Abstract
Background
Mutations in the GRIN2A gene, which encodes the GluN2A subunit of the N-methyl-D-aspartate receptor, have been identified in patients with epilepsy-aphasia spectrum disorders, idiopathic focal epilepsies with centrotemporal spikes, and epileptic encephalopathies with severe developmental delay. However, thus far, mutations in this gene have not been associated with a non-epileptic neurodevelopmental disorder with dystonia.
Objectives
To identify the disease-causing gene in two siblings with a neurodevelopmental and movement disorder with no epileptiform abnormalities.
Methods
Targeted next-generation sequencing panel for neuropediatric disorders and subsequent electrophysiological studies.
Results
The two siblings carry a novel missense mutation in the GRIN2A gene (p. Ala643Asp) that was not detected in genomic DNA isolated from blood cells of their parents, suggesting that the mutation is the consequence of germinal mosaicism in one progenitor. In functional studies, the GluN2A-A643D mutation increased the potency of the agonists L-glutamate and glycine and decreased the potency of endogenous negative modulators, including protons, Mg2+, and Zn2+, but reduced agonist-evoked peak current response in mammalian cells, suggesting that this mutation has a mixed effect on NMDAR function.
Conclusion
De novo GRIN2A mutations can give rise to a neurodevelopmental and movement disorder without epilepsy.
Keywords: Glutamate receptor, NMDA receptor, GRIN2A, GluN2A, movement disorder
INTRODUCTION
The N-methyl-D-aspartate receptor (NMDAR) is an ionotropic receptor that binds glutamate, the main excitatory neurotransmitter of the central nervous system. This receptor class plays important roles in development, synaptic plasticity, learning, and memory [1]. The NMDAR has a tetrameric structure, consisting of two GluN1 and two GluN2 subunits. The GluN1 subunit is encoded by the GRIN1 gene, while the GluN2 subunits are encoded by the genes GRIN2A, GRIN2B, GRIN2C, and GRIN2D, which have distinct spatial and temporal expression patterns [2–6]. Levels of GRIN2B and GRIN2D expression are high before birth and subsequently decline in most brain regions, whereas the expression of GRIN2A and GRIN2C increases after birth [7]. NMDARs are Ca2+-permeable, voltage-dependent receptors that mediate a slow synaptic current and are blocked by extracellular Mg2+ [8]. Overactivation of NMDARs has been implicated in the induction of seizures and cell death [9–13], whereas hypofunction of this receptor may contribute to schizophrenia [14, 15].
Alterations in GRIN2A were first described in 2010 [16] in three patients carrying deletions in chromosome 16p13, including the GRIN2A gene. These patients were intellectually impaired, with dysmorphic features and epilepsy involving the rolandic region. Three subsequent studies described point mutations and deletions in GRIN2A in patients with epilepsy-aphasia spectrum (EAS) and idiopathic focal epilepsies (IFE) with centrotemporal spikes, particularly in those with more severe phenotypes [17–19]. Lesca and coworkers [17] detected de novo or inherited GRIN2A deletions or mutations in about 20% of cases of Landau-Kleffner syndrome (LKS), epilepsy with continuous spikes and waves during slow-wave sleep (CSWSS), and atypical benign partial epilepsy (ABPE), often associated with speech impairment. Lemke et al. [18] reported genetic alterations in GRIN2A in 7.5% of IFE patients studied, and found that mutations were more common in more severe phenotypes: 4.9% (12/245) in benign epilepsy with centrotemporal spikes (BECTS) and 17.6% (9/51) in CSWSS patients. Moreover, they detected exon-disrupting microdeletions of the GRIN2A gene in 1% (3/286) of individuals screened for copy number variations (CNV). Finally, Carvill et al. [19] identified GRIN2A mutations in 9% (4/44) of patients with EAS. Alterations in GRIN2A have also been reported in patients with early-onset epileptic encephalopathy and severe developmental delay [1, 16–29]. To date, whole exome and genome sequencing have identified a substantial number of point mutations and deletions (>100) scattered across all domains of the GRIN2A gene [1, 30, 31]. The large number of mutations found in GRIN2A may reflect the postnatal increase in the expression of GluN2A, which prevents catastrophic preterm neurologic complications and allows patients to survive to full term, albeit with neurologic complications that appear later in life as GRIN2A expression increases.
Encephalopathies caused by mutations in genes encoding the other NMDAR subunits (GRIN1, GRIN2B, and GRIN2D) are considered part of a phenotypic spectrum that encompasses patients with a constellation of symptoms, including developmental delay, intellectual disability, autism spectrum disorders, epilepsy, and movement disorder [20, 26, 32–34]. Here, we describe a neurodevelopmental and movement disorder without seizures in two siblings with the same GRIN2A mutation.
METHODS
Consent and approval
Research protocols were approved by the Ethics Committee of the Hospital Santiago de Compostela (Spain), and families provided informed consent. All in vitro experiments were conducted in accordance with the guidelines of Emory University.
Targeted next-generation sequencing
Targeted enrichment was performed using an in-solution hybridization technology (Sure Select XT; Agilent Technologies), and the Miseq platform (Illumina) was used for subsequent sequencing. One custom Sure Select probe library was designed to capture the exons and exon-intron-boundaries of 428 genes associated with neuropediatric diseases, including all corresponding transcripts. The Sure Design web-based probe design tool was used for this purpose (https://earray.chem.agilent.com/suredesign/). To ensure optimal representation of all regions of interest, we designed different subgroups of probes according to guanine-cytosine content and location with respect to repetitive sequences. Sequence capture, enrichment, and elution were performed according to the manufacturer’s instructions. Captured fragments were sequenced in pair-end 100-base mode using the Miseq platform. Image analysis and processing of fluorescence intensity in sequences (“base calling”) was performed using Real Time Analysis (RTA) software v.1.8.70 (Illumina), and the FastQC v0.10.1 program (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/) was used for data quality control. Reads were aligned to the reference genome GRCh37 with BWA v0.7.9a [35]. NGSrich v0.7.5 software (http://ngsrich.sourceforge.net) was used as a control prior to variant detection, and BEDTools 2.17.0 (http://bedtools.readthedocs.org/en/latest/#) and Picard 1.114 (http://picard.sourceforge.net) for the intermediate steps.
Mutagenesis and electrophysiology studies
cDNAs encoding the wild-type (WT) human NMDA receptor subunit GluN1-1a (hereafter GluN1; GenBank accession number: NP_015566) and GluN2A (GenBank accession number: NP_000824) were subcloned into pCI-neo (Promega, Madison, Wisconsin) to express recombinant NMDARs. GluN2A-A643D was generated by site-directed mutagenesis using the QuikChange protocol (Stratagene, La Jolla, CA). The mutagenesis product was confirmed by DNA sequencing (Eurofins, Louisville, KY). cRNA was synthesized in vitro and co-injected into Xenopus laevis oocytes (Ecocyte Bio Science, Austin TX) as previously described [28]. Two-electrode voltage-clamp (TEVC) recordings from oocytes were performed at room temperature (23°C) to measure NMDAR-mediated inward currents in response to stimulation with the co-agonists glutamate and glycine. The recording solution contained (in mM) 90 NaCl, 1 KCl, 10 HEPES, 0.5 BaCl2, 0.01 EDTA (23°C, pH 7.4 unless otherwise stated). The membrane potential was held at −40 mV for all TEVC recordings unless otherwise stated. The agonist concentration-response data were fitted using equation 1:
| Equation 1 |
where N is the Hill slope and EC50 is the concentration of the agonist that produces a half-maximal effect. Experiments evaluating the sensitivity of the channel to extracellular zinc were performed in the presence of tricine (10 mM) at pH 7.3 (VHOLD, −20 mV). ZnCl2 solutions (10 mM) were prepared fresh daily and added directly to the recording solution (with no EDTA) to obtain the desired nominal free concentration of Zn2+ [36]. Experiments evaluating the sensitivity of the channel to Mg2+ blockade were performed in the recording solution with no EDTA at a holding potential of −60 mV. Glutamate (100 μM) and glycine (100 μM) were used in all oocyte experiments unless otherwise stated. The concentration-response curves for Mg2+ and Zn2+ inhibition were fitted using equation 2:
| Equation 2 |
where IC50 is the concentration of the inhibitor that produces a half-maximal effect and minimum is the degree of residual response at a saturating concentration of the inhibitor.
The whole cell voltage clamp current recordings were performed on transiently transfected HEK293 cells (ATCC CRL-1573) with plasmid cDNAs encoding wild type human GluN1/GluN2A or GluN1/GluN2A-A643D with a solution containing (in mM) 150 NaCl, 3 KCl, 10 HEPES, 0.01 EDTA, 0.5 CaCl2, and 11 D-mannitol, with the pH adjusted to 7.4 by addition of NaOH (23°C) [28]. The recording electrodes were prepared a vertical puller (Narishige P-10, Tokyo, Japan) using thin-walled filamented borosilicate glass (TW150F-4; World Precision Instruments, Sarasota, FL, USA) and filled with the internal solution (in mM: 110 D-gluconic acid, 110 CsOH, 30 CsCl, 5 HEPES, 4 NaCl, 0.5 CaCl2, 2 MgCl2, 5 BAPTA, 2 Na2ATP, 0.3 NaGTP; pH 7.35 with 300–310 mOsmol/kg of the osmolality). The current responses to the saturated concentrations of glutamate (1000 μM) and glycine (100 μM) were recorded using an Axopatch 200B patch-clamp amplifier (Molecular Devices, Sunnyvale, CA, USA) with the holding potential of −60 mV at room temperature (23°C). The current responses were digitized at 20 kHz using a Digidata 1440A data acquisition system (Molecular Devices, Sunnyvale, CA, USA), filtered at 8 kHz (−3 dB, 8 pole Bessel filter, Frequency Devices, Ottawa, IL, USA), and normalized to cell capacitance.
All reagents were purchased from Sigma (unless otherwise stated). Data were expressed as the mean ± SEM and the statistical difference was determined by unpaired t-test using the log EC50 and IC50 values. Current amplitudes from transfected HEK293 cells were compared using Mann Whitney test. Significance for all tests was set at p < 0.05.
RESULTS
Case report
Patient 1 is a boy (aged 8 years at the time of writing) born to non-consanguineous, healthy parents at week 35 of gestation by vaginal delivery following an uneventful pregnancy. His birth weight and length were 2,860 g and 48 cm, respectively, and his Apgar score was 9/10. He displayed retarded psychomotor development, with delayed acquisition of language and motor milestones (sitting at 19 months, walking at 20 months, two-word phrases at 5 years, and three-word phrases at 6–7 years). From the first months of life, he experienced generalized dystonic attacks and hypertonia predominantly affecting the lower limbs. Physical examination at 19 months revealed generalized dystonia with upper limb predominance, fluctuating muscular tone with a predominant extensor pattern in the lower limbs, truncal hypotonia, positive bilateral plantar grip, and an intense bilateral Galant reflex, and a diagnosis of dystonic cerebral palsy was established. A complete metabolic study of blood, urine, and cerebrospinal fluid, including screening for glucose transporter type 1 (GLUT-1) deficiency, yielded normal results. Magnetic resonance imaging (MRI) with spectroscopy, auditory and visual evoked potentials, and video-electroencephalogram (EEG) revealed no findings of relevance. The patient exhibited convergent strabismus and hypermetropia, which were treated with botulinum toxin and corrective lenses, as well as insomnia with frequent awakenings. In addition, the patient has obstructive sleep apnea syndrome, which required adenoidectomy and continuous positive airway pressure (CPAP) for one year, and makes a near continuous snoring-like breathing noise while awake, although for reasons unknown this is absent during sleep. Cognitive symptoms include moderate intellectual disability, reduced vocabulary and difficulties using expressive language. The patient has never experienced a seizure, and repeated EEG recordings have revealed an absence of epileptiform abnormalities. Recent physical examinations detected dystonic movements and postures of the head, neck, trunk, and upper limbs (right predominance), severe problems in fine motor coordination, and an altered gait with dystonic postures.
At 6 years of age, to establish a diagnosis, targeted next-generation sequencing (NGS) analysis of a panel of 428 genes associated with neuropediatric diseases was performed. The heterozygous variant c.C1928A, which promotes the amino-acid change p.Ala643Asp, was identified in exon 10 of the GRIN2A gene (reference sequence NM_000833.3). This variant was not registered in the 1000 Genomes Project database, the Single Nucleotide Polymorphism database (dbSNP), or the gnomAD database curated by the Broad Institute [37], nor has it been detected in 400 chromosomes analyzed in a Spanish population (Fernández-Marmiesse et al., unpublished data). An in silico pathogenicity assessment performed with CONDEL software [38] predicted that this variant was deleterious. The variant was not present in either parent.
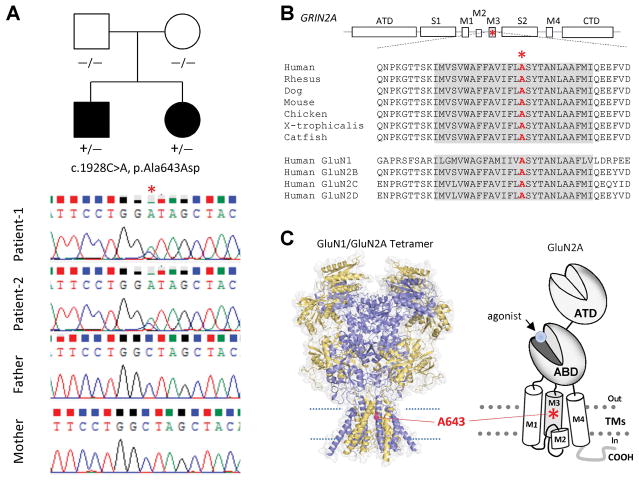
Patient 2 (aged 4 years at the time of writing) is the younger sister of Patient 1, and was born at week 37 of gestation via vaginal delivery. The pregnancy was complicated by preterm labor from week 30. The patient’s birth weight and length were 3,430 g and 50 cm, respectively, and her Apgar score was 9/10. Like her brother, her psychomotor development was retarded with delayed acquisition of language and motor milestones (walking at 20 months, first words at 17 months, and two-word phrases at 25 months). At 2 years of age, physical examination revealed mild dystonia of the upper limbs, dyskinesic movements, intentional and postural tremor, and major difficulties in fine manipulation, apraxic gait and lack of motor coordination. Additionally, she presented convergent strabismus, which was treated with lens occlusion, as well as sleep disturbances and the same breathing noise as her brother. Blood tests and MRI with spectroscopy revealed no findings of note, and EEG results were normal across multiple tests. Currently, she displays mild cognitive impairment, occasionally omitting words, repeating two-word phrases, and displaying serious difficulties in articulation, but has better verbal comprehension abilities than her brother. A genetic study using cycle sequencing after PCR amplification of exon 10 of the GRIN2A gene revealed that Patient 2 carries the same Ala643Asp variant as her brother. Since this variant is not present in the genomes of either parent but is present in two children born at different times, we conclude that it is the result of germline mosaicism of one of the parents (Fig. 1A).
Figure 1. Identification of a GRIN2A mutation in two siblings born to healthy, non-consanguineous parents.
(A) Family pedigree and whole-exome sequencing results of generations 1 and 2 confirming the presence in the patients, but not their parents, of the c.1928C>A mutation, which results in the amino acid change p.Ala643Asp. (B) Alignment of the GluN2A protein sequence across multiple species and of the GluN2A sequence with those of other NMDA subunits. The M3 transmembrane domain is highly conserved across vertebrate species and within the NMDA receptor family. The alanine residue (*) that is altered in the two patients is also highly conserved. ATD, amino terminal domain; S1 and S1, polypeptide chains that form the agonist-binding domain; M1, M2, M3, and M4, transmembrane domain helices 1, 3, and 4, and the membrane re-entrant loop 2; CTD, carboxy-terminal domain. (C) Schematic showing the fully formed GluN1/GluN2A receptor (left) and GluN2A monomer (right). GluN2A-Ala643 is located near the center of the channel pore (*).
The alanine residue at position 643 in GluN2A lies in the M3 transmembrane domain helix, immediately before the conserved SYTANLAAF motif, which is critically important for normal channel function. This alanine is highly conserved across vertebrate species and other GluN subunits (Fig. 1B, C).
Functional studies
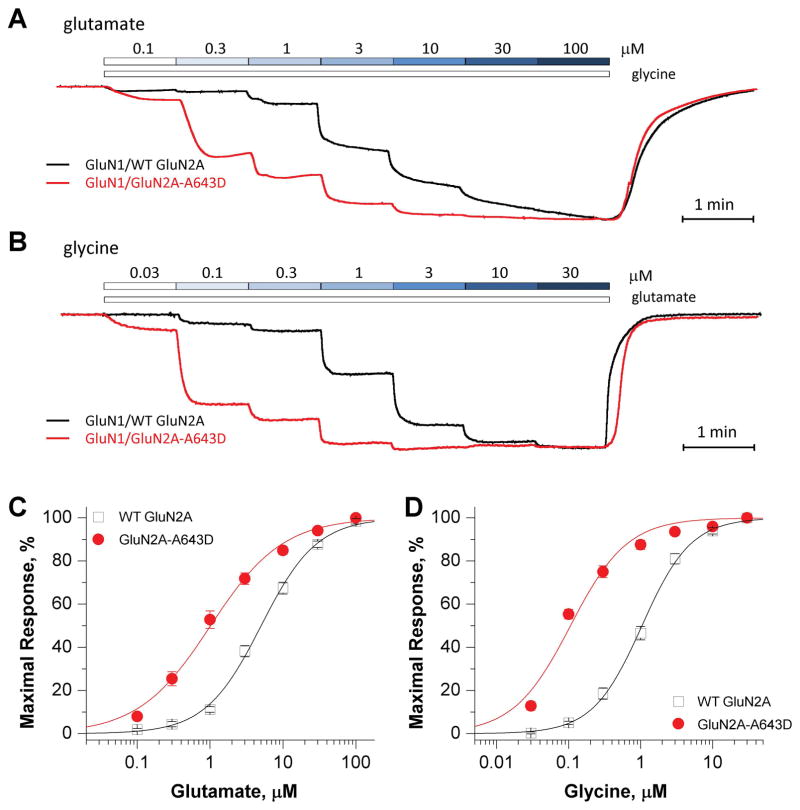
To examine the functional effects of the A643D variant, site-directed mutagenesis was used to introduce p.Ala643Asp into human WT GluN2A cDNA. Expression of GluN1/GluN2A-A643D in Xenopus laevis oocytes allowed functional evaluation of mutant and WT NMDARs using TEVC current recordings, as previously described [28]. We first evaluated the effects of the mutation on the potency of the co-agonists L-glutamate and glycine. Analysis of concentration-response relationships (Fig. 2A, B) showed that GluN2A-A643D increased glutamate potency by 5-fold (EC50, 1.0 μM vs. 5.0 μM for WT receptors; p < 0.0001, unpaired t-test) and increased glycine potency by 10-fold (EC50, 0.11 μM vs. 1.1 μM for WT receptors; p < 0.0001, unpaired t-test) (Fig. 2C, D; Table 1). These data suggest that NMDARs carrying the GluN1/GluN2A-A643D mutation can be activated by a lower concentration of agonist than WT receptors.
Figure 2. GluN2A-A643D increases agonist potency.
(A, B) Superimposed representative concentration-response TEVC traces for increasing concentrations of glutamate (A; in the presence of 30 μM glycine) and glycine (B; in the presence of 100 μM glutamate) for WT (wild-type) GluN2A- (black), and GluN2A-A643D- (red) expressing Xenopus laevis oocytes. (C, D) Fitted composite glutamate and glycine concentration-response curves for WT GluN2A- and GluN2A-A643D-expressing Xenopus laevis oocytes. The percentage of maximal current response is plotted as a function of agonist concentration in μM. TEVC recordings were conducted at a holding potential of −40 mV.
Table 1.
Summary of pharmacological data
| Measurement | WT GluN2A | GluN2A-A643D | p value | ||||
|---|---|---|---|---|---|---|---|
| Mean | 95% CI | n | Mean | 95% CI | n | ||
| Glutamate EC50, μM | 5.0 | (4.1, 6.1) | 18 | 1.0 | (0.7, 1.4) | 23 | <0.0001 |
| Glycine, EC50, μM | 1.1 | (0.9, 1.5) | 10 | 0.11 | (0.09, 0.13) | 13 | <0.0001 |
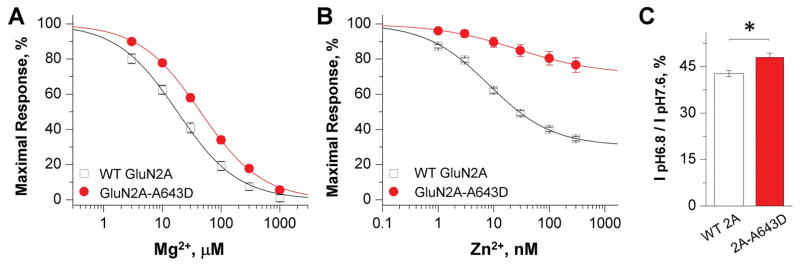
| Mg2+, IC50, μM | 18.6 | (12.9, 27.0) | 13 | 39.4 | (27.5, 56.2) | 12 | 0.004 |
| Proton, % A | 42.8 | (40.5, 45.0) | 8 | 48.0 | (45.1, 50.9) | 13 | 0.010 |
| Zn2+, IC50, nM | 9.1 | (6.6, 12.4) | 12 | 30.3 | (12.7, 72.3) | 11 | 0.007 |
| Zn2+, Residual Current (%)B | 30.9 | (24.3, 37.4) | 12 | 67.3 | (60.7, 73.9) | 11 | <0.0001 |
% current remaining measured at pH 6.8 compared to pH 7.6 at maximal L-glutamate and glycine activation
current remaining in maximum Zn2+ concentration (expressed as % of maximal receptor activation in the absence of Zn2+)
p value determined by unpaired t-tests (GraphPad Prism 5.0) comparing WT vs. mutant receptors. For EC50 and IC50 measurements log values were used for statistical comparison.
n = the number of oocytes evaluated for the measured endpoint.
CI = confidence interval
It is well known that NMDAR function can be negatively regulated by a number of endogenous negative modulators, including Mg2+, protons, and Zn2+ [8]. Using TEVC, we evaluated the effects of the mutation of on Mg2+ inhibition of NMDAR function in oocytes. Concentration-response data revealed a 2-fold reduction in the potency of Mg2+ in mutant NMDARs (IC50, 39 μM vs. 19 μM for WT receptors; p = 0.004, unpaired t-test) (Fig. 3A; Table 1). We subsequently evaluated the effects of GluN2A-A643D on sensitivity to extracellular protons and zinc. GluN2A-A643D reduced sensitivity to zinc by 3-fold (IC50, 30 nM vs. 9.1 nM for WT receptors; p = 0.007, unpaired t-test), and more than doubled the residual current measured in the presence of a saturating concentration (300 nM) of zinc (67% vs. 31% residual current for WT receptors; p < 0.0001, unpaired t-test) (Fig. 3B; Table 1). We further analyzed the effects of GluN2A-A643D on proton sensitivity by measuring current amplitude at pH 6.8 versus pH 7.6. The decrease in current amplitude at pH 6.8 versus pH 7.6 was slightly reduced in mutant receptors (48%) than WT receptors (43%; Figure 3C; Table 1), suggesting tonic proton inhibition may be reduced in the mutant receptors.
Figure 3. GluN2A-A643D decreases NMDAR sensitivity to endogenous negative modulators.
(A, B) Fitted concentration-response curves for endogenous antagonists. The percentage of maximal response is plotted against antagonist concentration in μM. TEVC recordings were conducted at a holding potential of −60 mV for magnesium and −20 mV for zinc. (C) Percentage current response at pH 6.8 vs. 7.6 of WT GluN2A- and GluN2A-A643D-expressing Xenopus laevis oocytes. GluN2A-A643D-containing NMDARs show decreased current attenuation in the presence of increased proton concentrations (i.e. low pH).
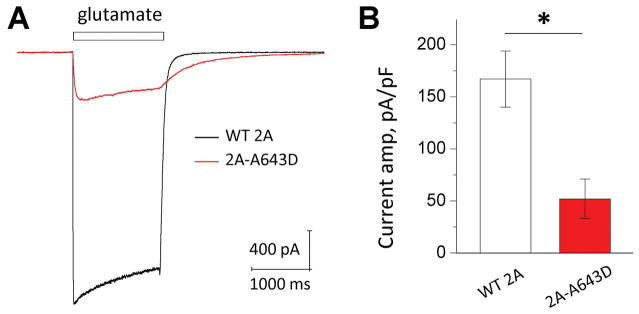
We then measured and compared the current response of the mutant GluN2A-A643D to the WT receptors from transiently transfected HEK293 cells. Surprisingly, the mutant showed a significantly decreased peak current amplitude to a maximal concentration of agonists when co-expressed with the GluN1 subunit (GluN1/GluN2A-A643D: 52 ± 19 pA/pF, n = 15 vs. WT GluN1/GluN2A: 167 ± 29 pA/pF, n = 8; p = 0.004, Mann Whitney test; Fig. 4A, B). Taken together, Our electrophysiological data suggest a mixed effects of this potation on NMDAR function, including an increased receptor activation in response to low concentrations of agonist and the attenuated inhibitory effect of endogenous inhibitors (magnesium, protons and zinc), but a decreased agonist-evoked current amplitude.
Figure 4. GluN2A-A643D changes current response.
(A) Current response was recorded on transiently transfected HEK293 cells by using the whole cell voltage patch clamp recordings. (B) The peak current amplitude to a maximal concentration of agonists normalized to cell capacitance was significant decreased in the GluN2A-A643D mutant receptors (*p = 0.004, Mann Whitney test).
DISCUSSION
Although GRIN2A mutations have been associated with a wide spectrum of neurodevelopmental disorders, including speech disorder without epilepsy in families with EAS [39], schizophrenia [40], autism spectrum disorders [40,41], and intellectual disability [42], mutations in this gene have not to date been implicated in movement disorders, in contrast to mutations in GRIN1 and GRIN2B [32–33]. Encephalopathies caused by GRIN1 and GRIN2B mutations share common features: they are characterized by developmental delay, intellectual disability, autism spectrum disorder, hypotonia, spasticity, epilepsy, hyperkinetic movement disorders (dystonia, dyskinetic movements, and/or chorea), sleep disturbances, cortical visual impairment, and cerebral atrophy. The clinical presentations of Patients 1 and 2 share some features with the phenotypes associated with GRIN1 and GRIN2B mutations, but with a less severe clinical course and without epilepsy, which is a component of all GRIN-associated phenotypes to date. However, both siblings present with movement disorder (predominantly with dystonia, but also with motor coordination disorder, dysmetria and tremor), sleep disturbances, abnormal breathing noise, and impaired ocular motility.
An association between a GRIN2A mutation and anti-NMDAR encephalitis has been proposed [43]. Both clinical entities present with epilepsy, and are similar in terms of seizure semiology and origination, as well as language impairment. Two other features of anti-NMDAR encephalitis, movement disorder and sleep disturbances, are also present in both our patients [43].
The p.Ala643Asp GluN2A variant is a non-conservative amino-acid substitution that likely affects secondary protein structure, as these residues differ in polarity, charge, size, and other properties. Moreover, in silico analysis predicts that this variant is probably deleterious to protein structure/function. This substitution is located in the M3 transmembrane domain of the ionotropic glutamate receptor family, a region devoid of variation in the healthy population [44]. This position is highly conserved across species, and reports have linked missense mutations in nearby residues (L649V, F652V) with GRIN2A-related disorders. While the de novo mutation L649V has been identified in a patient with severe intellectual disability, dysplastic corpus callosum, delayed myelination, epilepsy, severe feeding problems, hypothyroidism, and mild facial dysmorphism, functional analyses have not been performed in the patient in question [42]. The F652V variant has been detected in a patient with CSWSS [17]. Co-expression of GluN1 with WT GluN2A or GluN2A-F652V generates functional NMDA receptors, as demonstrated using single-channel recordings in cell-attached patches with altered single channel properties [17], further supporting the functional importance of the protein region in which the A643 residue lies.
Further studies will be necessary to determine why mild encephalopathy with dystonia, with no epileptic abnormalities, is the primary clinical feature of our patients. Our functional analysis of the GluN2A-A643D variant indicates a mixed effect on NMDAR function. More in-depth studies (i.e. in vivo study on transgenic animals) are required to elucidate the molecular mechanism underlying these patients’ phenotypes.
CONCLUSIONS
Our findings expand the phenotypic spectrum of GRIN2A-related disorders to include neurodevelopmental and movement disorder (dystonia) without seizures. The data reported here reinforce the association between GRIN2A mutations and speech development and/or cognitive impairment, and emphasize the importance of molecular analyses not only for genetic counseling purposes but also for the identification of potential treatments.
Supplementary Material
Normal waking electroencephalogram (EEG) for Patients 1 and 2.
Normal magnetic resonance imaging (MRI) findings for Patients 1 and 2. Images: A1 and B1 sagittal T1-weighted, A2 axial fluid-attenuated inversion recovery (FLAIR), B2 axial T2-weighted, A3 and B3 coronal T2-weighted.
Acknowledgments
Funding Resource: This work was supported by NIH - the Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD) [grant R01HD082373 to H.Y.], NIH- National Institute of Neurologic Disorders and Stroke (NINDS) [grants R01NS036654, R01NS065371, and R24NS092989 to S.F.T.], and the Instituto de Salud Carlos III Spain (FIS-PI13/02177 and JR13/0019). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
The authors thank Ellen Hess for her constructive criticism and Gil Shaulsky for outstanding technical assistance.
Footnotes
Competing Financial Interests: S.F.T. is a consultant for Janssen Pharmaceuticals, Inc., Boehringer Ingelheim Pharma GmbH & Co. KG, a member of the Scientific Advisory Board of Sage Therapeutics, and a co-founder of NeurOp Inc.
AUTHOR CONTRIBUTIONS
AFM performed the genetic analyses and wrote the paper.
SR and LGS were responsible for patient care and wrote the paper.
IR performed the bioinformatic analyses.
MLC reviewed the patients’ clinical histories and wrote the paper.
HK, JZ, and SJM conducted the functional studies and wrote the paper.
HY and SFT designed the experiments, analyzed the data, and wrote the paper.
References
- 1.Yuan H, Low C-M, Moody OA, Jenkins A, Traynelis SF. Ionotropic GABA and Glutamate Receptor Mutations and Human Neurologic Diseases. Mol Pharmacol. 2015;88:203–217. doi: 10.1124/mol.115.097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 3.Dunah AW, Luo J, Wang YH, Yasuda RP, Wolfe BB. Subunit composition of N-methyl-D-aspartate receptors in the central nervous system that contain the NR2D subunit. Mol Pharmacol. 1998;53:429–437. doi: 10.1124/mol.53.3.429. [DOI] [PubMed] [Google Scholar]
- 4.Dunah AW, Standaert DG. Subcellular segregation of distinct heteromeric NMDA glutamate receptors in the striatum. J Neurochem 2003. 2003;85:935–943. doi: 10.1046/j.1471-4159.2003.01744.x. [DOI] [PubMed] [Google Scholar]
- 5.Lopez de Armentia M, Sah P. Development and subunit composition of synaptic NMDA receptors in the amygdala: NR2B synapses in the adult central amygdala. J Neurosci. 2003;23:6876–6883. doi: 10.1523/JNEUROSCI.23-17-06876.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salter MG, Fern R. NMDA receptors are expressed in developing oligodendrocyte processes and mediate injury. Nature. 2005;438:1167–1171. doi: 10.1038/nature04301. [DOI] [PubMed] [Google Scholar]
- 7.Akazawa C, Shigemoto R, Bessho Y, Nakanishi S, Mizuno N. Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in the cerebellum of developing and adult rats. J Comp Neurol. 1994;347:150–160. doi: 10.1002/cne.903470112. [DOI] [PubMed] [Google Scholar]
- 8.Traynelis SF, Wollmuth LP, McBain CJ, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi DW. Glutamate receptors and the induction of excitotoxic neuronal death. Prog Brain Res. 1994;100:47–51. doi: 10.1016/s0079-6123(08)60767-0. [DOI] [PubMed] [Google Scholar]
- 10.Rothman SM, Olney JW. Excitotoxicity and the NMDA receptor—still lethal after eight years. Trends Neurosci. 1995;18:57–58. doi: 10.1016/0166-2236(95)93869-y. [DOI] [PubMed] [Google Scholar]
- 11.Obrenovitch TP, Hardy AM, Zilkha E. Effects of L-701,324, a high-affinity antagonist at the N-methyl-D-aspartate (NMDA) receptor glycine site, on the rat electroencephalogram. Naunyn Schmiedebergs Arch Pharmacol. 1997;355:779–786. doi: 10.1007/pl00005013. [DOI] [PubMed] [Google Scholar]
- 12.Dirnagl U, Iadecola C, Moskowitz MA. Pathobiology of ischaemic stroke: an integrated view. Trends Neurosci. 1999;22:391–397. doi: 10.1016/s0166-2236(99)01401-0. [DOI] [PubMed] [Google Scholar]
- 13.Yurkewicz L, Weaver J, Bullock MR, Marshall LF. The effect of the selective NMDA receptor antagonist traxoprodil in the treatment of traumatic brain injury. J Neurotrauma. 2005;22:1428–1443. doi: 10.1089/neu.2005.22.1428. [DOI] [PubMed] [Google Scholar]
- 14.Coyle JT. NMDA receptor and schizophrenia: a brief history. Schizophr Bull. 2012;38:920–926. doi: 10.1093/schbul/sbs076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menniti FS, Lindsley CW, Conn PJ, Pandit J, Zagouras P, Volkmann RA. Allosteric modulators for the treatment of schizophrenia: targeting glutamatergic networks. Curr Top Med Chem. 2013;13:26–54. doi: 10.2174/1568026611313010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reutlinger C, Helbig I, Gawelczyk B, et al. Deletions in 16p13 including GRIN2A in patients with intellectual disability, various dysmorphic features, and seizure disorders of the rolandic region. Epilepsia. 2010;51:1870–1873. doi: 10.1111/j.1528-1167.2010.02555.x. [DOI] [PubMed] [Google Scholar]
- 17.Lesca G, Rudolf G, Bruneau N, et al. GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nat Genet. 2013;45:1061–1066. doi: 10.1038/ng.2726. [DOI] [PubMed] [Google Scholar]
- 18.Lemke JR, Lal D, Reinthaler EM, et al. Mutations in GRIN2A cause idiopathic focal epilepsy with rolandic spikes. Nat Genet. 2013;45:1067–1072. doi: 10.1038/ng.2728. [DOI] [PubMed] [Google Scholar]
- 19.Carvill GL, Regan BM, Yendle SC, et al. GRIN2A mutations cause epilepsy-aphasia spectrum disorders. Nat Genet. 2013;45:1073–1076. doi: 10.1038/ng.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endele S, Rosenberger G, Geider K, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010;42:1021–1026. doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- 21.Pierson TM, Yuan H, Marsh ED, et al. GRIN2A mutation and early-onset epileptic encephalopathy: personalized therapy with memantine. Ann Clin Transl Neurol. 2014;1:190–198. doi: 10.1002/acn3.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venkateswaran S, Myers KA, Smith AC, Beaulieu CL, Schwartzentruber JA. Whole-exome sequencing in an individual with severe global developmental delay and intractable epilepsy identifies a novel, de novo GRIN2A mutation. Epilepsia. 2014;55:e75–79. doi: 10.1111/epi.12663. [DOI] [PubMed] [Google Scholar]
- 23.Singh D, Lau M, Ayers T, et al. De novo heterogeneous mutations in SCN2A and GRIN2A genes and seizures with ictal vocalizations. Clin Pediatr (Phila) 2016;55:867–870. doi: 10.1177/0009922815601060. [DOI] [PubMed] [Google Scholar]
- 24.Allen NM, Conroy J, Shahwan A, et al. Unexplained early onset epileptic encephalopathy: Exome screening and phenotype expansion. Epilepsia. 2016;57:e12–e17. doi: 10.1111/epi.13250. [DOI] [PubMed] [Google Scholar]
- 25.Yuan H, Hansen KB, Zhang J, et al. Functional analysis of a de novo GRIN2A missense mutation associated with early-onset epileptic encephalopathy. Nat Commun. 2014;5:3251. doi: 10.1038/ncomms4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swanger SA, Chen W, Wells G, et al. Mechanistic Insight into NMDA Receptor Dysregulation by Rare Variants in the GluN2A and GluN2B Agonist Binding Domains. Am J Hum Genet. 2016;99:1261–1280. doi: 10.1016/j.ajhg.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao K, Tankovic A, Zhang Y, et al. A de novo loss-of-function GRIN2A mutation associated with childhood focal epilepsy and acquired epileptic aphasia. PLoS One. 2017;12:e0170818. doi: 10.1371/journal.pone.0170818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen W, Tankovic A, Burger PB, Kusumoto H, Traynelis SF, Yuan H. Functional Evaluation of a De Novo GRIN2A Mutation Identified in a Patient with Profound Global Developmental Delay and Refractory Epilepsy. Mol Pharmacol. 2017;91:317–330. doi: 10.1124/mol.116.106781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lal D, Steinbrücker S, Schubert J, et al. Investigation of GRIN2A in common epilepsy phenotypes. Epilepsy Res. 2015;115:95–99. doi: 10.1016/j.eplepsyres.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Burnashev N, Szepetowski P. NMDA receptor subunit mutations in neurodevelopmental disorders. Curr Opin Pharmacol. 2015;20:73–82. doi: 10.1016/j.coph.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Soto D, Altafaj X, Sindreu C, Bayés À. Glutamate receptor mutations in psychiatric and neurodevelopmental disorders. Commun Integr Biol. 2014;7:e27887. doi: 10.4161/cib.27887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Platzer K, Yuan H, Schütz H, Winschel A, Chen W, Hu C, et al. GRIN2B encephalopathy: novel findings on phenotype, variant clustering, functional consequences and treatment aspects. J Med Genet. 2017;54(7):460–470. doi: 10.1136/jmedgenet-2016-104509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemke JR, Geider K, Helbig KL, Heyne HO, Schütz H, Hentschel J, et al. Delineating the GRIN1 phenotypic spectrum: a distinct genetic NMDA receptor encephalopathy. Neurology. 2016;86(23):2171–2178. doi: 10.1212/WNL.0000000000002740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li D, Yuan H, Ortiz-Gonzalez XR, Marsh ED, Tian L, McCormick EM, et al. GRIN2D Recurrent De Novo Dominant Mutation Causes a Severe Epileptic Encephalopathy Treatable with NMDA Receptor Channel Blockers. Am J Hum Genet. 2016;99(4):802–816. doi: 10.1016/j.ajhg.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010 Mar 1;26(5):589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Traynelis SF, Burgess MF, Zheng F, Lyuboslavsky P, Powers JL. Control of voltage-independent zinc inhibition of NMDA receptors by the NR1 subunit. J Neurosci. 1998;18:6163–6175. doi: 10.1523/JNEUROSCI.18-16-06163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.González-Pérez A, López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88:440–9. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner SJ, Mayes AK, Verhoeven A, Mandelstam SA, Morgan AT, Scheffer IE. GRIN2A: an aptly named gene for speech dysfunction. Neurology. 2015;84:586–593. doi: 10.1212/WNL.0000000000001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarabeux J, Kebir O, Gauthier J, et al. Rare mutations in N-methyl-D-aspartate glutamate receptors in autism spectrum disorders and schizophrenia. Transl Psychiatry. 2011;1:e55. doi: 10.1038/tp.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Roak BJ, Deriziotis P, Lee C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Ligt J, Willemsen MH, van Bon BW, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 43.Dalmau J, Lancaster E, Martinez-Hernandez E, Rosenfeld MR, Balice-Gordon R. Clinical experience and laboratory investigations in patients with anti-NMDAR encephalitis. Lancet Neurol. 2011;10:63–74. doi: 10.1016/S1474-4422(10)70253-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogden KK, Chen C, Swanger SA, McDaniel MJ, Fan LZ, Hu C, Tankovic A, Kusumoto H, Kosobucki GJ, Schulien AJ, Su Z, Pecha J, Bhattacharya S, Cohen AE, Aizenman E, Traynelis SF, Yuan H. Molecular mechanism of disease-associated mutations in the pre-M1 helix of NMDA receptors and potential rescue pharmacology. PLoS Genet. 2017;13(1):e1006536. doi: 10.1371/journal.pgen.1006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Normal waking electroencephalogram (EEG) for Patients 1 and 2.
Normal magnetic resonance imaging (MRI) findings for Patients 1 and 2. Images: A1 and B1 sagittal T1-weighted, A2 axial fluid-attenuated inversion recovery (FLAIR), B2 axial T2-weighted, A3 and B3 coronal T2-weighted.