Abstract
Increasing evidence indicates that human cytomegalovirus (HCMV) populations under the influence of host environment, can either be stable or rapidly differentiating, leading to tissue compartment colonization. We isolated previously from a 30-years old pregnant woman, a clinical isolate of HCMV, that we refered to as the HCMV-DB strain (accession number KT959235). The HCMV-DB clinical isolate demonstrated its ability to infect primary macrophages and to upregulate the proto-oncogene Bcl-3. We observed in this study that the genome of HCMV-DB strain is close to the genomes of other primary clinical isolates including the Toledo and the JP strains with the later having been isolated from a glandular tissue, the prostate. Using a phylogenetic analysis to compare the genes involved in virus entry, we observed that the HCMV-DB strain is close to the HCMV strain Merlin, the prototype HCMV strain. HCMV-DB infects human mammary epithelial cells (HMECs) which in turn display a ER−/PR−/HER2− phenotype, commonly refered to as triple negative. The transcriptome of HCMV-DB-infected HMECs presents the characteristics of a pro-oncogenic cellular environment with upregulated expression of numerous oncogenes, enhanced activation of pro-survival genes, and upregulated markers of cell proliferation, stemcellness and epithelial mesenchymal transition (EMT) that was confirmed by enhanced cellular proliferation and tumorsphere formation in vitro. Taken together our data indicate that some clinical isolates could be well adapted to the mammary tissue environment, as it is the case for the HCMV-DB strain. This could influence the viral fitness, ultimately leading to breast cancer development.
Introduction
Breast cancer, the most common cancer diagnosed among women, exhibits heterogeneous molecular characteristics. Several types of breast cancer have been identified based on the differential gene expression patterns. These groups include among others the normal breast epithelial-like, the luminal epithelial type A and type B, the basal-like and the claudin low groups1. Genetic risk factors and environmental risk factors are some of the etiologic factors involved in breast cancer2. Of all worldwide cancers close to one-fifth could involve infectious agents including viruses, as part of the environmental risk factors3–5.
The Betaherpesviridae human cytomegalovirus (HCMV) is know to cause, in the immunocompetent host, an infection that usually range from asymptomatic to mild. Serious complications can however be the result of the infection of immunocompromised hosts6. In opposition to laboratory strains of HCMV, which growth appears to be restricted to fibroblasts only, clinical isolates are able to infect and grow in several types of cells including epithelial cells, endothelial cells, monocytes, macrophages, fibroblasts, stromal cells, hepatocytes, smooth muscle cells, and neural stem/progenitor cells7–10. Recently, a detailed in vivo evolutionary map of HCMV was built by combining population genetics methods and high throughput sequencing. This map provided evidence that viral populations under the influence of host environment can either be stable or rapidly differentiating, leading to tissue compartment colonization11.
Several research groups focusing on inflammatory diseases and on cancer addressed the role played by HCMV in these diseases12–14. Tumor tissues from several cancers, including brain, colon, prostate, liver and breast cancer, have been found positive for HCMV DNA or antigens15–20. In the paradigm of oncomodulation, oncogenesis mechanisms could be amplified by HCMV acting as a cofactor, when infecting the tumor tissue21. In addition, monocytes and macrophages, respectively located in the blood and in the tissues, may act as sites for the establishment of latency, given their role as HCMV cellular reservoirs which are responsible for the dissemination of the virus8,22–24. Finally, in breast carcinomas and glioblastomas, tumor-associated macrophages (TAM) are a marker of poor prognosis, and their development might be influenced by macrophage-tropic HCMV strains25–28. Thus the quest of new HCMV isolates which target monocytes/macrophages and thereby might play a role in oncogenesis has shown increased interest.
In the present study, we found that the genomic sequence of the HCMV-DB strain isolated from a cervical swab specimen is close to the sequence of other primary clinical isolates, especially the Toledo strain, originally isolated in the urine of a child presenting a congenital infection by HCMV, and the JP strain, isolated from a glandular tissue, the prostate21,29. Based on the analysis of genes involved in virus entry, our results indicate that the HCMV-DB strain is close to the Merlin strain. Given the scarcity of evidence for a direct role of HCMV in the cellular transformation of epithelial cells, we studied the transcriptome profile of HCMV-DB infected human mammary epithelial cells (HMECs). We observed that the transcriptome of HCMV-DB infected HMECs displays a triple negative ER−/PGR−/HER2− phenotype, presents some oncogenic traits, favors cell cycling and cell proliferation, and modulate angiogenesis and proteolysis. All these phenomena are potentially involved in tumor development. Finally, the infection of HMECs with the HCMV strain DB resulted in enhanced proliferation and tumorsphere formation in vitro.
Results
Genomic profile of the HCMV-DB strain
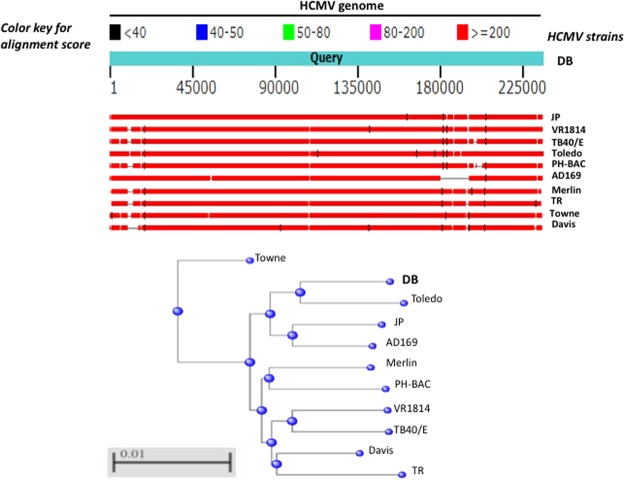
We previously isolated a novel HCMV strain, that we characterized and named HCMV-DB8. We compared its genomic sequence to that of ten HCMV strains, including two laboratory adapted strains (AD169, Towne) and eight clinical isolates with low passages in culture (Merlin, Toledo, TR, PH, VR1814, Davis, JP, TB40/E)30,31. The complete genomic sequence of HCMV-DB has a total length of 235,512 bp (Fig. 1 and Table 1). In comparison the sequence of the Merlin strain has approximately the same size (235,645 bp), the laboratory adapted strain AD169 has the shortest sequence (229,354 bp) and the clinical strain TB40/E the longest sequence (237,683 bp) (Fig. 1 and Table 1). Clinical isolates with low passages in culture have a total length ranging from 229,700 bp for PH to 237,683 bp for TB40/E (Fig. 1 and Table 1).
Figure 1.
Comparison of the genomic sequence of HCMV-DB with the genomic sequences of other clinical and laboratory adapted HCMV strains. The upper panel shows the genomic sequence of HCMV-DB as compared to two laboratory adapted strains (AD169 and Towne) and eight low passages clinical isolates (Merlin, Toledo, TR, PH, VR1814, Davis, JP, and TB40/E). The lower panel shows the alignment tree related to the whole viral genome with comparison between HCMV-DB genome and the genome of ten other HCMV strains.
Table 1.
Genomic comparison of HCMV-DB with other HCMV strains.
| HCMV strain | Clinical | Geographical | DNA | Passages | Nucleotide length | Genes Mutated | Max. score (e + 05) | Total score (e + 05) | Query cover | Identity | Accession |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DB | Pregnant women | France | Clinical material on macrophage cultures |
low passage on fibroblasts | 235512 | RL13, UL9 | 100 | 100 | KT959235 | ||
| Towne | Urine from a congenitally infected enfant | USA | Fibroblast culture cells | many | 235147 | many: RL13, UL1, UL40, UL130, US1 | 1.316 | 3.929 | 95 | 99 | FJ616285 |
| AD169 | Adenoid tissue | USA | Fibroblast culture | many | 229354 | 1.238 | 4.036 | 92 | 98 | X17403.1 | |
| DAVIS | Liver biopsy from a congenitally infected infant | USA | Fibroblast culture | many | 229768 | RL5A, RL12, RL13, UL1, UL2, UL4, UL5, UL6, UL99, UL130 | 1.29 | 3.936 | 95 | 98 | JX512198.1 |
| JP | Post mortem prostate tissue from an AIDS patient | UK | Clinical material | low passage on fibroblasts | 236375 | 2.838 | 4.129 | 98 | 99 | GQ221975.1 | |
| Merlin | Urine from a congenitally infected infant | UK | Clinical material | low passage on fibroblasts | 235646 | RL13 | 2.841 | 4.034 | 97 | 98 | AY446894 |
| PH | Bone marrow transplant recipient | USA | Clinical material | low passage on fibroblasts | 229700 | 1.545 | 3.856 | 95 | 98 | AC146904 | |
| TB40/E | Throat wash of a bone marrow transplant recipient | Germany | Fibroblast culture | Few | 237683 | RL13, UL128, IRS1, US1, US2 | 1.555 | 3.962 | 96 | 98 | KF297339 |
| Toledo | Urine from a congenitally infected infant | USA | Fibroblast culture | several | 235404 | RL13, UL9, UL128 | 1.548 | 4.171 | 99 | 98 | GU937742 |
| TR | Vitreous humor from eye of HIV-positive male | USA | Fibroblasts | several | 235681 | 1.539 | 3.955 | 96 | 98 | KF021605.1 | |
| VR1814 (FIX) | Cervical secretions of a pregnant woman with a primary HCMV infection | Italy | Clinical material | low passage on fibroblasts | 235233 | 1.564 | 3.995 | 97 | 98 | GU179289 |
Based on the sequence similarity between HCMV strains, the highest total scores by Blast were measured for JP and Toledo strains when compared to the DB strain (Table 1). The DB strain shares around 99% identity with JP and Toledo strains, by linear full genome alignment (Table 1). The other clinical isolates analyzed share 98% identity with the DB strain (Table 1). HCMV-DB strain is mutated in RL13 and UL9 genes, but not in the ULb’ region (Table 1). Altogether our analyses indicate that the genomic sequence of HCMV-DB is highly similar to the genomic sequences of the clinical strains JP and Toledo.
Phylogenetic classification of the HCMV-DB strain based on genes involved in virus entry
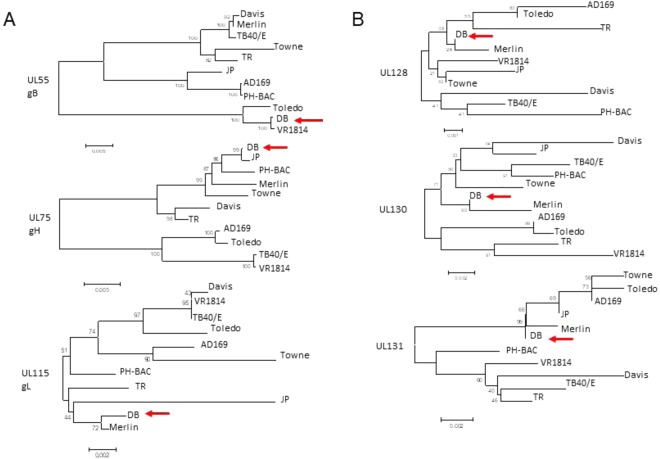
Since HCMV-DB is a highly macrophage-tropic strain8, we performed a phylogenetic analysis to compare the genes coding for the viral envelop that are involved in tropism (Fig. 2, Suppl. Table 1)32–34. For entry into fibroblasts, HCMV only requires the minimal complex of gB:gH/gL32,35. For entry into monocytes, macrophages, epithelial cells, and endothelial cells, HCMV requires, in addition to gB, a pentameric complex formed by glycoproteins gH and gL and proteins pUL128, pUL130 and pUL1317,36–41. We observed that the gB:gH/gL sequences from DB are close to those of VR1814, JP and Merlin, respectively (Fig. 2A). The HCMV-DB pentameric complex was close to that of JP for gH, and to that of Merlin for gL, UL128, UL130 and UL131 (Fig. 2A,B). For gB genotype, the DB strain is close to the VR1814 strain (Fig. 2A). Taking into account the sequences of the genes involved in virus entry, altogether our phylogenetic analysis indicates that the HCMV-DB strain is close to the Merlin strain. The UL144 gene, situated in the ULb’ region of the viral genome, is commonly used for genotyping HCMV and we previously reported that HCMV-DB belongs to the UL144 genotype C29.
Figure 2.
Phylogenetic analyses comparing HCMV-DB with several HCMV strains for genes involved in viral entry. (A) Phylogenetic analysis of genes coding for viral glycoproteins required for the entry into cells: UL55 (gB), UL75 (gH), and UL115 (gL). (B) Phylogenetic analysis of genes coding for UL128, UL130, and UL131 required in addition to the envelop glycoproteins for the viral entry into monocytes, macrophages, epithelial cells, and endothelial cells.
HCMV-DB infects HMECs in vitro
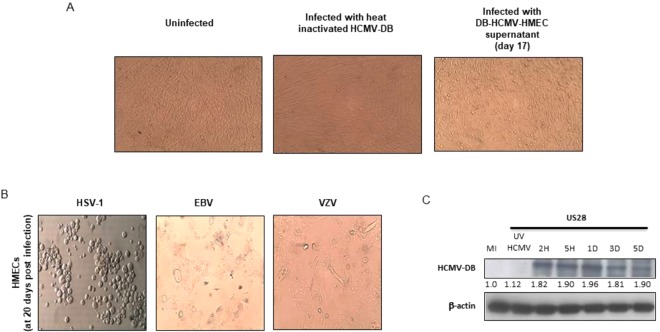
To determine whether HMECs are permissive to HCMV-DB, HMECs were infected with HCMV-DB. We harvested the supernatants of infected HMECs 17 days post infection and added it to MRC5 cells. We observed a cytopathic effect (CPE) typical of HCMV in MRC5 cultures infected with these supernatants (Fig. 3A). We did not detect any CPE in HMECs directly infected with Epstein-Barr virus (EBV) and varicella zoster virus (VZV) (Fig. 3B). In contrast, direct infection of HMECs with herpes simplex virus type 1 (HSV-1) resulted in the appearance of clusters of round refringent cells (Fig. 3B). To further confirm the direct infection of HMECs by HCMV-DB we detected the early protein pUS28 using western blotting in HCMV-DB infected HMECs after 2 hours and up to 5 days post infection (Fig. 3C). Our results confirm our previously published data that indicate a full replicative viral cycle of HCMV-DB in HMECs29.
Figure 3.
HCMV-DB infects HMECs. (A) HCMV cytopathic effect (CPE) in MRC5 cultures infected with supernatants harvested from HCMV-DB infected HMECs. Heat inactivated HCMV-DB is used as a negative control. (B) Clusters of round cells in HMECs infected with HSV-1. No CPE detected with the direct infection of HMECs with EBV and VZV. (C) Western blotting showing the time-course expression of pUS28 in HCMV-DB infected HMECs.
The transcriptome of HCMV-DB infected HMECs displays a triple negative basal-like phenotype
Uninfected HMECs and HCMV-DB-infected HMECs at low and high MOIs (1 or 10) were used one day after infection for the screening with oncogenes/tumor suppressor genes and human breast cancer genes RT2 profiler PCR assays (PAHS-502Z and PAHS-131Z, respectively). Positive controls included MCF-7 and MDA-MB231 breast cancer cell lines as luminal and basal-like phenotypes, respectively. Each of the RT2 Profiler PCR arrays profiles the expression of 84 key genes -which overlap sometimes in the two assays- commonly involved in tumor classification, signal transduction, and other commonly affected pathways such as angiogenesis, adhesion, proteolysis, cell cycle, and apoptosis (Suppl. Tables 2 and 3).
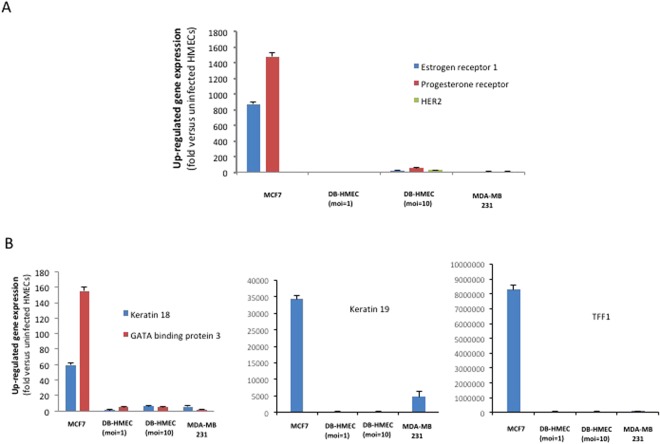
In regard to the expression of ER/PGR/HER2 transcripts, HCMV-DB infected HMECs and MDA-MB231 cells showed a similar transcriptome that was clearly distinct from the transcriptome of MCF-7 cells (Fig. 4A). Although the triple negative ER−/PGR−/HER2− phenotype was present in both infected and uninfected HMECs, the levels of estrogen receptor (ESR1), progesteron receptor (PGR) and HER2 (ERBB2) transcripts were higher in HCMV-DB infected HMECs in comparison to uninfected HMECs (Fig. 4A). The gene expression of luminal markers (KRT19, KRT18, GATA3, TFF1) was low in HCMV-DB-infected HMECs, similar to the phenotype of MDA-MB231 cells (Fig. 4B, Suppl. Tables 2 and 3). In contrast, MCF-7 cells expressed high levels of luminal markers transcripts (Fig. 4B).
Figure 4.
Transcriptome analysis of HMECs infected with HCMV-DB displays a triple negative basal-like phenotype. (A) HMECs infected with HCMV-DB and MDA-MB-231 cells show a similar pattern of ER−/PGR−/HER2− transcripts, in contrast to the ER+/PGR+/HER2− transcripts detected in MCF-7 cells. (B) Low levels of gene expression of luminal markers (KRT18, KRT19, GATA3, TFF1) in both HCMV-DB infected HMECs and MDA-MB-231 cells as compared to the luminal MCF-7 cells.
The transcriptome of HMECs infected with HCMV-DB presents oncogenic traits with enhanced cellular proliferation
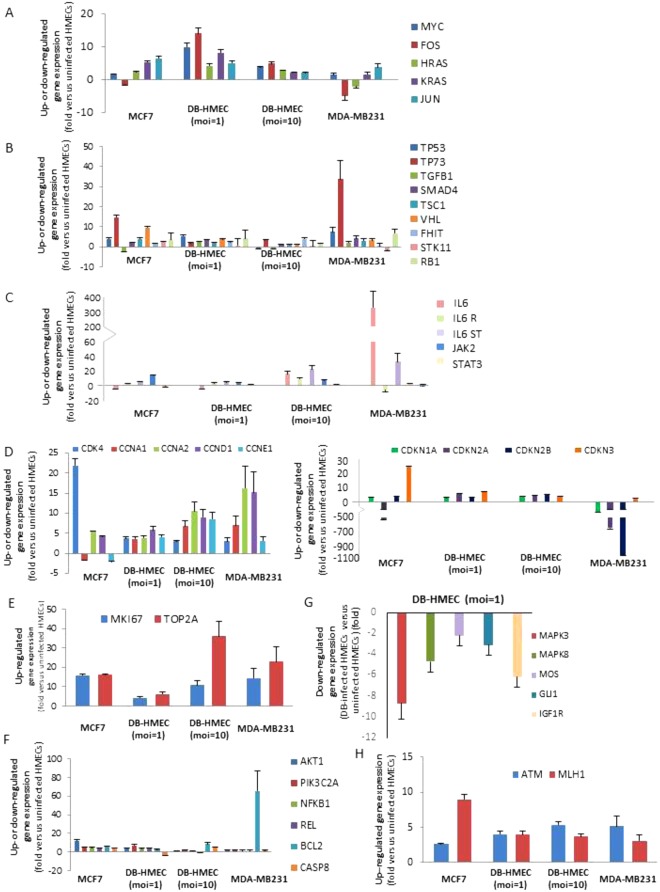
The gene expression of the oncogenes Myc (MYC), Fos (FOS), Jun (JUN), KRas (KRAS), HRas (HRAS) and NRas (NRAS) is upregulated in HCMV-DB-infected HMECs in comparison to uninfected HMECs (Fig. 5A). We also observed the upregulation of transcripts of numerous other oncogenes (KITLG, MCL1, MET, MYB, NFKBIA, PIK3CA, PML, PRKCA, RAF1, RARA, ROS1, RET, ABL1, ETS1, RUNX1, RUNX3) (Suppl. Tables 2 and 3). The gene expression of the tumor suppressor genes coding for the retinoblastoma protein (RB1) and the p53 protein (TP53) was upregulated in HCMV-DB-infected HMECs, as reported previously for basal-like triple negative cell lines such as MDA-MB231 cells (Fig. 5B and Suppl. Table 3)42,43. Similarly, the expression of several other tumor suppressor genes (TP73, FHIT, VHL, SMAD4, TGFB1, STK11, TSC1) is also upregulated in HCMV-DB-infected HMECs, mostly at low moi (moi = 1) compared to uninfected HMECs (Fig. 5B).
Figure 5.
The transcriptome of HMECs infected with HCMV-DB displays oncogenic traits. (A) Upregulation of the gene expression of oncogenes: Myc (MYC), Fos (FOS), Jun (JUN), KRas (KRAS), HRas (HRAS) and NRas (NRAS), (B) Upregulation of the gene expression of tumor suppressor genes: TP53, TP73, FHIT, VHL, SMAD4, TGFB1, STK11, TSC1, RB1. (C) Expression of genes from the IL-6/JAK-STAT3 axis. (D) Left panel, Upregulation of genes coding for the cyclins: CCNA1, CCNA2, CCND1, CCNE1 and the cyclin dependent kinase 4 CDK4. Right panel, Upregulation of genes coding for the cyclin dependent kinase inhibitors: CDKN 1 A, CDKN 2 A, CDKN 2B and CDKN 3. (E) Upregulation of the expression of genes coding for proliferation markers, the Ki67 antigen (MKI67) and the topoisomerase 2 (TOPO2A). (F) Upregulation of the expression of genes involved in cell survival (NFKB1, REL, AKT1, PIK3C2A, BCL-2) and downregulation of the expression of caspase 8 (CASP8). (G) Downregulation of gene expression of the members of the MAPK cascade (MAPK3, MAPK8, MOS, GLI1, IGF1R). (H) Upregulation of the expression of genes involved in DNA reparation: the ataxia telangiectasia mutated (ATM) and human MutL homolog (MLH1). The up- and down-regulation were measured in HMECs infected with HCMV-DB (moi, 1 and 10) as compared to uninfected HMECs.
The IL-6/JAK-STAT3/Cyclin D1 axis is activated in biopsies from breast cancer patients44. We observed the upregulation of IL-6, IL-6 receptor and JAK2 gene expression in HCMV-DB infected HMECs compared to uninfected HMECs especially at high moi (moi = 10), with stable levels of STAT3 gene expression (Fig. 5C). The gene expression of cyclin dependent kinase 4 (CDK4) and of several cyclins (CCNA1, CCNA2, CCND1, CCNE1) especially cyclin D1 which function as regulator of CDKs is up-regulated in HCMV-DB-infected HMECs (moi = 10) compared to uninfected controls (Fig. 5D, left panel). The gene expression of CDK inhibitors (CDKN) 1A (p21), 2A (p16), 2B (p15), and 3 (CDKN1A, CDKN2A, CDKN2B, CDKN3) is up-regulated by 5 folds in HCMV-DB-infected HMECs compared to uninfected cells (Fig. 5D, right panel).
In agreement with activation of the IL6/JAK/cyclin D1 pathway in HCMV-DB-infected HMECs, we observed the upregulation expression of proliferation marker genes such as the Ki67 antigen gene (MKI67) and the topoisomerase 2 gene (TOPO2A) when compared to uninfected HMECs (Fig. 5E). The expression of genes involved in cell survival (NFKB1, REL, AKT1, PIK3C2A, BCL-2) is increased in HCMV-DB-infected HMECs compared to uninfected HMECs, indicating a prosurvival signal in infected cells (Fig. 5F). In agreement with the induction of several prosurvival genes in infected cells compared to uninfected controls, the down-regulation of caspase 8 transcript (CASP8) is observed in HCMV-DB-infected HMECs at moi 1 compared to uninfected cells (Fig. 5F).
The transcriptome of HCMV-DB infected HMECs displays modifications in cell signaling, angiogenesis and proteolysis
The gene expression of the members of the MAPK cascade (MAPK3, MAPK8, MOS, GLI1, IGF1R) is down-regulated in HCMV-DB-infected HMECs (Fig. 5G). The gene expression for the transcription factors, tumor suppressor gene hypermethylated in cancer 1 (HIC1) and forkhead box D3 (FOXD3) is up-regulated in HCMV-DB-infected HMECs at high moi (Suppl. Figure 1). In HCMV-DB-infected HMECs, we observed the upregulation of the expression of the ataxia telangiectasia mutated (ATM) and human MutL homolog (MLH1) genes, both involved in DNA reparation (Fig. 5H).
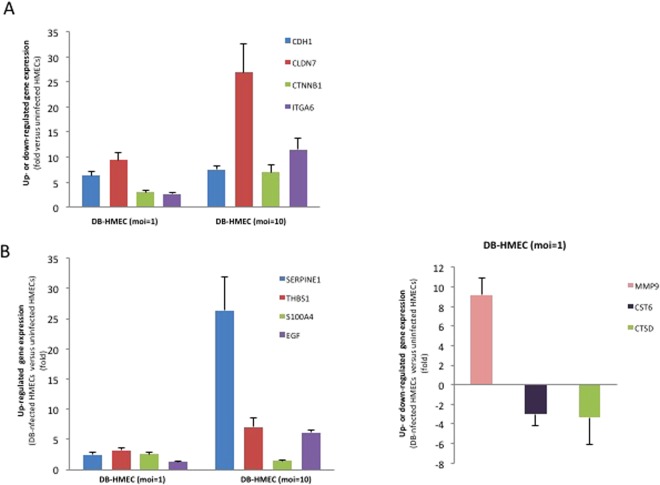
The gene expression of E-cadherin (CDH1), claudin 7 (CLDN7), beta-catenin (CTNNB1) and alpha 6 integrin (ITGA6) is upregulated in HCMV-DB-infected HMECs at high moi (moi = 10) (Fig. 6A). In HCMV-DB-infected HMECs, we observed both the up- and downregulation of genes involved in angiogenesis (upregulation: IL-6, SERPINE1, THBS1, S100A4, EGF; downregulation: SLIT2) and proteolysis (upregulation: MMP9; downregulation: CST6, CTSD) (Fig. 6B, Suppl. Table 3).
Figure 6.
Modification of the transcriptome of genes involved in cell adhesion, angiogenesis and proteolysis in HMECs infected with HCMV-DB. (A) Upregulated gene expression of E-cadherin (CDH1), claudin 7 (CDN7), beta-catenin (CTNNB1), alpha 6 integrin (ITGA6); (B) Upregulated expression of genes involved in angiogenesis (SERPINE1, THBS1, S100A4, and EGF) and proteolysis (MMP9), and downregulated expression of genes involved in proteolysis: CST6 and CTSD. The up- and down-regulation were measured in HMECs infected with HCMV-DB (moi, 1 and 10) as compared to uninfected HMECs.
The transcriptome of HMECs infected with HCMV-DB reveals a global hypomethylation state
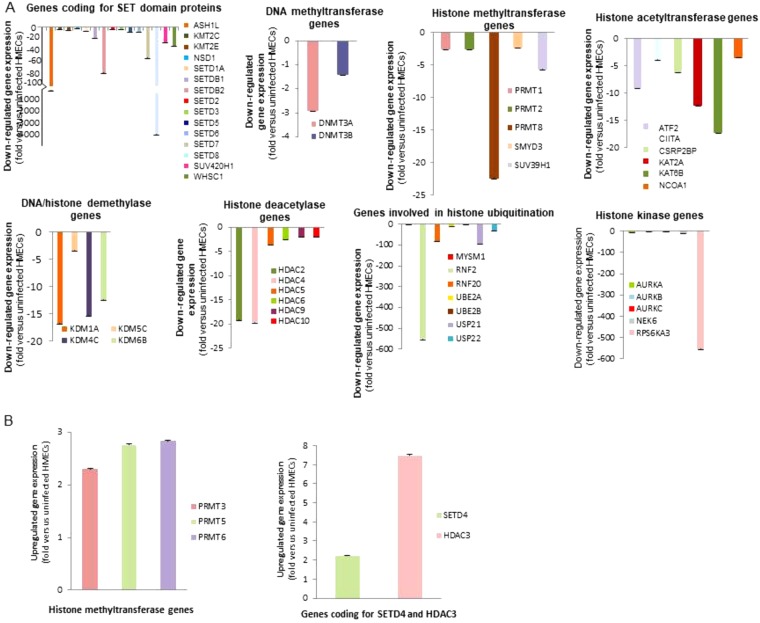
Since epigenetic regulation of chromatin has been associated with breast cancer classification and prognosis, we performed a Human Epigenetic Chromatin Modification Enzymes RT2 Profiler PCR Array (PAHS-085A) which profiles the expression of 84 key genes encoding enzymes known or predicted to regulate chromatin accessibility, and therefore gene expression, by modifying genomic DNA and histones (Suppl. Tables 4 and 5). Considering a standard two-fold increase or decrease of RNA expression between HCMV-DB-infected HMECs and uninfected HMECs to be biologically significant, performed analysis indicated the down-regulation of most (n = 50) of the genes including DNA methyltransferase genes (DNMT3A, DNMT3B), histone methyltransferase genes (PRMT1, PRMT2, PRMT8, SMYD3, SUV39H1), genes coding for SET domain proteins with a histone methyltransferase activity (ASH1L, MLL3 (KMT2C), MLL5 (KMT2E), NSD1, SETD1A, SETDB1, SETDB2, SETD2, SETD3, SETD5, SETD6, SETD7, SETD8, SUV420H1, WHSC1), histone acetyltransferase genes (ATF2, CIITA, CSRP2BP, KAT2A, KAT6B, NCOA1), histone kinase genes (AURKA, AURKB, AURKC, NEK6, RPS6KA3), genes involved in histone ubiquitination (MYSM1, RNF2, RNF20, UBE2A, UBE2B, USP21, USP22), DNA/histone demethylase genes (KDM1A, KDM5C, KDM4C, KDM6B) and histone deacetylase genes (HDAC2, HDAC4, HDAC5, HDAC6, HDAC9, HDAC10) (Fig. 7A, Suppl. Table 4), and the upregulation of few genes (n = 5) including histone methyltransferase genes (PRMT3, PRMT5, PRMT6), a gene coding for SET domain proteins with a histone methyltransferase activity (SETD4) and the histone deacetylase HDAC3 (Fig. 7B, Suppl. Table 4). Altogether our results indicate an overall down-regulation of transcripts involved in methylation of DNA and histones in HCMV-DB-infected HMECs. In addition, transcripts of histone acetyltransferase and deacetylase genes are mostly downregulated in HCMV-DB-infected HMECs compared to uninfected controls.
Figure 7.
A general hypomethylation state was observed in HMECs infected with HCMV-DB. (A) Downregulation of the gene expression of DNA methyltransferase genes (DNMT3A), histone methyltransferase genes (PRMT1, PRMT2, PRMT8, SMYD3, SUV39H1), genes coding for SET domain proteins with a histone methyltransferase activity (ASH1L, MLL3 (KMT2C), MLL5 (KMT2E), NSD1, SETD1A, SETDB1, SETDB2, SETD2, SETD3, SETD5, SETD6, SETD7, SETD8, SUV420H1, WHSC1), histone acetyltransferase genes (ATF2, CIITA, CSRP2BP, KAT2A, KAT6B, NCOA1), histone kinase genes (AURKA, AURKB, AURKC, NEK6, RPS6KA3), genes involved in histone ubiquitination (MYSM1, RNF2, RNF20, UBE2A, UBE2B, USP21, USP22), DNA/histone demethylase genes (KDM1A, KDM5C, KDM4C, KDM6B) and histone deacetylase genes (HDAC2, HDAC4, HDAC5, HDAC6, HDAC9, HDAC10). (B) Upregulation of the gene expression of histone methyltransferase genes (PRMT3, PRMT5, PRMT6), genes coding for SET domain proteins with a histone methyltransferase activity (SETD4) and HDAC3. The up- and down-regulation were measured in HMECs infected with HCMV-DB (moi, 1 and 10) as compared to uninfected HMECs.
Formation of tumorspheres from HMECs infected with HCMV-DB
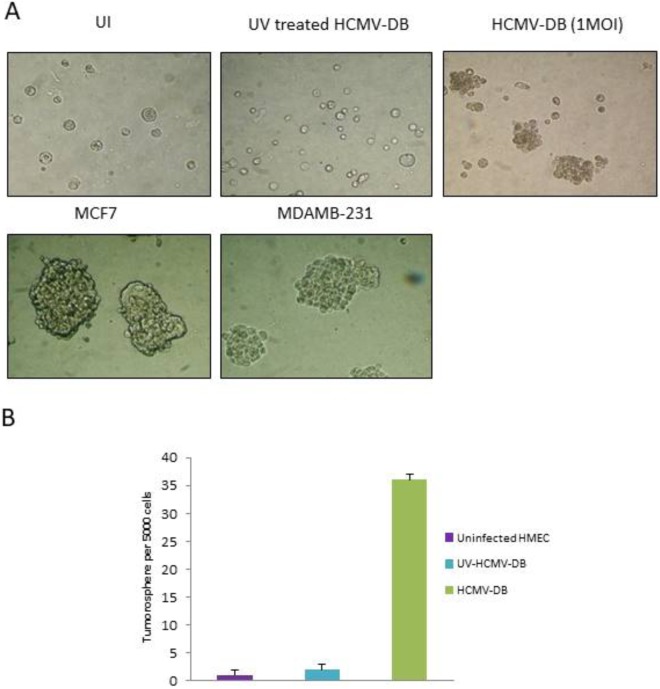
Since it was previously showed that the activation of the IL-6/STAT3 axis signaling in cancer stem cells (CSC) can enhances proliferation and survival of cells and in turn favor the growth of tumors in mice, we decided to detect the presence of CSC in HCMV-DB-infected HMECs using a tumorsphere (mammosphere) formation assay45,46. To determine whether the induction of CSC expansion could be the result of the HCMV-DB infection of HMECs, we infected them for 1 day and evaluated the proportion of stem-like cells by sphere formation assay. We found that HCMV-DB infected cultures formed tumorspheres (Fig. 8A,B). As expected MCF-7 cells and MDA-MB231 cells formed tumorspheres (Fig. 8A). In contrast, the use of UV-inactivated HCMV-DB to infect HMECs did not result in tumorspheres formation in cultures, neither did the use of uninfected HMECs (Fig. 8A,B).
Figure 8.
HMECs infected with HCMV-DB show tumorspheres formation. (A) Tumorsphere formation in HMECs infected with HCMV-DB as compared to uninfected HMECs (UI) and HMECs infected with UV-treated HCMV-DB. MCF-7 and MDA-MB-231 cells are used as positive controls. (B) The histogram represents quantification of tumorsphere formation in the cultures of HMECs infected with HCMV-DB. Uninfected HMECs and HMECs infected with UV-inactivated HCMV-DB were used as negative controls. Results are means (±SD) of three independent experiments.
Discussion
Our results indicate that, based on its genomic profile, the HCMV-DB strain is similar to clinical HCMV strains especially the JP and Toledo strains. Based on phylogenetic analysis of genes involved in virus entry, the HCMV-DB strain is close to the Merlin strain. In addition, we observed that HCMV-DB infects HMECs which display a triple negative phenotype. The transcriptome of HCMV-DB-infected HMECs shows the characteristics of a pro-oncogenic cellular environment with upregulated expression of numerous oncogenes, enhanced activation of pro-survival genes, and upregulated markers of cell proliferation, stemcellness and EMT that was confirmed by enhanced tumorsphere formation in vitro.
HCMV clinical isolates can infect among others epithelial cells, endothelial cells, monocytes, macrophages, fibroblasts, stromal cells, hepatocytes, smooth muscle cells, and neural stem/progenitor cells. In opposition to this broad cellular tropism, HCMV laboratory strains can only grow on fibroblasts7. Some clinical HCMV isolates infect the blood monocytes and tissue macrophages, induce a distinct macrophage polarization and establish latency in monocytes/macrophages8,22,24,47. The tropism of HMCV strains could have a major role in M1/M2 macrophage activation and enhance the viral fitness, ultimately favoring breast cancer promotion26,48,49. Previously, we isolated the HCMV-DB strain (KT959235)8. The HCMV-DB strain, given its characterization as highly macrophage-tropic, induces an M2 polarization of the macrophages and the upregulation of the proto-oncogene Bcl-38. The ULb’ region of the HCMV-DB strain is intact29 and HCMV-DB show genetic similarities to other primary clinical isolates such as the JP strain which has been isolated post mortem from the prostate of an HIV-infected patient30,31. Interestingly, DB and JP strains infect mammary and prostate glands respectively which show numerous similarities with regard to appearance, physiology and pathology50.
The HMECs infected with HCMV-DB exhibit a triple negative ER−/PR−/HER2− phenotype with low expression of luminal markers (KRT19, KRT18, GATA3, TFF1). Oncogenic conversion of HMECs requires a limited number of changes leading towards growth deregulation51. These changes include the inactivation of tumor suppressor pathways, mainly p53 and retinoblastoma protein (Rb)52, the establishment of telomere maintenance53,54, the activation of the Akt pathway and activation of mitogenic signal delivered by oncogenes such as Myc and Ras51,55,56. We observed the upregulation of Rb and p53 transcripts in HCMV-DB-infected HMECs. Increased p53 transcript and protein expressions have been reported in fibroblasts infected with several HCMV strains57. The functional inactivation of p53 required for cell transformation, could be explained by the previously reported potential inhibition of the transcriptional activity of p53 by HCMV-IE2-8658. In agreement with the upregulation of the p53 transcript, we previously reported that, in HCMV-DB infected HMECs, the p53 protein expression was enhanced, as well as its binding to IE2 and therefore could explain its functional inactivation29. The gene expression of retinoblastoma protein (pRb) is 7 fold higher in HCMV-DB-infected HMECs in comparison to uninfected HMECs at day 1 post infection. We also observed the upregulation of the Rb transcript in MDA-MB231 cells in agreement with the upregulation of the pRb protein in MDA-MB231 cells reported previously42. Although the expression of the gene Rb is increased in HCMV-DB-infected HMECs, the post translational phosphorylation of pRb has been reported to play a critical role in its inactivation56. The HCMV UL97 protein (pUL97) exhibits similar activities to that of cellular cyclin-dependant kinase (CDK) complexes; thus it phosphorylates and inactivates the pRb protein. The pUL97 protein lacks the amino acid residues conserved in CDKs that allow for the attenuation of kinase activity, and is not inhibited by the CDK inhibitor p21. Therefore the activity of pUL97 is immune from normal CDK control mechanisms59,60. Also the human cytomegalovirus pp71, encoded by UL82, has a role in the degradation of the retinoblastoma family of proteins61. In agreement with such an hypothesis, we reported previously the upregulation of UL82 transcript and protein (pp71) at day 1 and 3 post infection in HMECs infected with HCMV-DB29. Thus, in HCMV-DB infected HMECs, both p53 and pRb proteins could be inactivated, most likely blocking cellular senescence, and therefore promoting unchecked cell division51. We observed, in infected cells, the activation of IL-6/STAT3 axis with expression of the pUS28 protein, which is in agreement with other studies that confirm the role of HCMV pUS28 in mediating proliferation through this axis activation62,63.
The acquisition of a mitogenic signal, delivered by proto-oncogenes including c-Myc and Ras, could further allow the complete transformation of HCMV-DB-infected HMECs in vitro. In agrement with the enhanced expression of Myc and Ras transcripts in HCMV-DB-infected HMECs, we reported previoulsy enhanced expression of Myc and Ras proteins starting at day 1 postinfection as measured by western bloting29. Overexpression of other oncogene transcripts such as Fos and Jun could also participate in HMEC transformation induced by HCMV-DB64,65. Interestingly, the Myc region on chromosome 8q24.21 is a known site of frequent human papillomavirus (HPV) integration and Myc is overexpressed in cervical carcinoma55,66,67. In agreement with the upregulation of the Myc gene expression in HMECs infected with HCMV-DB, the activation of Fos and Myc is induced by the IE1 and IE2 proteins of HCMV68. In addition, MYB gene is induced in HCMV-DB-infected HMECs similar to enhanced MYB gene expression in HPV-induced carcinoma69. Altogether, the upregulation of oncogenes observed in HCMV-DB-infected HMECs is in part similar to the transcriptomic profile observed in HPV-induced carcinoma.
The PI3K/Akt pathway is activated in breast cancer and its activation is critical for the emergence of anchorage-independent growth, eventually leading to the transformation of HMECs70,71. We observed enhanced expression of genes of the PI3K/Akt pathway, in agreement with phosphorylation of Akt in HCMV-DB-infected HMECs29. In addition increased levels of transcripts of pro-survival genes such as Bcl-2, Akt, and NF-kB was observed parallel to decreased levels of the caspase-8 transcript in HCMV-DB-infected HMECs at low moi (moi = 1). By contrast, at high moi (moi = 10), we observed increased levels of the caspase-8 transcript in HMECs infected with HCMV-DB. This apparent discrepancy between low and high moi, could be explains as follows. At early stages of the infection after viral entry, only a limited number of virions are present within the infected cell. At this initital step of the viral life cycle anti-apoptotic mechanisms have to be activated to favor cell survival and to fuel viral replication within the infected cell. Thus it is critical that early after infection (low moi) the infected cell survives to favor the completion of the viral life cycle and to allow the production of a maximum of new progeny virions. Later during infection, when numerous virions have been produced within the infected cells (high moi), the pro-apoptotic signals e.g. enhanced caspase-8 gene expression are activated and result in the lyse of the infected cells with release of the newly produced virions. Altogether, these results indicate that the transcriptome of HCMV-DB-infected HMECs displays a flexible adaptation to the viral life cycle and can be modulated differentially depending on the intracellular viral load and/or the amount of viral input.
We observed the upregulation of gene expression of proliferation markers such as the Ki67 antigen, the topoisomerase 2 and the transcription factor E2F1 in HCMV-DB-infected HMECs compared to uninfected HMECs. Our results are in agreement with the increased proliferation previously observed in cultures of HCMV-DB-infected HMECs29. Parallel to enhanced cellular proliferation in HCMV-DB-infected HMECs, we noted the dysregulation of the cell cycling. In HCMV-DB-infected HMECs, we observed enhanced expression of cyclins genes (cyclin A1, D1, D2, E1) including cyclin D1, but also to a lesser extent of cell cycling inhibitors (CDKNs). In agreement with the induction of CDKN2A gene expression in HMECs infected with HCMV-DB, CDKN2A gene is strongly induced in HPV infection and is widely used as a surrogate marker for HPV carcinoma69,72. Although we observed enhanced proliferation of HCMV-DB-infected HMECs, we also noticed a dysregulation of cell cycling which could lead to the inhibition of the cell cycle G1 progression and to the block of G1/S transition, as previoulsy reported by other groups in fibroblasts infected with various HCMV strains73. In agreement with an inhibition of the cell cycle proliferation in HMECs infected with HCMV-DB, the Ras association domain-containing protein 1 (RASSF1) is upregulated in infected cells and was reported to inhibit the accumulation of cyclin D1 (Suppl. Table 3). An exquisite balance between enhanced cell cycling (enhanced cyclins gene expression) and enhanced cell cycling inhibition (enhanced CDKN gene expression), might ultimately result in enhanced cell proliferation as measured by increased Ki67Ag and TOP2A transcripts and enhanced Ki67 antigen detection in HMECs infected with HCMV-DB.
Decreased MAPK activation has been reported as pejorative in triple negative breast cancers74. In agreement with this observation, decreased gene expression of members of the MAPK cascades (MAPK3, MAPK8) is observed in HCMV-DB-infected HMECs which exhibit a ER−/PR−/HER2− phenotype.
Several herpesviruses have been reported to lead to genome instability with activation of the DNA reparation pathway with increased expression of the ATM gene75–77. We found that the ATM gene expression is upregulated in HCMV-DB-infected HMECs as reported previously in fibroblasts infected with other HCMV strains77. Enhanced ATM expression could favor the reparation of DNA damages such as chromosomal breaks already reported in fibroblasts infected with HCMV78,79, but could also lead to genome instability and thereby favor cellular transformation80.
Several studies, including a comprehensive study by The Cancer Genome Atlas (TCGA) Network, have shown that, in opposition to luminal estrogen receptor-positive cancers, which exhibit the highest degree of hypermethylation, the triple negative breast cancers (TNBCs) are characterized by the most extensive hypomethylation81,82. The expression of numerous histone methyltransferase genes and of genes coding for SET domain proteins with a histone methyltransferase activity is downregulated in HCMV-DB infected HMECs in comparison to uninfected HMECs. The gene expression of DNA methyltransferase 3 A (DMT3A) is also downregulated in infected HMECs.
Regarding mammary tumor progression, Suzuki et al.83 reported that in the tumor progression from normal mammary epithelium to ductal carcinoma in situ (DCIS), there is a reduction of the histone acetylation levels. This change in histone acetylation was not present when comparing DCIS to invasive ductal carcinoma. A significant decrease in histone deacetylase (HDAC) protein levels during tumor progression was also described84. This suggests an early role for histone acetylation modifications in breast tumor progression. We observed decreased expression of genes involved in histone acetylation (histone acetyl transferases) in HCMV-DB infected HMECs in comparison to uninfected HMECs. In addition, we observed the decreased gene expression of several histone deacetylases, including HDAC2, 4, 5, 6, 9 and 10 in HCMV-DB-infected HMECs in comparison to uninfected HMECs. Altogether, our data indicate that HCMV-DB-infected HMECs at day 1 post infection display already alterations of histone acetylation which have been evoked as an early event in breast tumor progression.
The transcriptomic analysis of HCMV-DB-infected HMECs indicates that genes involved in angiogenesis and proteolysis, and thereby potentially participating in the remodelling of the tumor microenvironment, are modulated in HCMV-DB-infected HMECs85,86. We also observed the upregulation of IL-6-JAK gene expression in HCMV-DB-infected HMECs compared to uninfected HMECs which has been reported to play a role in the appearance of tumorspheres in cultures of hepatocytes infected with HCMV10. In addition, the gene expression of alpha 6 integrin (ITGA6), a marker of stemcellness, is upregulated in HCMV-DB-infected HMECs. Finally in HCMV-DB-infected HMECs, the proto-oncogene tyrosine-protein kinase Src87, a gene involved in EMT, is upregulated compared to uninfected HMECs. Altogether our results indicate a potential trend towards the appearance of cancer stem cells (CSCs) in HMECs infected with HCMV-DB. CSCs are required for sustained tumor growth, invasiveness and metastasis formation88. The expansion of CSCs can be measured by the formation of tumorspheres. In agreement with the activation of IL-6-JAK pathway, upregulation of alpha 6 integrin and src gene expression, we observed tumorsphere formation in cultures of HCMV-DB-infected HMECs.
In conclusion, we define here the genomic profile of a newly isolated clinical HCMV strain, namely HCMV-DB, and its close phylogenetic links with other HCMV strains, especially the JP, Toledo and Merlin strains. Further, we confirmed that the HCMV-DB strain infects HMECs which display a triple negative phenotype. Our data indicate that the transcriptome of HCMV-DB-infected HMECs presents traits of a pro-oncogenic cellular environment with upregulated expression of numerous oncogenes, enhanced activation of pro-survival genes, and upregulated markers of cell proliferation, stemcellness and EMT that was confirmed by enhanced tumorsphere formation in vitro.
Materials and Methods
Reagents
Anti-US28 and anti β-actin antibodies were respectively acquired from Santa Cruz Biotechnology (Santa Cruz, CA) and Sigma-Aldrich (St. Louis, MO).
Cell cultures
HMECs were purchased from Life Technologies (Carlsbad, CA, USA). MDA-MB231 and MCF-7 cells were provided by Institut Hiscia (Arlesheim, Switzerland). Cell culture and cell viability assay was performed as previously described8. Cultures were free of mycoplasma.
Infection of HMECs with HCMV
The clinical isolate HCMV-DB was previously isolated in our laboratory8. Cell-free virus stocks of HCMV-DB were grown in macrophages, as described previously8. Human fibroblasts (MRC5) were cultured as previously described6,8. Cells infection by HCMV, heat-inactivated HCMV, UV-treated HCMV was performed as previously described29. Virus titers were determined by plaque-forming assay in MRC5 as described previously8. Viral stocks purity and viral replication were assessed as previously described29. Quantification of viral titer was performed by qPCR on cell culture supernatants as previously described8.
Western blotting
pUS28 and β-actin expression was examined by western blotting as described previously8, using infected or uninfected HMECs cell lysates.
RT2 ProfilerTM PCR Arrays
Total RNA was extracted from uninfected HMECs and HMECs infected with HCMV-DB at MOI of either 1 or 10 at day 1 post infection using TRIzol reagent (Life Technologies, Grand Island, NY), and the first strand cDNA synthesis was achieved using the RT2 First Strand Kit (SABiosciences, Valencia, CA) following the manufacturer’s instructions. Three RT2 ProfilerTM PCR Arrays for human breast cancer (PAHS-131ZA), oncogenes/tumor suppressor genes (PAHS-502Z) and human epigenetic chromatin modification enzymes (PAHS-085A) (all three from SABiosciences) were applied on the Strategene Mx3005P real-time PCR system (Agilent Technology, Santa Clara, CA) as per manufacturer’s instructions. Housekeeping genes, contamination control, reverse transcription control and positive controls were included in each PCR according to manufacturer’s instructions. Data analyses were performed using the web based analysis software (http://pcrdataanalysis.sabiosciencescom/pcr/arrayanalysis.php).
Tumorsphere assay
Tumorsphere formation by uninfected HMECs or by HMECs infected using HCMV-DB or UV-inactivated HCMV-DB, was assayed as described previously10. MCF-7 and MDA-MB231 were used as positive controls. The number of tumorspheres larger than 60 microns was counted.
Genomic analysis of HCMV-DB
The analysis of HCMV-DB genome and its comparison to other HCMV strains: AD-169 and Towne (laboratory adapted strains) and Merlin, Toledo, TR, PH, VR1814, Davis, JP, and TB40/E (clinical strains with low passages in culture) was done using the NCBI nucleotide blast tool (https://blast.ncbi.nlm.nih.gov/BlastAlign.cgi).
Phylogenetic analysis
Phylogenetic proximity was calculated among various HCMV strains (described in Table 1) for UL55, UL75, UL115, UL128, UL130, and UL131 genes. The phylogenetic analysis was performed as previously reported89.
Statistical analysis
Numerical values are shown as the means and SD of independent experiments. Mann Whitney U test was performed for statistical significance and differences were considered significant at a value of P < 0.05. The plots were prepared using Microsoft Excel.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the University of Franche-Comté (UFC), the Région Franche-Comté (RECH-FON12-000013) and Europe (FEDER, Fonds Européens de Développement Régional N°2014-0045) to Georges Herbein. Amit Kumar is a recipient of a postdoctoral fellowship of the Region Franche-Comté (N° 2012C-06102). Sébastien Pasquereau is an assistant Engineer in FEDER project. Fatima Al Moussawi is a recipient of a doctoral scholarship from the Lebanese University.
Author Contributions
F.A.M., A.K., S.P. and M.K.T. performed research; G.H., W.K. and M.D.A. designed research; F.A.M. and G.H. wrote the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fatima Al Moussawi and Amit Kumar contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-30109-1.
References
- 1.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hüsing A, et al. Prediction of breast cancer risk by genetic risk factors, overall and by hormone receptor status. J. Med. Genet. 2012;49:601–608. doi: 10.1136/jmedgenet-2011-100716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alibek K, Kakpenova A, Mussabekova A, Sypabekova M, Karatayeva N. Role of viruses in the development of breast cancer. Infect. Agents Cancer. 2013;8:32. doi: 10.1186/1750-9378-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geder KM, Lausch R, O’Neill F, Rapp F. Oncogenic transformation of human embryo lung cells by human cytomegalovirus. Science. 1976;192:1134–1137. doi: 10.1126/science.179143. [DOI] [PubMed] [Google Scholar]
- 5.Zur Hausen H. The search for infectious causes of human cancers: where and why. Virology. 2009;392:1–10. doi: 10.1016/j.virol.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Coaquette A, et al. Mixed cytomegalovirus glycoprotein B genotypes in immunocompromised patients. Clin. Infect. Dis. 2004;39:155–161. doi: 10.1086/421496. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. USA. 2005;102:18153–18158. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan KA, Coaquette A, Davrinche C, Herbein G. Bcl-3-Regulated Transcription from Major Immediate-Early Promoter of Human Cytomegalovirus in Monocyte-Derived Macrophages. J Immunol. 2009;182:7784–7794. doi: 10.4049/jimmunol.0803800. [DOI] [PubMed] [Google Scholar]
- 9.Belzile J-P, Stark TJ, Yeo GW, Spector DH. Human cytomegalovirus infection of human embryonic stem cell-derived primitive neural stem cells is restricted at several steps but leads to the persistence of viral DNA. J. Virol. 2014;88:4021–4039. doi: 10.1128/JVI.03492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lepiller Q, Abbas W, Kumar A, Tripathy MK, Herbein G. HCMV Activates the IL-6-JAK-STAT3 Axis in HepG2 Cells and Primary Human Hepatocytes. PLOS ONE. 2013;8:e59591. doi: 10.1371/journal.pone.0059591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renzette N, et al. Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection. PLoS Genet. 2013;9:e1003735. doi: 10.1371/journal.pgen.1003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cobbs CS, et al. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62:3347–3350. [PubMed] [Google Scholar]
- 13.Söderberg-Nauclér C. Does cytomegalovirus play a causative role in the development of various inflammatory diseases and cancer? J. Intern. Med. 2006;259:219–246. doi: 10.1111/j.1365-2796.2006.01618.x. [DOI] [PubMed] [Google Scholar]
- 14.Lepiller Q, Aziz Khan K, Di Martino V, Herbein G. Cytomegalovirus and Tumors: Two Players for One Goal-Immune Escape. The Open Virology Journal. 2011;5:60–69. doi: 10.2174/1874357901105010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samanta M, Harkins L, Klemm K, Britt WJ, Cobbs CS. High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J. Urol. 2003;170:998–1002. doi: 10.1097/01.ju.0000080263.46164.97. [DOI] [PubMed] [Google Scholar]
- 16.Harkins LE, et al. Detection of human cytomegalovirus in normal and neoplastic breast epithelium. Herpesviridae. 2010;1:8. doi: 10.1186/2042-4280-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baryawno N, et al. Detection of human cytomegalovirus in medulloblastomas reveals a potential therapeutic target. J Clin Invest. 2011;121:4043–4055. doi: 10.1172/JCI57147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharjee B, Renzette N, Kowalik TF. Genetic Analysis of Cytomegalovirus in Malignant Gliomas. J. Virol. 2012;86:6815–6824. doi: 10.1128/JVI.00015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taher C, et al. High prevalence of human cytomegalovirus proteins and nucleic acids in primary breast cancer and metastatic sentinel lymph nodes. PLoS ONE. 2013;8:e56795. doi: 10.1371/journal.pone.0056795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee S, et al. Distinct microbiological signatures associated with triple negative breast cancer. Sci Rep. 2015;5:15162. doi: 10.1038/srep15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michaelis M, Doerr HW, Cinatl J. The Story of Human Cytomegalovirus and Cancer: Increasing Evidence and Open Questions. Neoplasia. 2009;11:1–9. doi: 10.1593/neo.81178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hargett D, Shenk TE. Experimental human cytomegalovirus latency in CD14+monocytes. Proc. Natl. Acad. Sci. USA. 2010;107:20039–20044. doi: 10.1073/pnas.1014509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noriega VM, et al. Human cytomegalovirus modulates monocyte-mediated innate immune responses during short-term experimental latency in vitro. J. Virol. 2014;88:9391–9405. doi: 10.1128/JVI.00934-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith MS, Bentz GL, Alexander JS, Yurochko AD. Human cytomegalovirus induces monocyte differentiation and migration as a strategy for dissemination and persistence. J. Virol. 2004;78:4444–4453. doi: 10.1128/JVI.78.9.4444-4453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dziurzynski K, et al. Glioma-associated cytomegalovirus mediates subversion of the monocyte lineage to a tumor propagating phenotype. Clin. Cancer Res. 2011;17:4642–4649. doi: 10.1158/1078-0432.CCR-11-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herbein G, Kumar A. The oncogenic potential of human cytomegalovirus and breast cancer. Front Oncol. 2014;4:230. doi: 10.3389/fonc.2014.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medrek C, Pontén F, Jirström K, Leandersson K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer. 2012;12:306. doi: 10.1186/1471-2407-12-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A, et al. The Human Cytomegalovirus Strain DB Activates Oncogenic Pathways in Mammary Epithelial Cells. EBioMedicine. 2018;30:167–183. doi: 10.1016/j.ebiom.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cunningham C, et al. Sequences of complete human cytomegalovirus genomes from infected cell cultures and clinical specimens. J. Gen. Virol. 2010;91:605–615. doi: 10.1099/vir.0.015891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy E, et al. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA. 2003;100:14976–14981. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fouts AE, et al. Mechanism for neutralizing activity by the anti-CMV gH/gL monoclonal antibody MSL-109. Proc. Natl. Acad. Sci. USA. 2014;111:8209–8214. doi: 10.1073/pnas.1404653111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sijmons S, Van Ranst M, Maes P. Genomic and functional characteristics of human cytomegalovirus revealed by next-generation sequencing. Viruses. 2014;6:1049–1072. doi: 10.3390/v6031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkinson GWG, et al. Human cytomegalovirus: taking the strain. Med. Microbiol. Immunol. 2015;204:273–284. doi: 10.1007/s00430-015-0411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer-König U, et al. Glycoprotein B genotype correlates with cell tropism in vivo of human cytomegalovirus infection. J. Med. Virol. 1998;55:75–81. doi: 10.1002/(SICI)1096-9071(199805)55:1<75::AID-JMV12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 36.Ciferri C, et al. Structural and biochemical studies of HCMV gH/gL/gO and Pentamer reveal mutually exclusive cell entry complexes. Proc. Natl. Acad. Sci. USA. 2015;112:1767–1772. doi: 10.1073/pnas.1424818112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li G, Nguyen CC, Ryckman BJ, Britt WJ, Kamil JP. A viral regulator of glycoprotein complexes contributes to human cytomegalovirus cell tropism. Proc. Natl. Acad. Sci. USA. 2015;112:4471–4476. doi: 10.1073/pnas.1419875112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murrell I, et al. The pentameric complex drives immunologically covert cell-cell transmission of wild-type human cytomegalovirus. Proc. Natl. Acad. Sci. USA. 2017;114:6104–6109. doi: 10.1073/pnas.1704809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryckman BJ, Chase MC, Johnson DC. HCMV gH/gL/UL128-131 interferes with virus entry into epithelial cells: evidence for cell type-specific receptors. Proc. Natl. Acad. Sci. USA. 2008;105:14118–14123. doi: 10.1073/pnas.0804365105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J. Virol. 2006;80:710–722. doi: 10.1128/JVI.80.2.710-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wille PT, Knoche AJ, Nelson JA, Jarvis MA, Johnson DC. A human cytomegalovirus gO-null mutant fails to incorporate gH/gL into the virion envelope and is unable to enter fibroblasts and epithelial and endothelial cells. J. Virol. 2010;84:2585–2596. doi: 10.1128/JVI.02249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson TJW, et al. RB1 status in triple negative breast cancer cells dictates response to radiation treatment and selective therapeutic drugs. PLoS ONE. 2013;8:e78641. doi: 10.1371/journal.pone.0078641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dimas-González J, et al. Overexpression of p53 protein is a marker of poor prognosis in Mexican women with breast cancer. Oncol. Rep. 2017;37:3026–3036. doi: 10.3892/or.2017.5553. [DOI] [PubMed] [Google Scholar]
- 44.Banerjee K, Resat H. Constitutive activation of STAT3 in breast cancer cells: A review. Int. J. Cancer. 2016;138:2570–2578. doi: 10.1002/ijc.29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manuel Iglesias J, et al. Mammosphere formation in breast carcinoma cell lines depends upon expression of E-cadherin. PLoS ONE. 2013;8:e77281. doi: 10.1371/journal.pone.0077281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lombardo, Y., de Giorgio, A., Coombes, C. R., Stebbing, J. & Castellano, L. Mammosphere formation assay from human breast cancer tissues and cell lines. J Vis Exp97, e52671 (2015). [DOI] [PMC free article] [PubMed]
- 47.Chan G, Nogalski MT, Yurochko AD. Activation of EGFR on monocytes is required for human cytomegalovirus entry and mediates cellular motility. Proc. Natl. Acad. Sci. USA. 2009;106:22369–22374. doi: 10.1073/pnas.0908787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teng MW, et al. An endogenous accelerator for viral gene expression confers a fitness advantage. Cell. 2012;151:1569–1580. doi: 10.1016/j.cell.2012.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKinney C, et al. Global reprogramming of the cellular translational landscape facilitates cytomegalovirus replication. Cell Rep. 2014;6:9–17. doi: 10.1016/j.celrep.2013.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.López-Otín C, Diamandis EP. Breast and prostate cancer: an analysis of common epidemiological, genetic, and biochemical features. Endocr. Rev. 1998;19:365–396. doi: 10.1210/edrv.19.4.0337. [DOI] [PubMed] [Google Scholar]
- 51.Dimri G, Band H, Band V. Mammary epithelial cell transformation: insights from cell culture and mouse models. Breast Cancer Res. 2005;7:171–179. doi: 10.1186/bcr1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Helt A-M, Galloway DA. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis. 2003;24:159–169. doi: 10.1093/carcin/24.2.159. [DOI] [PubMed] [Google Scholar]
- 53.Hahn WC, Meyerson M. Telomerase activation, cellular immortalization and cancer. Ann. Med. 2001;33:123–129. doi: 10.3109/07853890109002067. [DOI] [PubMed] [Google Scholar]
- 54.Strååt K, et al. Activation of telomerase by human cytomegalovirus. J. Natl. Cancer Inst. 2009;101:488–497. doi: 10.1093/jnci/djp031. [DOI] [PubMed] [Google Scholar]
- 55.Abba MC, Laguens RM, Dulout FN, Golijow CD. The c-myc activation in cervical carcinomas and HPV 16 infections. Mutat. Res. 2004;557:151–158. doi: 10.1016/j.mrgentox.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 56.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hannemann H, Rosenke K, O’Dowd JM, Fortunato EA. The presence of p53 influences the expression of multiple human cytomegalovirus genes at early times postinfection. J. Virol. 2009;83:4316–4325. doi: 10.1128/JVI.02075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hsu C-H, et al. HCMV IE2-mediated inhibition of HAT activity downregulates p53 function. EMBO J. 2004;23:2269–2280. doi: 10.1038/sj.emboj.7600239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hume AJ, et al. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science. 2008;320:797–799. doi: 10.1126/science.1152095. [DOI] [PubMed] [Google Scholar]
- 60.Iwahori S, Umaña AC, VanDeusen HR, Kalejta RF. Human cytomegalovirus-encoded viral cyclin-dependent kinase (v-CDK) UL97 phosphorylates and inactivates the retinoblastoma protein-related p107 and p130 proteins. J. Biol. Chem. 2017;292:6583–6599. doi: 10.1074/jbc.M116.773150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kalejta RF, Bechtel JT, Shenk T. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol. Cell. Biol. 2003;23:1885–1895. doi: 10.1128/MCB.23.6.1885-1895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Slinger E, et al. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci Signal. 2010;3:ra58. doi: 10.1126/scisignal.2001180. [DOI] [PubMed] [Google Scholar]
- 63.Soroceanu L, et al. Human cytomegalovirus US28 found in glioblastoma promotes an invasive and angiogenic phenotype. Cancer Res. 2011;71:6643–6653. doi: 10.1158/0008-5472.CAN-11-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boldogh I, AbuBakar S, Deng CZ, Albrecht T. Transcriptional activation of cellular oncogenes fos, jun, and myc by human cytomegalovirus. J. Virol. 1991;65:1568–1571. doi: 10.1128/jvi.65.3.1568-1571.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boldogh I, AbuBakar S, Albrecht T. Activation of proto-oncogenes: an immediate early event in human cytomegalovirus infection. Science. 1990;247:561–564. doi: 10.1126/science.1689075. [DOI] [PubMed] [Google Scholar]
- 66.Kübler K, et al. c-myc copy number gain is a powerful prognosticator of disease outcome in cervical dysplasia. Oncotarget. 2015;6:825–835. doi: 10.18632/oncotarget.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Samir R, Asplund A, Tot T, Pekar G, Hellberg D. High-risk HPV infection and CIN grade correlates to the expression of c-myc, CD4+, FHIT, E-cadherin, Ki-67, and p16INK4a. J Low Genit Tract Dis. 2011;15:280–286. doi: 10.1097/LGT.0b013e318215170c. [DOI] [PubMed] [Google Scholar]
- 68.Hagemeier, C., Walker, S. M., Sissons, P. J. & Sinclair, J. H. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-fos, c-myc and hsp70 promoters via basal promoter elements. J. Gen. Virol.73, 2385–2393 (1992). [DOI] [PubMed]
- 69.Tang X. Tumor-associated macrophages as potential diagnostic and prognostic biomarkers in breast cancer. Cancer Lett. 2013;332:3–10. doi: 10.1016/j.canlet.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 70.Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–7464. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- 71.Zhao JJ, et al. Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase. Cancer Cell. 2003;3:483–495. doi: 10.1016/S1535-6108(03)00088-6. [DOI] [PubMed] [Google Scholar]
- 72.Zhao R, Choi BY, Lee M-H, Bode AM, Dong Z. Implications of Genetic and Epigenetic Alterations of CDKN2A (p16(INK4a)) in Cancer. Ebio Medicine. 2016;8:30–39. doi: 10.1016/j.ebiom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dittmer D, Mocarski ES. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hashimoto K, et al. Activated PI3K/AKT and MAPK pathways are potential good prognostic markers in node-positive, triple-negative breast cancer. Ann. Oncol. 2014;25:1973–1979. doi: 10.1093/annonc/mdu247. [DOI] [PubMed] [Google Scholar]
- 75.Castillo JP, et al. Human cytomegalovirus IE1-72 activates ataxia telangiectasia mutated kinase and a p53/p21-mediated growth arrest response. J. Virol. 2005;79:11467–11475. doi: 10.1128/JVI.79.17.11467-11475.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.E X, et al. A novel DDB2-ATM feedback loop regulates human cytomegalovirus replication. J. Virol. 2014;88:2279–2290. doi: 10.1128/JVI.03423-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shirata N, et al. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J. Biol. Chem. 2005;280:30336–30341. doi: 10.1074/jbc.M500976200. [DOI] [PubMed] [Google Scholar]
- 78.Fortunato EA, Dell’Aquila ML, Spector DH. Specific chromosome 1 breaks induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA. 2000;97:853–858. doi: 10.1073/pnas.97.2.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Siew V-K, Duh C-Y, Wang S-K. Human cytomegalovirus UL76 induces chromosome aberrations. J. Biomed. Sci. 2009;16:107. doi: 10.1186/1423-0127-16-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Angèle S, et al. ATM protein overexpression in prostate tumors: possible role in telomere maintenance. Am. J. Clin. Pathol. 2004;121:231–236. doi: 10.1309/JTKGGGKURFX3XMGT. [DOI] [PubMed] [Google Scholar]
- 81.Holm K, et al. Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast Cancer Res. 2010;12:R36. doi: 10.1186/bcr2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Szyf M. DNA methylation signatures for breast cancer classification and prognosis. Genome Med. 2012;4:26. doi: 10.1186/gm325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suzuki J, et al. Protein acetylation and histone deacetylase expression associated with malignant breast cancer progression. Clin. Cancer Res. 2009;15:3163–3171. doi: 10.1158/1078-0432.CCR-08-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Linares A, Dalenc F, Balaguer P, Boulle N, Cavailles V. Manipulating protein acetylation in breast cancer: a promising approach in combination with hormonal therapies? J. Biomed. Biotechnol. 2011;2011:856985. doi: 10.1155/2011/856985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sevenich L, Joyce JA. Pericellular proteolysis in cancer. Genes Dev. 2014;28:2331–2347. doi: 10.1101/gad.250647.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Diaz N, et al. Activation of stat3 in primary tumors from high-risk breast cancer patients is associated with elevated levels of activated SRC and survivin expression. Clin. Cancer Res. 2006;12:20–28. doi: 10.1158/1078-0432.CCR-04-1749. [DOI] [PubMed] [Google Scholar]
- 88.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 89.Waters A, et al. Human cytomegalovirus UL144 is associated with viremia and infant development sequelae in congenital infection. J. Clin. Microbiol. 2010;48:3956–3962. doi: 10.1128/JCM.01133-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.