Abstract
Background
Radiomic Analysis is defined as computationally extracting features from radiographic images for quantitatively characterizing disease patterns. There has been recent interest in examining the use of MRI for identifying prostate cancer aggressiveness in patients on active surveillance (AS).
Purpose
To evaluate the performance of MRI-based radiomic features in identifying presence or absence of clinically significant prostate cancer in AS patients.
Study Type
Retrospective.
Subjects Model
MRI/TRUS fusion-guided biopsy was performed for 56 prostate cancer patients on AS who had underwent pre-biopsy
Field Strength/Sequence
3T, T2-weighted and Diffusion-weighted.
Assessment
A pathologist histopathologically defined the presence of clinically significant disease. A radiologist manually delineated lesions on T2w-MRs. Then, three radiologists assessed MRIs using PIRADS v2.0 guidelines. Tumors were categorized into four groups: MRI-negative-biopsy-negative (Group1, N=15), MRI-positive-biopsy-positive (Group2, N=16), MRI-negative-biopsy-positive (Group3, N=10), and MRI-positive-biopsy-negative (Group4, N=15). 308 radiomic features (First-order statistics, Gabor, Laws Energy, and Haralick) were extracted from within the annotated lesions on T2w images and ADC maps. The top 10 features associated with clinically significant tumors were identified using minimum-redundancy-maximum-relevance and used to construct three machine learning models which were independently evaluated in their ability to identify the presence and absence of clinically significant disease.
Statistical Tests
Wilcoxon rank-sum tests with p-value <0.05 considered statistically significant.
Results
7 T2w-based (First-order Statistics, Haralick, Laws, and Gabor) and 3 ADC-based radiomic features (Laws, Gradient and Sobel) exhibited statistically significant differences (p-value <0.001) between malignant and normal regions in the training groups. The 3 constructed models yielded overall accuracy improvement of 33, 60, 80% and 30, 40, 60% for patients in testing groups, when compared to PIRADS v2.0 alone.
Data Conclusion
Radiomic features could help in identifying the presence and absence of clinically significant disease in AS patients when PIRADS v2.0 assessment on MRI contradicted pathology findings of MRI-TRUS prostate biopsies.
Keywords: Active Surveillance, Prostate Cancer, MRI, Radiomics, Radiomic Features, Texture Features
INTRODUCTION
Active surveillance (AS) is considered a viable alternative to radical treatments such as surgery or radiation therapy, specifically for patients diagnosed with lower risk or indolent prostate cancer (PCa)1–4. However, an AS protocol requires patients to be closely monitored and evaluated regularly with MRI scans and prostate biopsies. Annual biopsies identify whether patients on AS exhibit upgrading or upstaging of their disease; a change that would warrant reverting to more radical therapy.
Biopsies have an increased risk of complications (e.g. infection, bleeding, urinary retention, and erectile dysfunction) in addition to causing patient anxiety. Even with imaging-guided biopsy procedures, there is the risk of not correctly targeting the lesion of interest or even potentially missing the whole tumor altogether. Consequently, there has been increasing interest in the use of non-invasive MRI for monitoring PCa patients on AS regimens1,2,5–7.
Prostate multiparametric-MRI (mpMRI) provides structural and functional imaging via T2-weighted (T2w), diffusion-weighted (DW), and dynamic contrast-enhanced (DCE) sequences, for lesion characterization6–8. In 2015, the European Society of Urogenital Radiology (ESUR) introduced the second version of the prostate imaging reporting and data system (PIRADS v2)9, a structured reporting system that defines clinically significant cancer. PIRADS allocates lesion scores ranging from 1 (highly unlikely to be malignant)) to 5 (highly likely to be malignant))9. However, PIRADS evaluations greatly depend on the experience level of the radiologist and his/her ability to interpret mpMRI of the prostate. Schimmoller et al.10 showed that evaluating PCa using PIRADS gave only moderate to good inter-reader agreement when performed by three blinded readers.
Recently, there has been increasing interest in employing Radiomics or computerized feature analysis of MR images to quantitatively describe tissue appearance and subtle structural details, and thus enable diagnosis and risk stratification of PCa on mpMRI11. Wang et al.12 showed that a Radiomics-based machine learning classifier could increase the predictive performance of PIRADS. There have been several studies examining the capability of Radiomics for characterizing PCa based on Gleason score (i.e. clinically significant disease). Fehr et al.6 correlated mpMRI Radiomics (Haralick features) to Gleason scores from repeated TRUS-guided biopsies and whole-mount evaluation of radical prostatectomy specimens. Litjens et al.13 showed that radiomic features extracted from mpMRI could distinguish not only low- from high-grade PCa, but also tumor confounders from noncancerous tissue. Ginsburg et al.14 found that Radiomics-based machine learning classifiers trained primarily for PCa diagnosis did not accurately distinguish PCa tumors based off Gleason scores.
One limitation with these previous studies is that all considered patients underwent radical prostatectomy, in other words the patients in these studies typically tended to have later-staged, larger tumors. To the best of our knowledge, no previous work has focused on machine learning, Radiomics-based characterization, and risk stratification of PCa within AS regimens. These cases are typically characterized with earlier-staged tumors and also tend to represent the more challenging cases for PIRADS-based characterization.
In this work, our goal was to identify radiomic features from bi-parametric MRI (T2w and Diffusion) that could accurately identify the presence or absence of clinically significant disease in highly challenging cases where there was a discordance between PIRADS and histopathologic findings for PCa patients on AS.
MATERIALS AND METHODS
Dataset Description
Approval for this retrospective study was granted by Institutional Review Board (SVH12/007), and written informed consents were obtained from all patients before MRI and biopsy7. Figure 1 depicts patient selection/exclusion criteria and the overall experimental design. A total of 344 men, all older than 40 years with elevated PSA levels or abnormal digital rectal examination (DRE) outcome and with no previous prostate MRI or biopsy were considered for this study. All participants underwent pre-screening 30-core trans-perineal template-guided mapping biopsy (TTMB) between April 2012 and March 2014 as described in7 while being on active surveillance. A floor-mounted, trans-perineal grid ultrasound (TRUS) platform (BK Medical, Herlev, Denmark) combined with BioJet rigid MRI/TRUS fusion software were utilized for all biopsies. Details of the patient cohort included in this study are listed in Table 1.
Figure 1.
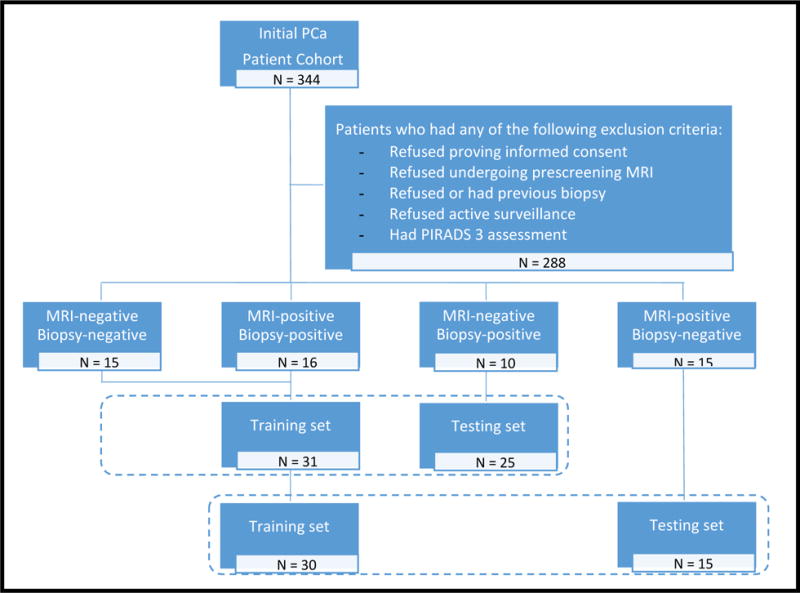
Dataset description and experimental design. 56-case dataset was divided to 4 groups. Cases from Groups 1 and 2 (true-negatives and true-positives, respectively) were used to train machine learning classifiers. Cases from Groups 3 and 4 (false-negatives and false-positives, respectively) were used as independent testing sets (for correctly identifying false-negatives identifying false-positives independently of histopathology findings). All cases with PIRADS 3 score were excluded from training and independent test sets.
Table 1.
Description of Study
| St Vincent’s Medical Center | |
|---|---|
| Number of Subjects | 56 |
| Age (mean ± SD) | 65.1 ± 6.4 |
| PSA (mean ± SD) | 6.9 ± 5.8 ng/ml |
| Lesion size (mean ± SD) | 1.30 ± 0.79 cm3 |
| Scanner | |
| Brand | Philips Achieva |
| Magnet strength | 3T |
| Coil type | Body coil |
| T2-weighted MRI | |
| Field-of-view | 220 × 220 mm2 |
| Matrix size | 444 × 332 |
| Slice thickness | 3 mm |
| Diffusion-Weighted MRI | |
| Field-of-view | 180 × 180 mm2 |
| Matrix size | 128 × 128 |
| Slice thickness | 3 mm |
| b-values | 0, 1500 s/mm2 |
| Gleason Scores | |
| Low (= 6) | 25 |
| High (7-9) | 26 |
After excluding men who refused MRI scans or biopsies, declined an AS regimen, had no sufficient diffusion parameters on imaging, or were assigned PIRADS score 3, of the 344, 56 remaining patients were considered for the study (Figure 1). All of these 56 patients, had previously undergone pre-biopsy 3 Tesla bi-parametric MRI (T2w, DW). Then, correlation of bi-parametric MRI with histopathological findings in TTMB was assessed. MRI-negative cases were defined as men who were assigned low scores on PIRADS assessments (1 or 2), while MRI-positive cases were those who were assigned high PIRADS scores (4 or 5). Based on PIRADS and biopsy findings of absence or presence of clinically significant PCa, the cohort was segregated into four patient groups; MRI-negative-biopsy-negative (Group 1: True-Negatives, N=15), MRI-positive-biopsy-positive (Group 2: True-positives, N=16), MRI-negative-biopsy-positive (Group 3: False-Negatives, N=10), and MRI-positive-biopsy-negative (Group 4: False-Positives, N=15). Table 2 summarizes the patient groups of this dataset.
Table 2.
Description of the patient groups
| Number of Patient Group | Number of Patients | Prostate Cancer-positive on | |
|---|---|---|---|
| MRI-positive (PIRADS score = 4-5) | Biopsy-positive (Gleason score 7-9) | ||
| 1 | 15 | No | No |
| 2 | 16 | Yes | Yes |
| 3 | 10 | No | Yes |
| 4 | 15 | Yes | No |
Experimental Design
Our study design comprised two experiments. Experiment 1 focused on finding radiomic features that discriminate patients who had confirmed clinically significant disease from those who did not. Experiment 2 evaluated the ability of these radiomic features to identify the presence and absence of clinically significant disease in the more challenging cases with discordance between PIRADS assessment and biopsy findings (Groups 3 and 4). Figure 2 shows the pipeline of modules that were developed and employed for addressing the aforementioned tasks. These individual modules are described in detail below.
Figure 2.
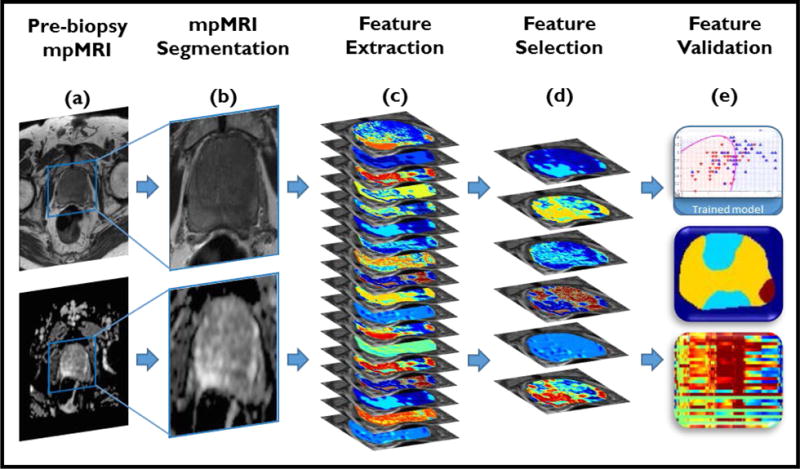
Radiomic analysis pipeline. a,b) Lesions on T2w images (top) and ADC maps (bottom) are manually annotated by an expert radiologist. c) 308 radiomic texture features are extracted from MRI. d) MRMR technique with feature voting is utilized for selecting the most relevant features. e) Validation strategies are applied to ensure robustness of the selected features.
Lesion Segmentation on MRI
All MRIs were manually annotated by an expert radiologist (R1: 7 years of experience) for regions suspicious for cancer (MRI-positive), then independently assessed by three more radiologists (R2, R3, and R4: 5, 9, and 11 years of experience, respectively) for all the 56 cases according to PIRADS v2.0 scoring system. Radiologist R1 delineated lesions and normal regions using the 3D Slicer open-source software (Kitware Inc., Carrboro, NC). Although MRI-negative (normal) regions existed in both Groups 1 and 2, annotations used for further processing were taken only from cases within Group 1. To ensure unbiased assessment, all MRI annotations were performed blinded to the biopsy findings. Furthermore, core locations on biopsy reports were matched to those on PIRADS reports (matching), then to radiologist’s delineations of lesions (validation). Only cases with matched locations (Biopsy, PIRADS reports, and radiologist’s delineations) were considered for inclusion in this study.
Preprocessing
For each patient in our cohort, T2w volume images and ADC maps were resampled to a uniform pixel spacing of 0.5 × 0.5 × 3 mm. Then, they were cropped to the lesion region of interest as delineated by R2, with 5 pixels (2.5 mm) padding along the x and y coordinates.
Inhomogeneity correction15 was applied to T2w images to account for presence of bias field artifacts. Next, T2w images and ADC maps were corrected for inherent acquisition-to-acquisition signal intensity variations (non-standardness) using scale-based standardization16. This procedure was applied to mitigate the inherent drift phenomenon that accompanies MRI intensities. This phenomenon results in MR intensities not having a tissue-specific numeric meaning, even within the same MRI protocol, for the same body region, and even for images of the same patient obtained on the same scanner16.
Radiomic Feature Extraction
The prostate capsules were manually segmented to ensure only lesions completely localized within the prostate contours are included. On a per-voxel basis, a total of 308 two-dimensional radiomic texture features (including gray-level statistical, steerable Gabor wavelets, Mean, Median, Sobel, Laws Energy, and Haralick) were extracted from within the prostate capsule regions on T2w images and ADC maps; 154 features from each sequence. A summary of these radiomic features and their relevance to PCa characterization is provided in Table 3. First order statistics (mean, median, standard deviation, skewness, and kurtosis) of each radiomic feature were calculated for each annotated lesion, and then feature values were normalized to values between 0 and 1. All features were implemented in MATLAB (R2017a, Mathworks, Natick, MA).
Table 3.
Description of Radiomic features extracted
| Radiomic Feature Category | Radiomic Feature Type/Param | Number of features extracted (total) | Relevance to PCa |
|---|---|---|---|
| Signal Intensity | T2w images, ADC maps | 1 × 2 (2) | Cancers are usually hypo-intense on MRI |
| First Order Statistics | Mean, Median, Sobel | 9 × 2 (18) | Intensity variability |
| Gabor | Frequency, Orientation | 80 × 2 (160) | Low-level oriented edges |
| Gray-level co-occurrence | Haralick | 3 × 13 × 2 (78) | Structural heterogeneity |
| Texture Energy | Laws’ texture energy | 25 × 2 (50) | Appearance of ROI |
Experiment 1: Radiomic Features that Discriminate Group 1 (PIRADS-/Bx-) from Group 2 (PIRADS+/Bx+)
Minimum redundancy, maximum relevance algorithm (MRMR)17 was used to identify and rank the top 10 features that could discriminate between the clinically significant lesions in Group 2 from normal regions in Group 1, using the studies within the training set. MRMR is a feature selection scheme that selects the most relevant features for prediction by maximizing the mutual information (MI) between the selected features and the lesions in Group 2. To minimize bias and ensure balanced sampling of tumor and normal regions, the ratio of the number voxels from the tumor to that of normal regions was maintained at 1:1 before invoking MRMR. Following MRMR, the top 10 most relevant features were identified.
In order to evaluate, the selected features, unsupervised hierarchical clustering of the top 10 selected features18 was performed within the training set to assess how well a selected set of features inherently clusters the patients into distinct groups without a priori knowledge of the corresponding class labels.
Experiment 2: Evaluating Ability of Radiomic Features Identified in Experiment 1 to Identify PCa Biopsy-Positive Cases in Group 3 and PCa Biopsy-Negative Cases in Group 4
Radiomic features identified in Experiment 1 were used to train three machine learning classifiers, Quadratic Discriminant Analysis (QDA), Random Forests (RF), and Support Vector Machine (SVM), to associate voxel-wise presence of clinically significant PCa. Output of the classification process was voxel-based probability maps reflecting the likelihood of clinically significant disease occurrence at every spatial location on the MR images.
Post-processing was applied to the probability maps in order to convert the continuous-valued image into a tri-level map representing areas of high, low, and very-low likelihood regions of clinically significant PCa. This was accomplished via Markov Random Fields (MRF)19,20 algorithm which provides tractable means for incorporating contextual information into a Bayesian framework. This contextual information is modeled using local conditional probability density functions which the MRF framework implicitly combines into a single joint probability density function. In our approach, MRF was employed in conjunction with the PCa probability maps obtained from the machine learning classifiers, providing a smoothed, filtered version of the original PCa likelihood maps. Recalling the PIRADS21,9,22 definition of clinically significant cancer (GS ≥ 7, and/or volume ≥ 0.5 cm3, and/or is or has an extra prostatic extension), all lesions with volume < 0.5 cm3 (lesion volume was calculated by multiplying the number of voxels comprising its voxel cluster by the voxel size) were considered clinically insignificant and filtered out.
The three machine learning classifiers were trained on data only from Groups 1 & 2. This involved a randomized, patient-stratified (to ensure patients in training are separated from those in testing folds i.e. voxels from one case would only be used either for training or testing models), 3-fold-100-run cross-validation scheme. For each cross-validation run, the 31 studies within Groups 1 and 2 were divided into three randomized subsets; two of which were used for training machine learning classifiers while the third subset was used for testing.
Then, classification was performed and the process was repeated until all of the patients within each of the three subsets had been classified. Each cross validation run was evaluated via AUC to identify the best performing classifier. This was then independently applied to every dataset in Groups 3 and 4 for voxel-wise identification of presence or absence of clinically significant disease. The resulting probability maps for Groups 3 and 4 were then denoised via MRF as described above. Finally, for every dataset in Groups 3 & 4, we assigned a label of clinically significant disease (PCa-positive or PCa-negative) based on the presence of any high-likelihood voxel clusters in the denoised MRF maps.
Both experiments were performed using in-house developed MATLAB (R2017a, MathWorks, Natick, MA) routines on a standard 3.4 GHz Intel Core i7-3770 (Quad-core processor with 8MB of on-chip L2 cache memory) Windows-based computer with 32 GB RAM. Average processing times broken down for individual modules are listed in Table 4.
Table 4.
Processing time break down for proposed Radiomic analysis pipeline
| Pipeline Step | Average Per-Study Processing Time |
|---|---|
| Image Preprocessing | 14 min |
| Feature Extraction | 17 min |
| Training Classifier | 24 min |
| Post processing | 1.7 min |
RESULTS
Experiment 1: Radiomic Features that Discriminate Group 1 from Group 2
The top ten radiomic features obtained by using MRMR for the primary PCa cohort (i.e discriminating Group 1 from Group 2) are shown in Table 5 and are qualitatively represented in Figures 3 and 4. These features include Gabor, first-order statistics, and grey-level co-occurrence-based texture features. Average feature values for features found to be the most relevant in identifying clinically significant disease exhibited statistically significant differences across MRI-negative-biopsy-negative and MRI-positive-biopsy-positive cases (p < 0.01). Of the ten selected features found to exhibit differential expression between biopsy-positive and biopsy-negative cases, three were extracted from DW ADC maps (First-order statistics and Laws) and seven were extracted from T2w MR images (Gabor, Haralick, and First-order statistics). Original intensities on T2w images or values on ADC maps were not picked by MRMR in the top set of features.
Table 5.
Top 10 radiomic features extracted from MRI that demonstrated better performance compared to PIRADS (Ranked according to relevance statistical significance of differential expression - p-value < 0.05 for all selected features)
| Rank | Feature (Parameters) | Protocol |
|---|---|---|
| 1 | Gabor (λ-2, ϴ=0.5) | T2w |
| 2 | Mean (win. size = 3) | T2w |
| 3 | Median (win. size = 3) | T2w |
| 4 | Std. Dev (win. size = 3) | T2w |
| 5 | Sobel (direction: x) | T2w |
| 6 | Laws (1) | T2w |
| 7 | Haralick (Energy) | T2w |
| 8 | Laws (8) | ADC |
| 9 | Gradient (direction: y) | ADC |
| 10 | Sobel (direction: y) | ADC |
Figure 3.
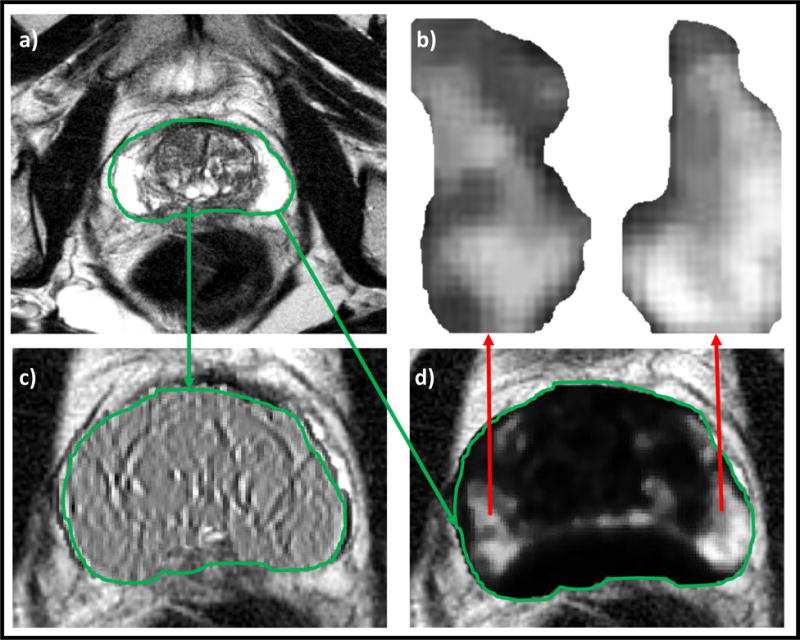
Low- and high-performing features for MRI-negative-biopsy-positive (PIRADS 2, GS 4+3) case. a) T2w MRI, b) Low-performance feature, c) High-performance feature, d) Markedly different texture appearance within biopsy-positive regions, not picked up via PIRADS.
Figure 4.
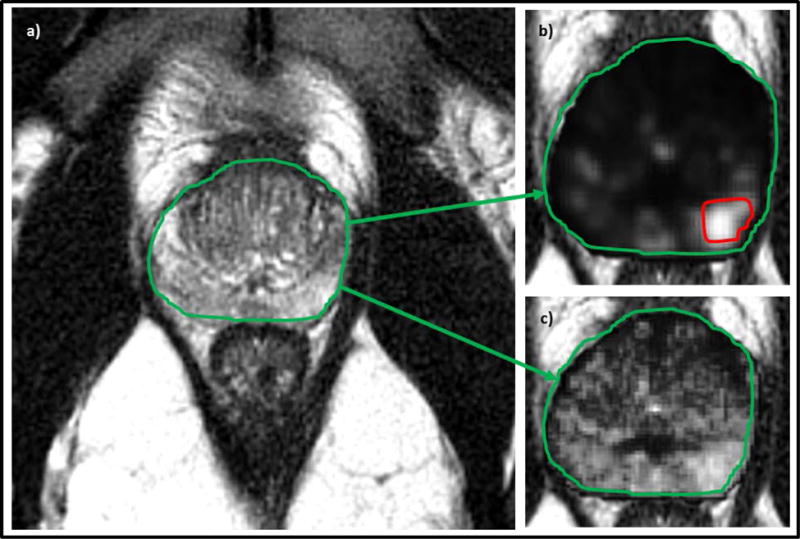
Low- and high-performing features for a MRI-positive-biopsy-negative (PIRADS 4) case: a) T2w MRI, b) Low-performance radiomic feature with the red region denoting “erroneous” PIRADS-based annotation, c) High-performance radiomic feature which demonstrates no clear signature for prostate cancer in this region (agreeing with biopsy findings).
Figure 3 illustrates an MRI-negative-biopsy-positive (PIRADS 2, GS 4+3) case where a radiomic feature showed differential expression of texture appearance within biopsy-positive regions. Similarly, Figure 4 illustrates an MRI-positive-biopsy-negative case for which a radiomic feature was identified that was under-expressing (or muted) for the biopsy-negative region despite that region have been assigned a PIRADS score of 4. Table 5 shows the top ten radiomic features found to be most relevant to identifying clinically significant disease.
Figure 5 shows an unsupervised hierarchical clustering dendrogram of the top ten radiomic features. Each unit is the mean absolute deviation of a radiomic feature extracted for a single patient. Patients are displayed across rows and features are displayed across columns. In this dendrogram, the Gabor feature was identified as the one exhibiting the most differential expression between the PCa-positive and PCa-negative regions (clusters) compared to the rest of features.
Figure 5.
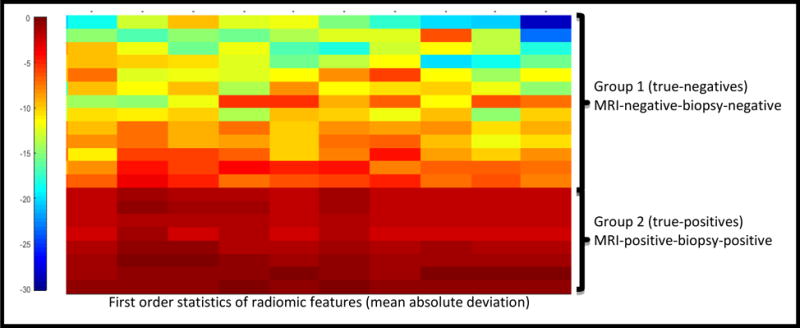
Unsupervised hierarchical clustering of mean absolute deviation of selected radiomic feature parameters (i.e. each feature was reduced to one value using a first order statistic). The x-axis corresponds to the individual patients and features are displayed across columns. Radiomic features separate patients into two distinct clusters: true-positives (red) and true-negatives (blue). Labels on the right indicate the group to which each individual case belongs. Shading indicates expression of the radiomic feature, where red and blue represent high and low expression, respectively.
Experiment 2: Evaluating Ability of Radiomic Features Identified in Experiment 1 to Identify Bx+ Cases in Group 3 and Bx- Cases in Group 4
Table 6 shows the results of each machine learning classifier for the patients in Groups 3 and 4. Among all the trained classifiers, QDA yielded the highest accuracy, correctly identifying 11 out of 15 cases in Group 3 as having a clinically significant and 7 out of 10 in Group 4 as not having clinically significant PCa. For the three machine learning models used (QDA, SVM, and RFs), an overall accuracy improvement of 80, 60, 33 % and 60, 40, 30 % for Groups 3 and 4, respectively, was observed when compared to PIRADS alone.
Table 6.
Description of the validation results for patient Groups 3 and 4 using PIRADS v2.0
| Group | Type | Total | PIRADS v2 scores |
QDA maps |
SVM maps |
RF maps |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1,2 | 4,5 | − | + | − | + | − | + | |||
| 3 | MRI-negative-Biopsy-positive | 15 | 15 | 0 | 3 | 12 | 6 | 9 | 10 | 5 |
| 4 | MRI-positive-Biopsy-negative | 10 | 0 | 10 | 6 | 4 | 4 | 6 | 3 | 7 |
The top radiomic features identified to be associated with clinically significant disease on MRI were Gabor, Haralick and first-order statistic features, all of which showed differential expression between the biopsy-positive and biopsy-negative regions in Groups 3 and 4.
Figure 6 shows the output probability maps of the QDA classifier before and after de-noising using MRF for patients in Groups 3 and 4. Detection accuracy for the MRI-negative-biopsy-positive patients was not largely different between classifiers, between 9 to 11 out of the 15 cases of Group 3 were correctly identified. However, only between 4 to 7 out of 10 cases of Group 4 were correctly identified by the radiomic classifiers (Table 6).
Figure 6.
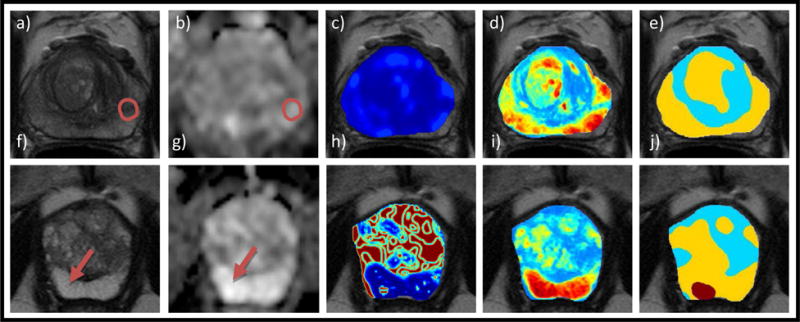
Top: MRI-negative-biopsy-positive case indicating correctly detected tumor presence: (a) PCa-positive region reported from biopsy indicated by red arrow on T2w image, (b) ADC map, (c) Gabor feature showing no differential expression within the prostate region (d) Computed heatmap with red representing high PCa probability and blue representing low PCa probability, (e) Denoised PCa heatmap segmented by Markov random fields with red for high, yellow for low, and cyan for very-low likelihood of disease occurrence, respectively. Bottom: MRI-positive-biopsy-negative case indicating correctly detected tumor absence: (f) T2w image with red region denoting the “erroneous” PIRADS-based annotation, (g) ADC map, (h) Haralick feature showing a high PCa probability spot although bright on ADC (i) Computed heatmap of PCa probability, (j) Denoised PCa heatmap with cyan representing low significance which shows no clear signature for PCa in this region (agreeing with biopsy).
DISCUSSION
In this study, we identified and evaluated the role of Radiomics on biparametric-MRI (bpMRI) for prostate cancer (PCa) characterization in patients on active surveillance (AS). Previous studies, which focused on Radiomics-based characterization of PCa, have typically employed cases where men were either sent to surgery (i.e. higher grade and higher stage disease) or where the histopathologic findings have been concordant with the imaging interpretations (i.e. PIRADS scores)6,12–14. Our work, on the other hand, focuses on the more radiographically confounding cases in patients undergoing AS where there was discordance between PIRADS and histopathology assessments of tumors.
Our experiments were designed to determine if radiomic features could be beneficial in identifying clinically significant disease, in cases where there was ambiguity between PIRADS scores and biopsy findings of clinically significant PCa. Specifically, we sought to identify the added contribution of radiomic features in the context of those MRI scans where (a) there was clinically significant disease and PIRADS scores were low (MRI-negative-biopsy-positive), and where (b) there was no clinically significant disease and PIRADS scores were high (MRI-positive-biopsy-negative).
According to PIRADS v2 guidelines9, scores of 4 or 5 are assigned to circumscribed, homogenous, moderately hypointense focuses/masses. However, a mixture of signal intensities may occur due to benign prostatic hyperplasia (BPH) nodules; predominantly stromal nodules that exhibit hypointensity on T2w images, especially if they are not clearly encapsulated. Decreased signals can also result from prostatitis, which is a known confounder for low grade PCa13. Radiology-based interpretation is known to be severely affected in scenarios where these benign confounders are not well circumscribed, or they exhibit hypointensity either on T2w images or ADC maps. Although this work focused only on clinically significant PCa, we posit that cases incorrectly identified by PIRADS as clinically significant disease in Group 4 appeared to have been on account of the presence of benign confounders (i.e. benign hyperplasia (BPH)).
We identified a set of ten radiomic features that had statistically significant differential expression between normal and clinically significant PCa regions for AS patients. These features were primarily steerable Gabor and co-occurrence Haralick texture features. By modeling localized frequency characteristics, Gabor wavelets appeared to capture differences between homogeneous texture of tumors and normal tissue when both pathologies were hypointense on T2w images. On the other hand, Haralick features appeared to pick the heterogeneity within lesions and differentiate normal hyper- from cancer hypointense regions when lesions were not clearly encapsulated. Interestingly, Ginsburg et al14 identified similar features in distinguishing PCa from benign prostate tissue, in patients who underwent radical prostatectomy.
Out of the 10 identified radiomic features, only three were extracted from ADC maps, while seven were extracted from T2w images. T2w signal intensity and ADC values were not highly ranked. All the 7 T2w-extracted radiomic features were highly ranked (1-7) by MRMR when compared to the ADC-extracted attributes (8-10). Although a substantial number of recent publications reference the value of diffusion weighted imaging for PCa grading23–25, our findings might be explained by the lower initial resolution of the available ADC maps compared to the corresponding T2w scans.
From among the three machine learning classifiers employed, QDA was identified as the best performing. QDA identified 80 % of the cases in Group 3 as having clinically significant PCa. Similarly, for cases in Group 4, QDA correctly identified 60 % of the patients as not having a focus of clinically significant disease. The classifier in conjunction with the Gabor and Haralick features appeared to distinguish benign tumor confounders from PCa, better than radiologic interpretations based off PIRADS v2 for patients in Group 4. We do acknowledge that this suggestion is somewhat speculative, since histopathologic confirmation for the false positive errors on account of PIRADS v2 was not available.
It was also found that the larger the tumor volume, the more likely it was to be identified on MRI. As 75 % of tumors in our cohort were < 1 cm3 in volume, most of them were missed on PIRADS assessment despite being clinically significant. Nevertheless, 70% of them were detected by the Radiomics-based QDA classifier. In terms of tumor locations, only 3 cases in Group 3 had tumors in the transition zone while the rest were in the peripheral zone of the prostate. Our QDA classifier correctly identified 100% of the tumors in the peripheral zone and 67% of the tumors in the transition zone. Transition zone tumors are notoriously more difficult to identify compared to peripheral zone tumors26. Additionally, the transition zone tumors in our study had an average volume of 1.1 cm3 compared to 1.4 cm3 for the peripheral zone tumors.
Our study did have its limitations. Firstly, we deliberately excluded cases with PIRADS score 3, since a consensus of interpretation could not be established between readers for these cases. Lesions assigned PIRADS score of 3 (indeterminate) pose a major clinical management challenge5,7,27,28. An additional improvement would also be to turn our Radiomics-based classifier into a spatially-aware one. Previous studies have shown that radiomic features used for prostate cancer detection differ between transition (TZ) and peripheral zones (PZ)29,30.
However, for this study we found that the majority of cases comprised PZ lesions (only 3 TZ were found out of 56 cases (5 %)), and since they were only in the test set, we had no training data to construct spatially-aware machine learning classifiers on. One of our future directions is training spatially-aware classifiers once we acquire balanced cohorts with lesions in both PZ and TZ.
A second limitation of this study was the limited cohort size (N = 56) and the fact that all the cases came from a single healthcare institution. In future work we will seek to further validate our findings and conclusions from this preliminary study on a larger population drawn within a multi-site setting with different scanners and vendor platforms.
A bottleneck for this procedure was the manual segmentation of the prostate capsules and benign lesions on MR images by the expert radiologist. There has been a lot of published work on automated segmentation of prostate capsule31–34, prostate zones35–38 and the individual lesions39. One of the avenues for future work will be to automate the entire processing pipeline. Another possible future direction will be to investigate correlations between the radiomic findings on the biopsy-naïve MRI with histopathologic findings on the surgical specimens; since it is well known that stage and Gleason scores can be substantially discordant between the initial diagnostic biopsy and surgically resected specimens27,40.
In conclusion, radiomic features were identified on biopsy-naïve T2w and DW MRI that were found to be associated with the presence of clinically important PCa. On a limited number of patients, these features appeared to be more relevant compared to the PIRADS v2 assessment of the tumors. Our findings appear to suggest a role for Radiomics-based assessment of clinically significant prostate cancer for patients on active surveillance and being followed via MRI. Employing Radiomics with MRI in patient management could potentially (1) improve risk stratification which could help in identifying which patients are candidates for active surveillance and (2) could allow for non-invasive monitoring of disease grade and stage for patients already on active surveillance, obviating the need for annual biopsies.
Acknowledgments
Grant Support: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers 1U24CA199374-01, R01CA202752-01A1, R01CA208236-01A1, R21CA179327-01, R21CA195152-01, and the DOD Peer Reviewed Cancer Research Program W81XWH-16-1-0329
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The U.S. Army Medical Research Acquisition Activity, 820 Chandler Street, Fort Detrick MD 21702-5014 is the awarding and administering acquisition office. This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs, through the Peer Reviewed Cancer Research Program, under Award No. W81XWH-16-1-0329. Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense.
References
- 1.Moore CM, Allen C, Emberton M. The Role of MRI in Active Surveillance. In: Klotz L, editor. Act Surveill Localized Prostate Cancer. Totowa, NJ: Humana Press; 2012. pp. 67–80. Available from: http://link.springer.com/10.1007/978-1-61779-912-9_6. [Google Scholar]
- 2.Recabal P, Ehdaie B. The role of MRI in active surveillance for men with localized prostate cancer. Curr Opin Urol. 2015 Nov;25(6):504–509. doi: 10.1097/MOU.0000000000000221. [DOI] [PubMed] [Google Scholar]
- 3.Somford DM, Hoeks CM, Hulsbergen-van de Kaa CA, Hambrock T, Fütterer JJ, Witjes JA, Bangma CH, Vergunst H, Smits GA, Oddens JR, van Oort IM, Barentsz JO. Evaluation of Diffusion-Weighted MR Imaging at Inclusion in an Active Surveillance Protocol for Low-Risk Prostate Cancer. Invest Radiol. 2013 Mar;48(3):152–157. doi: 10.1097/RLI.0b013e31827b711e. [DOI] [PubMed] [Google Scholar]
- 4.Chen RC, Rumble RB, Loblaw DA, Finelli A, Ehdaie B, Cooperberg MR, Morgan SC, Tyldesley S, Haluschak JJ, Tan W, Justman S, Jain S. Active Surveillance for the Management of Localized Prostate Cancer (Cancer Care Ontario Guideline): American Society of Clinical Oncology Clinical Practice Guideline Endorsement. J Clin Oncol. 2016 Jun 20;34(18):2182–2190. doi: 10.1200/JCO.2015.65.7759. [DOI] [PubMed] [Google Scholar]
- 5.Liddell H, Jyoti R, Haxhimolla HZ. mp-MRI Prostate Characterised PIRADS 3 Lesions are Associated with a Low Risk of Clinically Significant Prostate Cancer - A Retrospective Review of 92 Biopsied PIRADS 3 Lesions. Curr Urol. 2014;8(2):96–100. doi: 10.1159/000365697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fehr D, Veeraraghavan H, Wibmer A, Gondo T, Matsumoto K, Vargas HA, Sala E, Hricak H, Deasy JO. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci. 2015 Nov 17;112(46):E6265–E6273. doi: 10.1073/pnas.1505935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson JE, van Leeuwen PJ, Moses D, Shnier R, Brenner P, Delprado W, Pulbrook M, Böhm M, Haynes AM, Hayen A, Stricker PD. The Diagnostic Performance of Multiparametric Magnetic Resonance Imaging to Detect Significant Prostate Cancer. J Urol. 2016 May;195(5):1428–1435. doi: 10.1016/j.juro.2015.10.140. [DOI] [PubMed] [Google Scholar]
- 8.Schimmöller L, Quentin M, Arsov C, Hiester A, Buchbender C, Rabenalt R, Albers P, Antoch G, Blondin D. MR-sequences for prostate cancer diagnostics: validation based on the PI-RADS scoring system and targeted MR-guided in-bore biopsy. Eur Radiol. 2014 Oct;24(10):2582–2589. doi: 10.1007/s00330-014-3276-9. [DOI] [PubMed] [Google Scholar]
- 9.Weinreb JC, Barentsz JO, Choyke PL, Cornud F, Haider MA, Macura KJ, Margolis D, Schnall MD, Shtern F, Tempany CM, Thoeny HC, Verma S. PI-RADS Prostate Imaging – Reporting and Data System: 2015, Version 2. Eur Urol. 2016 Jan;69(1):16–40. doi: 10.1016/j.eururo.2015.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schimmöller L, Quentin M, Arsov C, Lanzman RS, Hiester A, Rabenalt R, Antoch G, Albers P, Blondin D. Inter-reader agreement of the ESUR score for prostate MRI using in-bore MRI-guided biopsies as the reference standard. Eur Radiol. 2013 Nov;23(11):3185–3190. doi: 10.1007/s00330-013-2922-y. [DOI] [PubMed] [Google Scholar]
- 11.Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep. 2015 Oct;5(1) doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Wu C-J, Bao M-L, Zhang J, Wang X-N, Zhang Y-D. Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. Eur Radiol. 2017 Oct;27(10):4082–4090. doi: 10.1007/s00330-017-4800-5. [DOI] [PubMed] [Google Scholar]
- 13.Litjens GJS, Elliott R, Shih NN, Feldman MD, Kobus T, Hulsbergen-van de Kaa C, Barentsz JO, Huisman HJ, Madabhushi A. Computer-extracted Features Can Distinguish Noncancerous Confounding Disease from Prostatic Adenocarcinoma at Multiparametric MR Imaging. Radiology. 2016 Jan;278(1):135–145. doi: 10.1148/radiol.2015142856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginsburg SB, Algohary A, Pahwa S, Gulani V, Ponsky L, Aronen HJ, Boström PJ, Böhm M, Haynes A-M, Brenner P, Delprado W, Thompson J, Pulbrock M, Taimen P, Villani R, Stricker P, Rastinehad AR, Jambor I, Madabhushi A. Radiomic features for prostate cancer detection on MRI differ between the transition and peripheral zones: Preliminary findings from a multi-institutional study: Radiomic Features for Prostate Cancer Detection on MRI. J Magn Reson Imaging. 2017 Jul;46(1):184–193. doi: 10.1002/jmri.25562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juntu J, Sijbers J, Dyck D, Gielen J. Bias Field Correction for MRI Images. In: Kurzyński M, Puchała E, Woźniak M, Żołnierek A, editors. Comput Recognit Syst. Berlin, Heidelberg: Springer Berlin Heidelberg; 2005. pp. 543–551. Available from: http://link.springer.com/10.1007/3-540-32390-2_64. [Google Scholar]
- 16.Palumbo D, Yee B, O’Dea P, Leedy S, Viswanath S, Madabhushi A. Interplay between bias field correction, intensity standardization, and noise filtering for T2-weighted MRI. IEEE. 2011:5080–5083. doi: 10.1109/IEMBS.2011.6091258. Available from: http://ieeexplore.ieee.org/document/6091258/ [DOI] [PubMed]
- 17.Auffarth B, López M, Cerquides J. Comparison of Redundancy and Relevance Measures for Feature Selection in Tissue Classification of CT Images. In: Perner P, editor. Adv Data Min Appl Theor Asp. Berlin, Heidelberg: Springer Berlin Heidelberg; 2010. pp. 248–262. Available from: http://link.springer.com/10.1007/978-3-642-14400-4_20. [Google Scholar]
- 18.Szekely GJ, Rizzo ML. Hierarchical Clustering via Joint Between-Within Distances: Extending Ward’s Minimum Variance Method. J Classif. 2005 Sep;22(2):151–183. doi: 10.1007/s00357-005-0012-9. [DOI] [Google Scholar]
- 19.Monaco J, Tomaszewski JE, Feldman MD, Moradi M, Mousavi P, Boag A, Davidson C, Abolmaesumi P, Madabhushi A. Pluim JPW, Dawant BM, editors. Probabilistic pairwise Markov models: application to prostate cancer detection. 2009:725903. doi: 10.1117/12.812462. Available from: http://proceedings.spiedigitallibrary.org/proceeding.aspx?doi=10.1117/12.812462. [DOI] [PMC free article] [PubMed]
- 20.Kato Z, Pong T-C. A Markov random field image segmentation model for color textured images. Image Vis Comput. 2006 Oct;24(10):1103–1114. doi: 10.1016/j.imavis.2006.03.005. [DOI] [Google Scholar]
- 21.Barentsz JO, Richenberg J, Clements R, Choyke P, Verma S, Villeirs G, Rouviere O, Logager V, Fütterer JJ. ESUR prostate MR guidelines 2012. Eur Radiol. 2012 Apr;22(4):746–757. doi: 10.1007/s00330-011-2377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan EJ, Fiske C, Zagoria RJ, Westphalen AC. Evaluating the performance of PI-RADS v2 in the non-academic setting. Abdom Radiol. 2017 Nov;42(11):2725–2731. doi: 10.1007/s00261-017-1169-5. [DOI] [PubMed] [Google Scholar]
- 23.Nezzo M, Di Trani MG, Caporale A, Miano R, Mauriello A, Bove P, Capuani S, Manenti G. Mean diffusivity discriminates between prostate cancer with grade group 1&2 and grade groups equal to or greater than 3. Eur J Radiol. 2016 Oct;85(10):1794–1801. doi: 10.1016/j.ejrad.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Helfrich O, Puech P, Betrouni N, Pinçon C, Ouzzane A, Rizk J, Marcq G, Randazzo M, Durand M, Lakroum S, Leroy X, Villers A. Quantified analysis of histological components and architectural patterns of gleason grades in apparent diffusion coefficient restricted areas upon diffusion weighted MRI for peripheral or transition zone cancer locations: Low ADC and Malignant Glands Density. J Magn Reson Imaging. 2017 Dec;46(6):1786–1796. doi: 10.1002/jmri.25716. [DOI] [PubMed] [Google Scholar]
- 25.Waseda Y, Yoshida S, Takahara T, Kwee TC, Matsuoka Y, Saito K, Kihara K, Fujii Y. Utility of computed diffusion-weighted MRI for predicting aggressiveness of prostate cancer: cDWI for Predicting Aggressiveness. J Magn Reson Imaging. 2017 Aug;46(2):490–496. doi: 10.1002/jmri.25593. [DOI] [PubMed] [Google Scholar]
- 26.Ginsburg SB, Viswanath SE, Bloch BN, Rofsky NM, Genega EM, Lenkinski RE, Madabhushi A. Novel PCA-VIP scheme for ranking MRI protocols and identifying computer-extracted MRI measurements associated with central gland and peripheral zone prostate tumors: Quantitative MRI Features of CG and PZ Tumors. J Magn Reson Imaging. 2015 May;41(5):1383–1393. doi: 10.1002/jmri.24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babaian RJ, Grunow WA. Reliability of Gleason grading system in comparing prostate biopsies with total prostatectomy specimens. Urology. 1985 Jun;25(6):564–567. doi: 10.1016/0090-4295(85)90280-8. [DOI] [PubMed] [Google Scholar]
- 28.Steiger P, Thoeny HC. Prostate MRI based on PI-RADS version 2: how we review and report. Cancer Imaging. 2016 Dec;16(1) doi: 10.1186/s40644-016-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viswanath S, Bloch BN, Genega E, Rofsky N, Lenkinski R, Chappelow J, Toth R, Madabhushi A. A comprehensive segmentation, registration, and cancer detection scheme on 3 Tesla in vivo prostate DCE-MRI. Med Image Comput Comput-Assist Interv MICCAI Int Conf Med Image Comput Comput-Assist Interv. 2008;11(Pt 1):662–669. doi: 10.1007/978-3-540-85988-8_79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viswanath SE, Bloch NB, Chappelow JC, Toth R, Rofsky NM, Genega EM, Lenkinski RE, Madabhushi A. Central gland and peripheral zone prostate tumors have significantly different quantitative imaging signatures on 3 tesla endorectal, in vivo T2-weighted MR imagery. J Magn Reson Imaging. 2012 Jul;36(1):213–224. doi: 10.1002/jmri.23618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Luca S, Passera R, Cattaneo G, Manfredi M, Mele F, Fiori C, Bollito E, Cirillo S, Porpiglia F. High prostate cancer gene 3 (PCA3) scores are associated with elevated Prostate Imaging Reporting and Data System (PI-RADS) grade and biopsy Gleason score, at magnetic resonance imaging/ultrasonography fusion software-based targeted prostate biopsy after. BJU Int. 2016 Nov;118(5):723–730. doi: 10.1111/bju.13504. [DOI] [PubMed] [Google Scholar]
- 32.Toth R, Madabhushi A. Multifeature Landmark-Free Active Appearance Models: Application to Prostate MRI Segmentation. IEEE Trans Med Imaging. 2012 Aug;31(8):1638–1650. doi: 10.1109/TMI.2012.2201498. [DOI] [PubMed] [Google Scholar]
- 33.Toth R, Tiwari P, Rosen M, Reed G, Kurhanewicz J, Kalyanpur A, Pungavkar S, Madabhushi A. A magnetic resonance spectroscopy driven initialization scheme for active shape model based prostate segmentation. Med Image Anal. 2011 Apr;15(2):214–225. doi: 10.1016/j.media.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reda I, Shalaby A, Elmogy M, Aboulfotouh A, Khalifa F, El-Ghar MA, Gimelfarb G, El-Baz A. Image-Based Computer-Aided Diagnostic System for Early Diagnosis of Prostate Cancer. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W, editors. Med Image Comput Comput-Assist Interv – MICCAI 2016. Cham: Springer International Publishing; 2016. pp. 610–618. Available from: http://link.springer.com/10.1007/978-3-319-46720-7_71. [Google Scholar]
- 35.Reda I, Shalaby A, Khalifa F, Elmogy M, Aboulfotouh A, El-Ghar MA, Hosseini-Asl E, Werghi N, Keynton R, El-Baz A. Computer-aided diagnostic tool for early detection of prostate cancer. IEEE. 2016:2668–2672. doi: 10.1109/ICIP.2016.7532843. Available from: http://ieeexplore.ieee.org/document/7532843/ [DOI]
- 36.Clark T, Zhang J, Baig S, Wong A, Haider MA, Khalvati F. Fully automated segmentation of prostate whole gland and transition zone in diffusion-weighted MRI using convolutional neural networks. J Med Imaging. 2017 Oct 17;4(04):1. doi: 10.1117/1.JMI.4.4.041307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egger J. PCG-Cut: Graph Driven Segmentation of the Prostate Central Gland. In: Culig Z, editor. PLoS ONE. 10. Vol. 8. 2013. Oct 11, p. e76645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toth R, Ribault J, Gentile J, Sperling D, Madabhushi A. Simultaneous segmentation of prostatic zones using Active Appearance Models with multiple coupled levelsets. Comput Vis Image Underst. 2013 Sep;117(9):1051–1060. doi: 10.1016/j.cviu.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Litjens G, Debats O, van de Ven W, Karssemeijer N, Huisman H. A Pattern Recognition Approach to Zonal Segmentation of the Prostate on MRI. In: Ayache N, Delingette H, Golland P, Mori K, editors. Med Image Comput Comput-Assist Interv – MICCAI 2012. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012. pp. 413–420. Available from: http://link.springer.com/10.1007/978-3-642-33418-4_51. [DOI] [PubMed] [Google Scholar]
- 40.Mills SE, Fowler JE. Gleason histologic grading of prostatic carcinoma. Correlations between biopsy and prostatectomy specimens. Cancer. 1986 Jan 15;57(2):346–349. doi: 10.1002/1097-0142(19860115)57:2<346::aid-cncr2820570226>3.0.co;2-e. doi: 10.1002/1097-0142(19860115)57:2<346::AID-CNCR2820570226>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]


