Abstract
Our recent studies have shown that crosstalk between histone deacetylase 5 (HDAC5) and lysine-specific demethylase 1 (LSD1) facilitates breast cancer progression. In this work, we demonstrated that regulatory activity at −356 to −100 bp promoter element plays a critical role in governing HDAC5 transcription. By using DNA affinity precipitation and mass spectrometry, we identified a group of factors that bind to this element. Among these factors, Upstream Transcription Factor 1 (USF1) was shown to play a critical role in controlling HDAC5 transcription. Through screening a panel of epigenetic modifying drugs, we showed that a natural bioactive HDAC inhibitor, sulforaphane, downregulated HDAC5 transcription by blocking USF1 activity. Sulforaphane facilitated LSD1 ubiquitination and degradation in an HDAC5 dependent manner. A comparative microarray analysis demonstrated a genome wide cooperative effect of HDAC5 and LSD1 on cancer related gene expression. shRNA knockdown and sulforaphane inhibition of HDAC5/LSD1 exhibited similar effects on expression of HDAC5/LSD1 target genes. We also showed that coordinated crosstalk of HDAC5 and LSD1 is essential for the antitumor efficacy of sulforaphane. Combination treatment with sulforaphane and a potent LSD1 inhibitor resulted in synergistic growth inhibition in breast cancer cells, but not in normal breast epithelial cells. Furthermore, combined therapy with sulforaphane and LSD1 inhibitor exhibited superior inhibitory effect on MDA-MB-231 xenograft tumor growth. Taken together, our work demonstrates that HDAC5-LSD1 axis is an effective drug target for breast cancer. Inhibition of HDAC5-LSD1 axis with sulforaphane blocks breast cancer growth and combined treatment with LSD1 inhibitor improved the therapeutic efficacy of sulforaphane.
Keywords: reast cancer, HDAC5, LSD1, USF1, sulforaphane, HCI-2509, combination therapy
INTRODUCTION
Epigenetic alterations include posttranslational histone modifications such as acetylation or methylation as well as abnormal DNA methylation in important genes.1, 2 Histone acetylation is typically associated with transcriptionally active chromatin and is a result of a dynamic balance between activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs). Abnormally high expression of HDACs in breast cancer cells may lead to the anomalous loss of expression of genes that are important in curbing tumor growth.3, 4 Our recent work has provided novel insights into molecular mechanisms by which histone deacetylase and demethylase interact in breast cancer cells. We identified a unique feature of HDAC5 in facilitating protein stabilization of FAD-dependent histone demethylase 1 (LSD1), leading to a dysregulated chromatin landscape which functions as an antibraking system in breast cancer development.5 Coordinated overexpression of HDAC5 and LSD1 proteins was observed in breast cancer cell lines and clinical patient samples. By gain- and loss-of-function studies using a breast tumor progression model, we have observed that HDAC5 possesses a critical oncogenic function in driving MCF10A transformation via blocking LSD1 protein degradation and reshaping epigenetic landscape.5 Our findings have revealed an important mechanism about the epigenetic regulation of LSD1 activity by HDAC5 that may lead to an alternative treatment approach against breast cancer.
HDAC5 is a key member of class II HDAC family. The class II HDAC isozymes, HDAC4, 5, 7, and 9, are unique due to their ability to shuttle between the nucleus and cytoplasm.6, 7 HDAC5 has been found to play critical roles in development of many pathogenic conditions including cancer.8, 9 Ozdag et al reported that overexpression of HDAC1, 5, and 7 may serve as a molecular biomarker of malignant versus normal tissues.10 Both HDAC5 and HDAC9 are overexpressed in tumors from high-risk medulloblastoma patients, suggesting a close linkage between their expression and poor survival of patients.11 These findings also imply that HDAC5 overexpression may act as an effective prognostic marker as well as a potential therapeutic target for cancer. However, little is known about the specific roles of HDAC5 in cancer initiation and progression. Therefore, it is important to fully elucidate the changes of molecular events leading to HDAC5 overexpression in cancer.
As the first identified histone demethylase, LSD1 has shown great potential as an effective target in cancer therapy.12–17 The activity of the LSD1 complex has been implicated in tumorigenesis of various cancers.18–20 Thus, there has been increasing interest in testing known compounds or designing new chemical entities that can inhibit LSD1 activity and function as novel therapeutic agents for cancer. Our previous studies have reported that LSD1 activity can be successfully inhibited by specific inhibitors in colorectal and breast cancer cells.12, 13, 21, 22 The rapid development of LSD1 inhibitors has led to the evaluation of several novel LSD1 inhibitors in early phase clinical trials.23–25
In this study, we have investigated the potential effect of targeting crosstalk between HDAC5 and LSD1 as a novel strategy for breast cancer treatment. The data presented here suggest that a natural HDAC inhibitor sulforaphane (SFN) suppresses HDAC5 expression, which in turn destabilizes LSD1 protein. SFN in combination with a novel LSD1 inhibitor has shown improved antineoplastic activity both in vitro and in vivo.
EXPERIMENTAL PROCEDURES
Reagents and cell culture
Sulforaphane was purchased from LKT Laboratories (Minneapolis, MN), Vorinostat (SAHA) was obtained from Cayman Chemical (Ann Arbor, MI). Trichostatin A (TSA) and MG-132 were obtained from Sigma-Aldrich (St. Louis, MO). MC-1568, LBH-589, Belinostat (PXD-101), MS-275 and Romidepsin were from Selleckchem (Houston, TX). HCI-2509 was purchased from Xcess Biosciences Inc (San Diego, CA). MDA-MB-231, MDA-MB-468, BT-474 and MCF-7 cell lines were obtained from ATCC. MCF10A and MCF10A-CA1a lines were provided by Dr. Saraswati Sukumar (Johns Hopkins University). Cells were cultured under the conditions as previously described.26
Plasmid construction
HDAC5 promoter elements were amplified by PCR with primers indicated in Supporting Information Table 1. The PCR products were engineered into the pGL2-Enhancer plasmid. Plasmids pcDNA3.1(+)-FLAG and pcDNA3.1(+)-FLAG-HDAC5 were purchased from Addgene (Cambridge, MA). pReceiver-FLAG-LSD1 was from Gene Copoeia (Rockville, MD). pcDNA3.1(+)-FLAG-Jade-2 was purchased from GenScript (Piscataway, NJ).
RNAi transfection
Pre-designed hUSF1 siRNA (ThermoFisher, Boston, MA), THOC1 siRNA (Santa Cruz) or scramble control siRNA were transfected into cells according to the manufacturer’s protocol. Cells were harvested 48 h post-transfection for further analysis.
Real time qPCR
Quantitative real-time PCR was carried out using the StepOne real-time PCR system as previously described5. All the probes for TaqMan® Gene Expression Assays were predesigned and provided by Life Technologies.
Immunoblotting
Whole cell lysate and nuclear proteins were extracted using methods described previously.26–29 Antibodies sources: H3K4me2, H3K4me1, and acetyl-H3K9 (Millipore, Billerica, MA); USP28 (Abcam, Cambridge, MA); LSD1 and HA (Cell Signaling Technology, Beverly, MA); Flag antibody (Sigma Aldrich); PARP1 (Active Motif, Carlsbad, CA); HDAC5, USF1, THOC1, PCNA and β-actin (Santa Cruz Biotechnology). Membranes were scanned using Li-Cor BioScience Odyssey Infrared Imager (Lincoln, NE).
Luciferase assays
2×105 cells per well were seeded into 24 well plates and 250 ng plasmid DNA were transiently co-transfected with 2.5 ng pRL-TK, a Renilla Luciferase Control Reporter Vector, (Promega, Madison, WI) using Lipofectamine 3000 (ThermoFisher). Cells were harvested 48 h post-transfection and luciferase activity was measured on a GLOMAX® 20/20 luminometer (Promega). Luciferase values (relative light units, RLUs) were normalized to Renilla luciferase activity and expressed as the fold change relative to pGL2-Enhancer transfected wells.
Cellular growth inhibition and drug combination index (CI) analysis
Crystal violet assays were performed as previously described.5, 30 The median effects (IC50) were determined using CalcuSyn software from Biosoft (Cambridge, UK). Effects of synergy, additivity, or antagonism of combination treatment were determined using the Chou-Talalay median effect/combination index (CI) model.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as described previously.21 Primary antibodies against USF1 protein were used for immunoprecipitation of protein–DNA complexes. Forward primer 5’-CATGCTAGCCTCGGCCGAACCCTGTGC-3' and reverse primer 5’-CATAAGCTTACCCCTCCCCTGCCTCT-3' were used for PCR amplification. Sheared genomic DNA was used as a positive control (Input).
Ubiquitination assays
The assay was performed as previously described5. Whole cell lysate was obtained using RIPA lysis buffer. After pre-clearing with Protein G-Plus Agarose beads (Santa Cruz Biotechnology), LSD1 or IgG antibody (Abcam) was added to whole cell lysate and rotated overnight at 4 °C. Then, 40 µl of Protein G-Plus Agarose beads was added for another 2 h. The agarose beads were washed and then subjected to immunoblotting.
In vitro DNA affinity precipitation assays (DAPA) and mass spectrometry analysis
Biotinylated primers (Integrated DNA Technologies, Coralville, IA) were used to generate double-stranded biotinylated HDAC5 promoter probes. For HDAC5 promoter −356 to −100 probe, forward primer 5’-biotin-CATGCTAGCACGATTGCACCATCCACGTTTTG-3' and reverse primer 5’-biotin-CATAAGCTTACCCCTCCCCTGCCTCT-3' were used for PCR amplification. The non-relevant biotinylated probes, sense: 5’-biotin-AGAGTGGTCACTACCCCCTCTG-3’, antisense: 5’-biotin-CAGAGGGGGTAGTGACCACTCT-3’, were also synthesized as a negative control. Streptavidin-agarose bead suspension was added to a mixture of nuclear proteins with double-strand biotinylated oligonucleotides. Mixture was placed on a gently rocking platform for 2 h and was centrifuged. DNA-protein complexes were washed and 2x protein sample buffer (Invitrogen) was added to the avidin-precipitated DNA-protein complex, which was then boiled for 5 min to dissociate the complexes. The proteins were fractionated on SDS acrylamide gels, and silver stained. The silver-stained bands were excised from the gel, and digested in gel with sequence-grade trypsin. The mass spectrometry analysis was performed at Biomedical Mass Spectrometry Center of University of Pittsburgh.
Microarray Analysis of Gene Expression
Total RNA from three independent biological replicates was extracted using QIAgen RNeasy kit (Valencia, CA). cRNA was hybridized to HG U133A 2.0 arrays (Affymetrix, Inc., Santa Clara, CA). The data were processed as RMA files (Affymetrix Robust Multi-Array Average). The raw intensity data were background corrected, log2 transformed, and quantile normalized according to Affymetrix recommendations. Differential gene expression was performed using BRB array tools (NCI). Refer to the Supporting Information material for more detailed methods for microarray processing and analysis.
Animal studies
4–5-week-old female BALB/c nu/nu athymic nude mice (Harlan Labs, Indianapolis, IN) were implanted with 4×106 human breast cancer MDA-MB-231 cells into the mammary fat pad. Five days after implantation, mice were randomly assigned into groups of vehicle control (10% DMSO), SFN (50 mg/kg), HCI-2509 (30 mg/kg), or combination treatment. Mice were treated by intraperitoneal injection once a day for 27 days. Tissues were processed into paraffin sections, and then subjected to hematoxylin-eosin staining at the Histology and Micro-Imaging Core at Magee Womens Research Institute. After staining, samples were examined and photographed by microscope (Nikon, Eclipse 90i).
Statistical analysis
Data were shown as the mean ± s.d. of three independent experiments. The quantitative variables were analyzed by Student’s t-test or One-way ANOVA. p-value <0.05 was considered statistically significant for all tests. GraphPad Prism 6 program (GraphPad Software Inc., La Jolla, CA) was used for statistical analyses.
RESULTS
1. Characterization of transcriptional regulatory activity at HDAC5 promoter
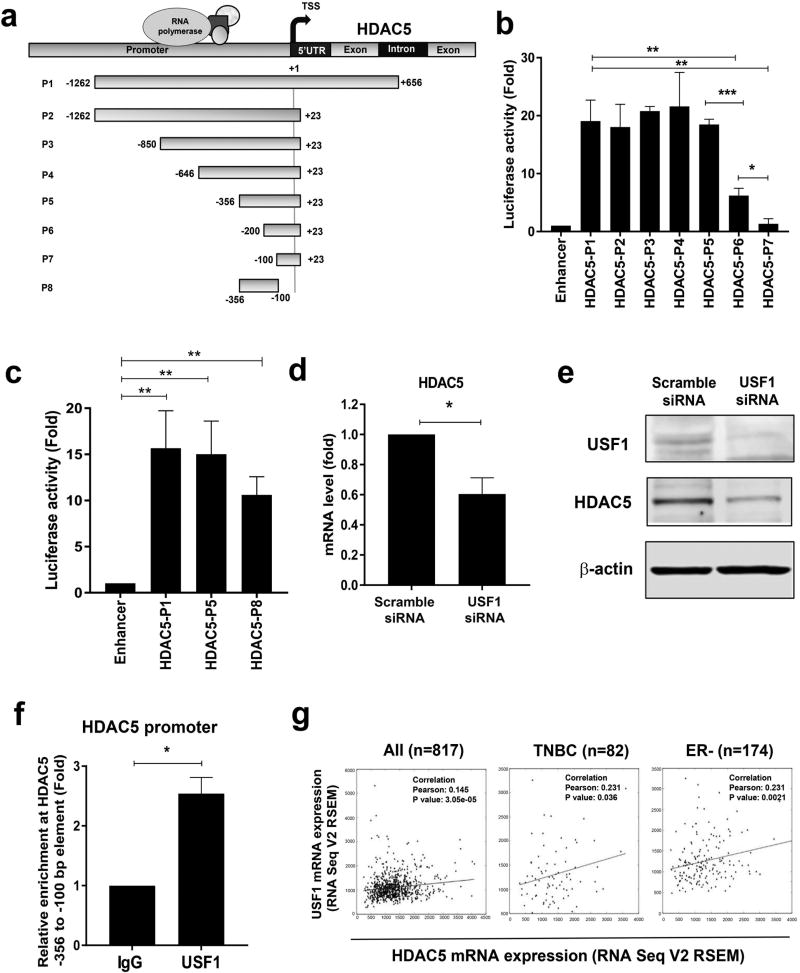
To better understand how changes of HDAC5 transcriptional regulation lead to HDAC5 overexpression in breast tumors, we engineered a series of deletion constructs of the HDAC5 5' flanking promoter elements into luciferase reporter pGL2-Enhancer vector (Fig. 1a). The plasmids were transfected into MDA-MB-231 cells followed by quantitative luciferase activity assays. While deletion of downstream element +24 to +656 bp (P2) or additional truncation of upstream elements from −1262 to −356 bp (P2, P3, P4, P5) exerted no obvious effect on luciferase reporter activity, extra truncation of −356 to −200 bp (P6) significantly attenuated and depletion of −356 to −100 bp (P7) nearly abolished luciferase reporter activity (Fig. 1b). To characterize the role of the −356 to −100 bp element in regulation of HDAC5 transcription, we engineered −356 to −100 bp element (P8) into the pGL2-Enhancer vector and showed that transfection of P8 element could generate significant luciferase activity when compared with the full length P1 or P5 elements (Fig. 1c). The results indicate that regulatory activity from −356 to −100 bp at HDAC5 promoter is essential for transcription regulation of HDAC5.
Figure 1. Analysis of transcriptional activity at HDAC5 promoter.
(a) Map of deletion constructs of HDAC5 promoter and coding region. TSS, transcription start site. (b) MDA-MB-231 cells were co-transfected with pGL2-Enhancer or pGL2-Enhancer-HDAC5 promoter elements and pRL-TK. Reporter gene activities were measured 48 h post-transfection. The relative luciferase activity of fragments P2-P7 was compared with that of full length P1. Transfection of pGL2-Enhancer plasmids was used as negative control. (c) MDA-MB-231 cells were co-transfected with pGL2-Enhancer or constructs of pGL2-Enhancer-HDAC5 promoter elements P1, P5 or P8 and pRL-TK. Reporter gene activities were measured 48 h post-transfection. Transfection of pGL2-enhancer was used as negative control. (d) MDA-MB-231 cells were transiently transfected with scramble or USF1 siRNA for 48 h. Effect of USF1 knockdown on HDAC5 mRNA expression was measured by quantitative PCR with β-actin as an internal control. (e) MDA-MB-231 cells were transfected with scramble or USF1 siRNA for 48 hr. Effect of USF1 knockdown on HDAC5 protein expression was examined by immunoblots with β-actin as loading control. (f) Quantitative ChIP analysis was used to determine the occupancy of USF1 protein at −356 to −100 bp element of HDAC5 promoter. (g) The Pearson correlation between mRNA expression of USF1 (y-axis) and HDAC5 (x-axis) in clinical TNBC or ER negative breast cancer specimens. Bars represent the mean of three independent experiments ± s.d. p<0.05 *, p<0.01 **, p<0.001 ***, Student’s t-test.
To identify coregulatory proteins that are associated with the −356 to −100 bp element (P8), in vitro DNA affinity precipitation assays (DAPA) and mass spectrometry analysis were performed. Biotin end-labeled sense and antisense oligonucleotides corresponding to P8 promoter element were custom synthesized. Non-denatured nuclear proteins from MDA-MB-231 cells were extracted and incubated with P8 oligonucleotides, and SDS-PAGE was performed, followed by silver staining. This experiment showed multiple protein complexes bound to P8 element, but absent from the negative scramble probe (Supporting Information Fig. 1a). Mass spectrometry analysis identified a group of potential binding proteins from recovered samples (Supporting Information Fig. 1b). Through functional analysis, several factors that play a role in chromatin remodeling and transcriptional regulation were selected to validate their physical association with the P8 element, such as polycomb protein SUZ12 (SUZ12), THO complex subunit 1 (THOC1) and upstream stimulatory factor 1 (USF1). Western blot results indicated that these factors exhibited stronger binding ability to P8 probe (Supporting Information Fig. 1c). To determine if activity of these factors is involved in regulation of HDAC5 transcription, MDA-MB-231 cells were transiently transfected with siRNA against these factors. Among these factors, we found that depletion of USF1 exhibits most significant inhibitory effect on HDAC5 mRNA expression (Fig. 1d). Depletion of SUZ12 or THOC1 in MDA-MB-231 cells exerted only marginal effect on the HDAC5 transcription activity (Supporting Information Fig. 2a & 2b). Western blots indicated that USF1 siRNA significantly decreased HDAC5 protein expression (Fig. 1e). To further confirm the binding ability of these factors in living cells, ChIP study was carried out and showed that USF1 is capable of physically binding to the P8 promoter region (Fig. 1f). Taken together, these studies identified USF1 as an important regulatory factor of HDAC5 transcription.
2. USF1 is overexpressed and positively associated with HDAC5 expression in TNBC/ER-breast tumors
In silico analysis using TCGA data (downloaded from GSE62944) indicates a significantly elevated mRNA level of USF1 in breast tumor specimens (n=1095) compared with normal tissue samples (n=113). Among these breast cancer tumors, estrogen receptor (ER) negative tumors (n=237) express significantly higher USF1 than ER positive counterparts (n=808) (Supporting Information Fig. 3a & 3b). Study of USF1 expression across all molecular subtypes of breast cancer showed that USF1 mRNA level is higher in basal-like tumors as compared to other breast cancer subtypes, such as Her2+, Luminal A, Luminal B, or normal-like (Supporting Information Fig. 3c & 3d). Analysis of TCGA database also showed that clinical TNBC specimens express significantly higher level of USF1 mRNA when compared with non-TNBC tissues (p=0.0013) (Supporting Information Fig. 3e). By assessing Pearson correlation coefficient, positively correlated mRNA expressions between USF1 and HDAC5 were observed. Triple negative breast cancer (TNBC) or ER negative tumors exhibit a stronger positive correlation than ER or Her2 positive subtypes (Fig. 1g & Supporting Information Fig. 3f). These data suggest that USF1 is positively correlated with HDAC5 expression in more aggressive subtypes of breast cancer which may warrant further investigation into its role in aggressive phenotypes of breast cancer.
3. Sulforaphane suppresses transcriptional activity of HDAC5 in breast cancer cells
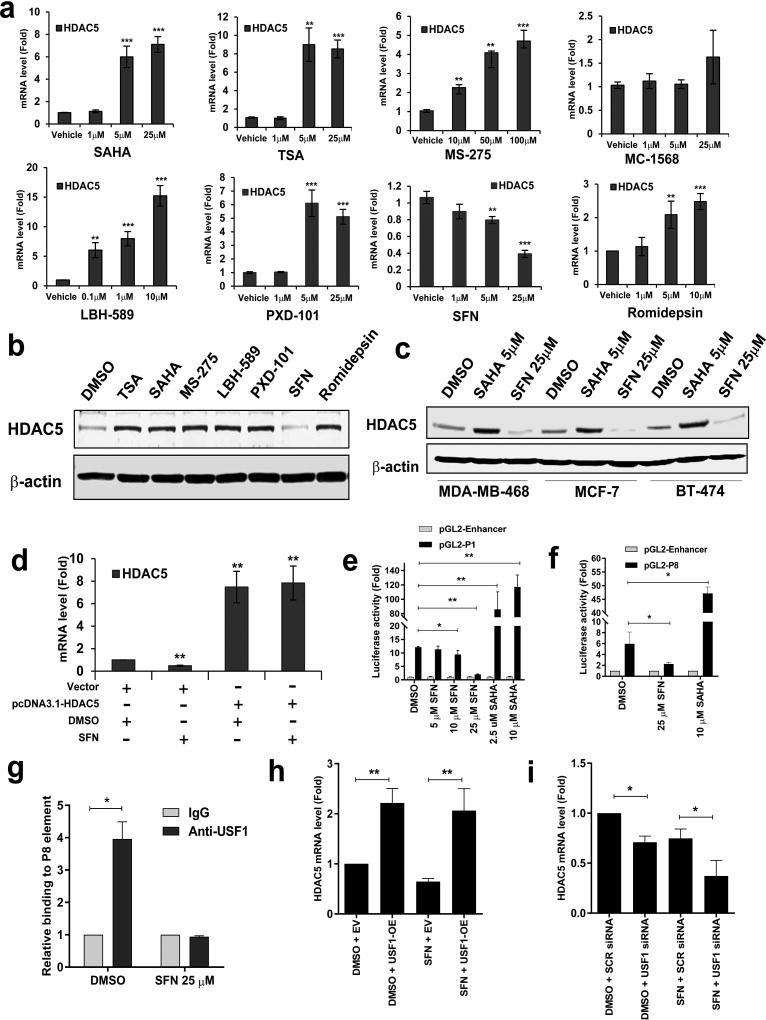
Next, we tested the effect of clinically relevant HDAC inhibitors (HDACi) on HDAC5 expression. The selected HDACi compounds include: hydroxamic acid derivatives SAHA (Vorinostat), TSA (Trichostatin A), LBH-589 (Panobinostat), and PXD-101 (Belinostat); a benzamide analog MS-275 (Entinostat); a selective class II HDAC inhibitor MC-1568; a natural product from the bacterium Chromobacterium violaceum Romidepsin and a natural bioactive HDACi sulforaphane (SFN). In MDA-MB-231 cells, treatment with most of the HDACis led to a significant increase of HDAC5 mRNA expression in a dose-dependent style. However, exposure of cells to SFN significantly inhibited mRNA expression of HDAC5 (Fig. 2a). While protein expression of HDAC5 was induced by TSA, SAHA, MS-275 LBH-589, PXD-101 and Romidepsin, SFN suppressed HDAC5 protein expression in MDA-MB-231 cells (Fig. 2b). A similar effect of SAHA and SFN on HDAC5 protein expression was also found in additional breast cancer cell lines including TNBC MDA-MB-468 and MCF10A-CA1a, ER+ MCF-7 and HER2+ BT474 cells (Fig. 2c; Supporting Information Fig. 4a). These results suggest that SFN, which is different from most other classes of HDACi, exhibited potent inhibitory effect against HDAC5 expression in breast cancer cells.
Figure 2. Effect of HDAC inhibitors on expression of HDAC5 in human breast cancer cells.
(a) After MDA-MB-231 cells were exposed to a variety of HDAC inhibitors for 24 h, mRNA expression of HDAC5 was quantitatively measured by real-time PCR. β-actin gene was used as an internal control. (b) MDA-MB-231 cells were treated with TSA (5 µM), SAHA (5 µM), MS-275 (50 µM), LBH-589 (1 µM), PXD-101 (5 µM), SFN (25 µM) and Romidepsin (5 µM) for 24 h. Whole cell lysates were extracted analyzed for protein expression of HDAC5 through Western blotting. β-actin was used as a loading control. (c) MDA-MB-468, MCF-7 or BT-474 cells were treated with 5 µM SAHA or 25 µM SFN for 24 h. Whole cell lysates were analyzed for protein expression of HDAC5 by Western blotting. β-actin was used as a loading control. (d) MDA-MB-231 cells transfected with pcDNA3.1 or pcDNA3.1-HDAC5 plasmids were treated with 25 µM SFN for 24 h. mRNA expression of HDAC5 was analyzed by quantitative RT-PCR with β-actin as an internal control. (e) pGL2-Enhancer plasmids or pGL2-P1 construct (–1262 to +656 bp) were transfected into MDA-MB-231 cells followed by treatment with indicated concentrations of SFN or SAHA for 24 h. Reporter gene activities were then measured. (f) pGL2-Enhancer or pGL2-P8 construct (–356 to −100 bp) were transfected into MDA-MB-231 cells followed by treatment with 25 µM SFN or 10 µM SAHA for 24 h. Reporter gene activities were then measured. (g) MDA-MB-231 cells were treated with DMSO or 25 µM SFN for 24 h. Quantitative ChIP analysis was used to determine the occupancy of USF1 at P8 element of HDAC5 promoter. (h) MDA-MB-231 cells were transiently transfected with USF1 expression plasmid followed by treatment with 25 µM SFN for 48 h. mRNA expression of HDAC5 was measured by qPCR. (i) MDA-MB-231 cells were transiently transfected with USF1 siRNA followed by treatment with 25 µM SFN for 48 h. mRNA expression of HDAC5 was measured by qPCR. β-actin was used as an internal control. All experiments were performed three times and showed similar results. Bars represent the mean of three independent experiments ± s.d. p<0.05 *, p<0.01 **, p<0.001 ***, Student’s t-test.
4. Sulforaphane suppresses transcriptional activity of −356 to −100 bp element at HDAC5 promoter
SFN failed to suppress exogenous HDAC5 mRNA expression driven by CMV promoter in MDA-MB-231 cells transfected with pcDNA3.1-HDAC5 plasmid (Fig. 2d), suggesting that SFN inhibited HDAC5 mRNA expression through repression of the transcriptional activity at its natural promoter. While SFN significantly inhibited the luciferase reporter activity in MDA-MB-231 cells or MDA-MB-468 cells, exposure of both cell lines to SAHA promoted reporter gene activity of the full-length construct (P1) (Fig. 2e & Supporting Information Fig. 4b). An opposite effect of SFN and SAHA on luciferase activity of P8 element was also detected in MDA-MB-231 cells (Fig. 2f). Treatment with SFN inhibited mRNA and protein expression of USF1 in a dose dependent manner (Supporting Information Fig. 4c & 4d). ChIP study showed that binding of USF1 to P8 element was abolished by SFN (Fig. 2g). A rescue study indicated that USF1 overexpression prevented downregulation of HDAC5 mRNA expression by SFN in MDA-MB-231 cells (Fig. 2h). Moreover, knockdown of USF1 by siRNA significantly enhanced SFN-induced suppression of HDAC5 transcription (Fig. 2i). These results suggest that USF1 plays an important role in regulating SFN-mediated HDAC5 transcription activity.
5. Sulforaphane destabilizes LSD1 protein in an HDAC5 dependent manner
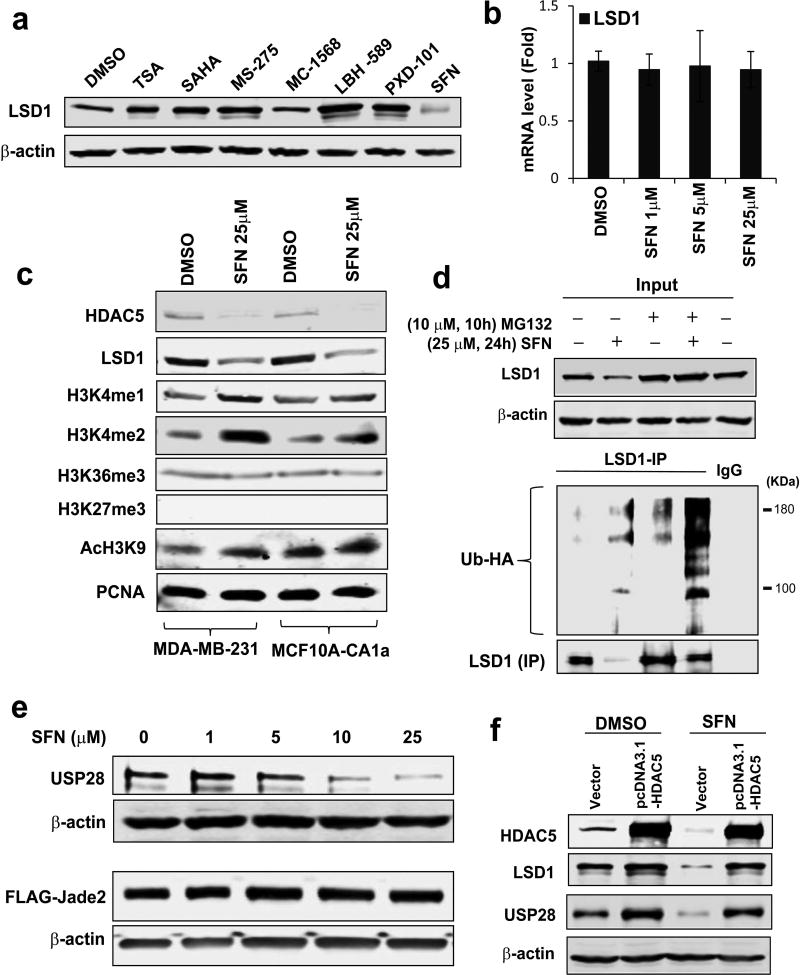
We further addressed whether SFN could disrupt activity of the HDAC5-LSD1 axis. Immunoblotting studies showed that unlike other HDAC inhibitors, SFN significantly inhibited expression of LSD1 protein (Fig. 3a). Exposure of MDA-MB-231 to SFN resulted in similar inhibition of HDAC5 and LSD1 protein in a dose-dependent manner (Supporting Information Fig. 5). qPCR studies demonstrated that LSD1 mRNA level was not affected by SFN treatment in MDA-MB-231 cells (Fig. 3b). SFN treatment increased nuclear levels of H3K4me1/2 and AcH3K9 without altering methylation of H3K36 and H3K27 in MDA-MB-231 or MCF10A-CA1a cells which is a transformed malignant line of MCF10A (Fig. 3c). We recently demonstrated that HDAC5 stabilizes LSD1 protein through down-regulation of LSD1 ubiquitination and degradation.5 To decipher whether SFN destabilizes LSD1 protein by regulating its ubiquitination modification, protein ubiquitination assays were conducted and showed that exposure to SFN led to a profound increase of LSD1 polyubiquitination in MDA-MB-231 cells (Fig. 3d). Next, we assessed the potential effect of SFN on protein expression of LSD1 E3 ubiquitin ligase and deubiquitinase, Jade-2 or USP28. Since the absence of specific antibody against Jade-2, plasmids expressing Jade-2-FLAG fusion protein were used as an alternative approach. Whereas depletion of HDAC5 did not alter Jade-2 protein expression, treatment with SFN significantly decreased USP28 protein levels (Fig. 3e). We performed a rescue expression of HDAC5 in MDA-MB-231 cells through transfection of pcDNA3.1-HDAC5 vector and observed that SFN had no effect on exogenous HDAC5 protein. SFN-mediated downregulation of LSD1 protein and USP28 was apparently reversed (Fig. 3f). These results demonstrate that SFN downregulates LSD1 protein stability through affecting LSD1-associated ubiquitination activity, which is largely dependent on HDAC5 activity.
Figure 3. SFN downregulates LSD1 protein stability through HDAC5 modulated LSD1 ubiquitination system.
(a) MDA-MB-231 cells were treated with TSA (5 µM), SAHA (5 µM), MS-275 (50 µM), MC-1568 (25 µM), LBH-589 (1 µM), PXD-101 (5 µM) and SFN (25 µM) for 24 h. Whole cell lysates were extracted and Western blotting was performed to analyze the expression of LSD1 protein. β-actin was used as a loading control. (b) MDA-MB-231 cells was treated with 25 µM SFN for 24 h. mRNA expression of LSD1 was measured by qPCR. β-actin was used as an internal control. (c) MDA-MB-231 and MCF10A-CA1a cells were exposed to 25 µM SFN for 24 h and analyzed for expression of indicated chromatin marks by immunoblots. PCNA was used as a loading control. (d) MDA-MB-231 cells were treated with 25 µM SFN for 24 h followed by treatment with proteasome inhibitor 10 µM MG132 for 10 h. IP was carried out using LSD1 antibody and immunoblots with anti-HA or LSD1 antibodies. (e) MDA-MB-231 cells were treated with increasing concentrations of SFN for 24 h. Whole cell lysates were analyzed for protein levels of USP28 and FLAG-Jade2. β-actin was used as loading control to normalize target protein levels. (f) MDA-MB-231 cells were transfected with empty or HDAC5 expression vectors for 48 h followed by treatment with 25 µM SFN for 24 h. Immunoblotting was performed for expression of HDAC5, LSD1 and USP28. All experiments were performed three times. Bars represent the mean of three independent experiments ± s.d. *p<0.05, **p<0.01, ***p<0.001, Student’s t-test.
6. Effect of sulforaphane on genome-wide transcription targets of the HDAC5/LSD1 complex
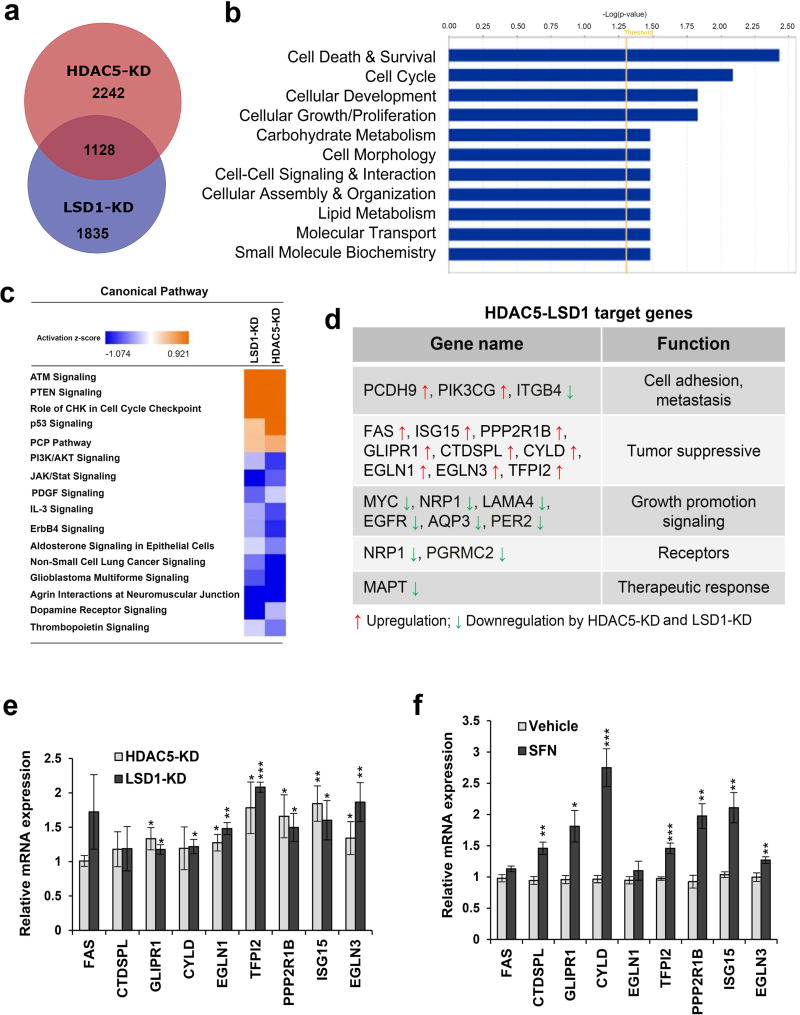
To define a comprehensive profile of genes whose expression is regulated by HDAC5/LSD1 complex, we performed genome-wide gene expression analysis in MDA-MB-231 cells with stable knockdown of HDAC5 or LSD1 by shRNA. We identified 3370 and 2963 genes whose expression was significantly changed by inhibition of HDAC5 and LSD1, respectively (Fig. 4a). Data has been deposited into Gene Expression Omnibus as GSE72687. Strikingly, more than 30% of genes in each group were overlapping and regulated by HDAC5-KD and LSD1-KD, reflecting a comprehensive genome wide cooperative effect of HDAC5 and LSD1 on target gene expression. A functional pathway analysis using the Ingenuity Pathway Analysis application showed that there are multiple important cellular networks whose activities are significantly altered by depletion of HDAC5 or LSD1 (Fig. 4b). These networks are associated with cell death & survival, cell cycle, cellular development, cellular growth/proliferation, carbohydrate metabolism, cell morphology, cell-cell signaling & interaction, cellular assembly & organization, lipid metabolism, molecular transport, small molecule biochemistry, etc. Figure 4c lists the top canonical pathways that are regulated by HDAC5-LSD1 complex. Depletion of HDAC5 or LSD1 activates several key tumor suppressive signaling pathways such as ATM, PTEN, p53, etc, and downregulates multiple tumor promoting signaling pathways including PI3K/AKT. JAK/Stat, PDGF, etc. Among the differentially expressed genes (DEGs) potentially regulated by HDAC5/LSD1 complex, we identified a subset of genes whose activities are associated with critical cellular processes in cancer - cell adhesion, metastasis, tumor suppression and cell growth, receptors, therapeutic response, etc (Fig. 4d). Among these genes, a group of important tumor suppressor genes (TSGs) was shown to be induced by either HDAC5 or LSD1 inhibition. These TSGs include FAS, CTDSPL, ISG15, GLIPR1, CYLD, EGLN1, TFPI2, PPP2R1B, EGLN3, etc. Induction of most of these TSGs was validated by qPCR (Fig. 4e). Furthermore, the effect of SFN on expression of these TSGs was examined in MDA-MB-231 cells. SFN treatment significantly induced expression of most of the TSG genes tested (Fig. 4f), showing a similar effect on activation of TSGs by SFN or HDAC5/LSD1 inhibition.
Figure 4. Genome-wide microarray analysis.
(a) Venn diagram illustration of gene expression similarity between HDAC5-KD and LSD1-KD microarray data. Numbers shown depict genes whose expression was significantly altered in the knockdown cell line compared to scramble cell line and the union of both datasets shows the number of genes significantly altered in both knockdown cell lines. (b) Functional analysis of genes whose expression is modulated by stable knockdown of both HDAC5 and LSD1 in MDA-MB-231 cells. The bar graphs were identified by Ingenuity Pathway Analysis (IPA). The statistically significant biological functions changed by knockdown of both genes are shown. Fisher’s exact test was used to calculate the significance (p<0.05). The threshold line shows the cutoff for significance. (c) Top canonical pathways affected by stable knockdown of HDAC5 in MDA-MB-231 cells. Ingenuity Pathway Analysis “Core analysis” enriched top canonical pathways are shown here. Straight orange vertical line running through the bars is threshold for p-value for the particular pathway’s enrichment. Bars represent overlap of genes from dataset with genes from that canonical pathway. (d) List of representative target genes of HDAC5-LSD1 complex which have critical roles in cellular processes of breast cancer. (e) Relative expression levels of HDAC5 target genes identified by HDAC5-KD microarray were validated in MDA-MB-231 cells depleted of HDAC5 or LSD1 using qPCR. (f) MDA-MB-231 cells were treated with 25 µM SFN or vehicle DMSO for 24 hr. mRNA expression was measured by qPCR for a group of tumor suppressor genes (TSGs) regulated by HDAC5-KD/LSD1-KD. β-actin was used as an internal control. Bars depict mRNA level as a fold change compared to that in vehicle treated cells. All experiments were performed three times. Student’s t-test was performed to assess significance. p<0.05 *, p<0.01 **, p<0.001 ***.
7. The HDAC5-LSD1 axis is essential for breast cancer cell sensitivity to sulforaphane
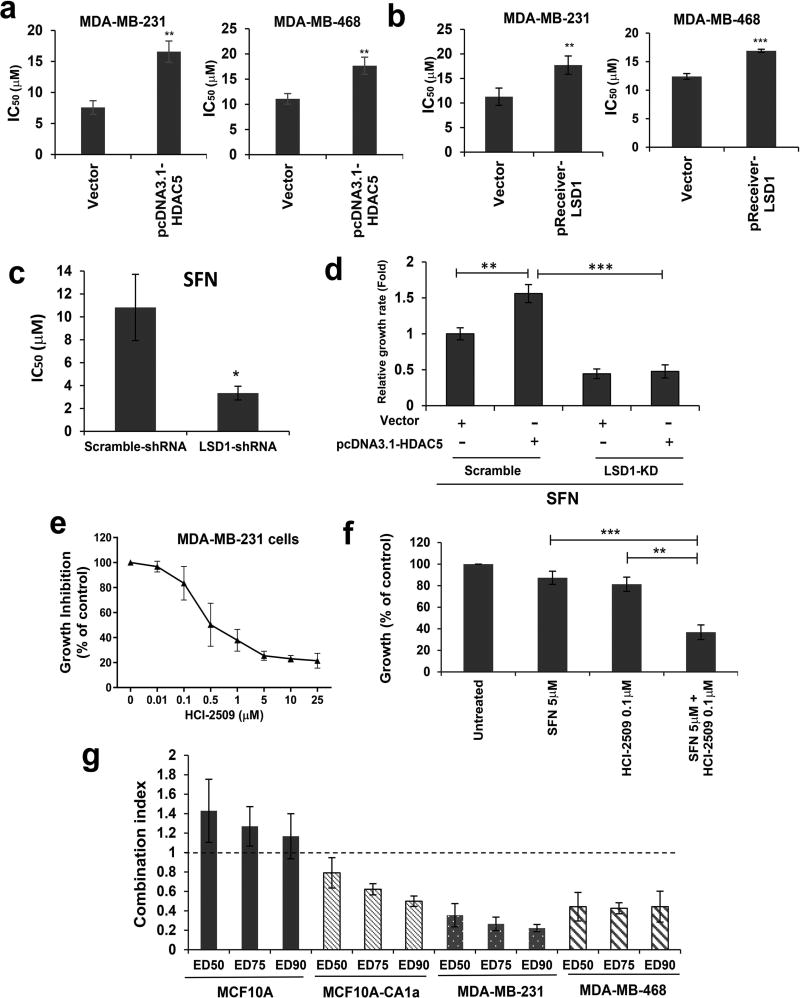
To determine whether HDAC5-LSD1 axis plays a role in regulating breast cancer sensitivity to SFN, MDA-MB-231 and MDA-MB-468 cells were transfected with pcDNA3.1-HDAC5 or pReceiver-LSD1 plasmids. In both cell lines, overexpression of HDAC5 or LSD1 protein increased resistance to SFN-mediated growth inhibition which was indicated by significantly increased IC50 values (Fig. 5a & 5b). shRNA-mediated depletion of LSD1 sensitized MDA-MB-231 cells to SFN which was evidenced by significantly reduced IC50 value (Fig. 5c). A rescue experiment showed that HDAC5 overexpression attenuated cellular sensitivity to SFN in MDA-MB-231-Scramble cells but was obviously reversed by LSD1 depletion in MDA-MB-231-LSD1-KD cells (Fig. 5d). Taken together, these results demonstrate that LSD1 acts as an important downstream effector of HDAC5 signaling in regulating cellular sensitivity to SFN in breast cancer.
Figure 5. LSD1 inhibitor sensitizes breast cancer cells to SFN-induced growth inhibition.
(a) MDA-MB-231 and MDA-MB-468 cells were transiently transfected with pcDNA3.1-HDAC5 flag plasmid followed by treatment with a series of concentrations of SFN for 48 h. Cell proliferation was analyzed by crystal violet assays. The median effects (IC50) were determined by CalcuSyn software. (b) MDA-MB-231 and MDA-MB-468 cells were transiently transfected with pReceiver-LSD1-flag plasmid followed by treatment with a series of concentrations of SFN for 48 h. Cell proliferation was analyzed by crystal violet assays. The median effects (IC50) were determined by CalcuSyn software. (c) Scramble or LSD1-shRNA transfected MDA-MB-231 cells were treated with 25 µM SFN for 72 h. Cell proliferation was analyzed by crystal violet assays. (d) MDA-MB-231-Scramble or MDA-MB-231-LSD1-KD cells were transfected with control vector pcDNA3.1 or pcDNA3.1-HDAC5 followed by treatment with 25 µM SFN for 72 h. Crystal violet assays for cell proliferation were carried out. (e) MDA-MB-231 cells were treated with increasing concentrations of HCI-2509 for 72 h followed by crystal violet growth assays. (f) MDA-MB-231 cells were treated with 5 µM SFN, 0.1 µM HCI-2509 or both drugs for 72 h. Cell proliferation was analyzed by crystal violet assays. (g) Effect of combination therapy on growth of breast cancer cells. Synergy was defined as any CI < 1, additivity as CI = 1 and antagonism as any CI > 1. Shown are means ± s.d. of three independent experiments. Student’s t-test was performed to assess significance. p<0.05 *, p<0.01 **, p<0.001 ***.
HCI-2509 is a highly potent, specific, non-MAOA and MAOB inhibitor of LSD1 which binds the FAD pocket of the enzyme.31 Proliferation assay showed that MDA-MB-231 cells were susceptible to HCI-2509 induced growth inhibition with a low IC50 value of 0.5 µM. (Fig. 5e). Combined treatment with low dose of SFN (5 µM) and HCI-2509 (0.1 µM) generated great synergistic effect on growth inhibition in MDA-MB-231 cells (Fig. 5f & Supporting Information Fig. 6a). We determined the growth inhibitory effect of combined treatment with HCI-2509 with SFN in multiple breast cancer cell lines and normal breast epithelial MCF10A cells using the combination index (CI) of growth inhibition via the Chou-Talalay model.32, 33 While an antagonistic effect of SFN and HCI-2509 was obviously seen in MCF10A cells (CI > 1 indicates antagonism), combination therapy exhibited significant synergy in hindering growth of breast cancer cell lines MCF10A-CA1a, MDA-MB-231 or MDA-MB-468 (CI < 1 indicates synergy) (Fig. 5g). In addition, combination therapy resulted in a robust increase of nuclear H3K4me2 level (Supporting Information Fig. 6b), indicating an apparent synergy between SFN and HCI-2509 against LSD1 activity.
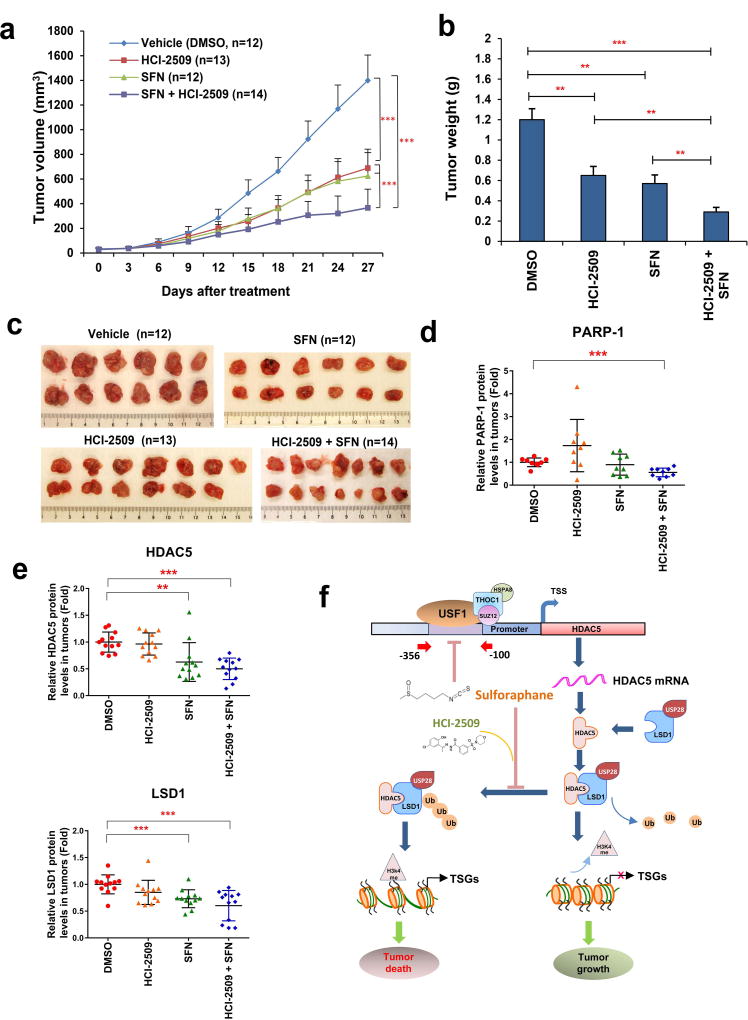
8. Sulforaphane in combination with LSD1 inhibitor profoundly inhibits growth of breast tumor xenografts in mice
To evaluate whether the above promising in vitro results may translate into in vivo therapeutic efficacy, we investigated the antineoplastic effect of combination therapy in athymic nude mice bearing MDA-MB-231 xenografts. While treatment with either SFN or HCI-2509 alone significantly inhibited the proliferation of MDA-MB-231 xenografts, the combined treatment displayed superior inhibitory effect against tumor growth (Fig. 6a). Average tumor weights were significantly lower in mice receiving combination treatment compared to those treated with either SFN or HCI-2509 alone (Fig. 6b). Statistical analysis between each group is shown in Supporting Information Table 2. At the end of experiments, xenograft tumors were extracted for further analysis of expression of key markers (Fig. 6c). To determine whether combination therapy promotes tumor cell apoptosis, PARP-1 cleavage was analyzed. Quantitative analysis showed that only the combination treatment significantly reduced the level of full-length PARP-1, which is cleaved in smaller fragments during apoptosis (Fig. 6d). The in vivo effect of drug treatment on protein expression of HDAC5 and LSD1 in xenograft tumors was also evaluated. Immunoblots showed that expression of both HDAC5 and LSD1 was attenuated in tumors treated with SFN alone or in combination with HCI-2509 (Fig. 6e). During the entire course of experiment, drug toxicity was acceptable as demonstrated by modest weight loss observed in animals with combination treatment, and no animal lethality occurring in any treatment group (Supporting Information Fig. 7a). Statistical analysis of animal weight between groups is shown in Supporting Information Table 3. To further evaluate in vivo toxicity, we performed hematoxylin-eosin (H&E) staining of animal livers and kidneys, and found no apparent changes in these tissues treated with SFN or HCI-2509 alone, or in combination (Supporting Information Fig. 7b). Collectively, these results indicate that SFN monotherapy effectively inhibits growth of MDA-MB-231 xenografts in vivo and exhibits significantly enhanced growth inhibition when used in combination with an LSD1 inhibitor. Based on these findings, we proposed a model to summarize the role of HDAC5–LSD1 axis in mediating antineoplastic effect of natural HDAC inhibitor sulforaphane in human breast cancer (Fig. 6f).
Figure 6. LSD1 inhibitor sensitizes MDA-MB-231 xenografts to sulforaphane induced growth inhibition in nude mice.
(a) MDA-MB-231 cells were transplanted into the mammary gland fat pads of athymic nude mice. Seven days after implantation, Sulforaphane (50 mg/kg, n=12), HCI-2509 (30 mg/kg, n=13), combination (SFN 50 mg/kg + HCI-2509 30 mg/kg, n=14) or vehicle (DMSO, n=12) were delivered via i.p. injection once a day for 5 days/week × 4 weeks. The vertical bars indicate mean tumor size (mm3) ± s.d. One-way ANOVA was performed to determine statistical significance between groups, with p < 0.05 considered significant. (b) Weight of animal tumors was measured at the end of experiment. (c) Images of xenograft tumors from each treatment group were taken at the end of experiment. (d) Protein was extracted from tumor samples of MDA-MB-231 xenografts treated with vehicle, HCI-2509, SFN or combination (n=12 for each group). Quantitative immunoblotting was used to determine the expression of full-length PARP-1 protein. β-actin was used as loading control. (e) Protein was extracted from tumor samples of MDA-MB-231 xenografts treated with vehicle, HCI-2509, SFN or combination (n=12 for each group). Quantitative immunoblotting was used to determine the expression of LSD1 and HDAC5 proteins. β-actin was used as loading control. (f) Proposed model of the role of HDAC5–LSD1 axis in mediating antineoplastic effect of SFN in human breast cancer. A complex of multiple factors (USF1, SUZ12, THOC1, HSPA8, etc.) were identified to be associated with −356 to −100 bp element at HDAC5 promoter. Among these factors, USF1 was shown to play an important role in governing HDAC5 transcription. A natural bioactive HDACi, sulforaphane (SFN), downregulates HDAC5 transcription by blocking USF1 activity. SFN facilitates LSD1 degradation through enhancing protein stability of LSD1 deubiquitinase USP28 in an HDAC5 dependent manner. SFN increases H3K4me2 level at promoter of tumor suppressor genes (TSGs) and promoted expression of TSGs. Combination treatment with SFN and a potent LSD1 inhibitor HCI-2509 results in synergistic growth inhibition of breast cancer proliferation in cell culture and xenograft models. TSGs, tumor suppressor genes; Ub, ubiquitination. HSPA8, Heat shock cognate 71 kDa protein.
DISCUSSION
Class II HDACs have been intensively investigated for their ability to regulate gene transcription and shape the epigenetic landscape. This class of HDAC enzymes have been recognized as important regulators of numerous cellular processes in human diseases. The increasing knowledge on regulatory signaling pathways of class II HDACs has provided novel targets and approaches for potential clinical intervention in cancer. HDAC5 is a key class II HDAC isozyme that has been shown to possess critical roles in many diseases including cancers.8, 9, 34, 35 Our recent tissue microarray study found that breast tumors expressed overall higher levels of HDAC5 protein compared to the matched adjacent normal tissues.5 Importantly, our analysis showed that elevated HDAC5 protein expression is positively correlated with higher stages of clinical breast cancer.5 These findings suggest that elevated expression of HDAC5 may serve as a potential novel prognostic marker as well as a possible therapeutic target for aggressive breast cancer. However, little is known about the regulatory roles of HDAC5 or other class II HDACs in human breast cancer. In this study, we explored the molecular mechanisms by which HDAC5 mRNA expression is upregulated during breast cancer progression. Through engineering a series of HDAC5 5' flanking promoter deletion elements in luciferase reporter plasmids, we showed that activity of an element at −356 to −100 bp of the HDAC5 promoter is essential in mediating its transcriptional activity. Further use of in vitro DAPA and mass spectrometry analysis identified USF1 as an important regulatory factor that binds to −356 to −100 bp element at HDAC5 promoter. USF1 is a member of the basic helix-loop-helix leucine zipper family and functions as a cellular transcription factor to activate transcription through pyrimidine-rich initiator elements and E-box motifs.36, 37 The dysfunction of USF1 has been reported to be linked with multiple human diseases and disorders, such as lipid metabolism, atherosclerosis, and acute cardiovascular events.38 The precise roles of USF1 activity in breast initiation and progression are still unclear. In silico data analysis indicates a significant elevation of USF1 expression in aggressive basal-like or ER negative breast tumors versus other breast cancer subtypes or normal adjacent tissues, suggesting a positive correlation between USF1 overexpression and aggressive phenotypes of breast cancer. Continuous exploration of the underlying mechanisms would aid in understanding how USF1 upregulates HDAC5 expression and the clinical impact of elevated USF1 expression on the risk stratification of breast cancer patients.
In our recent study, we have showed for the first time that HDAC5 stabilizes LSD1 protein through regulation of LSD1 associated ubiquitin-proteasome enzyme.5 To further explore whether the HDAC5-LSD1 axis has potential to be a novel therapeutic target for breast cancer, we surveyed a panel of clinically relevant HDAC inhibitors for their ability to alter the activity of the HDAC5-LSD1 axis. While most of the clinically relevant HDAC inhibitors significantly upregulated transcriptional level of HDAC5 which in turn led to increased expression of LSD1 protein, we found that SFN potently inhibited HDAC5 transcription in multiple breast cancer cell lines. SFN is generated by the hydrolytic conversion of glucoraphanin after ingestion of cruciferous vegetables, particularly broccoli and broccoli sprouts. Since Myzak et al. first reported that SFN inhibited in vitro HDAC activity by its two major metabolites, SFN-cysteine and SFN-N-acetylcysteine, numerous studies have demonstrated that SFN exhibits inhibitory effect against HDAC activity in many types of cancers including breast cancer.39–43 We have reported that SFN blocked growth, activated apoptosis, inhibited HDAC activity, and decreased the expression of key proteins involved in breast cancer proliferation.41 But the mechanisms of the inhibitory effect of SFN on HDAC activity in breast cancer has not been fully elucidated. In this study, we obtained new evidence to show that SFN downregulated HDAC5 mRNA expression, largely through inhibiting the transcriptional activities at the −356 to −100 bp element of gene promoter, therefore identifying this element and its associated factors as important targets for SFN. Future clarification of the function of key coregulatory proteins/complexes associated with this element would aid in elucidating the precise mechanism of SFN-induced downregulation of HDAC5 transcription in breast cancer. Our recent study showed that suppression of active histone marks H3K4 methylation and H3K9 acetylation mediated by enhanced activity of HDAC5-LSD1 signaling at promotes of tumor suppressor genes (TSGs) are important chromatin signature contributing to silencing of key TSGs in breast cancer cells.5 SFN significantly increases levels of both H3K4me and Acetyl H3K9, suggesting that SFN may act as an important epigenetic modulator to reactivate expression of TSGs through inhibiting crosstalk between LSD1 and HDAC5 in breast cancer cells.
We found that the suppressive effect of SFN on transcriptional activity was unique among the tested panel of HDAC inhibitors (HDACi), including clinically approved SAHA and Romidepsin. Several studies indicated that inherent resistance of HDACi was commonly observed in clinical trials of breast cancer patients.44–48 However, the mechanism of HDACi resistance in breast cancer is still unclear. Based on the findings from our work, we speculate that enhanced HDAC5 expression in response to treatment with conventional HDACi could contribute to refractoriness to HDACi therapy. Additional evidence supporting this hypothesis includes the findings that overexpression of HDAC5 or LSD1 in breast cancer cells significantly reduced sensitivity to growth inhibition mediated by several HDAC inhibitors. Future work is needed to explore the potential strategy of combining reagents targeting the HDAC5-LSD1 axis with clinically approved HDAC inhibitors to improve their therapeutic efficacy in breast cancer.
We recently demonstrated that HDAC5 promotes LSD1 protein stability through inhibition of the LSD1 associated ubiquitin-proteasome system, suggesting that the modulation of LSD1 protein stability by HDAC5 is a post-translational activity.5 In the current study, we showed that SFN suppresses HDAC5 expression, subsequently leading to degradation of LSD1 protein. SFN destabilizes LSD1 protein through inhibition of the LSD1 deubiquitinase, USP28, without affecting LSD1 mRNA expression. These results were further validated by a rescue strategy with overexpression of exogenous HDAC5 cDNA lacking a native promoter, showing that SFN treatment indeed leads to degradation of LSD1 protein in an HDAC5-dependent manner. Treatment with SFN or LSD1 inhibitor alone significantly inhibited the growth of MDA-MB-231 xenograft tumors in nude mice, but the greatest inhibition of tumor growth was observed when these two drugs were used in combination. These data clearly suggest that inhibition of HDAC5-LSD1 pathway by SFN in combination with a potent LSD1 inhibitor may serve as an effective approach to reduce non-specific side effects of SFN in breast cancer. Given that the inherent resistance to HDACi develops as a result of combined multi-factorial epigenetic abnormalities, our findings provide a rational basis for clinical trials combining agents targeting these dysregulated epigenetic targets in breast cancer.
As summarized in Fig. 6f, we have demonstrated that HDAC5-LSD1 axis is an effective drug target in breast cancer. Inhibition of HDAC5-LSD1 axis with sulforaphane suppresses breast cancer growth in vitro and in vivo. Notably, combined treatment with a novel and potent LSD1 inhibitor improves the anticancer efficacy of sulforaphane in breast cancer cells.
Supplementary Material
Novelty and Impact.
Our studies shed new light on regulatory mechanisms of HDAC5 transcription and identify that sulforaphane, a natural bioactive HDAC inhibitor, inhibited HDAC5 transcription through downregulation of USF1 that in turn destabilizes LSD1 protein in breast cancer cells. These novel findings suggest that targeting HDAC5-LSD1 axis by sulforaphane in combination with LSD1 inhibitor may serve as an effective strategy to enhance antineoplastic efficacy and overcome the nonspecific side effects of epigenetic reagents in breast cancer treatment.
Acknowledgments
This work is supported by US Army Breast Cancer Research Programs (W81XWH-14-1-0237 to YH; W81XWH-14-1-0238 to NED/SO), Breast Cancer Research Foundation (to NED and SO). This project used the University of Pittsburgh Hillman Cancer Center Cancer Bioinformatics, Cancer Proteomics and Cancer Genomics Services that are supported in part by NIH grant P30CA047904. The authors also gratefully acknowledge the animal and histological core facilities of Magee Womens Research Institute. We thank Lin Chen for technical support. Dr. Hao Wu was supported by China Scholarship Council and Project of Science and Technology Department of Qinghai Province of China (No. 2015-ZJ-751).
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
References
- 1.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–16. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Stearns V, Zhou Q, Davidson NE. Epigenetic regulation as a new target for breast cancer therapy. Cancer Invest. 2007;25:659–65. doi: 10.1080/07357900701719234. [DOI] [PubMed] [Google Scholar]
- 4.Connolly R, Stearns V. Epigenetics as a therapeutic target in breast cancer. J Mammary Gland Biol Neoplasia. 2012;17:191–204. doi: 10.1007/s10911-012-9263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao C, Vasilatos SN, Bhargava R, Fine JL, Oesterreich S, Davidson NE, Huang Y. Functional interaction of histone deacetylase 5 (HDAC5) and lysine-specific demethylase 1 (LSD1) promotes breast cancer progression. Oncogene. 2017;36:133–45. doi: 10.1038/onc.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang XJ, Gregoire S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol Cell Biol. 2005;25:2873–84. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin M, Kettmann R, Dequiedt F. Class IIa histone deacetylases: regulating the regulators. Oncogene. 2007;26:5450–67. doi: 10.1038/sj.onc.1210613. [DOI] [PubMed] [Google Scholar]
- 8.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–11. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin M, Kettmann R, Dequiedt F. Class IIa histone deacetylases: conducting development and differentiation. The International journal of developmental biology. 2009;53:291–301. doi: 10.1387/ijdb.082698mm. [DOI] [PubMed] [Google Scholar]
- 10.Ozdag H, Teschendorff AE, Ahmed AA, Hyland SJ, Blenkiron C, Bobrow L, Veerakumarasivam A, Burtt G, Subkhankulova T, Arends MJ, Collins VP, Bowtell D, et al. Differential expression of selected histone modifier genes in human solid cancers. BMC Genomics. 2006;7:90. doi: 10.1186/1471-2164-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milde T, Oehme I, Korshunov A, Kopp-Schneider A, Remke M, Northcott P, Deubzer HE, Lodrini M, Taylor MD, von Deimling A, Pfister S, Witt O. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res. 2010;16:3240–52. doi: 10.1158/1078-0432.CCR-10-0395. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y, Marton LJ, Woster PM, Casero RA. Polyamine analogues targeting epigenetic gene regulation. Essays Biochem. 2009;46:95–110. doi: 10.1042/bse0460007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM, Casero RA., Jr Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc Natl Acad Sci U S A. 2007;104:8023–8. doi: 10.1073/pnas.0700720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, Nayak S, Jankowitz R, Davidson NE, Oesterreich S. Epigenetics in breast cancer: what’s new? Breast Cancer Res. 2011;13:225. doi: 10.1186/bcr2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Marton LJ, Woster PM. The design and development of polyamine-based analogues with epigenetic targets. Royal Society of Chemistry Drug Discovery Series No 17, Thomas Graham House. 2012:238–56. [Google Scholar]
- 17.Katz TA, Huang Y, Davidson NE, Jankowitz RC. Epigenetic reprogramming in breast cancer: from new targets to new therapies. Ann Med. 2014;46:397–408. doi: 10.3109/07853890.2014.923740. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, Rose DW, Glass CK, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–18. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R, Kirfel J. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–20. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 20.Metzger E, Wissmann M, Yin N, Muller J, Schneider R, Peters A, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y, Stewart TM, Wu Y, Baylin SB, Marton LJ, Perkins B, Jones RJ, Woster PM, Casero RA., Jr Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes. Clin Cancer Res. 2009;15:7217–28. doi: 10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Q, Huang Y, Marton LJ, Woster PM, Davidson NE, Casero RA. Polyamine analogues modulate gene expression by inhibiting Lysine-Specific Demethylase 1 (LSD1) and altering chromatin structure in human breast cancer cells. Amino Acids. 2011 doi: 10.1007/s00726-011-1004-1. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maiques-Diaz A, Somervaille TC. LSD1: biologic roles and therapeutic targeting. Epigenomics. 2016 doi: 10.2217/epi-2016-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, Schneck JL, Carson JD, Liu Y, Butticello M, Bonnette WG, Gorman SA, et al. A DNA Hypomethylation Signature Predicts Antitumor Activity of LSD1 Inhibitors in SCLC. Cancer Cell. 2015;28:57–69. doi: 10.1016/j.ccell.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Cui S, Lim KC, Shi L, Lee M, Jearawiriyapaisarn N, Myers G, Campbell A, Harro D, Iwase S, Trievel RC, Rivers A, DeSimone J, et al. The LSD1 inhibitor RN-1 induces fetal hemoglobin synthesis and reduces disease pathology in sickle cell mice. Blood. 2015;126:386–96. doi: 10.1182/blood-2015-02-626259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasilatos SN, Katz TA, Oesterreich S, Wan Y, Davidson NE, Huang Y. Crosstalk between lysine-specific demethylase 1 (LSD1) and histone deacetylases mediates antineoplastic efficacy of HDAC inhibitors in human breast cancer cells. Carcinogenesis. 2013;34:1196–207. doi: 10.1093/carcin/bgt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Vasilatos SN, Boric L, Shaw PG, Davidson NE. Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res Treat. 2012;131:777–89. doi: 10.1007/s10549-011-1480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katz TA, Vasilatos SN, Harrington E, Oesterreich S, Davidson NE, Huang Y. Inhibition of histone demethylase, LSD2 (KDM1B), attenuates DNA methylation and increases sensitivity to DNMT inhibitor-induced apoptosis in breast cancer cells. Breast Cancer Res Treat. 2014;146:99–108. doi: 10.1007/s10549-014-3012-9. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, Vasilatos SN, Qin Y, Katz TA, Cao C, Wu H, Tasdemir N, Levine KM, Oesterreich S, Davidson NE, Huang Y. Functional characterization of lysine-specific demethylase 2 (LSD2/KDM1B) in breast cancer progression. Oncotarget. 2017;8:81737–53. doi: 10.18632/oncotarget.19387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, Hager ER, Phillips DL, Dunn VR, Hacker A, Frydman B, Kink JA, Valasinas AL, Reddy VK, Marton LJ, Casero RA, Jr, Davidson NE. A novel polyamine analog inhibits growth and induces apoptosis in human breast cancer cells. Clin Cancer Res. 2003;9:2769–77. [PMC free article] [PubMed] [Google Scholar]
- 31.Sankar S, Bell R, Stephens B, Zhuo R, Sharma S, Bearss DJ, Lessnick SL. Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene. 2013;32:5089–100. doi: 10.1038/onc.2012.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou TC, Talaly P. A simple generalized equation for the analysis of multiple inhibitions of Michaelis-Menten kinetic systems. J Biol Chem. 1977;252:6438–42. [PubMed] [Google Scholar]
- 33.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 70:440–6. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 34.He P, Liang J, Shao T, Guo Y, Hou Y, Li Y. HDAC5 promotes colorectal cancer cell proliferation by up-regulating DLL4 expression. Int J Clin Exp Med. 2015;8:6510–6. [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh TH, Hsu CY, Tsai CF, Long CY, Wu CH, Wu DC, Lee JN, Chang WC, Tsai EM. HDAC inhibitors target HDAC5, upregulate microRNA-125a-5p, and induce apoptosis in breast cancer cells. Mol Ther. 2015;23:656–66. doi: 10.1038/mt.2014.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shieh BH, Sparkes RS, Gaynor RB, Lusis AJ. Localization of the gene-encoding upstream stimulatory factor (USF) to human chromosome 1q22-q23. Genomics. 1993;16:266–8. doi: 10.1006/geno.1993.1174. [DOI] [PubMed] [Google Scholar]
- 37.Gregor PD, Sawadogo M, Roeder RG. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 1990;4:1730–40. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- 38.Kristiansson K, Ilveskoski E, Lehtimaki T, Peltonen L, Perola M, Karhunen PJ. Association analysis of allelic variants of USF1 in coronary atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:983–9. doi: 10.1161/ATVBAHA.107.156463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tortorella SM, Royce SG, Licciardi PV, Karagiannis TC. Dietary Sulforaphane in Cancer Chemoprevention: The Role of Epigenetic Regulation and HDAC Inhibition. Antioxid Redox Signal. 2015;22:1382–424. doi: 10.1089/ars.2014.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho E, Clarke JD, Dashwood RH. Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. J Nutr. 2009;139:2393–6. doi: 10.3945/jn.109.113332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pledgie-Tracy A, Sobolewski MD, Davidson NE. Sulforaphane induces cell type-specific apoptosis in human breast cancer cell lines. Mol Cancer Ther. 2007;6:1013–21. doi: 10.1158/1535-7163.MCT-06-0494. [DOI] [PubMed] [Google Scholar]
- 42.Atwell LL, Zhang Z, Mori M, Farris PE, Vetto JT, Naik AM, Oh KY, Thuillier P, Ho E, Shannon J. Sulforaphane Bioavailability and Chemopreventive Activity in Women Scheduled for Breast Biopsy. Cancer Prev Res (Phila) 2015;8:1184–91. doi: 10.1158/1940-6207.CAPR-15-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–74. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 44.Traynor AM, Dubey S, Eickhoff JC, Kolesar JM, Schell K, Huie MS, Groteluschen DL, Marcotte SM, Hallahan CM, Weeks HR, Wilding G, Espinoza-Delgado I, et al. Vorinostat (NSC# 701852) in patients with relapsed non-small cell lung cancer: a Wisconsin Oncology Network phase II study. J Thorac Oncol. 2009;4:522–6. doi: 10.1097/jto.0b013e3181952478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luu TH, Morgan RJ, Leong L, Lim D, McNamara M, Portnow J, Frankel P, Smith DD, Doroshow JH, Wong C, Aparicio A, Gandara DR, et al. A phase II trial of vorinostat (suberoylanilide hydroxamic acid) in metastatic breast cancer: a California Cancer Consortium study. Clin Cancer Res. 2008;14:7138–42. doi: 10.1158/1078-0432.CCR-08-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Modesitt SC, Sill M, Hoffman JS, Bender DP. A phase II study of vorinostat in the treatment of persistent or recurrent epithelial ovarian or primary peritoneal carcinoma: a Gynecologic Oncology Group study. Gynecol Oncol. 2008;109:182–6. doi: 10.1016/j.ygyno.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 47.Blumenschein GR, Jr, Kies MS, Papadimitrakopoulou VA, Lu C, Kumar AJ, Ricker JL, Chiao JH, Chen C, Frankel SR. Phase II trial of the histone deacetylase inhibitor vorinostat (Zolinza, suberoylanilide hydroxamic acid, SAHA) in patients with recurrent and/or metastatic head and neck cancer. Invest New Drugs. 2008;26:81–7. doi: 10.1007/s10637-007-9075-2. [DOI] [PubMed] [Google Scholar]
- 48.Mithraprabhu S, Khong T, Spencer A. Overcoming inherent resistance to histone deacetylase inhibitors in multiple myeloma cells by targeting pathways integral to the actin cytoskeleton. Cell Death Dis. 2014;5:e1134. doi: 10.1038/cddis.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.