Abstract
OBJECTIVE
Somatic variants are a recognized cause of epilepsy-associated focal malformations of cortical development (MCD). We hypothesized that somatic variants may underlie a wider range of focal epilepsy, including non-lesional focal epilepsy (NLFE). Through genetic analysis of brain tissue, we evaluated the role of somatic variation in focal epilepsy with and without MCD.
METHODS
We identified somatic variants through high-depth exome and ultra-high-depth candidate gene sequencing of DNA from epilepsy surgery specimens and leukocytes from 18 individuals with NLFE and 38 with focal MCD.
RESULTS
We observed somatic variants in five cases in SLC35A2, a gene associated with glycosylation defects and rare X-linked epileptic encephalopathies. Nonsynonymous variants in SLC35A2 were detected in resected brain, and absent from leukocytes, in 3/18 individuals (17%) with NLFE, one female and two males, with variant allele frequencies (VAFs) in brain-derived DNA of 2–14%. Pathologic evaluation revealed focal cortical dysplasia type Ia (FCD1a) in two of the three NLFE cases. In the MCD cohort, nonsynonymous variants in SCL35A2 were detected in the brains of two males with intractable epilepsy, developmental delay, and MRI suggesting FCD, with VAFs of 19–53%; evidence for FCD was not observed in either brain tissue specimen.
INTERPRETATION
We report somatic variants in SLC35A2 as an explanation for a substantial fraction of NLFE, a largely unexplained condition, as well as focal MCD, previously shown to result from somatic mutation but until now only in PI3K-AKT-mTOR pathway genes. Collectively, our findings suggest a larger role than previously recognized for glycosylation defects in the intractable epilepsies.
INTRODUCTION
Epilepsy affects approximately 1% of the world’s population and causes substantial morbidity and mortality.1 Approximately one-third of individuals with epilepsy do not respond to anti-epileptic medications, and many patients with medically intractable epilepsy ultimately undergo surgical resection of a seizure focus.2, 3 Intractable focal epilepsy poses a particular challenge when pre-operative brain magnetic resonance imaging (MRI) does not reveal a seizure-related structural abnormality, referred to conventionally as “non-lesional focal epilepsy” (NLFE). While germline genetic variants in a small number of genes—including KCNT1, LGI1, RELN, NPRL2, NPRL3, GRIN2A, and DEPDC5—have been associated with familial and sporadic focal epilepsy,4–12 the genetic basis for the majority of focal epilepsies remain unknown.
A number of human epilepsy syndromes associated with early onset seizures and intellectual disability have been associated with de novo variants in genes involved in early brain development and epileptogenesis.10,13–17 Recent studies have also identified brain-specific somatic variants in focal epilepsy due to malformations of cortical development (MCD), including hemimegalencephaly and the more common, radiographically evident focal cortical dysplasia (FCD) type II,18–21 characterized pathologically by the presence of dysmorphic neurons.22 These variants arise in the post-zygotic state, creating a mosaic of variant-carrying cells intermixed with variant-negative cells.23 Pathological evaluation of surgical epilepsy specimens often reveals FCD, even in cases in which a malformation was not evident on preoperative neuroimaging.23 In part on the basis of these observations, it has been suspected that patients with focal epilepsy without clear radiographic malformation to which seizures can be attributed may also be caused by somatic variants in focal regions of the brain, resulting in localized network disruption.24, 25
We hypothesized that somatic mutation, likely during corticogenesis, results in variants that would be detectable in brain tissue resected during epilepsy surgery, even in patients without explanatory lesions detected with modern neuroimaging. To test this hypothesis, we analyzed data from exome sequencing and targeted deep sequencing of a broad list of epilepsy- and malformation-related genes of paired DNA from resected brain and leukocytes in 18 individuals with NLFE. We identified low-level somatic variants in SLC35A2 only in brain from 3/18 individuals with NLFE (17%), two of whom had pathological findings consistent with FCD type Ia, defined by disordered cortical lamination without dysmorphic neurons.22 Through additional investigation of a MCD-related epilepsy cohort (n=38), we identified two additional cases harboring somatic SLC35A2 variants in brain, each with focal epilepsy and radiographic lesions suggestive of FCD. We thus provide compelling evidence for an association of somatic variants in SLC35A2 with intractable focal epilepsy, both with and without radiographic evidence of a seizure-associated lesion on neuroimaging.
METHODS
Participants
For all cases included in research and sequenced, clinical determination of localization of the epileptogenic region and candidacy for focal resection was determined by standard clinical practice at each site, incorporating seizure semiology, EEG data, structural MRI, functional imaging (PET, SPECT), and consensus at epilepsy surgery conferences.
At each center, review of clinical data was performed by that center’s research team, with input from involved clinicians, to identify eligible participants prior to enrollment. To be eligible for the study enrolling patients with NLFE, it was required that the participant had no lesion identified on MRI that was felt to be responsible for seizures and that the patient had a planned or completed surgical resection. Focal features on other modalities, such as nuclear imaging studies, were not considered criteria for exclusion. Eighteen individuals with NLFE undergoing focal resection as treatment for intractable epilepsy were enrolled from Columbia University Medical Center (CUMC), New York University Langone Medical Center (NYULMC), and University of California, San Francisco (UCSF) (Table S2). To be eligible for the study enrolling patients with MCD, it was required that the patient have MRI findings consistent with FCD, polymicrogyria, or hemimegalencephaly and have a planned or completed surgical resection. Thirty-eight individuals with intractable epilepsy and MCD were enrolled from Boston Children’s Hospital, Duke University Medical Center, and Lucille Packard Children’s Hospital at Stanford (Table S2). All patients were consented for research approved by the respective institutional review boards.
For cases described in detail here with somatic variants identified in SLC35A2, epilepsy and general medical history, neuroimaging, and neuropathological data were reviewed in detail by a team of board-certified physicians, including epileptologists (MRW, PBC, AHP), a neuroradiologist (EY), and neuropathologists (DZ, HGWL, PC) in addition to the referring neurologists and neurosurgeons for each case. Seizure semiology and post-operative Engel outcome scores were characterized according to International League Against Epilepsy (ILAE) terminology.26 Neuropathological analysis assessed specimens for features of FCD based on established ILAE criteria.22
Next-generation sequencing
We extracted DNA from one or more regions of resected brain specimens and from leukocytes from the same individuals using a DNeasy Blood & Tissue Kit (Qiagen). High-depth exome sequencing was performed for all 18 cases with NLFE as well as the 38 cases with MCDs using a Nimblegen SeqCap EZ V3.0 Exome Enrichment Kit using established methods.14 To increase sensitivity to detect variants present at low allele frequency, we performed ultra-high-depth targeted sequencing of 2.5 megabases (Mb) of genomic sequence encompassing 838 genes (Table S1), including 274 genes related to the PI3K-AKT-mTOR pathway, 308 genes implicated in neurogenesis, a subset of epilepsy-associated genes that included SLC35A2, and the remaining genes included for other genetic studies unrelated to this work (Roche SeqCap EZ). Targeted sequencing was performed on 17/18 NLFE samples and 30/38 MCD samples (Table S2). Alignment and variant calling were performed using established methods.14
Somatic variant calling
Somatic single-nucleotide variants (sSNVs) and somatic insertion-deletion variants (sindels) were called from aligned BAM files using Mutect2 in all cases and controls.27 In addition to sSNVs and sindels passing recommended filters, we included short tandem repeat (STR) variants that failed the Mutect2 contraction filter. Variants were annotated with SnpEff 4.2. After annotation, qualifying variants, defined as variants predicted to be functional [i.e., missense (rated possibly or probably damaging by Polyphen-2), splice site, stop-gained, codon deletions/insertions, and frameshift variants)] and rare [(i.e., absent from 13,472 in-house sequenced controls, the ExAC database, and the gnomAD database)], were prioritized for analysis. Qualifying variants were required to have a variant allele frequency (VAF) of 1% or higher in brain, with at least one read with an R1 orientation and at one read with an R2 orientation. In six individuals, we performed sequencing twice on the same DNA sample. Variants were eliminated from the list of qualifying variants if they were not present in the second sequencing run despite sufficient coverage to detect the variant.
Test for enrichment of somatic variants in individual genes
To ensure we had the sensitivity to detect somatic variants present at low allele frequency, we limited these analyses the protein coding regions (defined by CCDS version 14) and the two-base pair highly conserved splice sites flanking exons included on the ultra-high-depth targeted sequencing gene panel. Among the qualifying calls, we observed a high rate of G-T substitutions (84% of single nucleotide substitutions), which are a well-recognized Illumina sequencing error and can also arise from oxidative DNA damage28, 29 This overrepresentation was less pronounced in variants with higher VAF (>2%) for variants called in the targeted sequencing data (VAF) (69%) compared to those with a VAFs between 1–2% (88%). To address this artifact, we restricted enrichment analyses to somatic calls with VAFs ≥2%. We then compiled all qualifying variants calls (defined above) in these regions with a VAF ≥2%. Assuming no sequencing error and that each allele is equally likely to be sequenced, when the variant site was sequenced to at least 150-fold and the true frequency at least 2%, we have a 95% chance of calling at least one variant allele. We therefore calculated the ‘callable real estate’ across the 2.5 MB of genomic sequence for each individual sequenced to 150-fold in both brain and leukocyte DNA. We then calculated the individual’s mutability across each gene on the custom capture sufficiently sequenced in that individual by summing the trinucleotide mutability for each possible single base substitution across the ‘callable real estate’.
Using these data, we tested for enrichment of somatic calls in individual genes using a previously published method30. In brief, we compared the observed number of somatic variants within each gene to a null rate in which somatic variants are randomly placed within the individuals ‘callable-real estate’ in accordance with the sequence-context-dependent mutation rate. We randomly distributed the somatic calls in this way 1,000,000 times to establish the null. This observed vs. expected contrast was calculated for each individual, thereby explicitly accounting for inter-individual differences in number of variants that may arise from other sources (e.g., environmental exposure, DNA quality, etc.). To calculate the gene-level p-value, we computed the proportion of simulated statistics that are as extreme or more extreme than that computed from the observed somatic variants.
Variant confirmation
Somatic variants in SLC35A2 were confirmed to be present in DNA derived from brain tissue and absent in DNA from leukocytes using digital droplet polymerase chain reaction PCR (ddPCR, BioRad QX-200 Droplet Digital PCR system) or conventional PCR (Sanger sequencing), depending on the level and type of variant.
Control cohorts
Variant frequencies were evaluated in a control dataset of 13,472 individuals sequenced from genetic studies at the Columbia Institute for Genomic Medicine. Approximately 6900 were neuropsychiatrically normal, and the remaining individuals had conditions without known co-morbidities with epilepsy or MCD. We also used control data available in ExAC and gnomad31 to ascertain population allele frequencies in the germline.
To evaluate the frequency of somatic variants in SLC35A2 in brain, we evaluated paired next-generation sequencing data (from the high-depth targeted sequencing panel) from DNA from 20 autopsy-collected cortical brain specimens and matched leukocyte-derived DNA collected pre-mortem from individuals with Alzheimer’s disease (AD) or dementia from the New York Brain Bank at Columbia University.
Phosphorylated ribosomal S6 immunohistochemistry
Representative fixed sections of each from the five cases and age-matched post-mortem neocortical fixed sections were probed with antibodies recognizing phosphorylated ribosomal S6 protein (P-S6; Ser 240/244, mouse monoclonal, Cell Signaling; 1:250) to assess the extent of mTOR signaling and NeuN (1:100, mouse monoclonal; Novus).
RESULTS
Identification of somatic variants in SLC35A2 present in brain
We first collated all qualifying somatic variants (Methods) from exome sequencing and targeted deep sequencing of 838 genes (Table S1) of paired DNA from resected brain and leukocytes in the 18 individuals with NLFE and 38 with MCD (Table S2). Exome and targeted sequenced DNA pairs were sequenced across protein-coding regions to an average depth of 137-fold +/− 41 and 492-fold +/− 121, respectively, with an average of 12 and 4 qualifying somatic variants called from exome and targeted sequencing, respectively (Table S3).
We observed variants in SLC35A2, which is located on the X chromosome, present in brain and absent in leukocytes, in 3/18 cases with NLFE (Table 1). These variants are present only in a subset of cells, consistent with having been acquired in the post-zygotic state. The brain specimen from Case 1 (female) harbored the variant Ser304Pro predicted to be protein-altering (Polyphen2-probably damaging, SIFT-deleterious, CADD-26.7) with variant allele frequency of 3.3% and 6.5%, from exome and targeted sequencing. This indicates that between 6.6% and 13% of brain cells in the tested specimens have the variant since the patient is female (XX) and for each cell with the variant we would expect there to be one variant allele and one reference allele (heterozygous genotype). We were able to obtain DNA from a buccal swab from Case 1, from which no variant alleles were detected using ddPCR, suggesting that the mutation occurred after neuroectoderm formation. Case 2 (male) harbored an in-frame indel (codon duplication) Leu113dup in brain tissue, and Case 3 (male) a frameshift indel Ser212Leufs*9. VAFs from exome and target sequencing ranged between 2–14%, respectively. Since these were both male (XY) patients, we estimate that between 2–14% of cells in the assayed brain tissue specimens carry the variant (hemizygous genotype) (Table 1). Despite non-lesional MRIs, Cases 2 and 3 had pathological evidence of FCD type I (Table 2).
Table 1.
Somatic variants identified in SLC35A2 using NGS (exome and targeted sequencing) and confirmed with ddPCR.
| Sample /specimen(s) | SLC35A2 somatic variant * | Brain DNA VAF – exome [95% CI] | Leukocyte DNA VAF – exome | Brain VAF – targeted sequencing ****[95% CI] | Leukocyte DNA VAF – targeted sequencing | Average +/− SD Brain VAF – ddPCR **** [no. replicates] |
|---|---|---|---|---|---|---|
| Case 1/s3 | c.910T>C; p.(Ser304Pro) | 3.3% (2/61) ** [0.4–11.4%] |
0% (0/63) | 6.5 % (40/612) [4.7–8.8%] |
0.21% (1/477)*** | 6.2% +/− 0.6 [10] |
| Case 2/s1 | c.339_340insCTC;p.(Leu113dup) | 8.9% (4/45) [2.5–21.2%] |
0% (0/34) | 2.4% (6/246) [0.9–5.2%] |
0% (0/393) | 5.3% +/− 0.6 [8] |
| Case 3/s2 | c.634_635del;p.(Ser212Leufs*9) | 14.3% (5/35) [4.8–30.2%] |
0% (0/42) | 6.8% (9/133) [3.1–12.5%] |
0% (0/167) | 8.8% +/− 0.2 [6] |
| Case 4/s1 | c.164G>T; p.(Arg55Leu) | 52.6% (20/38) [36–69%] |
0% (0/71) | 50.8% (97/191) [43.8–57.8%] |
0% (0/226) | 54.6% +/− 0.2 [3] |
| Case 5/s1 | c.747_757dup; p.(Ala253Glyfs*100) | 18.8% (8/45) [8–32.1%] |
0% (0/95) | 26.5% (58/219) [20.8–34.8%] |
0% (0/160) | NA |
annotated based on NM_005660.1
not called using the calling algorithm but reads supporting the variant were detected
The one read supporting the variant could represent a sequencing artifact or very low levels of variant. No variant was detected in leukocyte or saliva DNA using ddPCR with a sensitivity to detect variant allele down to 0.1%, suggesting this one read supporting the variant is a sequencing artifact.
VAF as estimated by targeted sequencing varies compared to that estimated from exome sequencing. This is explained by the fact that DNA aliquots for sequencing were extracted from two different pieces of the same specimen and therefore the levels will also reflect the regional variability across the specimen.
Table 2.
Phenotypes of patients with somatic SLC35A2 mutations.
| Subject ID/ Sex | Epilepsy Type/ Seizures types | Age at first seizure /Age at surgery | Post-operative Engel score (time since surgery) | Preoperative MRI | Pathology | Neuropsych assessment | Localization | |
|---|---|---|---|---|---|---|---|---|
| Case 1 (Ser304Pro) /F | FE/FoS with impaired awareness | 13 yrs /17 and 18 yrs | Engel 1A after second surgery (20 mos) | 3T 1) Possible R MTS; 2) no lesion corresponding to epileptogenic focus* |
First procedure: no architectural abnormalities in GM or WM; patchy microgliosis Second procedure: CMS; subacute infarcts (likely due to electrode placement) |
Average intelligence; frontal dysfunction likely secondary to AEDs | R frontal: mid cingulate gyrus, R anterior cingulate gyrus, possible R SMA | |
| Case 2 (Leu113dup)/M | FE/FoS with impaired awareness, followed by secondary generalization | 2 yrs /13 yrs | Engel IV (12 mos) | 1.5T 1) Possible L MTS; 2) Slight ventricular enlargement; 3) No lesion corresponding to epileptogenic focus |
FCD Ia; CMS; Mild patchy microgliosis | Difficulty with auditory naming and contextual verbal learning/memory; Mild bilateral frontal dysfunction. | Posterior L cingulate and precuneate region | |
| Case 3 (Ser212 Leufs*9) /M | Fe/ FoS with impaired awareness, followed by secondary generalization | 17 yrs /23 yrs | Engel 1A (19 mos) | 3T normal |
Hypercellular with FCD Ia; reactive changes | Nondominant posterior hemisphere dysfunction | right posterior temporal-occipital lobe | |
| Case 4 (Arg55Leu) /M | Infantile spasms | 4 mo /2 yrs | Engel IV (24 mos) | 3T (4 mos) normal 3T (23 mos) (1) R fronto-temporal lesion, FCD vs gliosis; (2) R>L fronto-temporal dysmyelination |
Gliosis | At 28 months: at or below 3-month level in all domains | R frontal and temporal | |
| Case 5 (Ala253 Glyfs*100) /M | Epileptic spasms | 14 mo /7 yrs | Engel 1A (24 mos) | 3T (3 yrs) R frontal FCD |
Gliosis | At 7 years: delayed in all domains but ambulatory; conversant with behavioral dysregulation | R frontal |
FE = focal epilepsy, FoS = Focal seizures, CMS = Chaslin’s ; marginal sclerosis, LRE = localization-related epilepsy, AED = antiepileptic drug, FCD = focal cortical dysplasia MTS = Mesial Temporal Sclerosis, SMA = supplemental motor area, R=Right L=Left, WM = White matter, GM = White matter
a small periventricular signal abnormal abnormality was identified in post-hoc analysis (Figure 2).
From 38 individuals with MCDs, we identified somatic variants in SLC35A2 in brain tissue from two male individuals, both with FCD suggested by MRI (Table 2). Case 4 harbored the putatively damaging missense variant Arg55Leu (Polyphen2-probably damaging, SIFT-deleterious, CADD-34), and Case 5 the frameshift indel Ala253Glyfs*100. The VAFs for these cases ranged from 19–53%, reflecting 19–53% of cells carrying the variant (Table 1).
Confirmation of somatic variants in cases and absence in controls
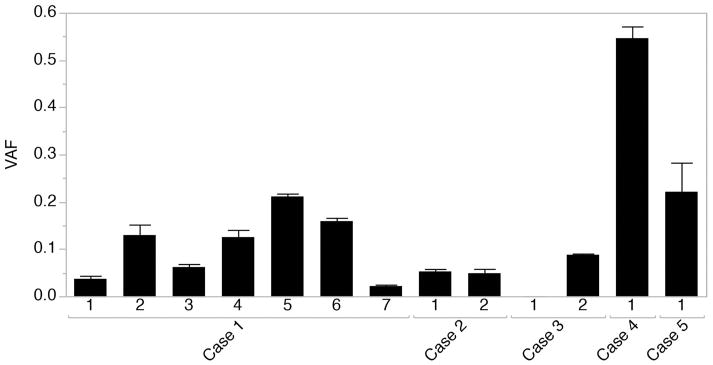
The four somatic variants observed in Cases 1–4 were confirmed with ddPCR, and the variant for Case 5 (Ala253Glyfs*100) with Sanger sequencing. The percentage of variant alleles was estimated using ddPCR highly correlated with that detected with next-generation sequencing results above (Table 1). Comparing the VAF across all available specimens per case, we observed highly variable levels within cases (Fig 1). All five variants are absent from local control cohorts and public variant databases (listed in Methods). In 20 control brains sequenced to the same depth (or more) on average as cases and subjected to identical calling algorithms, no somatic variants in SLC35A2 were found Table S3)
Figure 1.
Variant allele frequency across all available specimens as detected with digital droplet PCR (Cases 1,2,3, and 5) or next-generation sequencing (Case 4). Data are presented mean +/− standard deviation. Case 1 (two separate surgeries): two specimens (1 and 2) were collected from a right frontal resection within the supplementary motor area, and 5 specimens (3, 4, 5, 6, and 7) were collected from the right frontal premotor region anterior to the precentral sulcus. Case 2: two specimens were collected from a parietal precuneate and posterior two thirds cingulate resection. Case 3: Two specimens were collected from a right posterior temporal-occipital resection. Case 4: A temporal lobe specimen was collected as part of a functional hemispherectomy. Case 5: A frontal lobe specimen was collected as part of a right frontal resection.
We next assessed whether the somatic variants observed were selectively enriched in SLC35A2 across individuals with nonlesional and lesional epilepsy using a published method30. The uncorrected p-value associated with observing four qualifying variants in SLC35A2 in a cohort of 56 individuals, using the criteria described above in the methods, was <1 x 10−6. The fifth somatic variant in SLC35A2 did not reside in the ‘callable real estate’ since the site was only covered 133-fold (Table 1), not meeting the minimum coverage requirement of 150-fold in the targeted sequence data. Correcting for the 838 protein-coding genes sequenced on the targeted sequencing panel using a Bonferroni correction, SLC35A2 is significantly enriched for somatic variants. No other gene was significantly enriched for somatic variants.
Clinical, neuroimaging, and neuropathological features of the mosaic SLC35A2 cases
While all five patients harboring mosaic variants in SLC35A2 had intractable epilepsy leading to surgical resection, they represent a wide phenotypic range (Table 2). In keeping with the brain-localized presence of the SLC35A2 variants, no patients were reported to have any symptoms or signs classically associated with a glycosylation defect, such as inverted nipples or cerebellar atrophy32.
All three cases with NLFE presented with focal epilepsy in childhood or adolescence and brain MRI without overt structural abnormality identified as responsible for their epilepsy. Case 1 is a young woman with onset of focal intractable epilepsy at 13 years, normal intelligence, and a normal neurological examination. MRI was interpreted clinically as not having a structural abnormality that explained the focal epilepsy; a small periventricular signal abnormal abnormality (Fig 2), was identified during post-hoc review. Epilepsy surgery at age 17 resulted in seizure freedom for 7–8 months, but seizures recurred, necessitating a second resection in the same area, performed at age 18. She has remained seizure-free since her second surgery. Histopathological examination of resected brain tissue from the first surgery revealed patchy gliosis but no architectural abnormalities, while tissue from the second surgery showed Chaslin’s marginal sclerosis (and subacute infarcts in the setting of electrode placement) (Table 2, Fig 3). Case 2 is a boy who had mild language delay and onset of focal seizures with impaired awareness and secondarily generalized seizures at age 2 years. He had a right homonymous quadrantanopia on pre-operative ophthalmological examination, contralateral to the EEG seizure focus. MRI did not reveal a lesion to explain the focal epilepsy or visual field defect. Focal resection of an electrographically defined seizure focus, limited by the presence of adjacent eloquent cortex, did not result in sustained seizure control. Pathology of surgically resected brain revealed FCD type Ia, Chaslin’s marginal sclerosis, and mild patchy microgliosis. Case 3 is a young man who developed intractable epilepsy with focal seizures with impaired awareness beginning at age 17 years, normal intelligence, and a normal neurological examination. Pathology in Case 2 and Case 3 demonstrated findings consistent with FCD type Ia (Table 2, Fig 3).
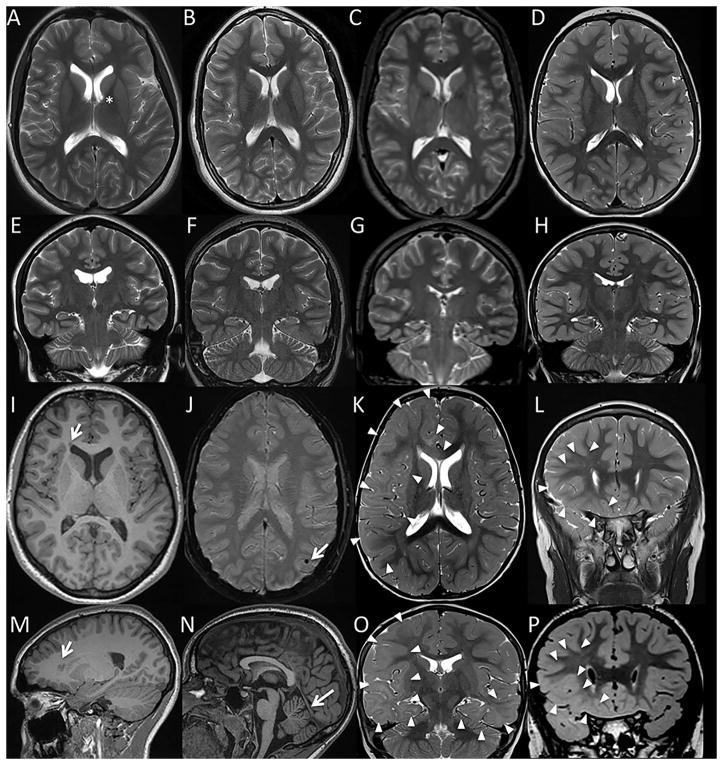
Figure 2.
Representative brain MR images from patients with somatic mutations in SLC35A2: Case 1 (A, E, I, M), Case 2 (B, F, J, N), Case 3 (C,G), Case 4 (K,O), and Case 5 (D, H, L, P). Axial T2 (A–D, K) and coronal T2 (E–H, L) images demonstrated grossly normal cerebral volume and preserved size of the hippocampi in all 5 subjects, allowing for some minimal prominence of the lateral ventricles and variant underrotation of the left hippocampus in Case 1 (E). For subjects Case 1, Case 2, and Case 3, there was no evidence of a malformation of cortical malformation apart from a tiny probable right frontal periventricular gray matter heterotopion in Case 1 (arrows in axial and sagittal T1 weighted images, I and M). For Cases 1, 2, and 3 there was also no evidence of a significant parenchymal injury or a metabolic/neurodegenerative condition, apart from nonspecific findings which were not thought to be related to seizure: an incidental left caudo-thalamic groove germinolytic cyst in Case 1 (asterisk in A); punctate mineralization versus hemosiderin deposition in the left temporo-parietal cortex of Case 2 (arrow in susceptibility weighted imaging, J); and some minimal cerebellar vermis volume loss in Case 3 (arrow sagittal T1 series, N). By comparison, subjects Case 4 and Case 5 demonstrated moderate to extensive parenchymal signal abnormality. In Case 4, T2 weighted imaging demonstrated diffuse haziness of the cerebral white matter concentrated in the right-greater-than-left frontotemporal lobes and right forceps minor (outlined by arrowheads in axial and coronal T2 weighted sequences, K and O). This pattern was consistent with cortical dysplasia and brain dysmyelination/gliosis; cortical dysplasia was thought unlikely to solely explain the findings given extent of signal abnormality, and tumor was thought unlikely given lack of mass effect. While T1 and T2 weighted imaging appeared grossly normal for subject Case 5, volumetric FLAIR imaging demonstrated loss of gray-white matter differentiation throughout the lower half of the right frontal lobe (arrowheads in coronal volumetric FLAIR, P; corresponding image slice on coronal T2, L). Although lacking a transmantle sign specific for cortical dysplasia, the regional blurring of gray-white matter differentiation in Case 5 was most suggestive of a focal cortical dysplasia; however, regional dysmyelination/gliosis could conceivably also have this appearance.
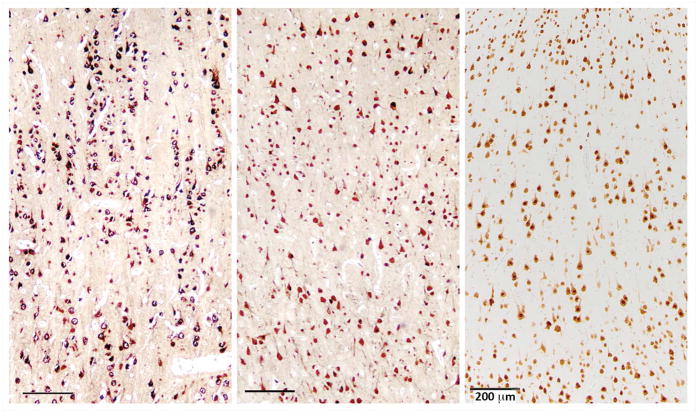
Figure 3.
Representative histopathological findings from the surgical specimens. Immunoperoxidase stains for the neuronal marker NeuN (brown) were performed to assess the cytoarchitecture of the resected cortex. Left panel (case 2) shows prominent radial arrangement of neurons with numerous microcolumns containing more than 8 tightly aligned neurons, characteristic of FCD type 1A. The middle panel (case 1) and right panel (case 5) show normal cortical architecture and reactive changes with patchy neuronal loss. Scale bars = 200 microns.
Both SLC35A2 variant-carrying cases from the MCD cohort had intractable epilepsy with localization consistent with a visible lesion suspected to be FCD based on MRI. Case 4 is a boy who presented with infantile spasms at 4 months and severe intellectual disability. MRI showed a right fronto-temporal lesion suspicious for FCD vs. gliosis and bilateral white matter abnormalities (Fig 2). He continues with intractable epilepsy after functional hemispherectomy. Case 5 is a boy who had onset of epileptic spasms at age 14 months and mild developmental delay, with mild intellectual disability diagnosed on assessment at age 7 years. His MRI revealed a lesion consistent with FCD, and there was good concordance between EEG and MRI pre-operatively; he remains seizure-free 2 years after focal resection. Both cases, despite pre-operative MRI suggestion of MCD, showed only gliosis on neuropathological review; for Case 4, available tissue was limited as the therapeutic procedure was primarily a disconnection rather than a resection. Both had a mild excess of neurons in the white matter and a few scattered SMI31-positive neurons but did not have evidence for FCD based on ILAE criteria.22
The presence of MRI appearance or pathological evidence of FCD in Cases 2–5 prompted assessment of increased mTOR activation since classically FCDs have been associated with mTOR-related genetic abnormalities.11,21,33–42 Consistent with previous reports, we found low levels of baseline P-S6 immunoreactivity in control brain sections within scattered cortical neurons across all layers in the subpial region of layer I (Fig 4).43,44 A similarly low level of baseline P-S6 labeling was observed the SLC35A2-associated cases (Fig 4), suggesting that SLC35A2 haploinsufficiency does not result in activation of mTOR signaling.
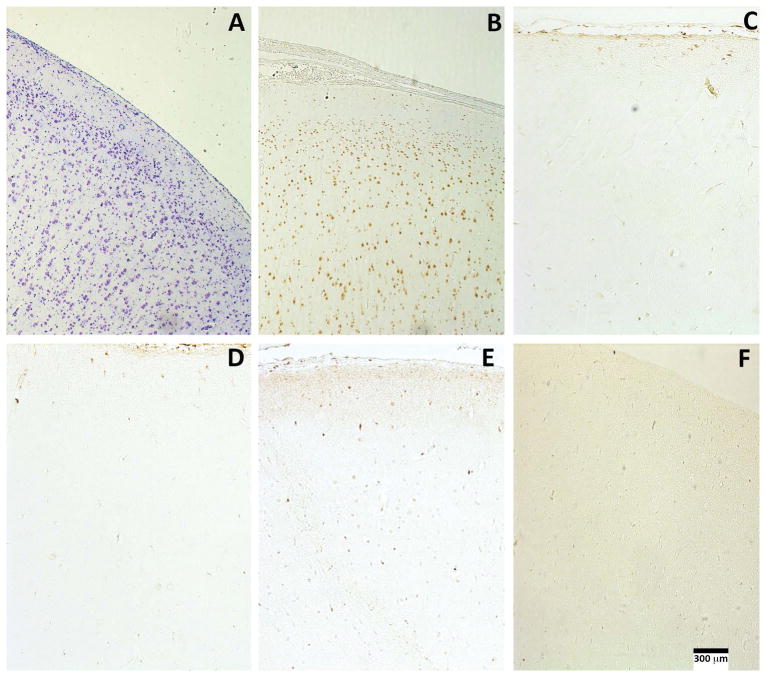
Figure 4.
No evidence for mTOR pathway activation in any of the five SLC35A2-associated cases. Cresyl violet staining (A) and NeuN immunoreactivity (B) in case 5 show normal laminar cytoarchitecture. There are scattered cortical neurons labeled with P-S6 antibodies in representative sections of cases 1, 4, 5, (D, E, F, respectively) associated with somatic SLC35A2 variants. The distribution and P-S6 labeling intensity in three cases was similar to that seen in control cortex from an unaffected individual (C). Scale bar = 300 microns.
DISCUSSION
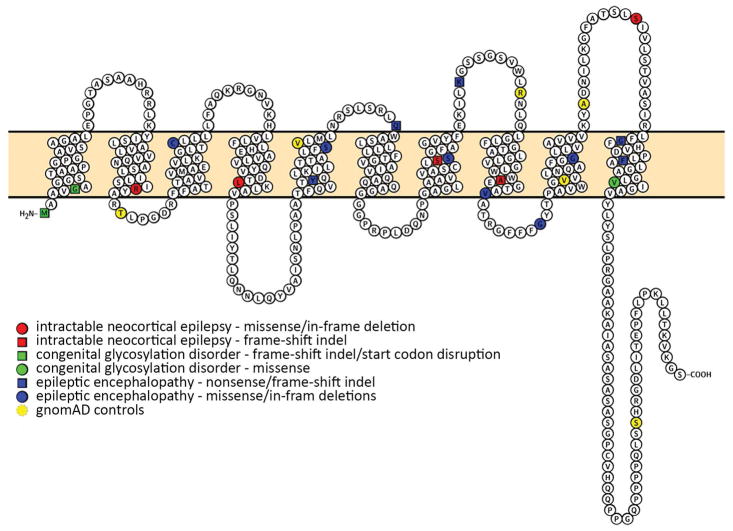
By sequencing brain specimens from 56 individuals undergoing epilepsy surgery for intractable neocortical epilepsy, with and without MCD on MRI, we identified five individuals with mosaic variants in SLC35A2 that are predicted to be deleterious. Variants in SLC35A2 were not present in DNA from leukocytes from the same individuals, confirming somatic (post-zygotic) rather than germline origin of the variants, consistent with the lack of other clinical features suggesting a more generalized disorder of glycosylation in these cases. SLC35A2 is the only gene for which we observed more somatic variants than we would expect to observe in a single gene by chance, supporting the assertion that somatic SLC35A2 variants are associated with intractable focal epilepsy. Nonsynonymous germline variants in SLC35A2 are uncommon in the population, indicating that such variants are under strong negative selection and likely to result in a severe phenotype. Specifically, there are no loss-of function variants in the canonical transcript of SLC35A2 reported in the more than 130,000 individuals in the gnomAD database, and likewise only six Polyphen2 probably damaging missense alleles are reported in this population (Fig 5).
Figure 5.
Novel and previously reported rare functional variation in SLC35A2. Somatic mutations identified in this study are shown in red, those previously implicated in congenital disorders of glycosylation are shown in green, and those previously implicated in epileptic encephalopathies in blue. Six Polyphen2 probably damaging ultra-rare missense mutations identified in gnomAD in the canonical transcript are shown in yellow.
SLC35A2, located at Xp11.23, encodes a UDP-galactose transporter (UGT), a member of the nucleotide-sugar transporter family that transports galactose from the cytosol or nucleus into Golgi vesicles. The encoded protein is a multi-pass membrane protein that permits transport of galactose necessary for glycosylation. The first evidence for a role for SLC35A2 in disease came from a report of two male patients with mosaic variants and one female patient with a de novo germline variant in SLC35A2, all with apparent glycosylation disorders.45 Based on these findings, SLC35A2 was included as a cause of type IIm congenital disorders of glycosylation (CDGIIm), multi-system disorders associated with seizures. Subsequently, germline heterozygous putative loss-of-function de novo variants, including missense, nonsense, and frameshift indels, in SLC35A2 have been identified in multiple girls with epileptic encephalopathy14,46–48,51 (Fig 4). The lack of males with germline variants in this gene likewise supports the assertion that an intact allele, in at least some tissues of the body, is required for survival. Across all previously and our newly reported associated phenotypes, there are both protein-truncating and missense variants observed, suggesting that the missense variants are likely acting through a loss-of-function mechanism (Fig 4).
In this study, we identified mosaic SLC35A2 variants from the resected brain tissue of two boys with early onset epileptic encephalopathy in the form of intractable epileptic spasms associated with developmental delay. Both had MRI-identified lesions and higher VAFs than the NLFE cases, corresponding to a higher fraction of cells in the brain containing the variant allele; this higher burden of variant SLC35A2 may explain why their presentation more closely resembles the central nervous system features of female cases with germline de novo variants and epileptic encephalopathy. Interestingly, one of the somatic mutation sites identified in this study (c.164G>T;p.Arg55Leu in Case 4) was found to be affected, but with a different amino acid substitution (c.164G>C;p.Arg55Pro), in a fraction of cells in the blood (9% VAF in leukocyte DNA) of a girl with epileptic encephalopathy from a previously published trio sequencing study.14 This variant was not highlighted in the original report since mosaic variants at this level in leukocytes had been excluded,14 but it has since been confirmed to be mosaic and absent in parents (unpublished data). This patient exhibited infantile spasms at age four months, developmental delay, and normal MRI, similar to other reported cases with germline SLC35A3 variants.14,46–48,51 In contrast to the two cases with mosaic SLC35A2 variants in brain tissue who presented with epileptic encephalopathy and abnormal MRI, the NLFE cases with mosaic SLC35A2 variants in brain tissue did not present with epileptic encephalopathy or infantile seizure onset.
Pathology in these five cases showed a range of findings. In the three individuals in the NLFE cohort with identified variants in SLC35A2, for whom preoperative MRI did not identify a structural cause responsible for the patients’ focal epilepsy, two had findings that met criteria for FCD1a, while the other had only peri-electrode changes (Table 2). Of the two individuals in the MCD cohort found to have variants in SLC35A2, one had only gliosis on pathology and the other gliosis with an excess of neurons in white matter but not sufficient to meet criteria for FCD (Table 2). Given this range of findings, and the possibility that FCD cannot always be definitively diagnosed by neuropathological assessment because of limited sample size and within-specimen variability, we cannot conclude that a single neuropathological phenotype is associated with mosaic SLC35A2-related dysfunction. Enhanced P-S6 staining was not observed in the resected specimens and thus, the SLC35A2 variants do not appear to be acting through activation of mTOR signaling. We acknowledge, however, that to sampling constraints and the variability observed across specimens, we cannot definitively say that mTOR-related processes are not contributing to the epileptogenic lesions in these cases.
Consistent with the presence of SLC35A2 variants localized to brain in our five cases, we did not observe any systemic signs of a glycosylation defect48 and only one case (Case 4), with the highest burden of mosaic SLC35A2, has severe intellectual disability. The bilateral MRI white matter abnormalities seen on MRI of this case may, in retrospect, be consistent with a defect in glycosylation48 that is more widespread in the brain, but of course we do not have data from other brain regions. Our cases represent a wide range of findings, with presence or absence of MRI lesions, pathological findings suggestive of FCD, and associated intellectual disability. This wide phenotypic range is perhaps not surprising given recent observations in epilepsy genetics. For example, recently a large exome sequencing study of leukocyte DNA from NLFE patients with a family history of epilepsy reported more than expected ultra-rare variants in genes previously associated with epileptic encephalopathies in cases compared to controls, suggesting that some of these variants contribute to disease risk.8 While SLC35A2 was not specifically implicated in this previously published study,8 our findings likewise demonstrate that the same gene may underlie phenotypic extremes with the added aspect that variant burden in the brain may also govern aspects of the clinical presentation.
In terms of the potential impact of SLC35A2 mosaic variants in non-lesional epilepsy, we found that 16.7% (3/18) of our sequenced NLFE cohort had an SLC35A2 somatic variant identified in brain tissue, allowing us to estimate, based on the calculated 95% CI, that somatic variants in SLC35A2 could explain between 3.5% to 42% of cases of NLFE undergoing resection for focal epilepsy. Overall, our data suggest that SLC35A2 plays a larger role in epilepsy than previously recognized.
Somatic mutation has been identified in major structural epileptogenic malformations including hemimegalencephaly, and in FCD type II, which includes dysmorphic neurons in addition to cortical disorganization.18–20,41 However, genes associated with isolated FCD type I, with its more subtle cortical disorganization without dysmorphic neurons, have not been previously described, though rare patients with gremline genetic syndromes have been described as having FCD type I on pathology.49,50 Furthermore, the relatively new field of post-zygotic mutation in human focal cortical malformation has thus far implicated only genes in the mTOR pathway;18,20,41,52 thus, our findings represent a novel, non-mTOR-related genetic cause of somatic mosaic focal epileptic brain lesions. Our findings may consequently have important treatment implications. First, despite growing numbers of cases of FCD type II being explained by somatic variants in the PI3K-AKT-mTOR pathway, raising the possibility that mTOR modulation might be a treatment possibility, our data suggest that not all FCD can summarily be classified as uniquely related to this pathway, though a downstream effect on the mTOR pathway cannot be conclusively ruled out. Second, two previously reported patients with SLC35A2 variants were reported to have a positive response to treatment with exogenous galactose,47,53 raising an interesting possibility for treatment of epilepsy if variants in this gene can be identified early in patients with intractable epilepsy. Collectively, our findings advance our understanding of the pathophysiology of intractable focal epilepsy and the role of somatic mutation in both lesional and non-lesional epilepsy. Further, they present the possibility of innovative, rational therapy for one of the most difficult to treat and common types of epilepsy.
Supplementary Material
Acknowledgments
We thank all the study participants who generously enrolled into the study and shared biological samples to advance epilepsy research. We also thank Sylwia Misciewicz and Felicia Kuo who were instrumental in coordinating patient ascertainment, patient consent, and sample management in the nonlesional focal epilepsy cohort. We thank Michael Wilson and Joseph DeRisi for their involvement in the collaboration studying the genomic basis of nonlesional focal epilepsy, and specifically work evaluating possible viral etiologies, and George Zanazzi for his help acquiring control brain tissue. We also thank the Columbia University Alzheimer’s Disease Research Center, funded by NIH grant P50AG008702 to S.A. Small (P.I.), J.P. Vonsattel, and L.S. Honig, for providing biological samples from brain and blood.
This work was funded by National Institute for Neurological Diseases and Stroke through the following grants: R01-NS089552 (PIs: MW, PBC); R01-NS094596 (PIs: ELH, PBC; CO-Is: AHP, SAS, BEP, GAG, GMM, PC); The Epilepsy Phenome/Genome Project - NS053998, Epi4K—Administrative Core NS077274, Epi4K—Sequencing, Biostatistics and Bioinformatics Core NS077303, and Epi4K—Phenotyping and Clinical Informatics Core NS077276. This research was also supported in part by the Repository Core for Neurological Disorders, Department of Neurology, Boston Children’s Hospital, and the IDDRC (NIH P30HD018655). AHP is also funded by Translational Research Program, Boston Children’s Hospital, and Boston Children’s Hospital Department of Neurology Core for Neurological Diseases.
The collection of control samples and data was funded in part by: Biogen, Inc.; Gilead Sciences, Inc.; UCB; Bryan ADRC NIA P30AG028377; B57 SAIC-Fredrick Inc M11-074; National Institute of Neurological Disorders and Stroke (RC2NS070344; RC2MH089915; U01NS077303; U01NS053998, U54NS078059, P01HD080642); National Human Genome Research Institute (Yale Mendelian Genomics Center - UM1HG006504, U01HG007672); National Institute of Mental Health (K01MH098126, R01MH097971, R01MH099216, RC2MH089915); National Institute of Diabetes and Digestive and Kidney Diseases (R01DK080099); National Institute of Allergy and Infectious Diseases (Division of Intramural Research, 1R56AI098588-01A1); National Institute of Allergy and Infectious Diseases Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (UM1AI100645, U19AI067854); National Center for Advancing Translational Sciences (UL1TR000040); Eunice Kennedy Shriver National Institute of Child Health and Human Development(R01HD048805); the Ellison Medical Foundation New Scholar award AG-NS-0441-08; the Duke Chancellor’s Discovery Program Research Fund 2014; Neil Molberger Brain Research Fund; Endocrine Fellows Foundation Grant; Bill and Melinda Gates Foundation; The Murdock Study Community Registry and Biorepository; The Stanley Institute for Cognitive Genomics at Cold Spring Harbor Laboratory; the Duke Genome Sequencing Clinic; New York-Presbyterian Hospital; Columbia University College Physicians and Surgeons; Columbia University Medical Center; The J. Willard and Alice S. Marriott Foundation; The Muscular Dystrophy Association; The Nicholas Nunno Foundation; The JDM Fund for Mitochondrial Research; The Arturo Estopinan TK2 Research Fund; and The Endocrine Fellows Foundation; Helaine B Allen and Emily Allen Wolff.
Data collection and sharing for the Washington Heights-Inwood Columbia Aging Project (WHICAP) project (used as controls in this analysis) was supported by the grants (WHICAP, PO1AG07232, R01AG037212, RF1AG054023) funded by the National Institute on Aging (NIA) and by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant Number UL1TR001873. This manuscript has been reviewed by WHICAP investigators for scientific content and consistency of data interpretation with previous WHICAP Study publications. We acknowledge the WHICAP study participants and the WHICAP research and support staff for their contributions to this study.
Footnotes
AUTHOR CONTRIBUTIONS
MRW, PBC, AHP, and ELH conceived and the designed the study. MRW, NGG, EY, PBC, AHP, CAS, and ELH drafted the manuscript and figures. MRW, NGG, JS, EHB, DR, SR, AZ, CAS, PD, MH, SAS, GMM, WKD, GAG, BEP, MAM, CRM, CDM, AMRB, JMP, DKM, AMP, CIA, CML, KMK, JRM, EY, PGWL, CS, ASA, PC, PBC, AHP, ELH were involved in acquisition and analysis of data.
POTENTIAL CONFLICTS OF INTEREST
Dr. Sheth reports personal fees from Boston Scientific, personal fees from Medtronic.
References
- 1.Hesdorffer DC, Logroscino G, Benn EK, Katri N, Cascino G, Hauser WA. Estimating risk for developing epilepsy: a population-based study in Rochester, Minnesota. Neurology. 2011 Jan 4;76(1):23–7. doi: 10.1212/WNL.0b013e318204a36a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dwivedi R, Ramanujam B, Chandra PS, et al. Surgery for Drug-Resistant Epilepsy in Children. N Engl J Med. 2017 Oct 26;377(17):1639–47. doi: 10.1056/NEJMoa1615335. [DOI] [PubMed] [Google Scholar]
- 3.Wiebe S, Blume WT, Girvin JP, Eliasziw M, EES T. A randomized, controlled trial of surgery for temporal-lobe epilepsy. New Engl J Med. 2001 Aug 2;345(5):311–8. doi: 10.1056/NEJM200108023450501. [DOI] [PubMed] [Google Scholar]
- 4.Picard F, Makrythanasis P, Navarro V, et al. DEPDC5 mutations in families presenting as autosomal dominant nocturnal frontal lobe epilepsy. Neurology. 2014 Jun 10;82(23):2101–6. doi: 10.1212/WNL.0000000000000488. [DOI] [PubMed] [Google Scholar]
- 5.Dibbens LM, de Vries B, Donatello S, et al. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nat Genet. 2013 May;45(5):546–51. doi: 10.1038/ng.2599. [DOI] [PubMed] [Google Scholar]
- 6.Ottman R, Winawer MR, Kalachikov S, et al. LGI1 mutations in autosomal dominant partial epilepsy with auditory features. Neurology. 2004 Apr 13;62(7):1120–6. doi: 10.1212/01.wnl.0000120098.39231.6e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heron SE, Smith KR, Bahlo M, et al. Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet. 2012 Oct 21;44(11):1188–90. doi: 10.1038/ng.2440. [DOI] [PubMed] [Google Scholar]
- 8.Epi4K Consortium, Epilepsy Phenome/Genome Project. Ultra-rare genetic variation in common epilepsies: a case-control sequencing study. Lancet Neurol. 2017 Feb;16(2):135–43. doi: 10.1016/S1474-4422(16)30359-3. [DOI] [PubMed] [Google Scholar]
- 9.Dazzo E, Fanciulli M, Serioli E, et al. Heterozygous reelin mutations cause autosomal-dominant lateral temporal epilepsy. Am J Hum Genet. 2015 Jun 4;96(6):992–1000. doi: 10.1016/j.ajhg.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvill GL, Regan BM, Yendle SC, et al. GRIN2A mutations cause epilepsy-aphasia spectrum disorders. Nat Genet. 2013 Sep;45(9):1073–6. doi: 10.1038/ng.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weckhuysen S, Marsan E, Lambrecq V, et al. Involvement of GATOR complex genes in familial focal epilepsies and focal cortical dysplasia. Epilepsia. 2016 Jun;57(6):994–1003. doi: 10.1111/epi.13391. [DOI] [PubMed] [Google Scholar]
- 12.Ricos MG, Hodgson BL, Pippucci T, et al. Mutations in the mammalian target of rapamycin pathway regulators NPRL2 and NPRL3 cause focal epilepsy. Ann Neurol. 2016 Jan;79(1):120–31. doi: 10.1002/ana.24547. [DOI] [PubMed] [Google Scholar]
- 13.Epi4K Consortium and Epilepsy Phenome/Genome Project. De novo mutations in epileptic encephalopathies. Nature. 2013 Sep 12;501(7466):217–21. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Euro Epinomics- R. E. S. Consortium, Epilepsy Phenome/Genome Project, Epi4k Consortium. De Novo Mutations in Synaptic Transmission Genes Including DNM1 Cause Epileptic Encephalopathies. Am J Hum Genet. 2014 Oct 2;95(4):360–70. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura K, Kodera H, Akita T, et al. De Novo Mutations in GNAO1, Encoding a Galphao Subunit of Heterotrimeric G Proteins, Cause Epileptic Encephalopathy. Am J Hum Genet. 2013 Sep 5;93(3):496–505. doi: 10.1016/j.ajhg.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veeramah KR, O’Brien JE, Meisler MH, et al. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am J Hum Genet. 2012 Mar 9;90(3):502–10. doi: 10.1016/j.ajhg.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers CT, Stong N, Mountier EI, et al. De Novo Mutations in PPP3CA Cause Severe Neurodevelopmental Disease with Seizures. Am J Hum Genet. 2017 Oct 05;101(4):516–24. doi: 10.1016/j.ajhg.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JH, Huynh M, Silhavy JL, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44(8):941–5. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poduri A, Evrony GD, Cai X, et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron. 2012 Apr 12;74(1):41–8. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffin NG, Cronin KD, Walley NM, et al. Somatic uniparental disomy of Chromosome 16p in hemimegalencephaly. Cold Spring Harb Mol Case Stud. 2017 Sep;3(5) doi: 10.1101/mcs.a001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim JS, Kim WI, Kang HC, et al. Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat Med. 2015 Apr;21(4):395–400. doi: 10.1038/nm.3824. [DOI] [PubMed] [Google Scholar]
- 22.Blumcke I, Muhlebner A. Neuropathological work-up of focal cortical dysplasias using the new ILAE consensus classification system - practical guideline article invited by the Euro-CNS Research Committee. Clin Neuropathol. 2011 Jul-Aug;30(4):164–77. doi: 10.5414/np300398. [DOI] [PubMed] [Google Scholar]
- 23.Porter BE, Judkins AR, Clancy RR, Duhaime A, Dlugos DJ, Golden JA. Dysplasia: a common finding in intractable pediatric temporal lobe epilepsy. Neurology. 2003 Aug 12;61(3):365–8. doi: 10.1212/01.wnl.0000076487.28227.6e. [DOI] [PubMed] [Google Scholar]
- 24.Lindhout D. Somatic mosaicism as a basic epileptogenic mechanism? Brain. 2008 Apr;131:900–1. doi: 10.1093/brain/awn056. [DOI] [PubMed] [Google Scholar]
- 25.Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013 Jul 05;341(6141):1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher RS, Cross JH, French JA, et al. Operational classification of seizure types by the International League Against Epilepsy: Position Paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017 Apr;58(4):522–30. doi: 10.1111/epi.13670. [DOI] [PubMed] [Google Scholar]
- 27.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013 Mar;31(3):213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costello M, Pugh TJ, Fennell TJ, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013 Apr 01;41(6):e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Liu P, Evans TC, Jr, Ettwiller LM. DNA damage is a pervasive cause of sequencing errors, directly confounding variant identification. Science. 2017 Feb 17;355(6326):752–6. doi: 10.1126/science.aai8690. [DOI] [PubMed] [Google Scholar]
- 30.Hildebrand MS, Griffin NG, Damiano JA, et al. Mutations of the Sonic Hedgehog Pathway Underlie Hypothalamic Hamartoma with Gelastic Epilepsy. Am J Hum Genet. 2016 Aug 04;99(2):423–9. doi: 10.1016/j.ajhg.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016 Aug 17;536(7616):285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sparks SE, Krasnewich DM. Congenital Disorders of N-Linked Glycosylation and Multiple Pathway Overview. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, et al., editors. GeneReviews((R)) Seattle (WA): 1993. [Google Scholar]
- 33.Baulac S, Ishida S, Marsan E, et al. Familial focal epilepsy with focal cortical dysplasia due to DEPDC5 mutations. Ann Neurol. 2015 Apr;77(4):675–83. doi: 10.1002/ana.24368. [DOI] [PubMed] [Google Scholar]
- 34.D’Gama AM, Geng Y, Couto JA, et al. Mammalian target of rapamycin pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Annals of neurology. 2015 Apr;77(4):720–5. doi: 10.1002/ana.24357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Gama AM, Woodworth MB, Hossain AA, et al. Somatic Mutations Activating the mTOR Pathway in Dorsal Telencephalic Progenitors Cause a Continuum of Cortical Dysplasias. Cell reports. 2017 Dec 26;21(13):3754–66. doi: 10.1016/j.celrep.2017.11.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansen LA, Mirzaa GM, Ishak GE, et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain : a journal of neurology. 2015 Jun;138(Pt 6):1613–28. doi: 10.1093/brain/awv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirzaa GM, Campbell CD, Solovieff N, et al. Association of MTOR Mutations With Developmental Brain Disorders, Including Megalencephaly, Focal Cortical Dysplasia, and Pigmentary Mosaicism. JAMA Neurol. 2016 Jul;73(7):836–45. doi: 10.1001/jamaneurol.2016.0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakashima M, Saitsu H, Takei N, et al. Somatic Mutations in the MTOR gene cause focal cortical dysplasia type IIb. Ann Neurol. 2015 Sep;78(3):375–86. doi: 10.1002/ana.24444. [DOI] [PubMed] [Google Scholar]
- 39.Aldred MA, Trembath RC. Activating and inactivating mutations in the human GNAS1 gene. Human mutation. 2000 Sep;16(3):183–9. doi: 10.1002/1098-1004(200009)16:3<183::AID-HUMU1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 40.Conti V, Pantaleo M, Barba C, et al. Focal dysplasia of the cerebral cortex and infantile spasms associated with somatic 1q21.1-q44 duplication including the AKT3 gene. Clinical genetics. 2014 Aug 5; doi: 10.1111/cge.12476. [DOI] [PubMed] [Google Scholar]
- 41.Lim JS, Gopalappa R, Kim SH, et al. Somatic Mutations in TSC1 and TSC2 Cause Focal Cortical Dysplasia. Am J Hum Genet. 2017 Mar 02;100(3):454–72. doi: 10.1016/j.ajhg.2017.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheffer IE, Heron SE, Regan BM, et al. Mutations in mTOR regulator DEPDC5 cause focal epilepsy with brain malformations. Annals of neurology. 2014 Mar 1; doi: 10.1002/ana.24126. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Reeves C, Michalak Z, et al. Evidence for mTOR pathway activation in a spectrum of epilepsy-associated pathologies. Acta Neuropathol Commun. 2014 Jul 8;2:71. doi: 10.1186/2051-5960-2-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talos DM, Jacobs LM, Gourmaud S, et al. Mechanistic target of rapamycin complex 1 and 2 in human temporal lobe epilepsy. Ann Neurol. 2018 Feb;83(2):311–27. doi: 10.1002/ana.25149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng BG, Buckingham KJ, Raymond K, et al. Mosaicism of the UDP-galactose transporter SLC35A2 causes a congenital disorder of glycosylation. Am J Hum Genet. 2013 Apr 04;92(4):632–6. doi: 10.1016/j.ajhg.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hino-Fukuyo N, Kikuchi A, Arai-Ichinoi N, et al. Genomic analysis identifies candidate pathogenic variants in 9 of 18 patients with unexplained West syndrome. Hum Genet. 2015 Jun;134(6):649–58. doi: 10.1007/s00439-015-1553-6. [DOI] [PubMed] [Google Scholar]
- 47.Dorre K, Olczak M, Wada Y, et al. A new case of UDP-galactose transporter deficiency (SLC35A2-CDG): molecular basis, clinical phenotype, and therapeutic approach. J Inherit Metab Dis. 2015 Sep;38(5):931–40. doi: 10.1007/s10545-015-9828-6. [DOI] [PubMed] [Google Scholar]
- 48.Kimizu T, Takahashi Y, Oboshi T, et al. A case of early onset epileptic encephalopathy with de novo mutation in SLC35A2: Clinical features and treatment for epilepsy. Brain Dev. 2017 Mar;39(3):256–60. doi: 10.1016/j.braindev.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Kurian M, Korff CM, Ranza E, et al. Focal cortical malformations in children with early infantile epilepsy and PCDH19 mutations: case report. Dev Med Child Neurol. 2018 Jan;60(1):100–5. doi: 10.1111/dmcn.13595. [DOI] [PubMed] [Google Scholar]
- 50.Barba C, Parrini E, Coras R, et al. Co-occurring malformations of cortical development and SCN1A gene mutations. Epilepsia. 2014 Jul;55(7):1009–19. doi: 10.1111/epi.12658. [DOI] [PubMed] [Google Scholar]
- 51.Kodera H, Nakamura K, Osaka H, et al. De Novo Mutations in SLC35A2 Encoding a UDP-Galactose Transporter Cause Early-Onset Epileptic Encephalopathy. Hum Mutat. 2013 Dec;34(12):1708–14. doi: 10.1002/humu.22446. [DOI] [PubMed] [Google Scholar]
- 52.Crino PB. The mTOR signalling cascade: paving new roads to cure neurological disease. Nat Rev Neurol. 2016 Jul;12(7):379–92. doi: 10.1038/nrneurol.2016.81. [DOI] [PubMed] [Google Scholar]
- 53.Demos M, Guella I, McKenzie MB, et al. Diagnostic Yield And Treatment Impact Of Targeted Exome Sequencing In Early-Onset Epilepsy. bioRxiv. 2017 doi: 10.1101/139329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.