Materials and Methods) (B), and BAL eosinophils (C). Cells were analyzed by immunofluorescent staining using mAb to PSGL1 and AF488-conjugated secondary antibody. Nuclei were stained with DAPI. Bar, 10 µm. Representative of two experiments (i.e., 2 subjects, these 2 subjects were the same as used in Figure 5 but were different from the 6 subjects used in Figures 1–4). (D) Quantitation of PSGL1 localization using Fiji software, peripheral PSGL1 staining as percentage of circumference in unactivated blood eosinophils (blood eos), activated blood eosinophils (blood eos+IL5), and BAL eosinophils (BAL eos), **p < 0.01, ***p < 0.001 versus unactivated blood eosinophils.
5. Discussion
Cells in BAL obtained 48 h after segmental lung allergen challenges were examined for Siglec-8 expression. BAL eosinophils had high unimodal Siglec-8 expression. BAL eosinophil Siglec-8 level was similar to that on blood eosinophils. BAL basophils had lower but detectable Siglec-8 expression. Other BAL cell populations, including monocytes, alveolar macrophages, neutrophils, and pDCs, did not express Siglec-8.
The staining pattern of human BAL cells differs from what has been reported in mice when comparing surface levels of Siglec-F, the closest mouse counterpart to Siglec-8. Mouse Siglec-F is expressed on eosinophils, but unlike human Siglec-8, Siglec-F is not expressed on basophils and is unexpectedly expressed on BAL macrophages [9,34]. Regardless, the finding that the level of Siglec-8 on human BAL eosinophils resemble that on their blood counterparts suggests that during recruitment to the lung these cells neither shed Siglec-8 nor encounter endogenous ligands capable of stimulating internalization of Siglec-8, as has been reported for Siglec-F and the airway mucin Muc5b [13,35]. The current data also suggest that Siglec-8 on human lung eosinophils is still available for therapeutic targeting [36].
Immunofluorescence confocal microscopy of unactivated blood eosinophils demonstrated that Siglec-8 was homogeneously distributed around the cell periphery, which is similar to what was recently shown [13]. On acutely IL-5-activated blood eosinophils, Siglec-8 was localized in patches at the nucleopod and at the opposite cell pole, indicating that the process of eosinophil activation includes a relocalization of Siglec-8 into patches. On BAL eosinophils, Siglec-8 was also localized in patches, although at different sites around the cell periphery. Quantitation confirmed that Siglec-8 covered a significantly smaller proportion of the cell periphery on BAL or IL-5-activated blood eosinophils than on unactivated blood eosinophils. PSGL1 staining was also performed, and as observed before [19], unactivated blood eosinophils had uniform PSGL1 distribution similar to the Siglec-8 distribution, and IL-5-activated blood eosinophils had PSGL1 concentrated at the nucleopod tip. The latter staining was thus more restricted than that of Siglec-8 on IL-5-activated cells. BAL eosinophils had heterogeneous PSGL1 localization ranging from concentrated at a tip over a crescent-like pattern to an even distribution. As with Siglec-8, PSGL1 covered a significantly smaller part of the cell periphery on BAL or activated blood eosinophils than on unactivated cells. The patchy distribution patterns of Siglec-8 and PSGL1 on BAL and IL-5-activated blood eosinophils indicate that signaling via these receptors may be intensified in cytokine-activated or airway eosinophils compared to unactivated eosinophils. In particular, Siglec-8-initiated signaling in airway eosinophils may be similar to that in cytokine-primed eosinophils, including amplified Siglec-8-dependent apoptosis.
Thus, Siglec-8 is highly expressed on airway eosinophils following allergen challenge and is expressed at a lower level on airway basophils. Siglec-8 is redistributed in the membrane of BAL eosinophils consistent with an activated phenotype for eosinophils in the airway following allergen challenge.
Finally, Siglec-8 expression and localization on airway eosinophils may have therapeutic implications. Human airway eosinophils, which are important for the clinical expression of asthma [1], retain a level of surface Siglec-8 similar to that on blood eosinophils. This is unlike what has been reported for the level of the IL-5 receptor, which is reduced on BAL eosinophils compared to their blood counterparts [27]. It has been shown that anti-IL-5 therapy leads to about a 50% reduction in asthma exacerbations [2] and only partially decreases eosinophils and eosinophil granular proteins in airway tissue [24,37]; the partial effects may be explained by the reduced levels of IL-5 receptor on airway eosinophils. In contrast, Siglec-8 on human lung eosinophils should still be available for therapeutic targeting [36] and thus targeting Siglec-8 may allow for depletion of eosinophils from the airway.
Acknowledgments
We thank Michele Wolff and Holly Eversoll for study coordination; Evelyn Falibene for subject recruitment; Loren Denlinger, Richard Cornwell and Keith Meyer for performing bronchoscopies; Arturo Guadarrama and Lei Shi for processing BAL and blood samples; Elizabeth Schwantes and Paul Fichtinger for the purification of blood and BAL eosinophils; Rustom Falahati, Nenad Tomasevic, Christopher Bebbington, Brad Youngblood, and Emily Brock for Siglec antibodies, and helpful suggestions and discussions; Elle Grevstad for advice on confocal microscopy; and Deane Mosher and Sameer Mathur for suggestions and discussions.
Disclosure statement
MWJ received a fee from Genentech for speaking and funds for research from Hoffmann-La Roche, and was a scientific advisory board member for Genentech, none of which is related to Siglec-8. NNJ has received honoraria from Teva and AstraZeneca for consultation with regards to new therapies for severe asthma, none of which is related to Siglec-8. BSB has current or recent consulting or scientific advisory board arrangements or has received honoraria from Sanofi-Aventis, Teva, GlaxoSmithKline, AstraZeneca, and Allakos, and owns stock in Allakos, Inc. He receives publication-related royalty payments from Elsevier and UpToDate™ and is a co-inventor on existing Siglec-8-related patents and thus may be entitled to a share of royalties received by Johns Hopkins University on the potential sales of such products. BSB is also a cofounder of Allakos, Inc., which makes him subject to certain restrictions under University policy. The terms of this arrangement are being managed by John Hopkins University and Northwestern University in accordance with their conflict of interest policies. EAK and CLN have no conflicts of interest to declare.
Funding source
This work was funded by Program Project grant P01 HL088594 (NNJ and DF Mosher) and Clinical and Translational Science Award grant UL1 RR025011 (MK Drezner) from the National Institutes of Health.
Footnotes
Statement of ethics
Informed written consent was obtained from each subject before participation. These studies were approved by the University of Wisconsin-Madison Health Sciences Institutional Review Board (protocol No. 2016-0021 for bronchoalveolar lavage study and No. 2013-1570 for eosinophil purification).
Author contributions
BSB conceived the study and participated in manuscript revision. MWJ and EAK designed the research, performed experiments, analyzed data, and wrote the manuscript. CLN conducted image analysis. NNJ led the research program on the role of eosinophils in airway inflammation and participated in manuscript revision.
References
- 1.McBrien CN, Menzies-Gow A. The biology of eosinophils and their role in asthma. Front Med. 2017;4:93. doi: 10.3389/fmed.2017.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, Marshall RP, Bradding P, Green RH, Wardlaw AJ, Pavord ID. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med. 2009;360:973–984. doi: 10.1056/NEJMoa0808991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efthimiadis A, Pizzichini E, Hargreave FE, O'Byrne PM. Mepolizumab for prednisone-dependent asthma with sputum eosinophilia. N Engl J Med. 2009;360:985–993. doi: 10.1056/NEJMoa0805435. [DOI] [PubMed] [Google Scholar]
- 4.Kay AB, Phipps S, Robinson DS. A role for eosinophils in airway remodelling in asthma. Trends Immunol. 2004;25:477–482. doi: 10.1016/j.it.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Nhu QM, Aceves SS. Tissue remodeling in chronic eosinophilic esophageal inflammation: Parallels in asthma and therapeutic perspectives. Front Med. 2017;4:128. doi: 10.3389/fmed.2017.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg HF, Dyer KD, Foster PS. Eosinophils: changing perspectives in health and disease. Nat Rev Immunol. 2013;13:9–22. doi: 10.1038/nri3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larose M-C, Archambault A-S, Provost V, Laviolette M, Flamand N. Regulation of eosinophil and group 2 innate lymphoid cell trafficking in asthma. Front Med. 2017;4:136. doi: 10.3389/fmed.2017.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson MW. Eosinophil activation status in separate compartments and association with asthma. Front Med. 2017;4:75. doi: 10.3389/fmed.2017.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Sullivan JA, Carroll DJ, Bochner BS. Glycobiology of eosinophilic inflammation: Contributions of siglecs, glycans, and other glycan-binding proteins. Front Med. 2017;4:116. doi: 10.3389/fmed.2017.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao SP, Ge XN, Sriramarao P. Regulation of eosinophil recruitment and activation by galectins in allergic asthma. Front Med. 2017;4:68. doi: 10.3389/fmed.2017.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kikly KK, Bochner BS, Freeman SD, Tan KB, Gallagher KT, D'Alessio KJ, Holmes SD, Abrahamson JA, Erickson-Miller CL, Murdock PR, Tachimoto H, Schleimer RP, White JR. Identification of SAF-2, a novel siglec expressed on eosinophils, mast cells, and basophils. J Allergy Clin Immunol. 2000;105:1093–1100. doi: 10.1067/mai.2000.107127. [DOI] [PubMed] [Google Scholar]
- 12.Nutku E, Aizawa H, Hudson SA, Bochner BS. Ligation of Siglec-8: a selective mechanism for induction of human eosinophil apoptosis. Blood. 2003;101:5014–5020. doi: 10.1182/blood-2002-10-3058. [DOI] [PubMed] [Google Scholar]
- 13.O'Sullivan JA, Carroll DJ, Cao Y, Salicru AN, Bochner BS. Leveraging Siglec-8 endocytic mechanisms to kill human eosinophils and malignant mast cells. J Allergy Clin Immunol. 2017 doi: 10.1016/j.jaci.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nutku-Bilir E, Hudson SA, Bochner BS. Interleukin-5 priming of human eosinophils alters siglec-8 mediated apoptosis pathways. Am J Respir Cell Mol Biol. 2008;38:121–124. doi: 10.1165/rcmb.2007-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Na HJ, Hudson SA, Bochner BS. IL-33 enhances Siglec-8 mediated apoptosis of human eosinophils. Cytokine. 2012;57:169–174. doi: 10.1016/j.cyto.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kano G, Bochner BS, Zimmermann N. Regulation of Siglec-8-induced intracellular reactive oxygen species production and eosinophil cell death by Src family kinases. Immunobiology. 2017;222:343–349. doi: 10.1016/j.imbio.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll DJ, O'Sullivan JA, Nix DB, Cao Y, Tiemeyer M, Bochner BS. Sialic acid-binding immunoglobulin-like lectin 8 (Siglec-8) is an activating receptor mediating beta2-integrin-dependent function in human eosinophils. J Allergy Clin Immunol. 2017 doi: 10.1016/j.jaci.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen HS, Chang AT, Tomasevic N, Bebbington C. A randomized, double-blind, placebo-controlled, ascending dose phase 1 study of AK002, a novel Siglec-8 selective monoclonal antibody, in healthy subjects. J Allergy Clin Immunol. 2018;141:AB403. (abstract) [Google Scholar]
- 19.Han ST, Mosher DF. IL-5 induces suspended eosinophils to undergo unique global reorganization associated with priming. Am J Respir Cell Mol Biol. 2014;50:654–664. doi: 10.1165/rcmb.2013-0181OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denlinger LC, Kelly EA, Dodge AM, McCartney JG, Meyer KC, Cornwell RD, Jackson MJ, Evans MD, Jarjour NN. Safety of and cellular response to segmental bronchoprovocation in allergic asthma. PLoS One. 2013;8:e51963. doi: 10.1371/journal.pone.0051963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson MW, Kelly EA, Busse WW, Jarjour NN, Mosher DF. Up-regulation and activation of eosinophil integrins in blood and airway after segmental lung antigen challenge. J Immunol. 2008;180:7622–7635. doi: 10.4049/jimmunol.180.11.7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esnault S, Johansson MW, Kelly EA, Koenderman L, Mosher DF, Jarjour NN. IL-3 upregulates and activates human eosinophil CD32 and alphaMbeta2 integrin causing degranulation. Clin Exp Allergy. 2017;47:488–498. doi: 10.1111/cea.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson MW, Gunderson KA, Kelly EA, Denlinger LC, Jarjour NN, Mosher DF. Anti-IL-5 attenuates activation and surface density of beta(2) -integrins on circulating eosinophils after segmental antigen challenge. Clin Exp Allergy. 2013;43:292–303. doi: 10.1111/j.1365-2222.2012.04065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly EA, Esnault S, Liu LY, Evans MD, Johansson MW, Mathur S, Mosher DF, Denlinger LC, Jarjour NN. Mepolizumab attenuates airway eosinophil numbers, but not their functional phenotype, in Asthma. Am J Respir Crit Care Med. 2017;196:1385–1395. doi: 10.1164/rccm.201611-2234OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansson MW, Mosher DF. Activation of beta1 integrins on blood eosinophils by P-selectin. Am J Respir Cell Mol Biol. 2011;45:889–897. doi: 10.1165/rcmb.2010-0402OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson MW, Han ST, Gunderson KA, Busse WW, Jarjour NN, Mosher DF. Platelet activation, P-selectin, and eosinophil beta1-integrin activation in asthma. Am J Respir Crit Care Med. 2012;185:498–507. doi: 10.1164/rccm.201109-1712OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu LY, Sedgwick JB, Bates ME, Vrtis RF, Gern JE, Kita H, Jarjour NN, Busse WW, Kelly EA. Decreased expression of membrane IL-5 receptor alpha on human eosinophils: I. Loss of membrane IL-5 receptor alpha on airway eosinophils and increased soluble IL-5 receptor alpha in the airway after allergen challenge. J Immunol. 2002;169:6452–6458. doi: 10.4049/jimmunol.169.11.6452. [DOI] [PubMed] [Google Scholar]
- 28.Han X, Jorgensen JL, Brahmandam A, Schlette E, Huh YO, Shi Y, Awagu S, Chen W. Immunophenotypic study of basophils by multiparameter flow cytometry. Arch Pathol Lab Med. 2008;132:813–819. doi: 10.5858/2008-132-813-ISOBBM. [DOI] [PubMed] [Google Scholar]
- 29.Johansson MW, Barthel SR, Swenson CA, Evans MD, Jarjour NN, Mosher DF, Busse WW. Eosinophil beta 1 integrin activation state correlates with asthma activity in a blind study of inhaled corticosteroid withdrawal. J Allergy Clin Immunol. 2006;117:1502–1504. doi: 10.1016/j.jaci.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 30.Barthel SR, Jarjour NN, Mosher DF, Johansson MW. Dissection of the hyperadhesive phenotype of airway eosinophils in asthma. Am J Respir Cell Mol Biol. 2006;35:378–386. doi: 10.1165/rcmb.2006-0027OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson MW, Khanna M, Bortnov V, Annis DS, Nguyen CL, Mosher DF. IL-5-stimulated eosinophils adherent to periostin undergo stereotypic morphological changes and ADAM8-dependent migration. Clin Exp Allergy. 2017;47:1263–1274. doi: 10.1111/cea.12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo CB, Liu MC, Galli SJ, Bochner BS, Kagey-Sobotka A, Lichtenstein LM. Identification of IgE-bearing cells in the late-phase response to antigen in the lung as basophils. Am J Respir Cell Mol Biol. 1994;10:384–390. doi: 10.1165/ajrcmb.10.4.7510984. [DOI] [PubMed] [Google Scholar]
- 33.Yu YR, Hotten DF, Malakhau Y, Volker E, Ghio AJ, Noble PW, Kraft M, Hollingsworth JW, Gunn MD, Tighe RM. Flow cytometric analysis of myeloid cells in human blood, bronchoalveolar lavage, and lung tissues. Am J Respir Cell Mol Biol. 2016;54:13–24. doi: 10.1165/rcmb.2015-0146OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng YH, Mao H. Expression and preliminary functional analysis of Siglec-F on mouse macrophages. J Zhejiang Univ Sci B. 2012;13:386–394. doi: 10.1631/jzus.B1100218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiwamoto T, Katoh T, Evans CM, Janssen WJ, Brummet ME, Hudson SA, Zhu Z, Tiemeyer M, Bochner BS. Endogenous airway mucins carry glycans that bind Siglec-F and induce eosinophil apoptosis. J Allergy Clin Immunol. 2015;135:1329–1340. e1329. doi: 10.1016/j.jaci.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bochner BS. "Siglec"ting the allergic response for therapeutic targeting. Glycobiology. 2016;26:546–552. doi: 10.1093/glycob/cww024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flood-Page PT, Menzies-Gow AN, Kay AB, Robinson DS. Eosinophil's role remains uncertain as anti-interleukin-5 only partially depletes numbers in asthmatic airway. Am J Respir Crit Care Med. 2003;167:199–204. doi: 10.1164/rccm.200208-789OC. [DOI] [PubMed] [Google Scholar]