Abstract
Leukocyte sensing of microbial genetic material often elicits beneficial proinflammatory cytokines, but dysregulated responses can cause severe pathogenesis. Genome-wide association studies have linked Phospholipase D3 (PLD3) to Alzheimer’s disease and PLD4 to rheumatoid arthritis and systemic sclerosis. PLD3 and PLD4 are endolysosomal proteins whose functions are obscure. PLD4-deficient mice were found to have an inflammatory disease, marked by elevated interferon-γ (IFN-γ) and splenomegaly. These phenotypes were traced to an altered responsiveness of PLD4-deficient dendritic cells to ligands of the single-stranded DNA (ssDNA) sensor Toll-like receptor 9 (TLR9). Macrophages from PLD3-deficient mice also had exaggerated TLR9 responses. Although PLD4 and PLD3 were presumed to be phospholipases, we found that they are 5′ exonucleases, likely identical to spleen phosphodiesterase, that break down TLR9 ligands. Mice deficient in both PLD3 and PLD4 developed lethal early life liver inflammation indicating that both enzymes are required to regulate inflammatory cytokine responses by degradation of nucleic acids.
Introduction
Leukocytes carry conserved sensors for endocytosed DNA and RNA that trigger the production of proinflammatory cytokines. The ensuing inflammation can be beneficial for host defense to microorganisms, viruses, and tumors, but can also be pathogenic. Host recognition of nucleic acids triggers production of the type I interferons (IFN), IFN-α and IFN-β, and inflammatory cytokines such as interleukin-6 (IL-6) and IL-121, 2. Viral, bacterial or host DNA and RNA can be recognized depending upon sequence motifs, concentration, intracellular localization, and rate of degradation. One class of sensors resides in endosomes and includes TLRs 3, 7, 8, 9, and 13, which recognize, respectively, double-stranded (ds) RNA, ssRNA, ssRNA, ssDNA carrying unmethylated CpG motifs, and bacterial 23S RNA. A second sensor class resides in the cytoplasm. The RIG-I-like receptors (RLRs) recognize uncapped dsRNA, whereas the cyclic GMP-AMP synthase (cGAS) and Absent in Melanoma 2 (AIM2) are cytoplasmic DNA sensors. These sensors are believed to preferentially recognize foreign genetic material because host DNA and RNA often have distinct sequence abundances and modifications, are localized away from the sensors, or are degraded in a more efficient or organized manner.
Evidence is mounting that regulation of nucleic acid abundance by nucleases prevents autoinflammatory and autoimmune diseases driven by nucleic acid sensors3. Deficiency in Dnase1 or Dnase1L3 leads to systemic lupus erythematosus in both humans and mice4,5,6. In mice, Dnase2a deficiency leads to a lethal IFN-mediated disease, while patients deficient for DNASE2a exhibit an autoinflammatory anemia, kidney and joint disease7,8. Patients with TREX1 deficiency develop Aicardi-Goutières syndrome, whilst Trex1-deficient mice develop a lethal cardiomyopathy inflammatory disease9,10,11. The prevention of abnormal accumulation of host nucleic acid in differing tissues and cellular compartments is thus performed by an array of enzymes with non-redundant functions.
Phospholipase D family proteins share a catalytic site sequence signature (HxxKxxxxD) that is found in PLD1 and PLD2 cytosolic phospholipases, but most family members have distinct functions and intracellular localization12, 13, 14. PLD3 and PLD4 are glycosylated type II transmembrane proteins localized in endolysosomes15, 16, 17, residing in subcellular compartments distinct from most other PLD family members. Studies have linked PLD4 polymorphisms to systemic sclerosis and rheumatoid arthritis18, 19, 20. Several works link PLD3 to Alzheimer’s disease21,22 with some controversy23, 24. The functions of PLD3 and PLD4 are unknown. Despite their names, PLD3 and PLD4 have not been demonstrated to have phospholipase activity25, 26. PLD4 is highly expressed in dendritic cells (DCs) and other myeloid cells, and in lower amounts in B cells. PLD3 is structurally similar to PLD4, but more broadly expressed. We find that PLD3 and PLD4 are 5′ exonucleases with properties strikingly similar to the enzymatic activity described many years ago as spleen acid exonuclease or phosphodiesterase27, 28. Mutants in Pld3 and Pld4 genes have heightened sensitivity to, and diminished turnover of, ligands of TLR9.
Results
PLD4-deficient mice display a TLR9-driven inflammatory syndrome
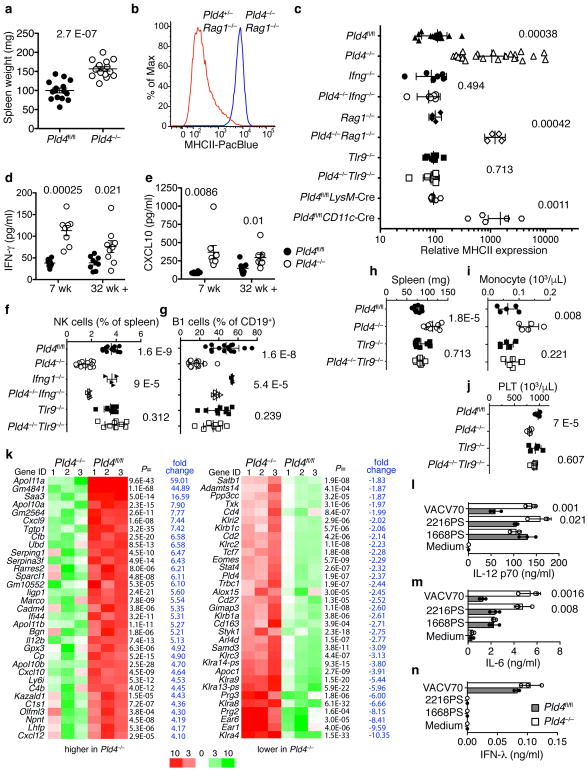
To assess effects of PLD4 deficiency, a conditional knockout allele (Pld4fl) was engineered, from which we generated homozygous knockout (Pld4−/−) and conditional strains (Pld4fl/fl) (Supplementary Fig. 1a–c); Pld4−/− mice lacked detectable PLD4 protein (Supplementary Fig. 1d) and exhibited a phenotype of chronic immune activation, with splenomegaly (Fig. 1a) and elevated major histocompatibility class II (MHCII) expression on resident peritoneal macrophages (Fig. 1b,c), whereas CD86 was not elevated (Supplementary Fig. 1g). The MHCII upregulation was driven by IFN-γ produced by innate immune cells, as it occurred in Pld4−/−Rag1−/− mice (Fig. 1b,c), which lack B and T lymphocytes, but not in IFN-γ-deficient Ifng−/−Pld4−/− mice (Fig. 1c). Expression of MHCII by macrophages was not elevated in Pld4fl/flLysM-Cre mice, in which PLD4 deficiency was limited to macrophages, but was evident in Pld4fl/flCD11c-Cre mice, suggesting that PLD4-deficient CD11c+ DCs indirectly promoted MHCII upregulation (Fig. 1c). Plasma of Pld4−/− mice had significantly elevated concentrations of IFN-γ and CXCL10 (Fig. 1d,e). Pld4−/− mice had altered leukocyte subsets, including fewer natural killer (NK) cells, peritoneal B1 lymphocytes, and platelets, and more marginal zone (MZ) B cells and blood monocytes (Fig. 1f–j, Supplementary Fig. 1h). However, splenic DC numbers were similar to those of PLD4-sufficient mice (Supplementary Fig. 1e,f). Consistent with elevated IFN-γ concentrations, altered B1 and MZ B cell numbers seen in Pld4−/− mice were partly reversed in Ifng−/−Pld4−/− mice (Fig. 1f,g and Supplementary Fig. 1i). Moreover, Pld4−/− but not Ifng−/−Pld4−/− mice, were resistant to the induction of experimental autoimmune encephalomyelitis (EAE) (Supplementary Fig. 2), in which IFN-γ is protective29. RNA sequencing comparison of Pld4−/−Rag1−/− and Rag1−/− splenocytes revealed an IFN signature in Pld4−/− animals, as >40 of the 109 genes whose expression was significantly elevated were known to be inducible by IFN-γ or type I IFNs (Fig. 1k, Supplementary Table 1). Expression of IL12b, Cxcl9 and Il10 were also significantly elevated. Of the 33 down-regulated genes, some were known to be suppressed by IFN, but most were NK-specific genes, consistent with their lower numbers in the spleens of Pld4−/− mice and Pld4−/−Rag1−/− mice.
Figure 1.
Splenomegaly, IFN-γ-driven MHCII upregulation in peritoneal macrophages, and other TLR9-dependent anomalies in Pld4−/− mice. (a) Spleen weights of Pld4fl/fl compared to age-matched Pld4−/− mice (n=15,15). (b) MHCII expression on F4/80+-gated resident peritoneal macrophages from Pld4+/−Rag1−/− compared to Pld4−/−Rag1−/− mouse. Summary data (n=4,4) shown in (c). (c) Cell type-, IFN-γ-, and TLR9-dependence of PLD4 deficiency on MHCII upregulation in peritoneal macrophages (n=13,15,6,6,4,4,9,9,8,5 independent mice in groups listed from top to bottom). Plasma cytokine concentrations of (d) IFN-γ and (e) CXCL10 (7-wk-old mice, n=8/group; 32+wk-old mice, n=9/group). (f–j) Effects of compound deficiency of Pld4 and Tlr9 or Pld4 and Ifng on (f) splenic NK cell frequency, (g) peritoneal B1 cell frequency, (h) spleen weight, (i) monocyte counts, (j) blood platelets. n=12,12,4,4,9,9 represent values obtained from independent mice in groups listed from top to bottom in f and g; n=9 for each group in h; n=5 for each group in i and j. In (a,d–g) error bars show mean and SEM, c shows geometric standard deviation and h–j show linear standard deviation with each data point representing the value obtained from an independent mouse and P values calculated using unpaired 2-tailed T-test. (k) mRNA sequencing analysis comparing splenocytes of Pld4+/+Rag1−/− versus Pld4−/−Rag1−/− mice. Expression of all genes shown was significantly different in the two groups (false discovery rate ≤ 0.05 calculated using edgeR, n=3/group). (l–n) Cytokine responses of sorted splenic CD8+ DCs to the indicated TLR9 ligands. (l) IL-12p70, (m) IL-6, (n) IFN-λ (mean is shown for l–n, triplicate cultures, repeated in at least two independent experiments, unpaired 2-tailed T test).
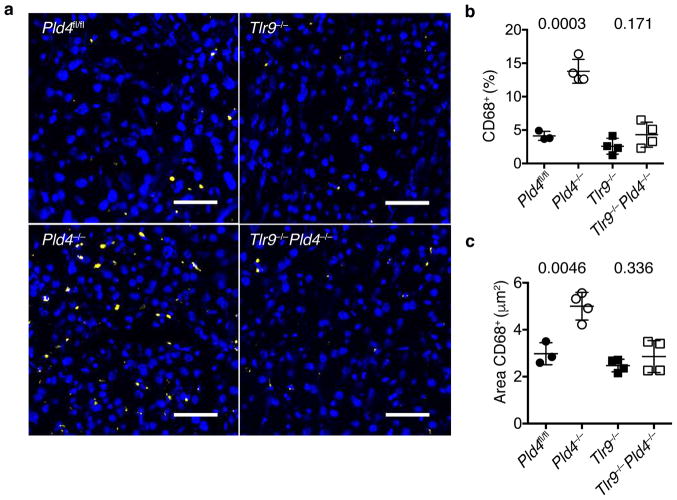
Certain features of the phenotype of Pld4−/− mice resembled the human disease Macrophage Activation Syndrome, which can be mimicked in mice by repeated challenge with ligands for TLR930. To assess the role of TLR9 in the PLD4-deficient phenotype, we bred Tlr9−/−Pld4−/− mice. Notably, analysis of these mice indicated that all of the abnormalities of PLD4-deficient mice tested were dependent upon TLR9, including splenomegaly, MHCII upregulation, and alterations in numbers of monocytes, platelets, NK, and B1 cells (Fig. 1c,f–j). In contrast, combined deficiency in both PLD4 and IFN-γ reversed only a subset of phenotypes associated with PLD4 deficiency (Fig. 1c,f,g), suggesting that IFN-γ production by Pld4−/− mice was secondary to TLR9 triggering. Histological analysis of Pld4−/− livers revealed a TLR9-dependent increase in the frequency and size of CD68+ macrophages, consistent with mild liver inflammation (Fig. 2). Overall, these results suggested that the major effects of PLD4 deficiency stemmed from enhanced or dysregulated TLR9 recognition of ssDNA in endolysosomes.
Figure 2.
Effect of Pld4 and Tlr9 deficiency on liver infiltration by CD68+ macrophages. (a) Frozen sections of liver from mice of the indicated genotypes were stained with Hoechst dye (blue) and antibody for CD68 (yellow). Scale bar indicates 50 μm. Analysis of (b) the percentages of liver cells staining with CD68 antibody relative to Hoechst 33258+ cells and (c) the area occupied by CD68+ cells in mice of the indicated genotypes. Each point in (b,c) represents the value measured in an independent mouse (mean and SD shown, n=3,4,4,4; P value calculated by unpaired 2-tailed T test).
That Pld4−/− mice had excessive TLR9-driven IFN-γ production and Il12b mRNA suggested that activation of CD8+ DCs might be dysregulated in these mice. Secretion of IL-12 by CD8+ DCs is known to stimulate IFN-γ production, particularly by NK cells31. PLD4-deficient sorted splenic DCs were found to produce more IL-12p70 and IL-6 in response to two TLR9 ligands, VACV70 and 2216PS (an A-type CpG oligodeoxynucleotide (ODN)), compared to PLD4-sufficient controls, however responses to the B-type CpG ODN PS, were similar (Fig. 1l,m). 1668PS is fully modified by phosphorothioate (PS) linkages, 2216PS is partially modified, and VACV70 is unmodified dsDNA containing CpG motifs (Table 1). In addition, VACV70, but not 1668PS and 2216PS, elicited IFN-λ responses in CD8+ DCs through the STING cytoplasmic DNA sensing pathway, but did not differ between wild-type and PLD4-deficient cells (Fig. 1n, Supplementary Fig. 3a,b). IFN-γ signals are known to promote IL-12 responses32, however sorted CD8+ DCs isolated from Pld4−/−Ifng−/− mice also showed elevated IL-12 responses to TLR9 stimuli (Supplementary Fig. 3c). CD8−CD11bhi splenic DCs also had enhanced IL-6 and IL-12 responses, especially to VACV70 (Supplementary Fig. 3d). These results supported the notion that Pld4−/− CD8+ DCs had enhanced cytokine responses to at least some TLR9 ligands.
Table 1.
Synthetic deoxyoligonucleotide ligands used in this study*
| 2216PS | ggGGGACGATGATCGTCgggggg | 2216 | GGGGGACGATGATCGTCGGGGGG |
| A5′11PS | ggGGGACGATC | A5′11 | GGGGGACGATC |
| A3′14PS | CGATGATCGTCgggggg | A3′14 | CGATGATCGTCGGGGGG |
| A3′13PS | GATGATCGTCgggggg | A3′13 | GATGATCGTCGGGGGG |
| A3′12PS | ATGATCGTCgggggg | A3′12 | ATGATCGTCGGGGGG |
| A3′11PS | TGATCGTCgggggg | A3′11 | TGATCGTCGGGGGG |
| A3′10PS | GATCGTCgggggg | A3′10 | GATCGTCGGGGGG |
| 1668PS | tccatgacgttcctgatgct | 1668 | TCCATGACGTTCCTGATGCT |
| 2006PS | tcgtcgttttgtcgttttgtcgtt | 2006 | TCGTCGTTTTGTCGTTTTGTCGTT |
| VACV70 | CCATCAGAAAGAGGTTTAATATTTTTGTGAGACCATCGAAGAGAGAAAGAGATAAAACTTTTTTACGACT GGTAGTCTTTCTCCAAATTATAAAAACACTCTGGTAGCTTCTCTCTTTCTCTATTTTGAAAAAATGCTGA |
||
UPPER CASE: phosphodiester linkage, lower case: phosphorothioate linkage
PLD4 and PLD3 are single-stranded 5′ exonucleases
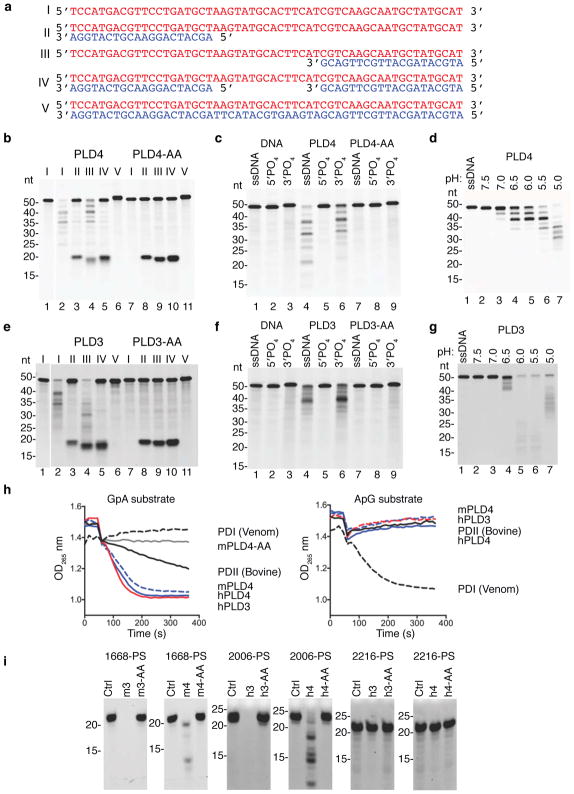
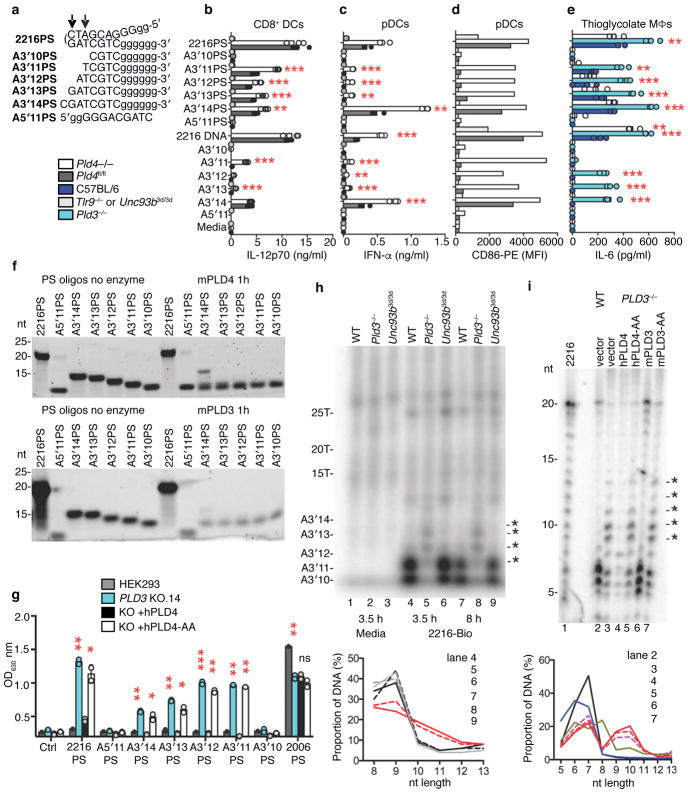
These results suggested a role for PLD4 in the processing of TLR9 ligands. To test for nuclease activity, recombinant soluble mouse PLD4 was expressed by substituting a secretion signal peptide for the N-terminal transmembrane and cytoplasmic tail. Using DNA substrates shown in Fig. 3a, the activity of purified recombinant mouse PLD4 was then assessed (Fig. 3b). PLD4 degraded a 55 nt ssDNA (lane 2), but not a dsDNA version of the same sequence (lane 6). When paired with shorter ssDNAs complementary with the 5′ or 3′ ends, PLD4 degraded only substrate with an unpaired 5′ end (lane 4), indicating 5′-to-3′, but not 3′-to-5′, ssDNA exonuclease activity, and demonstrating a lack of endonuclease activity (lane 5). PLD4 protein carrying H-to-A mutations in both HKD motifs (PLD4-AA) lacked detectable enzymatic activity (lanes 7–11). PLD4 only cleaved substrates that lacked phosphorylated 5′ ends and was unaffected by 3′ phosphorylation (Fig. 3c, lanes 4,6). As predicted for an endosomal nuclease, the reaction was most efficient at pH<6, but was still detectable at pH 7 (Fig. 3d). Parallel experiments with mouse PLD3 revealed a similar profile of substrate specificity and an acidic pH optimum (Fig. 3e–g). We conclude that PLD3 and PLD4 are 5′ exonucleases that are able to cleave ssDNA.
Figure 3.
Analysis of the nuclease activity of PLD4 and PLD3. (a) Roman numerals indicate each nucleic acid substrate tested. (b,c) Soluble recombinant mouse PLD4 or PLD4-AA (100 nM) was incubated with substrate (2.5 μM) in reaction buffer (50 mM MES pH 5.5, 100 mM NaCl) for 2 hours at 37°C, then analyzed by denaturing Tris-Borate-EDTA polyacrylamide gel electrophoresis. In b all oligonucleotides lacked 5′ or 3′ phosphate. (c) Substrate I that lacked phosphate (ssDNA) or carried phosphorylation on the 5′ or 3′ ends was left untreated (DNA) or was incubated with PLD4 or PLD4-AA. (d) Reactions using substrate I in buffers adjusted to the indicated pH. (e-g) Reactions were identical to b–d except that PLD3 or PLD3-AA proteins (10 nM) were used as indicated. In (b,e) photo is cropped between lanes 1 and 2 to eliminate additional undigested control lanes. (h) Assay of the release of adenosine by digestion of the indicated dinucleotides (left panel, GpA; right panel, ApG) was carried out as described34 using the indicated enzymes. PDI, snake venom phosphodiesterase; PDII (Bovine), calf spleen phosphodiesterase II (spleen exonuclease). (i) Digestion of the indicated PS-linked substrate ODNs (Table 1, 2.5 μM) with recombinant proteins (500 nM) in reaction buffer adjusted to pH 5.0 for PLD4 and pH 5.5 for PLD3, incubated at 37°C for 6 hours. Mouse and human PLD3 and PLD4 are abbreviated as m3, h3, m4, h4, respectively. nt, length in nucleotides. All experiments were repeated at least twice.
PLD3 and PLD4 appear to be classic spleen phosphodiesterases
The enzymatic properties of PLD4 and PLD3 that we observed were reminiscent of the activity previously described as “spleen acid exonuclease” or “spleen phosphodiesterase”28, 33 (Enzyme Commission registry number 3.1.16.1). We therefore directly compared PLD3 and PLD4 with a commercial preparation of bovine spleen phosphodiesterase as a positive control using a diagnostic assay that measured their abilities to digest GpA or ApG dinucleotides (Fig. 3h). Digestion was readily tracked spectroscopically by the release and loss of adenosine, which is deaminated in a coupled assay34. Mouse and human recombinant soluble PLD3 and PLD4, but not PLD4-AA, specifically cleaved the phosphodiester bond between the phosphate and the second nucleotide, matching the activity described of bovine spleen exonuclease. Snake venom phosphodiesterase served as a control for possible cleavage at the upstream phosphodiester bond, leaving 5′ phosphorylated mononucleotides. The results indicated that digestion by PLD3 and PLD4 generated 3′ mononucleotides and nucleosides from short oligonucleotide substrates. The commercial Phosphodiesterase II (Methods) was further purified by size exclusion to reduce complexity of the sample and the enzymatically active fractions were subjected to trypsin-Liquid Chromatography-Mass Spectrometry (LC-MS) analysis. Six peptides unique to bovine PLD3 were identified (Supplementary Fig. 4a,b). Other proteins, however, were also identified (Supplementary Fig. 4c), consistent with low specific activity of the commercial preparation in the dinucleotide cleavage assay (Fig. 3h). We conclude that PLD3 and PLD4 have activities indistinguishable from classical spleen acid exonuclease.
PLD3 and PLD4 can degrade PS-linked CpG agonists of TLR9 in vitro
Phosphorothioate (PS) linkages are often used in synthetic ODNs to hinder their digestion by nucleases. CpG-B-type ODNs 2006PS and 1668PS are fully PS-modified. They stimulate TLR9 preferentially through the NF-κB pathway to produce cytokines such as IL-6 and IL-1235. CpG-A-type ODN 2216PS is a potent inducer of IFN-α and has an internal palindromic sequence composed of phosphodiester-linked nucleotides flanked by strings of PS-modified G nucleotides known to form unusual secondary structures35. Human and mouse PLD3 and PLD4 were able to digest B-type CpG ODNs 2006PS and 1668PS despite their PS linkages (Fig 3i), albeit between 20–50 times less efficiently than versions with phosphodiester bonds (Supplementary Fig. 5a,c), but failed to digest 2216 or 2216PS (Fig. 2i, right; Supplementary Fig. 5b,d). We conclude that PLD3 and PLD4 digest ssODNs, including those with phosphorothioate linkages, but are blocked by features of A-type CpG ODNs, likely their secondary structure.
PLD3 and PLD4 limit cellular responses to exogenous TLR9 ligands
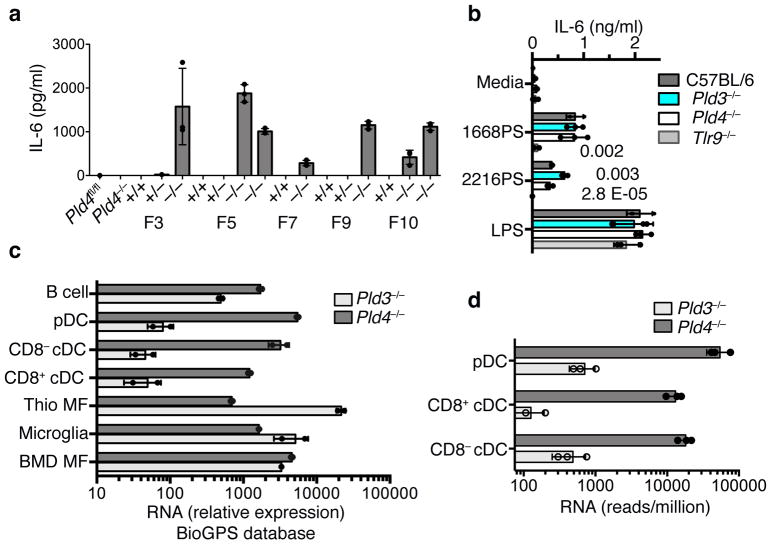
To assess PLD3 function and potential redundancy with PLD4, we used CRISPR/Cas9 mutagenesis of zygotes to generate Pld3 strains lacking PLD3 protein (Supplementary Fig. 1j,k). Thioglycolate-elicited Pld3−/− macrophages expressed more IL-6 in response to 2216PS, relative to wild-type or Pld4−/− thioglycolate-elicited macrophages (Fig. 4a). The enhanced response was observed using macrophages from multiple founder lines, but not from wild-type or Pld3+/− littermates. In contrast, Pld3−/− macrophage responses to the TLR4 agonist lipopolysaccharide (LPS) were comparable to wild-type and Pld4−/− macrophages (Fig. 4b). RNA expression analysis revealed that wild-type thioglycolate-elicited macrophages highly expressed Pld3 and poorly expressed Pld4, whereas wild-type DCs showed the opposite pattern of expression (Fig. 4c,d). Pld3−/− mice lacked other phenotypes observed in Pld4−/− mice and had similar spleen size, splenic NK proportions, and resident peritoneal macrophage MHCII surface expression as their heterozygous littermate controls (Supplementary Fig. 1l–n) and FLT3L-DC responses were unchanged (Supplementary Fig. 6f). Functional redundancy and distinct expression profiles are thus consistent with the altered responses of Pld3−/− thioglycolate-elicited macrophages and of Pld4−/− CD8+ DCs.
Figure 4.
Exaggerated responses of Pld3−/− thioglycolate-elicited macrophages to an A-type TLR9 agonist and analysis of relative RNA expression of Pld3 and Pld4 in selected cell types. (a) Elevated IL-6 responses of thioglycolate-elicited macrophages from independent Pld3−/− founder lines to in vitro incubation with ODN 2216PS. Founder lines 3, 5, 7, 9, and 10 were outcrossed to C57BL/6 then intercrossed to yield heterozygosity or homozygosity of the Pld3 mutations indicated in Fig. S1j, or homozygosity of the wild-type Pld3 allele. Results shown are from triplicate cultures performed once, showing mean and SD. (b) TLR9-dependence and elevated responses of Pld3−/− thioglycolate-elicited macrophages. PLD3-deficient macrophages of founder line 7 were challenged with the indicated stimuli and the IL-6 response compared to Pld4−/−, Tlr9−/−, or wild-type cells as indicated. Bars show mean and points show values obtained in 3 separate cultures. P values calculated by two-tailed T test. These experiments were repeated at least twice. (c,d) Analysis of Pld3 and Pld4 expression. Points show independent RNA samples and bars show geometric mean. (c) RNA expression data from the BioGPS MOE430 Gene Atlas Data set of the indicated mouse cell types: including dendritic cells (CD8α+ DCs, CD8α− DCs, and pDCs), B cells, bone marrow-derived macrophages (BMD MF), and thioglycolate-elicited macrophages (Thio MF). n=2,2 independent RNA samples. (d) RNA sequencing analysis of Pld3 and Pld4 expression in DC subsets. n=3,3; each data point was from sorted cells of independent pools of splenocytes from 10–14 mice.
Recombinant PLD3 was unable to degrade the TLR9 ligand 2216PS in vitro (Fig. 3i, Supplementary Fig. 5d), so it was surprising that thioglycolate-elicited Pld3−/− macrophages had an enhanced IL-6 response to 2216PS (Fig. 4a,b). TLR9 responses to 2216PS require cleavage by DNase II in the palindromic phosphodiester-linked central region36. As DNAse II cleaves dsDNA or palindromic ssDNA, creating fragments with unphosphorylated 5′ ends and 3′ phosphates, we hypothesized that DNAse II-cleaved 2216PS fragments should be naturally degraded by PLD3 or PLD4. We therefore tested a series of synthesized fragments of 2216 or 2216PS (Fig. 5a) for their ability to stimulate FLT3L-cultured DCs and thioglycolate-elicited macrophages deficient in PLD3 or PLD4 (Fig. 5b–e). Pld4−/− DCs and Pld3−/− thioglycolate-elicited macrophages showed enhanced responses to these shorter ligands compared to PLD3/4-sufficient controls (Fig. 5b–e). When packaged in the transfection reagent Lyovec, the same ODN ligands gave similar strong responses in both wild-type and PLD4-deficient DCs (Supplementary Fig. 6a,b), suggesting that ligand stability rather than TLR9 signaling was affected by PLD4 deficiency and could be bypassed using this reagent. Interestingly, PLD3-deficient, thioglycolate-elicited macrophages still had enhanced responses to Lyovec complexes of 2216 subfragments, indicating some cell type-specific differences (Supplementary Fig. 6c). In both DCs and macrophages, responses to ODNs 1668PS and 2216PS were dependent on TLR9 (Fig. 5b–d). The enhanced bioactivity of particular CpG fragments in PLD3- or PLD4-deficient cells correlated with their sensitivity to PLD3 and PLD4 in vitro (Fig. 5f).
Figure 5.
Stimulatory ability and stability of 2216 and its fragments in the presence or absence of PLD3 or PLD4. (a) Synthetic substrates tested; PS-linked nucleotides (lower case). Arrows indicate the ability of DNase II to cut within the palindrome of 2216. (b–e) Cytokine responses of the indicated cell types to ODNs, 1 μM. Cells derived from (b–d) FLT3L DC cultures or (e) thioglycolate-elicited macrophages. (b) IL-12p70 responses of CD8+ DCs. (c) IFN-α responses of FLT3L DCs. (d) CD86 upregulation in CCR9+/B220+-gated (pDC) cells analyzed in c. Here, cells from replicate cultures were pooled for flow cytometry analysis, yielding a single sample per condition. (e) IL-6 responses of thioglycolate-elicited macrophages. In (b,c,e) bars show mean and points show values obtained in 3 separate cultures. (f) In vitro digestion by PLD4 (100 nM, upper panel) or PLD3 (10 nM, lower panel) of the substrates listed in a. These experiments were carried out twice with similar results. (g) 293HEK-BluehTLR9 clones deficient for PLD3 were assessed for TLR9 responses to the indicated ODNs. PLD3-deficient clone #14 was reconstituted with PLD4 or PLD4-AA expression constructs and challenged with the indicated ODNs (1 μM). NF-κB reporter activation was then measured. Plots show mean of duplicate cultures. P values calculated by 2-tailed T test. In b,c,e,g *, **, *** denote P< 0.05, .01, .001, respectively. This experiment was repeated at least twice with similar results. (h) Analysis of in vivo stability of 3′-biotin-tagged-2216 in thioglycolate-elicited macrophages. After the indicated duration of incubation, ODNs were isolated and visualized on a TBE-Urea acrylamide gel. Asterisks indicate ODN species increased in abundance in Pld3−/− lysates. Macrophages were from C57BL/6 (WT), PLD3-deficient or Unc93b13d/3d mice. (i) Similar analysis of 2216-biotin recovered after 6 h with PLD3−/− #10 HEK293-BluehTLR9 cells or cells that had been reconstituted with PLD3, PLD4, or nuclease dead variants PLD3-AA or PLD4-AA. 2216-biotin was radiolabeled as a size marker (2216-bio). Electrophoresis performed in 50 mM histidine buffer for higher resolution of small fragments50. Panels below h,i quantitate relative intensity of bands in the 5–13 nt range of the indicated lanes above. Experiments in (h) were carried out twice and in (i) once.
To enable the analysis of the function of PLD3 and PLD4 in human cells, PLD3-deficient clones of the human cell line HEK-BluehTLR9 were generated by CRISPR/Cas9 treatment. Independent PLD3-deficient clones had greatly enhanced responses to 2216PS and its synthetic fragments, as well as a phosphodiester form of B-type CpG 2006 (Fig. 5g, Supplementary Fig. 6d–e). Reconstitution with PLD4 inhibited the response to baseline, whereas reconstitution with PLD4-AA did not (Fig. 5g), suggesting that nuclease activity was required to limit the response.
Enhanced stability of TLR9 ligands in cells lacking PLD3 and PLD4
To assess ODN degradation in vivo, we stimulated wild-type or mutant thioglycolate-elicited macrophages with 2216 carrying a 3′-biotin tag; after 3.5 or 8 hours, we recovered ODNs from cell lysates using streptavidin beads and visualized them by 32P end-labelling (Fig. 5h). The analysis revealed that the 2216-biotin fragments that demonstrated enhanced stimulatory activity in Pld3−/− macrophages and Pld4−/− DCs were less degraded in PLD3-deficient lysates than in those from control macrophages (Fig. 5h, asterisks), which had shorter species composed mainly of the poly-G 3′ end (<10 nt). The degradation pattern in Unc93b3d/3d macrophages, which lack signaling by TLR 3, 7 and 937, was identical to wild-type macrophages, indicating that a TLR9 signal is not required for enzymatic degradation by PLD3. Similar results were obtained in PLD3−/− HEK293 cells fed 2216-biotin, where DNA fragments of 10–15 nt were more abundant, and were specifically depleted in cells reconstituted with PLD4 or PLD3 but not PLD4-AA or PLD3-AA mutants (Fig. 5i). We conclude that PLD3 and PLD4 have redundant exonuclease function that can limit TLR9 responses to CpG ODNs, particularly those lacking phosphorothioate linkages, by reducing the concentration of ssDNA able to stimulate TLR9 and that these activities are similar in human and mouse.
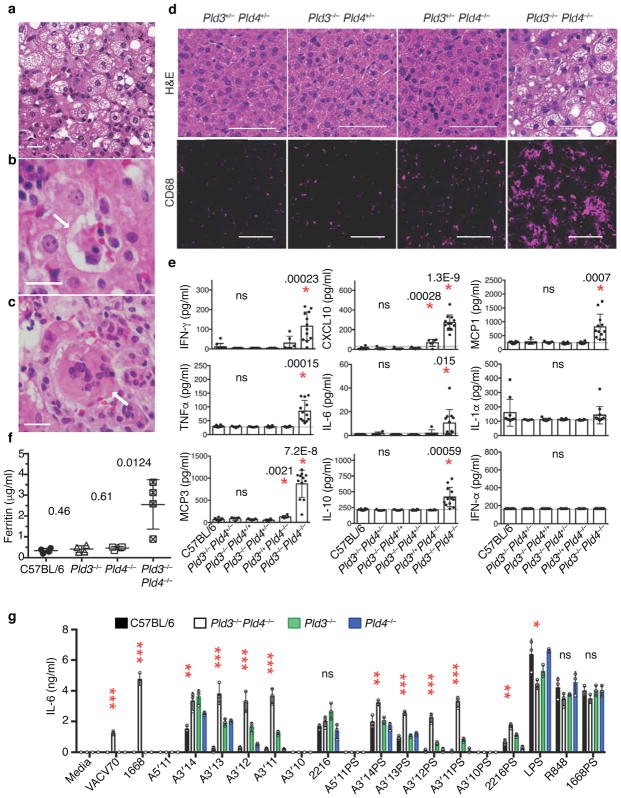
PLD3 and PLD4 prevent a lethal hepatic autoinflammatory disease
The overlapping enzymatic activities of PLD3 and PLD4 and their co-expression in macrophage subsets prompted us to interbreed these strains to generate animals deficient for both enzymes. No animals deficient for both genes survived to weaning age (Supplementary Fig. 7a). Pld3−/−Pld4−/− mice were identified of either sex, but these animals were undersized compared to littermate controls and died between 12 and 21 days of age with severe liver inflammation (Fig 6a–d, Supplementary Fig. 7b,c). The livers from Pld3−/−Pld4−/− animals showed macroscopic white areas of necrosis (Supplementary Fig. 7c), with regions of vesicular steatosis, hemophagocytosis, and multinucleated hepatocytes (Fig. 6a–c, respectively). Importantly, only a single allele of either Pld3 or Pld4 was required to allow survival and to prevent the substantial liver pathology (Fig. 6d and Supplementary Table 2). Although CD68+ myeloid cells in the livers from 16 day old Pld3+/−Pld4−/− animals were more prevalent than in Pld3+/−Pld4+/− or Pld3−/−Pld4+/− littermates, livers from Pld3−/−Pld4−/− animals demonstrated extensive infiltration of CD68+ myeloid cells (Fig. 6d). Sera from Pld3−/−Pld4−/− animals revealed elevated concentrations of ferritin, IL-6, TNF, IFN-γ, CXCL10, MCP3, MCP1, and IL-10 compared to controls of the same age, whereas IFN-α and IL-1α were not significantly elevated (Fig 6e,f). We infer that neonatal mice require PLD3 and PLD4 to prevent an inflammatory disease consistent with hemophagocytic lymphohistiocytosis (HLH).
Figure 6.
Liver inflammation and elevated inflammatory cytokine production in Pld3−/−Pld4−/− mice. (a–c) Hematoxylin/eosin (H&E) staining of paraffin-embedded sections from Pld3−/−Pld4−/− livers; scale bars are 50 μm. Pathogenic features of disease include (a) vesicular hepatopathy (steatosis), (b) hemophagocytosis (arrow), and (c) hepatic multinucleate cells (arrow). (d) H&E analysis of liver sections of littermates with the indicated genotypes (upper panels, scale bar 50 μm) or CD68 immunofluorescent staining of frozen liver sections derived from different lobes of the same liver (lower, scale bar 100 μm). These data are representative of at least 3 mice/group. (e) Serum cytokines measured in 16–19 day-old mice of the indicated genotypes. n=10, 5, 7, 6, 6, 12 for C57BL/6, Pld3+/−Pld4+/−, Pld3−/−Pld4+/+, Pld3−/−Pld4+/−, Pld3+/+Pld4−/−, Pld3−/−Pld4−/−, respectively. P values calculated by two-tailed T test; asterisks indicate significant differences. Dotted lines in (e) indicate assay background. (f) Serum ferritin concentrations from indicated genotypes n= four/group. (g) IL-6 secretion by bone marrow-derived macrophages isolated from 19 day-old C57BL/6, Pld3−/−, Pld4−/− or Pld3−/−Pld4−/− mice after stimulation with indicated TLR agonists. Asterisks refer to significant differences between C57BL/6 and Pld3−/−Pld4−/−. *, **, *** denote P< 0.05, .01, .001, respectively. Significance of IL-6 responses assessed by unpaired two-tailed T-test. All plots show mean and SD of triplicate cultures; this experiment was repeated a second time with similar results.
Analysis of TLR9 ligand responses of macrophages grown from Pld3−/−Pld4−/− bone marrow revealed elevated IL-6 secretion to 2216 subfragments, VACV70, and 1668 (DNA form), but responses to 1668PS, R848 and LPS were similar to wild-type or single deficient controls (Fig. 6g). As both Pld3 and Pld4 genes are expressed in bone marrow derived macrophages (Fig. 4c), the enhanced responses to particular TLR9 ligands occurred when both genes were deficient, but not when either gene was present (Fig. 6g). Pld3−/−Pld4−/− bone marrow failed to generate dendritic cells in FLT3L culture, suggesting that either the on-going inflammation was affecting DC precursors in the bone marrow or that both proteins are required for in vitro FLT3L-driven DC development.
PLD3 and PLD4 in hematopoietic cells prevent autoinflammation
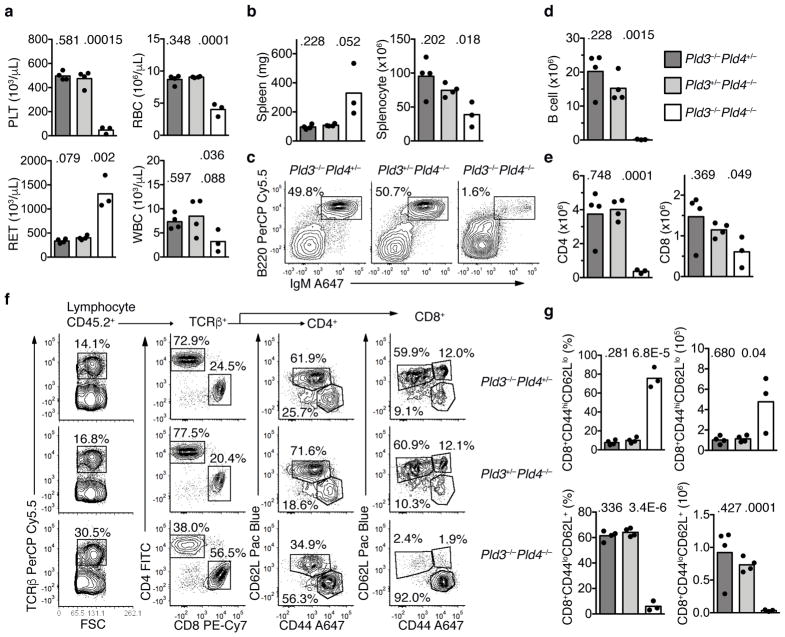
To determine whether the immune system itself was the driver of lethal inflammation in Pld3−/−Pld4−/− mice, bone marrow transplantation into lethally irradiated 6 week old wild-type CD45.1+ recipients was performed using 19 day littermate Pld3+/−Pld4−/−, Pld3−/−Pld4+/− or Pld3−/−Pld4−/− bone marrow as donors. Eight weeks post transfer, blood analysis revealed low platelet counts, anemia, elevated reticulocytes and leukopenia in the recipients receiving Pld3−/−Pld4−/− bone marrow, but not in recipients transplanted with littermate bone marrow containing a single wild-type allele of either Pld3 or Pld4 (Fig. 7a). Despite overt splenomegaly, the number of splenic leukocytes recovered after erythrocyte lysis was lower in Pld3−/−Pld4−/− bone marrow recipients compared to controls (Fig. 7b). Flow cytometric analysis of CD45.2+ splenocytes revealed a marked reduction in B cell proportions and numbers when both Pld3 and Pld4 were absent (Fig. 7c,d). The total numbers of TCRβ+ CD4+ and CD8+ T cells were also reduced in the spleens of Pld3−/−Pld4−/− bone marrow recipients compared to controls (Fig. 7e), however the majority of CD8+ T cells exhibited an activated phenotype (CD44hiCD62Llo) and very few naïve CD8+ T cells (CD44loCD62L+) were present (Fig. 7f,g). Consistent with disease observed in Pld3−/−Pld4−/− mice, recipients of Pld3−/−Pld4−/− bone marrow had elevated expression of MHCII (with CD86 expression similar to controls) on peritoneal macrophages, elevated blood monocytes, and elevated concentrations of serum cytokines CXCL10, IFN-γ, MCP3, IL-6, but IFN-α was not detected (Supplementary Fig. 7d–k). We conclude that both PLD3 and PLD4 limit autoinflammation, and that a single wild-type allele of either can prevent the development of the inflammatory disease transferred by hematopoietic stem cells.
Figure 7.
Transfer of an inflammatory disease by transplantation of Pld3−/−Pld4−/− bone marrow. (a) Eight weeks post transfer, whole blood was analyzed for platelet (PLT), red blood cell (RBC), reticulocyte (RET) and white blood cell (WBC) counts from B6.SJL-CD45.1 recipient mice receiving bone marrow of the indicated genotypes: Pld3−/−Pld4+/− (dark grey bar), Pld3+/−Pld4−/− (light grey bar), Pld3−/−Pld4−/− (white bar). Bar indicates mean and symbols indicate individual recipient values. (b) Spleen weight (mg) and splenocyte count after erythrocyte lysis was determined 8 weeks post transfer. (c) Representative flow cytometry plots indicating recipient splenic CD45.2+-gated B cell populations stained with B220 and IgM from the indicated bone marrow donors. Summary data shown in d. (d) Numbers of donor (CD45.2+) B220+IgM+ B cells identified in spleens as gated in (c). (e) Numbers of CD45.2+TCRβ+CD4+ (left) or CD8+ (right) T cells from the spleens of recipient mice transferred with donor bone marrow of the indicated genotypes. (f) Representative flow cytometry plots of splenic CD45.2+ cells from mice receiving bone marrow of the indicated genotypes. Panels show gating and proportions of TCRβ+, CD4 and CD8 T cells. Cells gated on either CD4 or CD8 cells were further analyzed by CD62L and CD44 staining. Summary data shown in g. (g) Frequencies and numbers of CD8 T cells staining CD44hiCD62Llo (upper panel) or CD44loCD62L+ (lower panel) in the spleens of recipients receiving bone marrow of the indicated genotypes. n=4, 4, 3 for Pld3−/−Pld4+/−, Pld3+/−Pld4−/−, and Pld3−/−Pld4−/−, respectively. Two-tailed T test was used to calculate P values. This experiment was performed once.
Discussion
We present evidence that the endolysosomal proteins PLD3 and PLD4 trim the 5′ end of ss oligodeoxynucleotides, thus degrading them and limiting their ability to stimulate TLR9. Consistent with this conclusion, PLD3 and PLD4 are most active at acidic pH (5–5.5) and are localized to endolysosomes15, 16, 17. Importantly, neither enzyme appears to have phospholipase activity25, 38.
Human GWAS studies have implicated polymorphisms of PLD3 and PLD4 in inflammatory diseases. In animals, spontaneous Pld4 null mutations have been linked to skin disease and runting. A mouse mutant identified on the BALB/c background called “Thin hair with small size” (Thss) appears to have abnormalities distinct from PLD4-deficiency on the C57BL/6 background that we have investigated here (http://www.informatics.jax.org/downloads/Reference_texts/J171492.pdf). A spontaneous PLD4-null mutation occurred in a herd of cattle, where homozygotes presented as runts with skin lesions, respiratory and digestive tract inflammation and mild anemia39. It will be of interest to determine if these phenotypes are also connected to TLR9 activation.
The similarities in enzymatic activities of PLD3 and PLD4 and their partly overlapping expression patterns suggest that they have redundant functions in key cell types. RNA profiling data imply that PLD4 has a narrow tissue distribution, with high levels in DCs and myeloid cells, whereas PLD3 has a broad distribution and appears to digest artificial substrates with higher efficiency than does PLD4. PLD3 enters the lysosome by an unusual ESCRT-regulated sorting and ubiquitination pathway17 and other PLD3-deficient mice reportedly showed no phenotype except for effects on the morphology of the lysosomal system16. The phenotype of our Pld3−/− mice also seems benign compared to that of Pld4−/− mice. This observation might be explained by the high levels of PLD4 normally present in cells expressing high levels of TLR9. Although Pld4−/− mice have a relatively mild form of Macrophage Activation Syndrome, residual PLD3 in myeloid cells likely provides protection from more severe disease as a single allele of either Pld3 or Pld4 protect against lethal inflammation early in life.
Nucleic acid sensors are endowed with a restricted fine specificity and affinity for ligands. Responses are limited by the quantity of ligands available and their intracellular localization. PLD3 and PLD4 provide ssDNAse activity in endolysosomes; it is likely no coincidence that they reside in the same compartment as active TLR9. Importantly, the DNA substrates of PLD3 and PLD4 require a non-phosphorylated 5′ end and the absence of secondary structure. Nucleic acids with such features are relatively unusual in nature, but are generated efficiently in endolysosomes by DNAse II, which cleaves dsDNA down to ssDNA fragments of ~10 nt carrying 3′ phosphate and 5′ hydroxyl groups36. DNAse II is also required to process DNA hairpins, such as A-type CpG ligands, for TLR9 recognition. Recent work indicates that TLR9 has two binding sites, one binding ssDNA with a CpG motif and the second binding short ssDNA carrying a 5′ hydroxyl40. Thus, the short stimulatory ligands that TLR9 is predicted to encounter should also be good substrates for PLD3 and PLD4. Endosomal build-up of such short ssDNA in the absence of PLD3 or PLD4 is unlikely to trigger STING/cGAS-mediated IFN responses as lengths of ssDNA greater than 21 nt are required and may be prevented by the presence of Trex1 exonuclease in the cytoplasm41, 42.
Although mild, the phenotype of Pld4−/− mice is reminiscent of human macrophage activation syndrome, which is associated with elevated IFN-γ, IL-12, CXCL9, CXCL10, low NK cell number, an increase of liver CD68+ cells, and can be mimicked in mice by repeated injection of a TLR9 ligand3, 30, 43, 44. Importantly, all of the phenotypes of Pld4−/− mice were absent in mice also lacking TLR9, consistent with the notion that defective degradation of ligands for TLR9 is central to the phenotype. This is further supported by evidence that Pld3−/− macrophages and HEK293 cells have exaggerated responses to particular ssDNA TLR9 ligands. Several features of the disease transferred by Pld3−/−Pld4−/− bone marrow into wild-type recipients mimic dysregulated TLR9 signaling45. Animals receiving stem cells expressing a mutated TLR9 that does not require proteolytic activation or traffic normally to endolysosomes, succumb to a DC-driven fatal inflammation characterised by profound anemia and reduced splenic B cells45. The lethality of Pld3−/−Pld4−/− animals in early life reflects the important role these enzymes play in limiting autoinflammation and although we predict that TLR9 signaling is required for the disease, this needs to be addressed by further breeding.
Presenting in infancy, livers from children with primary HLH show several features similar to those from Pld3−/−Pld4−/− animals, including giant cell hepatitis, steatosis, necrosis, and the name sake hemophagocytosis46. High serum ferritin and cytokine levels and extensive CD68+ cell hepatic infiltration in Pld3−/−Pld4−/− mice provide further similarities to primary HLH, although this disease is usually associated with mutations resulting in defective cytotoxic granule content or exocytosis from NK and T cells47.
PS-linkages are used in synthetic ODNs to hinder their digestion by nucleases. Fully phosphorothioated CpG-B type ODNs (1668PS) could drive TLR9-mediated IL-6 and IL-12 secretion from macrophages and dendritic cells independently of PLD3 or PLD4. However, PS linkages are by no means strictly artificial, and are found widely in nature in the DNA of bacteria48. That PLD3 and PLD4 digest ODNs carrying PS linkages or strings of Gs more slowly than host DNA might contribute to preferential innate recognition of bacteria or viruses.
We find that PLD3 and PLD4 are 5′ exonucleases with properties strikingly similar to the enzymatic activity described many years ago as spleen acid exonuclease27, 28. The bovine enzyme has a pH optimum of 5.5, and is 5′ single strand-specific; it also was characterized to be a non-processive nuclease, inhibited by 5′ phosphate, and generating 3′ nucleotide monophosphate products27, 49. Degradation of ssDNA substrates by PLD3 and PLD4 was similarly non-processive, blocked by 5′ phosphate, generated 3′ nucleotide monophosphates, and was able to digest to completion, generating mononucleotides and nucleosides as does the spleen exonuclease34. Although the gene encoding spleen exonuclease has never been cloned, the enzymatic activity has been described in human, rat, cow and pig, whose predicted PLD3 proteins are highly conserved (93–96% identical between species). The similarities in enzymatic features and the high amino acid identity between bovine, mouse and human PLD3, suggest that PLD3 may be identical to the spleen phosphodiesterase activity previously described, though PLD4 cannot be excluded, as it is also conserved among these species (>70% identical) and has a very similar enzymatic activity.
Methods
Mice
Targeting of Pld4 was carried out in 129/J strain embryonic stem cells by the TSRI Mouse Genetics Core. The neor cassette was removed by breeding to mice expressing Flp recombinase to generate Pld4fl/+ mice. Subsequent breeding to EIIa-Cre mice generated Pld4+/− mice, and both Pld4fl/+ and Pld4+/− were subsequently backcrossed to C57BL/6J for >20 generations. Pld4-mutant mice were genotyped by PCR using the following primers #32- TTTGCCATACCACCATAGCA, #33- GTTGGGCAGCAAATCTTAGG, #34- CTGGATTCCCCAAGAGAACA, and generating product sizes of 300 bp (wild-type), 350 bp (Flox), and 420 bp (ablated) for indicated alleles. The following mouse strains bred with Pld4−/− mice including B6.129S7-Rag1tm1Mom/J, B6.129P2-Lyz2tm1(cre)Ifo/J, B6.Cg-Tg(Itgax-cre)1–1Reiz/J, B6.129S7-Ifngtm1Ts/J, C57BL/6J-Tlr9M7Btlr/Mmjax (CpG11) were from the Jackson Laboratory. Unc93b3d/3d mice were kindly provided by B. Beutler, Univ. Texas Southwestern. Mutation of Pld3 was carried out by nuclear injection of C57BL/6J zygotes with plasmid eSpCas9(1.1)51 (a gift from Y. Doyon, Centre Hospitalier Universitaire de Québec, Addgene plasmid 79877) engineered to express the guide sequence GGACTGGCGGTCGCTGACCC targeting Pld3 exon 9, which encodes the first HKD motif. DNA (5 ng/μl) in Tris-HCl, pH 7.5, 10 mM, EDTA 0.1 mM, NaCl 50 mM was microinjected by the TSRI Mouse Genetics Core staff. Pld3−/− mice were genotyped by PCR using P3Ex5F-GCACATGCACACACACACAAAG and P3Ex6R- TACAATGAGGGCCAGGTAAGTG and the PCR product was digested with PshA1 restriction enzyme as the PshA1 site is only present in the wild-type allele. Experiments were performed as approved by the TSRI IACUC Committee.
Dendritic cell generation, isolation, and stimulation
DCs were expanded in vitro from bone marrow precursors using FLT3L as described52. Sorted splenic DCs were isolated essentially as described52. CD8α+ cDC (CD45RA−, CD11c+, CD11b+ and CD8α+) cells were plated at a density of 106/ml in round-bottom plates in media containing GM-CSF (10 ng/ml), mouse IL-4 (10 ng/ml) and rat IFN-γ (10 ng/ml). Stimuli were added and supernatant was collected 18–24 h later for cytokine analyses. VACV70 was purchased from InvivoGen and added to the culture media after being complexed with Lyovec at a concentration of 1 μg/ml. Oligonucleotides purchased from IDT or InvivoGen were also used directly; 2216PS (1 μM), 2216 (1 μM) 1668PS (0.25 μM) and 1668 (1 μM). 2216 subfragments were used at 1 μM. ELISA kits were used to measure the following cytokines: IL-12p70 (Duoset, R&D Systems), IL-6 (BD OptEIA, BD Biosciences), IFN-α (Lumikine, InvivoGen), IFN-λ (mouse IL-28A/B, R&D Systems).
Macrophage analysis
Macrophages were elicited into the peritoneal cavity by 2% thioglycolate broth treatment as described53 and stimulated essentially as described above. Ultrapure LPS from Salmonella minnesota R595 (InvivoGen) was used at 100 ng/ml. For the experiments described in Fig. 5h thioglycolate-elicited macrophages (2 × 106) were stimulated for either 3.5 or eight h with CpG-containing stimuli 2216-3′-biotin at 5 μM. Cells were washed twice with cold PBS before cell lysis with 0.5% NP40, 150 mM NaCl and 1 mM EDTA. Nuclei and cell debris were removed by centrifugation and the supernatant was incubated with 20 μl streptavidin magnetic beads (Invitrogen) for 40 min at 4°C with rotation. Beads were isolated using a magnet, washed twice with lysis buffer, and twice with T4 Polynucleotide Kinase (PNK) buffer (NEB). Bound DNA was labeled with γ32P-ATP (Perkin Elmer) with PNK for 30 min at 37°C and beads were washed twice with lysis buffer. Labeled DNA was then eluted with 95% formamide, 10 mM EDTA, heated to 70°C for 10 min and loaded onto 20% TBE/Urea Acrylamide gels. Radiolabeled DNA was exposed to a phosphorimager screen and the signal visualized using a Molecular Dynamics Typhoon phosphorimager and quantitated using ImageQuant software. In some cases, bone marrow derived macrophages were grown in 20% L929 supernatant containing medium over 7 days and stimulated as above.
Protein purification and nuclease assay
The N-terminal and transmembrane domain-deleted forms of mouse and human PLD3, PLD3-AA, PLD4, and PLD4-AA were cloned into vector pCEP4 (Thermo) and transfected into HEK293-EBNA cells (Invitrogen). The stable cell lines expressing appropriate proteins were generated by selection with G418 (0.25 mg/ml) and puromycin (0.5 μg/ml) in DMEM complete medium. After selection, cells were expanded in suspension culture in 293 Freestyle media (Life Technologies) containing G418 (0.25 mg/ml) and puromycin (0.5 μg/ml) in a humidified 37°C CO2 incubator (8%), rotating at 135 rpm at 1–2 × 106 cells/ml. Cells were harvested, centrifuged at 5,000g for 20 min and supernatant filtered using 0.2 μm filter (Millipore). Protino Ni-NTA Agarose beads (Macherey-Nagel) were added to the supernatants and rotated for 4 h at 4°C. After incubation, the beads were washed three times in buffer (Tris-HCl 20 mM, pH 7.5, NaCl 250 mM, Imidazole 10 mM) and eluted (Tris-HCl 20 mM, pH 7.5, NaCl 250 mM, Imidazole 200 mM). The eluted proteins were dialyzed overnight in PBS at 4°C and protein concentrations were determined using the BCA protein assay kit (Thermo). Proteins were analyzed for purity by SDS-PAGE stained with Coomassie Brilliant Blue. Nuclease assays for DNA and RNA digestion were carried out for 2 h at 37°C in 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) pH 5.5, 125 mM NaCl, with substrates at 2.5 μM and enzyme at 100 nM. For phosphorothioate-modified substrates, pH was pH 5.0, enzyme concentration 500 nM, and incubation time 6 h unless otherwise noted.
Dinucleotide substrate assay
Enzymatic activity assay on dinucleotide substrates was performed as described34. Lyophilized bovine spleen phosphodiesterase II (PDII) and snake venom phosphodiesterase I (PDI) were diluted to 2.5 U/ml and 100 U/ml in water, respectively. Adenosine deaminase (ADA) was resuspended in 1 ml water then diluted to 20 μg/ml in 0.5× PBS. All enzymes were purchased from Worthington. The dinucleotide substrates GpA and ApG were purchased from TriLink Biotechnologies. Final reaction condition for PLD3, PLD4, and bovine spleen PDII was 50 mM MES pH 6.1, 20 mM NaCl. Snake venom PDI enzyme reaction was performed in 50 mM Tris, pH 7.9, 20 mM NaCl. Each reaction consisted of dinucleotide substrate (60 μM), ADA enzyme (2 μg/ml), and either PLD3 (50 nM), PLD4 (50 nM), PDII (0.25 U/ml), or PDI (10 U/ml) in a total volume of 250 μl. The reaction was performed with the Nanodrop 3000C in a heated quartz cuvette (37 °C) and absorbance at 265 nm measured every 15 s. The tested phosphodiesterase was added after four initial absorbance readings (1 min) and absorbance measured for another 5 min. In the absence of an added phosphodiesterase, such as substrate alone, or ADA alone, no change in absorbance was observed throughout the 6 min of recording.
Studies with 293HEK cells
HEK293-BluehTLR9 cells (InvivoGen) were cultured and stimulated according to the manufacturer’s instructions. Briefly, 100 μl cells/well at a density of 106/ml were cultured in 96-well plates. Stimuli were added and culture supernatants were harvested 16 h later for SEAP1 detection using a kit (InvivoGen). For PLD3-deficient cells, HEK293-BluehTLR9 cells were transiently transfected with a CRISPR/Cas9 construct previously validated for efficacy that carried the guide RNA sequence TCCTCATTCTGGCGGTTGT. Single-cell clones were then isolated and individually screened by sequencing for homozygous mutation of PLD3. PLD3-deficient cells were transduced with lentiviruses encoding human PLD4, human PLD4-AA, mouse PLD3, and mouse PLD3-AA, respectively and cultured for several days before stimulation. ODN stimuli were purchased from InvivoGen or Integrated DNA Technologies. Cells were cultured with 2216-3′-biotin for 6 h at 37 °C then washed and lysed as for macrophages. Streptavidin precipitated DNA was 5′ radiolabeled using polynucleotide kinase and γ32P-ATP. To achieve higher resolution of smaller oligonucleotide fragments, samples in formamide were loaded onto a 20% acrylamide (19:1 Accugel acrylamide (National Diagnostics)) denaturing gel in 50 mM histidine as described50.
Blood analysis
Monocyte, platelet, reticulocyte, red blood cell, and white blood cell counts in whole blood from age-matched male and female animals were determined using an IDEX Procyte DX Machine for blood subset analysis. Plasma concentrations of IFN-γ and CXCL10 of adult Pld4fl/fl and Pld4−/− mice were determined using FlowCytomix beads (eBioscience). Serum from 16–19-day-old animals was analyzed for cytokine concentrations using a kit (Flowcytomix, eBiosciences)
Ferritin
Mouse Ferritin concentrations were determined from sera isolated from 12–21 day old animals as outlined by the manufacturer of the ELISA kit (Alpco).
Generation of Bone Marrow Chimeras
Bone marrow from 16-day-old littermates of CD45.2+ Pld3−/− Pld4+/−, Pld3+/− Pld4−/−, or doubly deficient Pld3−/−Pld4−/− genotypes were injected intravenously into lethally irradiated (1000 rad) 8 week old sex matched B6.SJL-CD45.1+ recipients. Eight weeks after transfer, blood and spleen populations and serum cytokines were analyzed.
Flow cytometry analysis of leukocyte compartments
Tissue suspensions were generated by mashing tissue between frosted glass slides. Bone marrow was released from tibia and femurs by a mortar and pestle crushing method. Red blood cells were lysed with ammonium chloride (0.83%) and cells filtered to generate single cell suspensions. Peritoneal cavity cells were isolated by lavage. Cells were pre-incubated with 1 μg/106 cells of Fc Block (2.4G2) prior to staining with antibodies. Splenic NK cell proportions were determined by flow cytometry of splenocytes from age- and sex-matched animals using the following gates (TCRβ− NK1.1+CD49b+). Flow cytometry of DCs was performed from spleens treated with collagenase and DNase I treatment prior to separation on light density gradients (Nycodenz). The following gating strategies were used to identify and quantify different leukocyte subsets. pDCs (CD11c+CD45RA+SiglecH+CD19−TCRβ−); CD8+ cDCs (CD19−TCRβ−CD11c+CD45RA−CD11bloCD8α+); CD11b+ cDCs (CD19−TCRβ−CD11c+CD45RA−CD11bhiCD8α+); CD4+ (TCRβ+CD4+CD8α−); CD8+ (TCRβ+CD4−CD8α+); CD19+ B cells (CD19+B220+); Transitional 1 (TI) B cells (B220+CD93+IgM+CD23−); Transitional 2 (T2) B cell (B220+CD93+IgM+CD23+); Follicular B2 cells (B2) (CD19+B220+CD93−IgM+CD23+); Marginal zone B cells (MZ) (CD19+ B220+CD93−CD23loIgM+CD21hi); Germinal Center (GC) (TCRβ−B220+FAS+ GL7+); Plasma cell (PC) (TCRβ−F480−IgD−CD138+); resident peritoneal B1 cells (B220loCD19+CD43+IgM+); resident peritoneal B2 (B220hiCD19+CD43−IgM+); resident peritoneal macrophages (size, F4/80+CD11b+). Antibodies used in this study are listed in Supplementary Table 3. Gating strategies used for dendritic cells, peritoneal macrophages, and bone marrow chimera B and T cells are shown in Supplementary Figure 8.
Experimental autoimmune encephalomyelitis induction and monitoring
EAE was induced by immunization with MOG35–55 peptide emulsified in complete Freund’s adjuvant followed by Pertussis toxin administration as outlined by the manufacturer (Hooke Laboratories) and scored according to the manufacturer’s instructions. Female animals over 8 weeks old were used. In some cases, C57BL/6 mice congenic for CD45.1 (B6.CD45.1) were lethally irradiated with two doses of 500 rads and were reconstituted with bone marrow from either Pld4fl/fl or Pld4−/− mice for 10 weeks prior to immunization.
CD68+ liver stains
Livers from age and sex matched mice were frozen in optimal cutting temperature (O.C.T., Sakura). Then 7 μm sections were cut, fixed with acetone for 10 min, and stained with anti-CD68 Alexa Fluor 647 (FA-11, BioLegend). After washing in PBS, nuclei were stained with Hoechst 33258 for 5 min prior to mounting in Prolong Diamond Antifade mounting medium (Invitrogen). Sections were imaged using the Keyence BZ-X710 fluorescence microscope. CD68+% and area was determined using FIJI software54. Briefly, color images were split into channels and a threshold set for each channel. The analyze particles tool was used to estimate number of nuclei in image (Hoechst blue channel). The analyze particle tool was then applied to the CD68+ channel (Red channel). The percentage was then calculated. The measure tool was used to calculate the average area of the particles above the threshold in the CD68+ channel.
Mass spectrometry
Phosphodiesterase II (Worthington Biochemical Corp. Cat#: LS003602, LOT #: 36m16915) was fractionated on a Superdex 200 column in PBS and two adjacent fractions with peak enzymatic activity were pooled. The solution sample was reduced (200 mM DTT), alkylated (200 mM iodoacetamide) and digested with trypsin overnight using an estimated 1:30 enzyme to substrate ratio before being analyzed by nano-LC-MS/MS at the TSRI core facility. The MS/MS raw data were searched against the custom sequence database, which included the sequence [NP_001071509.1 phospholipase D3 [Bos taurus]] and the NCBInr database. Bovine PLD3 protein was identified with 7 peptides and 17% sequence coverage and the sequence [XP_015324054.1 PREDICTED: phospholipase D4 isoform X2 [Bos taurus]] was not identified. Other proteins also identified with at least five different peptides are listed in Supplementary Fig. 4.
RNA expression analysis and sequencing
RNA expression data in Fig. 4c was from the BioGPS MOE430 Gene Atlas Data set55, 56. For the sequencing analysis in Fig. 1k, total RNA was isolated from splenocytes using a kit (RNA easy, Qiagen). RNA was prepared into RNAseq libraries using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina following the manufacturer’s recommended protocol. The libraries were then PCR amplified 15 cycles using barcoded PCR primers, purified and size selected using AMPure XP Beads before loading onto an Illumina NextSeq500 for 75 base single read sequencing. A total of 16.3–19.6 million “passed filter” reads were obtained for each of the 6 samples. Reads were compared to the mouse genome build mm10 and quantitated using the program Salmon. DC RNAseq data described in Fig. 4d were generated as follows. DC subsets were purified from spleens of 10–14 C57BL/6 mice/sample as described57. Briefly, cell sorting was performed on a BD Influx™ machine. RNA was extracted using RNeasy Mini Kit (QIAGEN) and residual genomic DNA removed using RNase-free DNase (QIAGEN). Sequencing libraries were prepared using the Illumina TruSeq Stranded mRNA kit and sequencing was carried out by Micromon using High-Output SBS on the Illumina NextSeq 500. RNAseq data analyses were performed in Degust (http://degust.erc.monash.edu/ Version: 3.1.0) by Monash Bioinformatics platform, David R Powell & Adele Barugahare.
Statistical analyses
Analysis of differential gene expression was carried out using the program Edge. Other data comparisons used the program Prism (Graph Pad), with correction for multiple comparisons. When two groups were compared, an unpaired, two-tailed T test was used. Actual P values are provided in Supplementary Table 4. Additional information is provided in the Life Sciences Reporting Summary.
Supplementary Material
Acknowledgments
This project was supported by grants from the National Institutes of Health R21AI126011, R21AI101692, R37AI059714. We thank our TSRI colleagues S. Kupriyanov and G. Martin (Genetics Core) and S. Head and P. Natarajan (NGS Core) for expert technical assistance.
Footnotes
Declaration of interests
The authors have no competing interests
Author contributions: A.L.G. participated in many of the studies, identifying anomalies in PLD4- and PLD3-deficient mice and their myeloid cell cytokine responses and in the preparation of PLD3 and PLD4 proteins. C.H. generated the Pld4fl/fl and Pld4−/− mutant mice and characterized many of their phenotypes. D.H. elucidated the enzymatic function of PLD3 and PLD4, carried out all studies in 293 cells, and contributed to other studies. A.M. cloned Pld4 cDNA and characterized Pld4 expression in the B cell lineage. V.T. and B.R.L. participated in analysis of EAE in Pld4−/− mice. P.D.S., T.R.B., and T.C.T. provided technical support. K.O. provided liver pathology histological analysis. F. K., H.S.C., A.Ze., R.L.S., P.K., M.B., S.R.S. were student interns who participated in generating CRISPR/Cas9 mutagenesis constructs and site-directed mutagenesis of Pld3 and Pld4. A.Za. provided assistance with blood analysis using the IDEX Procyte DX Machine. B.C. assisted C.H. in a search for potential phospholipase activity of PLD4. J.T. and J.C.dlT. provided advice and assistance with RNA virus studies. M.O., E.S.P. and H.H. provided advice and assistance with DC studies (Fig. 3e) and data in Extended Data Fig. 4. L.T. provided advice and assistance with PLD4 protein expression. M.D. assisted with PLD3 reagents and antibodies. D.N. and A.L.G. codirected these studies. D.N., A.L.G., and D.H. wrote the paper.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol. 2016;16:566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 3.Rigby RE, Leitch A, Jackson AP. Nucleic acid-mediated inflammatory diseases. Bioessays. 2008;30:833–842. doi: 10.1002/bies.20808. [DOI] [PubMed] [Google Scholar]
- 4.Al-Mayouf SM, et al. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nat Genet. 2011;43:1186–1188. doi: 10.1038/ng.975. [DOI] [PubMed] [Google Scholar]
- 5.Napirei M, et al. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- 6.Yasutomo K, et al. Mutation of DNASE1 in people with systemic lupus erythematosus. Nat Genet. 2001;28:313–314. doi: 10.1038/91070. [DOI] [PubMed] [Google Scholar]
- 7.Kawane K, et al. Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science. 2001;292:1546–1549. doi: 10.1126/science.292.5521.1546. [DOI] [PubMed] [Google Scholar]
- 8.Rodero MP, et al. Type I interferon-mediated autoinflammation due to DNase II deficiency. Nat Commun. 2017;8:2176. doi: 10.1038/s41467-017-01932-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morita M, et al. Gene-targeted mice lacking the Trex1 (DNase III) 3′-->5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee-Kirsch MA, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 11.Rice G, et al. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutieres syndrome. Am J Hum Genet. 2007;80:811–815. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491:279–283. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waite M. The PLD superfamily: insights into catalysis. Biochim Biophys Acta. 1999;1439:187–197. doi: 10.1016/s1388-1981(99)00094-3. [DOI] [PubMed] [Google Scholar]
- 14.McDermott M, Wakelam MJ, Morris AJ. Phospholipase D. Biochem Cell Biol. 2004;82:225–253. doi: 10.1139/o03-079. [DOI] [PubMed] [Google Scholar]
- 15.Otani Y, et al. PLD4 is involved in phagocytosis of microglia: expression and localization changes of PLD4 are correlated with activation state of microglia. PloS one. 2011;6:e27544. doi: 10.1371/journal.pone.0027544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fazzari P, et al. PLD3 gene and processing of APP. Nature. 2017;541:E1–E2. doi: 10.1038/nature21030. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez AC, et al. Unconventional Trafficking of Mammalian Phospholipase D3 to Lysosomes. Cell Rep. 2018;22:1040–1053. doi: 10.1016/j.celrep.2017.12.100. [DOI] [PubMed] [Google Scholar]
- 18.Terao C, et al. PLD4 as a novel susceptibility gene for systemic sclerosis in a Japanese population. Arthritis Rheum. 2013;65:472–480. doi: 10.1002/art.37777. [DOI] [PubMed] [Google Scholar]
- 19.Okada Y, et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet. 2012;44:511–516. doi: 10.1038/ng.2231. [DOI] [PubMed] [Google Scholar]
- 20.Chen WC, et al. rs2841277 (PLD4) is associated with susceptibility and rs4672495 is associated with disease activity in rheumatoid arthritis. Oncotarget. 2017;8:64180–64190. doi: 10.18632/oncotarget.19419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cruchaga C, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2014;505:550–554. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang DF, et al. PLD3 in Alzheimer’s Disease: a Modest Effect as Revealed by Updated Association and Expression Analyses. Mol Neurobiol. 2016;53:4034–4045. doi: 10.1007/s12035-015-9353-5. [DOI] [PubMed] [Google Scholar]
- 23.Hooli BV, et al. PLD3 gene variants and Alzheimer’s disease. Nature. 2015;520:E7–8. doi: 10.1038/nature14040. [DOI] [PubMed] [Google Scholar]
- 24.Heilmann S, et al. PLD3 in non-familial Alzheimer’s disease. Nature. 2015;520:E3–5. doi: 10.1038/nature14039. [DOI] [PubMed] [Google Scholar]
- 25.Yoshikawa F, et al. Phospholipase D family member 4, a transmembrane glycoprotein with no phospholipase D activity, expression in spleen and early postnatal microglia. PloS one. 2010;5:e13932. doi: 10.1371/journal.pone.0013932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osisami M, Ali W, Frohman MA. A role for phospholipase D3 in myotube formation. PloS one. 2012;7:e33341. doi: 10.1371/journal.pone.0033341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernardi A, Bernardi G. Studies on acid hydrolases. IV Isolation and characterization of spleen exonuclease. Biochim Biophys Acta. 1968;155:360–370. [PubMed] [Google Scholar]
- 28.Hilmoe RJ. Purification and properties of spleen phosphodiesterase. J Biol Chem. 1960;235:2117–2121. [PubMed] [Google Scholar]
- 29.Willenborg DO, Fordham SA, Staykova MA, Ramshaw IA, Cowden WB. IFN-gamma is critical to the control of murine autoimmune encephalomyelitis and regulates both in the periphery and in the target tissue: a possible role for nitric oxide. J Immunol. 1999;163:5278–5286. [PubMed] [Google Scholar]
- 30.Behrens EM, et al. Repeated TLR9 stimulation results in macrophage activation syndrome-like disease in mice. J Clin Invest. 2011;121:2264–2277. doi: 10.1172/JCI43157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferlazzo G, et al. Distinct roles of IL-12 and IL-15 in human natural killer cell activation by dendritic cells from secondary lymphoid organs. Proc Natl Acad Sci USA. 2004;101:16606–16611. doi: 10.1073/pnas.0407522101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X, et al. The interleukin 12 p40 gene promoter is primed by interferon gamma in monocytic cells. J Exp Med. 1996;183:147–157. doi: 10.1084/jem.183.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Razzell WE. Tissue and intracellular distribution of two phosphodiesterases. J Biol Chem. 1961;236:3028–3030. [PubMed] [Google Scholar]
- 34.Ipata PL, Felicioli RA. A convenient spectrophotometric assay for phosphodiesterases, using dinucleoside-monophosphates as substrates. Eur J Biochem. 1969;8:174–179. doi: 10.1111/j.1432-1033.1969.tb00511.x. [DOI] [PubMed] [Google Scholar]
- 35.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 36.Chan MP, et al. DNase II-dependent DNA digestion is required for DNA sensing by TLR9. Nat Commun. 2015;6:5853. doi: 10.1038/ncomms6853. [DOI] [PubMed] [Google Scholar]
- 37.Tabeta K, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 38.Pedersen KM, Finsen B, Celis JE, Jensen NA. Expression of a novel murine phospholipase D homolog coincides with late neuronal development in the forebrain. J Biol Chem. 1998;273:31494–31504. doi: 10.1074/jbc.273.47.31494. [DOI] [PubMed] [Google Scholar]
- 39.Langenmayer MC, et al. Zinc Deficiency-Like Syndrome in Fleckvieh Calves: Clinical and Pathological Findings and Differentiation from Bovine Hereditary Zinc Deficiency. J Vet Intern Med. 2018;32:853–859. doi: 10.1111/jvim.15040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohto U, et al. Toll-like Receptor 9 Contains Two DNA Binding Sites that Function Cooperatively to Promote Receptor Dimerization and Activation. Immunity. 2018;48:649–658. e644. doi: 10.1016/j.immuni.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 41.Erdal E, Haider S, Rehwinkel J, Harris AL, McHugh PJ. A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev. 2017;31:353–369. doi: 10.1101/gad.289769.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luecke S, et al. cGAS is activated by DNA in a length-dependent manner. EMBO Rep. 2017;18:1707–1715. doi: 10.15252/embr.201744017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Canna SW, Behrens EM. Not all hemophagocytes are created equally: appreciating the heterogeneity of the hemophagocytic syndromes. Curr Opin Rheumatol. 2012;24:113–118. doi: 10.1097/BOR.0b013e32834dd37e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bracaglia C, et al. Elevated circulating levels of interferon-gamma and interferon-gamma-induced chemokines characterise patients with macrophage activation syndrome complicating systemic juvenile idiopathic arthritis. Ann Rheum Dis. 2017;76:166–172. doi: 10.1136/annrheumdis-2015-209020. [DOI] [PubMed] [Google Scholar]
- 45.Mouchess ML, et al. Transmembrane mutations in Toll-like receptor 9 bypass the requirement for ectodomain proteolysis and induce fatal inflammation. Immunity. 2011;35:721–732. doi: 10.1016/j.immuni.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen JH, et al. Pathology of the liver in familial hemophagocytic lymphohistiocytosis. Am J Surg Pathol. 2010;34:852–867. doi: 10.1097/PAS.0b013e3181dbbb17. [DOI] [PubMed] [Google Scholar]
- 47.Sepulveda FE, de Saint Basile G. Hemophagocytic syndrome: primary forms and predisposing conditions. Curr Opin Immunol. 2017;49:20–26. doi: 10.1016/j.coi.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 48.Wang L, et al. DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc Natl Acad Sci USA. 2011;108:2963–2968. doi: 10.1073/pnas.1017261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernardi A, Cantoni GL. Action of spleen exonuclease on transfer ribonucleic acid. J Biol Chem. 1969;244:1468–1476. [PubMed] [Google Scholar]
- 50.Mandecki W, Hayden M. High-resolution polyacrylamide gel electrophoresis of oligonucleotides using L-histidine buffer. DNA. 1988;7:57–62. doi: 10.1089/dna.1988.7.57. [DOI] [PubMed] [Google Scholar]
- 51.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vremec D, Shortman K. The isolation and identification of murine dendritic cell populations from lymphoid tissues and their production in culture. Meth Molec Biol. 2008;415:163–178. doi: 10.1007/978-1-59745-570-1_10. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Prot Immunol/edited by John E. Coligan … [et al.] 2008;Chapter 14(Unit 14):11. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lattin JE, et al. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu C, Jin X, Tsueng G, Afrasiabi C, Su AI. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucl Acids Res. 2016;44:D313–316. doi: 10.1093/nar/gkv1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lauterbach H, et al. Mouse CD8αlpha+ DCs and human BDCA3+ DCs are major producers of IFN-lambda in response to poly IC. J Exp Med. 2010;207:2703–2717. doi: 10.1084/jem.20092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.