Abstract
Biogenic amines (BAs) have been reported to threaten the Douchi safety, while the BAs formation mechanism and corresponding control method have not been clarified for Douchi. The present study aims to investigate the microbial contribution to BAs in Douchi, and to find the beneficial strain for BAs control. Firstly, the BAs profiles of 15 Douchi samples were analyzed, and common 6 kinds of BAs were detected from different samples. All the samples showed the total BAs contents within the safe dosage range, while the histamine concentrations in 2 samples and β-phenethylamine in 6 samples were above the toxic level. Then, the bacterial and fungal communities were investigated by high-throughput sequencing analysis, and Bacillus and Candida were identified as the dominant bacteria and fungi genus, respectively. Furthermore, nineteen strains were selected from the dominant species of Douchi samples, including 14 Bacillus strains, 2 Staphylococcus strains, 1 Enterococcus strain and 2 Candida strains, and their BAs formation and degradation abilities were evaluated. B. subtilis HB-1 and S. pasteuri JX-2 showed no BAs producing ability, and B. subtilis GD-4 and Candida sp. JX-3 exhibited high BAs degradation ability. Finally, fermented soybean model analysis further verified that B. subtilis HB-1 and S. pasteuri JX-2 could significantly reduce BAs. This study not only contributed to understanding the BAs formation mechanism in Douchi, but also provided potential candidates to control the BAs in fermented soybean products.
Introduction
Biogenic amines (BAs) are organic nitrogenous compounds with low molecular weight, and they are formed during normal metabolic processes in various organisms1. BAs can be categorized into aliphatic (putrescine and cadaverine), aromatic (tyramine and β-phenethylamine) and heterocyclic amines (histamine and tryptamine) based on their chemical structures2,3. Recently, BAs have been broadly reported in various fermented foods, such as cheese, beer, wine and fermented soybeans, which shows potential harmful risk for human health4–10. BAs at low concentrations are essential for many physiological functions, while high concentrations of BAs can cause some deleterious effects7,11. Histamine can cause adverse neurological, gastrointestinal, circulatory and respiratory symptoms, such as headache, nausea, hot flushes, skin rashes and intestinal problems12. Tyramine, tryptamine and β-phenylethylamine have been confirmed to be implicated in hypertensive crises and dietary-induced migraine13. Therefore, evaluation and control of BAs are highly important to guarantee the safety of fermented food.
Douchi is a traditional fermented soybean product commonly consumed as flavouring agents in China, Japan, and other Asian countries14–16. The BAs have been reported as potential toxic substances in commercial Douchi17–19, thus evaluation and control of the BAs are valuable for the safety of Douchi. The BAs are mainly generated through decarboxylation of amino acids by various microbes, and putrescine, cadaverine, tyramine, β-phenethylamine, histamine and tryptamine were generated from ornithine, lysine, tyrosine, phenylethylamine, histidine and tryptophan, respectively20. Moreover, putrescine can also be formed through deimination of agmatine21. Different microbes may excrete different amino acid decarboxylases to produce specific BA, and some can also produce amine oxidases to degrade BAs22–25. Hence, the relations between BAs and microbes are complicated. Using the high BAs-degrading strains or the low BAs-producing strains as the starter cultures will be beneficial to control the Douchi BAs.
Generally, Douchi is produced by spontaneous fermentation with an uncontrolled manner, and various microbial communities are formed at different environment conditions14–16. The microbial diversities in commercial Douchi have been investigated using traditional culture method, denaturing gradient gel electrophoresis (DGGE) and high-throughput sequencing16,26–28, and the Bacillus species were identified as the dominant bacteria in Douchi11,14,15,27. However, the microbial contributions on BAs formation in Douchi have not been clarified, and the efficient starter cultures for BAs control have not developed. In this study, BAs contents and microbial communities in Douchi samples from different regions were investigated, and the BAs producing and degradation abilities of representative strains were evaluated to elucidate the microbial contribution and select beneficial candidate for BAs control.
Materials and Methods
Samples and media
Fifteen fresh Douchi samples were purchased from 15 different regions in China, named JXJJ (Jiangxi), GXWZ (Guangxi), GDYJ (Guangdong), SXHZ (Shanxi), YNKM (Yunnan), GZTR (Guizhou), HNLY (Hunan), HBYC (Hubei), CQ (Chongqing), GSLN (Gansu), SCCD (Sichuan), TJ (Tianjing), ZJJS (Zhejiang), JSXZ (Jiangsu) and HNLB (Henan). All samples were collected in three replicates, and stored at 4 °C for further analysis. The media used this study included LB medium (peptone 10 g/L, yeast extract 5 g/L, sodium chloride 10 g/L), MRS medium (peptone 10 g/L, beef extract 8 g/L, yeast extract 4 g/L, glucose 20 g/L, diammonium hydrogen citrate 2 g/L, sodium acetate 5 g/L, K2HPO4 2 g/L, MgSO4 0.2 g/L, MnSO4 0.04 g/L, Tween 80 1 g/L, pH = 5.7), and SDB medium (peptone 10 g/L, glucose 20 g/L, pH = 5.6), and 1.5% agar was added to prepare solid medium.
Illumina MiSeq sequencing
The total DNA was extracted from each Douchi sample by a SDS-based DNA extraction method previously described29. DNA quality was monitored by agarose gel electrophoresis, and stored at −20 °C for subsequent analysis. Primers of 515 F (GTGCCAGCMGCCGCGGTAA) and 806 R (GGACTACHVGGGTWTCTAAT) were designed according to the V4 region of bacterial 16S rRNA gene, and ITS1 (TCCGTAGGTGAACCTGCGG) and ITS2 (GCTGCGTTCTTCATCGATGC) were designed based on the ITS1 region of the fungal internal transcribed spacer (ITS).
The Miseq sequencing for bacteria and fungi followed the protocols described by Caporaso et al. and Kozich et al.30,31, respectively. The bacterial 16S rDNA and fungal ITS genes fragments were amplified using the total DNA, primers, and Thermo Scientific® Phusion High-Fidelity PCR Master Mix (New England Biolabs, UK), purified by the QIAquick PCR Purification Kit (QIAGEN, Germany). Then, purified amplicons were employed for DNA library construction using the TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina). At last, DNA libraries were evaluated using Qubit@ 2.0 Fluorometer (Thermo Scientific) and real-time PCR, then sequenced on the MiSeq platform at Beijing Novogene Bioinformatics Technology Co., Ltd. All experiments were conducted in three replicates.
Bioinformatics analysis
Raw sequences generated through MiSeq sequencing were merged using fast length adjustment of short reads (FLASH)32, and low-quality sequences were discarded using QIIME. Clean paired sequences retained for each sample were analyzed using the UPARSE pipeline to generate operational taxonomic units (OTUs) and select representative sequences at 97% similarity33. PyNAST alignment and ribosomal database project (RDP) assignment were carried out based on the latest Green genes database34. Resampling was performed according to the minimum sequence numbers across all samples before the downstream analysis. In addition, the community compositions were provided at different taxonomic levels.
Isolation of strains from Douchi samples
Douchi samples (5 g) were added into 45 mL sterile water, rotated at 37 °C and 140 rpm for 40 min. The liquid mixtures were diluted, and spread onto the LB plates, MRS plates added with 20 g/L CaCO3 and SDB plates added with 50 mg/L rifampicin, respectively. The LB and SDB plates were cultured at 37 °C and 30 °C to isolate the dominant bacteria and fungus strains, respectively, and the MRS plates were incubated at 37 °C in a vacuum bag to select the lactic acid bacteria.
Identification of strains
The bacterial genomic DNA was extracted by Gen-EluteTM Kit (Tiangen Biotech Co., Ltd, Beijing, China) following the manufacturer’s protocol, and the fungal genomic DNA was extracted using a SDS-based DNA extraction method previously described29. The 16S rDNA fragment was amplified through the universal primers of 27f (AGAGTTTGATCMTGGCTCAG) and 1492r (CTACGGCTACCTTGTTACGA), and the ITS fragment was amplified with the universal primers ITS1 (TCCGTAGGTGAACCTGCGG) and ITS4 (GCATATCAATAAGCGGA). The PCR protocol was set as: 95 °C, 5 min; 95 °C, 45 s, 55 °C, 1 min, 72 °C, 1 min, 32 cycles; 72 °C, 10 min; and 4 °C for 10 min. The PCR is conducted in the 25 μL reaction system consisting of 2 μL genomic template, 2.5 μL Easy-Taq Buffer, 2 μL primers, 2.5 μL dNTPs, 0.3 μL Easy-Taq enzyme and 15.7 μL nuclease-free water. The DNA fragments were sequenced by Tsingke Biological Technology Co., Ltd. (Wuhan, China), and the sequence identity was analyzed using the Blastn program (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Evaluation of BAs production and degradation ability
The BAs production ability was assessed by culturing the strains in 5 mL LB (Bacillus and Staphylococcus), MRS (Enterococcus) or SDB (Candida) medium added with 1 g/L of histidine, tyrosine, tryptophan, phenylalanine, ornithine monohydrochloride, lysine or agmatine sulfate salt. After incubating for 48 h, the resulting BAs were determined. To measure the BAs-degrading ability, the cells were cultured and collected by centrifugation at 6000 × g for 5 min. After washing with 0.05 mol/L phosphate buffer (pH = 7), the cell pellets were diluted to OD600 = 0.4 in phosphate buffer (0.05 mol/L) containing 100 mg/L of histamine, tyramine, tryptamine, β-phenethylamine, putrescine and cadaverine, cultured for 48 h to detect the residual BAs, and the blank phosphate buffer without cell pellets was applied as the control. The BA-degradation rate was calculated according to the equation i.e., M = [(A − B)/A] × 100%, where M means the BAs degradation percentage, A and B indicate the initial and residual contents of BAs, respectively20.
Fermented soybean product model analysis
The fermented soybean model was applied to compare the BAs-controlling capacities of different strains. The cells (OD600 = 2) were inoculated into the soaked soybeans (30 g) with the inoculum size of 5% (v/w), incubated for 7 days, and the total BAs contents were measured by HPLC. The soybeans were fermented by inoculating with B. subtilis HB-1, S. pasteuri JX-2, B. subtilis GD-4 and Candida sp. JX-3, respectively, and the soybeans added with equivalent sterile water was set as the control.
BAs determination
The BAs contents were detected based on a previous report20. Briefly, 1.5 mL of 0.4 M HClO4 was added into 0.5 g solid sample or 0.5 mL liquid sample, extracted for 1 h. After centrifugation at 12,000 × g for 10 min, the supernatant (250 μL) was added with 25 μL of 2 M NaOH and 75 μL of saturated NaHCO3, then reacted with 500 μL of 5 mg/mL dansyl chloride at 50 °C for 45 min. After that, the reactant was mixed with 25 μL of 25% NH4OH and incubated at 50 °C for 15 min to remove the residual dansyl chloride. Then the mixture was adjusted to 1.5 mL with acetonitrile and centrifuged at 2500 × g for 5 min. The supernatant was filtered through a 0.22 μm membrane for HPLC (high-performance liquid chromatography) analysis, which was carried out using an Agilent 1260 HPLC system with an Agilent column Zorbax Eclipse XDB-C18 (4.6 mm × 250 mm, 5 μm) at 30 °C. The separation was achieved using a linear gradient of mobile phase A (acetonitrile) and B (H2O) at a flow rate of 1 mL/min. The solvent gradient was as follows: 50% A (0–3 min), 50–90% A (3–20 min), 90% A (20–29 min), 90–50% A (29–32 min), 50% A (32–35 min). The detection was carried out at 254 nm. Each BA in the samples was quantified with a calibration curve generating by analyzing standard BA solution. Typical chromatograms are shown in Figure S1.
Statistical analysis
Alpha diversity including Chao 1, Shannon diversity indices and Goods coverage were subjected to statistical analysis using Qiime (Version 1.7.0). The community structure was statistically analyzed at different classification levels. The significance was analyzed by one-way ANOVA using the statistical software SPSS 20.0, and means were compared by Duncan’s multiple range test at 5%.
Results and Discussion
BAs contents in Douchi samples
BAs were widely detected in various soybean foods. Kim et al. detected six BAs in fermented soybean products, natto4, and Gong et al. reported that tryptamine, putrescine and cadaverine were detected in Douchi samples19. In this study, six common BAs with different contents were found among the Douchi samples (Table 1), including histamine (1.43–213.13 mg/kg), tyramine (1.37–64.33 mg/kg), tryptamine (0.86–99.31 mg/kg), β-phenethylamine (1.47–191.24 mg/kg), putrescine (2.65–171.00 mg/kg), and cadaverine (0.49–397.70 mg/kg), and the total BA contents ranged from 5.09 to 679.46 mg/kg. Generally, total BA contents in food are expected to be not more than 900 mg/kg3,35,36, and they may cause serious harm to human health when the total contents exceed 1000 mg/kg24. Our results indicated that no sample was above the harmful level (900 mg/kg) in the total content level, and as-detected samples were in the relative safe level on the whole. However, different BA showed different toxic level for human, thus each BA is needed to be further analyzed for comprehensive safety assessment.
Table 1.
Contents of BAs in Douchi samples.
| Douchi Sample | Histamine (mg/Kg) | Tyramine (mg/Kg) | Tryptamine (mg/Kg) | β-Phenethylamine (mg/Kg) | Putrescine (mg/Kg) | Cadaverine (mg/Kg) | Total (mg/Kg) |
|---|---|---|---|---|---|---|---|
| JXJJ | 75.16 ± 3.41 c | 54.80 ± 4.32 f | 14.71 ± 1.24 d | 191.24 ± 2.93 j | 97.08 ± 1.27 j | 246.47 ± 4.16 f | 679.46 ± 17.33 j |
| GXWZ | 1.43 ± 0.11 a | 64.33 ± 1.85 h | ND | 86.58 ± 0.91 i | 45.62 ± 0.48 h | 397.70 ± 4.35 g | 595.66 ± 7.70 i |
| GDYJ | 213.13 ± 2.37 d | 20.81 ± 1.31 d | 9.08 ± 1.79 bc | 69.44 ± 3.40 h | 171.00 ± 3.13 k | 25.08 ± 0.90 e | 508.54 ± 12.9 h |
| SXHZ | ND | 45.33 ± 3.72 e | 11.38 ± 0.48 cd | 38.57 ± 1.38 f | 71.66 ± 0.23 i | 2.69 ± 0.03 abc | 169.63 ± 5.84 g |
| YNKM | 11.54 ± 0.47 b | 59.94 ± 6.56 g | 0.86 ± 0.18 a | 69.11 ± 0.65 h | 12.68 ± 0.33 d | 5.62 ± 0.18 cd | 159.75 ± 8.37 fg |
| GZTR | ND | 2.09 ± 0.23 ab | 99.31 ± 6.41 e | 24.02 ± 2.05 e | 19.25 ± 1.31 f | 4.83 ± 0.57 bcd | 149.50 ± 10.57 f |
| HNLY | 11.84 ± 1.61 b | 11.30 ± 2.39 c | ND | 48.47 ± 4.79 g | 15.09 ± 2.86 e | 7.39 ± 0.91 d | 94.09 ± 12.56 e |
| HBYC | ND | 16.64 ± 2.94 d | 5.89 ± 0.21 b | 14.59 ± 0.54 d | 8.59 ± 0.38 c | 1.70 ± 0.16 ab | 47.41 ± 4.23 d |
| CQ | ND | 5.44 ± 0.38 b | ND | 4.90 ± 0.46 ab | 18.85 ± 0.05 f | 0.54 ± 0.00 a | 29.73 ± 0.89 c |
| GSLN | ND | ND | ND | ND | 23.33 ± 0.30 g | 1.93 ± 0.17 ab | 25.26 ± 0.47 c |
| SCCD | ND | 0.26 ± 0.09 a | ND | ND | 21.46 ± 0.68 g | 2.87 ± 0.14 abc | 24.53 ± 0.91 c |
| TJ | ND | 6.24 ± 0.84 b | ND | 7.19 ± 2.40 bc | 12.72 ± 1.24 d | ND | 26.15 ± 4.48 c |
| ZJJS | ND | 4.08 ± 0.13 ab | ND | 8.79 ± 0.25 c | 8.16 ± 0.41 c | ND | 21.03 ± 0.79 bc |
| JSXZ | ND | 1.37 ± 0.34 ab | ND | 1.47 ± 0.24 a | 5.49 ± 0.06 b | 0.49 ± 0.20 a | 8.82 ± 0.84 ab |
| HNLB | ND | 2.44 ± 0.44 ab | ND | ND | 2.65 ± 0.14 a | ND | 5.09 ± 0.58 a |
Data are presented as mean ± SDs of three replicates; ND means “not detected”. Different letters (a, b, c, etc.) indicate significantly different means at P < 0.05.
As shown in Table 1, histamine was detected in five Douchi samples, the histamine concentration of two samples were higher than the allowable limit (50 mg/kg) suggested by the US Food and Drug Administration3,13. And 100–800 mg/kg of tyramine and 30 mg/kg of β-phenylethylamine have been reported to be the toxic doses in food4,10,37. Although most of the Douchi samples contained tyramine, no sample showed the amount above the hazardous level (100 mg/kg). Tryptamine was detected in six samples, and their concentrations were under 100 mg/kg. Though no β-phenethylamine was detected in three samples, it was noteworthy that the concentrations of six products were higher than the toxic dose (30 mg/kg). While Gong et al. detected no histamine, tyramine and β-phenethylamine in Douchi samples19. The differences in distribution and content of BAs could be attributed to the variation of the raw materials, the microbiological compositions and the fermentation conditions.
Putrescine and cadaverine are regarded as two typical BA reported in soybean products including sufu, soy sauce, tempe, miso and natto4,7. Though putrescine was found in all Douchi samples, the concentrations were under 100 mg/kg in 14 Douchi samples. And the content of putrescine under 100 mg/kg has been usually attributed to natural occurrence of amine in soybean products7. Meanwhile, cadaverine was detected in twelve products, while the concentrations of cadaverine in two samples were higher than 200 mg/kg. The toxicity of putrescine and cadaverine is not obvious relatively, but they can interact with nitrite to form carcinogenic nitrosamine38.
Microbial communities among the Douchi samples
As described above, BAs contents in Douchi samples showed relatively large variations, which was probably due to the different microbiological compositions. Therefore, it is important to analysis the microbial communities among the Douchi samples. By high-throughput sequencing, 3,514,240 16S rRNA (V3–V4 regions) and 3,402,964 ITS2 reads were generated from 45 sequencing samples. After quality filtering and chimera removal, 62,603 bacterial and 74,327 fungal sequences were obtained from each sample, with the average read length of 426 bases for bacteria and 211 bases for fungi. Community richness and diversity were assessed using four alpha-diversity metrics (observed OTU, Chao1, Shannon and Good’s sample coverage) (Table 2). Chao1 is an estimator of species richness, and Chao1 values indicated that all the Douchi samples possessed good sample richness. The Shannon diversity index, a measurement of overall diversity, indicated the diversified microbiota throughout the samples, and the Good’s coverage result, an estimator of completeness of sampling39, highlighted good overall sampling, with levels of 99%.
Table 2.
Alpha-diversity metrics characterizing Douchi samples and inferred from the sequencing of 16S V3–V4 and ITS amplicons. Sequences were clustered at 97% identity.
| Douchi sample | Bacteria | Fungi | ||||||
|---|---|---|---|---|---|---|---|---|
| Observed species | Chao1 | Shannon | Good’s coverage (%) | Observed species | Chao1 | Shannon | Good’s coverage (%) | |
| JXJJ | 503 ± 52 c | 571.94 ± 106.20 bcd | 4.20 ± 0.15 cd | 99 | 115 ± 38 a | 136.41 ± 57.77 a | 1.06 ± 0.34 a | 99 |
| GXWZ | 360 ± 76 abc | 489.92 ± 165.10 abc | 4.12 ± 0.16 bcd | 99 | 121 ± 108 ab | 153.31 ± 117.03 ab | 2.08 ± 1.29 a | 99 |
| GDYJ | 283 ± 80 abc | 408.98 ± 108.89 abc | 2.35 ± 0.28 abc | 99 | 92 ± 52 a | 115.29 ± 61.53 a | 1.63 ± 1.10 a | 99 |
| SXHZ | 274 ± 85 abc | 400.36 ± 98.48 abc | 3.60 ± 0.32 abcd | 99 | 131 ± 20 ab | 156.35 ± 27.29 ab | 1.81 ± 0.35 a | 99 |
| YNKM | 555 ± 196 cd | 667.53 ± 231.33 cd | 4.72 ± 0.73 d | 99 | 126 ± 31 ab | 149.72 ± 28.19 a | 1.63 ± 1.35 a | 99 |
| GZTR | 149 ± 23 a | 191.62 ± 48.98 a | 2.07 ± 0.22 ab | 99 | 235 ± 103 cd | 274.10 ± 110.50 bcd | 3.44 ± 0.77 bc | 99 |
| HNLY | 389 ± 47 abc | 522.55 ± 84.83 abcd | 1.75 ± 0.70 a | 99 | 122 ± 82 ab | 141.24 ± 105.69 a | 1.65 ± 1.04 a | 99 |
| HBYC | 196 ± 66 ab | 269.26 ± 87.25 ab | 2.42 ± 0.19 abc | 99 | 246 ± 62 d | 284.65 ± 99.82 cd | 3.63 ± 0.39 c | 99 |
| CQ | 549 ± 76 cd | 625.56 ± 71.96 bcd | 3.98 ± 1.90 bcd | 99 | 174 ± 0 abcd | 197.38 ± 0 abc | 2.12 ± 0 a | 99 |
| GSLN | 362 ± 47 abc | 426.41 ± 44.12 abc | 4.00 ± 0.25 bcd | 99 | 220 ± 16 bcd | 273.77 ± 26.88 bcd | 3.49 ± 0.32 bc | 99 |
| SCCD | 481 ± 422 bc | 558.25 ± 535.29 bcd | 5.10 ± 1.69 d | 99 | 223 ± 0 bcd | 349.04 ± 0 d | 1.20 ± 0 a | 99 |
| TJ | 471 ± 66 bc | 581.94 ± 53.24 bcd | 3.58 ± 0.51 abcd | 99 | 98 ± 6 a | 119.90 ± 13.96 a | 2.26 ± 0.13 ab | 99 |
| ZJJS | 789 ± 291 d | 863.44 ± 275.59 d | 4.71 ± 1.52 d | 99 | 87 ± 4 a | 103.00 ± 6.77 a | 1.08 ± 0.32 a | 99 |
| JSXZ | 440 ± 62 abc | 562.34 ± 96.40 bcd | 2.04 ± 0.95 ab | 99 | 136 ± 50 abc | 193.42 ± 44.81 abc | 1.28 ± 1.00 a | 99 |
| HNLB | 444 ± 96 abc | 528.99 ± 67.78 abcd | 3.04 ± 2.51 abcd | 99 | 157 ± 50 abcd | 185.22 ± 60.82 abc | 1.65 ± 0.44 a | 99 |
Data are presented as mean ± SDs of three replicates. Different letters (a, b, c, etc.) indicate significantly different means at P < 0.05.
As to bacteria, Firmicutes was the predominant phylum in most Douchi samples, while Proteobacteria was only dominant in the sample of SCCD (Fig. 1a). Yang et al. found Firmicutes and Proteobacteria were major phyla in Douchi fermentation16. At the genus level, Bacillus was the dominant bacterial genus in most Douchi samples (Fig. 1a), and it was also reported as the predominant genus in various fermented soybean products40–42. In addition, the genera of Weissella, Enterococcus, Pantoea and Tetragenococcus were also the major genera in some Douchi samples, which was consistent with previous reports8,16,43,44. In many fermented soybean products, Bacillus and Tetragenococcus played the crucial role in production of nutrients and flavors26,45.
Figure 1.
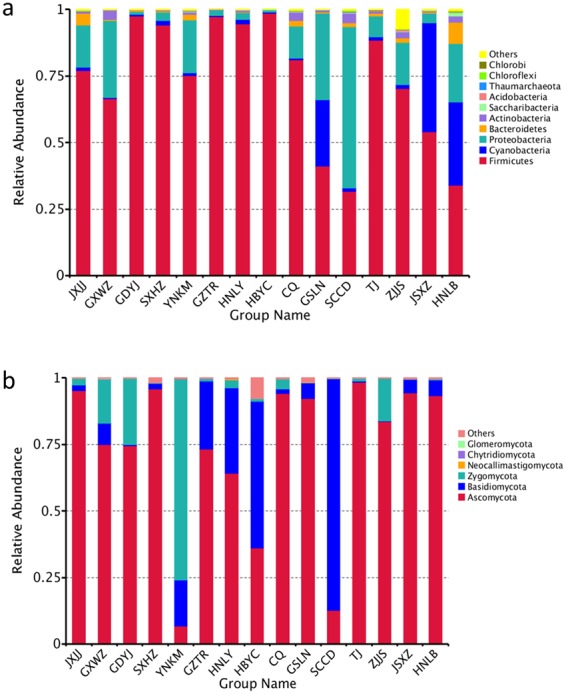
Microbial community in Douchi samples at the phylum level. (a) Bacteria, (b) Fungi.
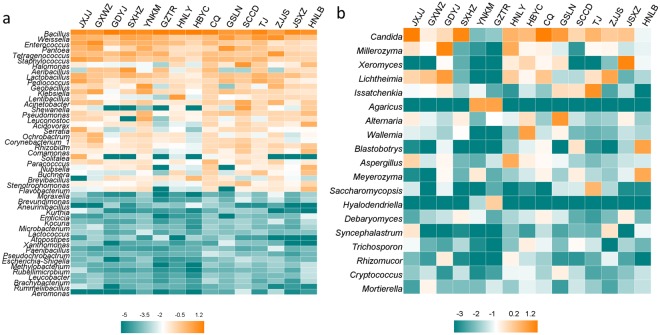
Figure 1b showed the difference in fungal abundance among samples at phylum level. Ascomycota was the most popular phylum in twelve Douchi samples, and this result was similar to previous studies16,44. The heatmap revealed differences in fungal genera among different Douchi samples (Fig. 2b). Candida was the most prevailing genus in JXJJ, SXHZ and CQ Douchi samples. Zhang et al. reported Candida was the dominant genus in Douchi44. During Douchi fermentation, Candida species grew more quickly than other fungi under high contents of organic acids and salts, then became the dominant fungal species46. Additionally, the genera of Millerozyma, Xeromyces, Lichtheimia, Issatchenkia, Agaricus, Alternaria, Wallemia and Blastobotrys were dominant in some Douchi samples. In a previous study, Lichtheimia was also detected as the major genus in Douchi16. Garcia et al. found that Lichtheimia produced β-glucosidase during solid-state fermentation47, which contributed to the release of glucose to improve the flavour of Douchi.
Figure 2.
Heatmap of Douchi samples at the genus level. The color intensity of each panel is proportional to the OTU abundance. (a) Bacteria, (b) Fungi.
Microbial contribution to BAs contents in Douchi
According to the results of microbial diversity in Douchi samples, the JXJJ, GXWZ, GDYJ, SXHZ, GZTR, HNLY and HBYC Douchi samples were used to isolate strains. Nineteen strains were isolated, named HB-1, HN-1, HN-2, HN-3, HN-4, GD-1, GD-2, GD-3, GD-4, SX-1, SX-2, GZ-1, GZ-2, GZ-3, JX-1, JX-2, JX-3, JX-4 and GX-1. The 16S rDNA or ITS sequences were analyzed to identify these strains, and sequences similarities of isolated strains with representative strains were shown in Table 3. According to the sequences similarities results, these strains were identified as Bacillus subtilis (5 isolates), Bacillus methylotrophicus (2 isolates), Bacillus amyloliquefaciens (2 isolates), Bacillus sonorensis (1 isolate), Bacillus licheniformis (4 isolates), Staphylococcus carnosus (1 isolate), Staphylococcus pasteuri (1 isolate), Enterococcus faecium (1 isolate) and Candida sp. (2 isolates), respectively (Table 3). Most of these strains belonged to the dominant genera in Douchi samples, and they were often reported in fermented soybean products17,44,48–52.
Table 3.
16S rDNA and ITS sequences similarities of isolated strains with representative microbe.
| Isolates | Closest strains | Identities (%) | Accession No. |
|---|---|---|---|
| HB-1 | B. subtilis CS10 | 99 | MH373531 |
| HN-1 | B. licheniformis L5 | 99 | MH373532 |
| HN-2 | B. subtilis L23 | 99 | MH373533 |
| HN-3 | B. sonorensis SXYC17 | 99 | MH373534 |
| HN-4 | B. subtilis yxw4 | 99 | MH373535 |
| GD-1 | B. methylotrophicus HB25 | 99 | MH373536 |
| GD-2 | B. amyloliquefaciens SXAU001 | 99 | MH373537 |
| GD-3 | B. licheniformis JD19 | 99 | MH373538 |
| GD-4 | B. subtilis NJ1 | 99 | MH373539 |
| SX-1 | B. licheniformis DC3-1 | 99 | MH373540 |
| SX-2 | B. subtilis SBE1 | 99 | MH373541 |
| GZ-1 | B. licheniformis JD18 | 99 | MH373542 |
| GZ-2 | B. amyloliquefaciens Y26 | 99 | MH373543 |
| GZ-3 | B. methylotrophicus HB26 | 99 | MH373544 |
| JX-1 | S. carnosus a23 | 99 | MH445557 |
| JX-2 | S. pasteuri HN-35 | 99 | MH445558 |
| JX-3 | Candida sp. 4 TMS-2011 | 98 | MH443337 |
| JX-4 | Candida sp. 4 TMS-2011 | 98 | MH443338 |
| GX-1 | E. faecium RK 204 | 99 | MH445559 |
To further investigate the microbial contribution to BAs accumulation in Douchi, we evaluated the BAs producing and degradation abilities of these isolated strains. The BAs producing abilities of all isolated strains were detected in medium supplemented with corresponding precursors, and the BAs produced by different strains were shown in Table 4. Most of the isolated strains (79%) could produce putrescine via agmatine, while none could synthesize putrescine from ornithine. Only a few strains could generate histamine, tyramine, tryptamine, β-phenethylamine or cadaverine from corresponding substrates. Thereinto, B. subtilis HB-1 and S. pasteuri JX-2 showed no ability to produce six common BAs, while E. faecium GX-1 produced the highest concentration of total BAs, especially the relative high level of tyramine. In addition, E. faecium also exhibited the ability of generating β-phenethylamine, which was consistent with a previous study53.
Table 4.
The BA-producing abilities of the isolated strains with corresponding precursor.
| Strain | Histamine (Histidine) (mg/L) | Tyramine (Tyrosine) (mg/L) | Tryptamine (Tryptophan) (mg/L) | β-Phenethylamine (Phenylalanine) (mg/L) | Putrescine (Ornithine monohydrochloride) (mg/L) | Putrescine (Agmatine sulfate salt) (mg/L) | Cadaverine (Lysine) (mg/L) | Total Bas (mg/L) |
|---|---|---|---|---|---|---|---|---|
| HB-1 | ND | ND | ND | ND | ND | ND | ND | ND |
| HN-1 | ND | ND | ND | 5.42 ± 0.54 a | ND | 3.11 ± 0.23 cd | ND | 8.53 ± 0.77 ab |
| HN-2 | ND | ND | ND | ND | ND | 8.20 ± 0.08 e | ND | 8.20 ± 0.08 ab |
| HN-3 | ND | ND | 30.98 ± 4.49 a | ND | ND | 1.64 ± 0.06 ab | ND | 32.62 ± 4.55 c |
| HN-4 | ND | ND | ND | ND | ND | 9.8 ± 0.11 f | ND | 9.80 ± 0.11 ab |
| GD-1 | ND | ND | ND | 7.11 ± 0.32 a | ND | 1.48 ± 0.14 a | ND | 8.59 ± 0.46 ab |
| GD-2 | ND | ND | ND | ND | ND | 7.88 ± 0.62 e | ND | 7.88 ± 0.62 ab |
| GD-3 | ND | ND | 59.63 ± 6.39 b | ND | ND | 1.72 ± 0.24 ab | ND | 61.35 ± 6.61 e |
| GD-4 | ND | ND | ND | ND | ND | 3.23 ± 0.16 cd | ND | 3.23 ± 0.16 a |
| SX-1 | 20.91 ± 1.21 a | ND | 24.24 ± 6.02 a | ND | ND | 2.44 ± 0.10 bc | ND | 47.59 ± 7.33 d |
| SX-2 | ND | ND | ND | ND | ND | 8.36 ± 0.13 e | ND | 8.36 ± 0.13 ab |
| GZ-1 | 17.60 ± 5.96 a | ND | 27.21 ± 5.98 a | ND | ND | 8.64 ± 1.64 e | ND | 53.45 ± 13.58 de |
| GZ-2 | ND | ND | ND | ND | ND | 8.04 ± 0.22 e | ND | 3.04 ± 0.22 a |
| GZ-3 | ND | ND | ND | ND | ND | 8.84 ± 0.52 e | ND | 8.84 ± 0.52 ab |
| JX-1 | ND | 34.76 ± 3.04 a | 120.18 ± 4.29 c | 378.42 ± 9.15 c | ND | 2.12 ± 0.04 ab | ND | 535.48 ± 16.52 g |
| JX-2 | ND | ND | ND | ND | ND | ND | ND | ND |
| JX-3 | ND | 364.34 ± 7.15 b | ND | ND | ND | ND | 2.86 ± 0.53 | 367.20 ± 7.68 f |
| JX-4 | 14.81 ± 0.80 a | ND | ND | ND | ND | 3.42 ± 0.11 d | ND | 18.23 ± 0.91 b |
| GX-1 | ND | 652.76 ± 11.52 c | ND | 40.69 ± 2.38 b | ND | ND | ND | 693.69 ± 13.98 i |
Data are presented as mean ± SDs of three replicates; ND means “not detected”. Different letters (a, b, c, etc.) indicate significantly different means at P < 0.05 (analysis of variance (ANOVA)).
The degrading abilities of the nineteen isolates were investigated. None of the strains was able to cause a complete disappearance of histamine, tyramine, tryptamine, β-phenethylamine, putrescine or cadaverine under the experimental conditions used (Table 5). Among the 19 strains tested, 53% of those strains were able to degrade histamine, 21% for tyramine, 58% for tryptamine, 84% for β-phenethylamine, 11% for putrescine and 11% for cadaverine. Some strains showed high histamine degradation ability (>20%), including B. licheniformis GD-3, B. subtilis GD-4, B. subtilis SX-2 and Candida sp JX-3. Moreover, Candida sp JX-3 also exhibited a high ability of degrading β-phenethylamine. Consistently, several B. licheniformis and B. subtilis strains were also reported to produce histamine in previous studies50,54.
Table 5.
Profiles of BA-degradation rates of selected strains.
| Strain | Histamine (%) | Tyramine (%) | Tryptamine (%) | β-Phenethylamine (%) | Putrescine (%) | Cadaverine (%) |
|---|---|---|---|---|---|---|
| HB-1 | 0.00 | 0.00 | 0.00 | 8.77 ± 0.95 cde | 0.00 | 0.00 |
| HN-1 | 0.00 | 0.00 | 3.29 ± 0.12 b | 10.05 ± 1.13 de | 0.00 | 0.00 |
| HN-2 | 0.00 | 0.00 | 0.00 | 9.98 ± 0.38 de | 0.00 | 1.93 ± 0.12 |
| HN-3 | 4.42 ± 0.64 a | 7.29 ± 0.42 b | 2.52 ± 0.20 b | 14.56 ± 1.25 f | 0.00 | 6.07 ± 0.54 |
| HN-4 | 0.00 | 0.00 | 1.14 ± 0.17 a | 5.34 ± 0.96 bc | 0.00 | 0.00 |
| GD-1 | 0.00 | 0.00 | 2.01 ± 0.13 ab | 1.43 ± 0.16 a | 0.00 | 0.00 |
| GD-2 | 0.00 | 0.00 | 2.61 ± 0.26 b | 5.93 ± 0.86 bc | 0.00 | 0.00 |
| GD-3 | 50.53 ± 2.64 d | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| GD-4 | 50.73 ± 4.06 d | 2.45 ± 0.12 a | 10.45 ± 1.50 d | 11.85 ± 1.20 ef | 9.78 ± 0.44 | 0.00 |
| SX-1 | 0.00 | 7.95 ± 0.58 b | 0.00 | 3.75 ± 0.40 ab | 7.59 ± 0.62 | 0.00 |
| SX-2 | 21.99 ± 2.81 c | 0.00 | 0.00 | 5.82 ± 0.39 bc | 0.00 | 0.00 |
| GZ-1 | 11.20 ± 1.49 b | 11.06 ± 1.22 c | 0.00 | 14.66 ± 0.66 f | 0.00 | 0.00 |
| GZ-2 | 0.00 | 0.00 | 3.00 ± 0.78 b | 1.61 ± 0.44 a | 0.00 | 0.00 |
| GZ-3 | 0.00 | 0.00 | 6.92 ± 1.00 c | 4.92 ± 0.32 ab | 0.00 | 0.00 |
| JX-1 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| JX-2 | 0.00 | 0.00 | 0.00 | 6.92 ± 0.40 bcd | 0.00 | 0.00 |
| JX-3 | 56.25 ± 5.98 d | 0.00 | 10.77 ± 0.25 d | 50.21 ± 4.98 g | 0.00 | 0.00 |
| JX-4 | 0.00 | 0.00 | 12.96 ± 1.34 e | 49.03 ± 5.44 g | 0.00 | 0.00 |
| GX-1 | 0.00 | 0.00 | 9.61 ± 0.22 d | 0.00 | 0.00 | 0.00 |
Data are presented as mean ± SDs of three replicates. Different letters (a, b, c, etc.) indicate significantly different means at P < 0.05 (analysis of variance (ANOVA)).
The BAs-controlling capacity of selected strains in fermented soybean model
For the 19 strains, B. subtilis HB-1 and S. pasteuri JX-2 showed no BAs producing ability (Table 4), and B. subtilis GD-4 and Candida sp. JX-3 revealed high degradation rates for BAs (Table 5). Those four strains were probably the beneficial candidates for controlling the BAs, and thus their BAs-controlling capacities were evaluated in a fermented soybean model. After fermentation for 7 days, the total BAs content of the control sample without inoculation reached 284.88 mg/Kg (Fig. 3), which might be due to the native microbes in soybean. After inoculation with B. subtilis HB-1 and S. pasteuri JX-2, the total BAs concentrations were much lower than that of control (Fig. 3). Meanwhile, the highest BAs content (432.72 mg/kg) was detected in the soybean inoculated with Candida sp. JX-3. These results might be due to their different BAs producing ability. Among the four strains, B. subtilis HB-1 and S. pasteuri JX-2 had no BAs formation ability, with Candida sp. JX-3 exhibiting the highest BAs formation ability (367.2 mg/L). The B. subtilis HB-1 and S. pasteuri JX-2 with no BAs producing ability were more beneficial to reduce the BAs accumulation in fermented soybean product, and their existence probably inhibited the growth of BAs-producing strains by competition for nutrients, which might be the main reason for the decreased BAs. Eom et al. also observed that B. subtilis isolated from a Korean traditional fermented food was able to inhibit BA production50. In addition, Staphylococcus xylosus was used as a starter culture to inhibit BA production in a salted and fermented anchovy55. Similarly, the B. subtilis HB-1 and S. pasteuri JX-2 isolated in this study can be used as potential candidates for BA control in fermented soybean.
Figure 3.
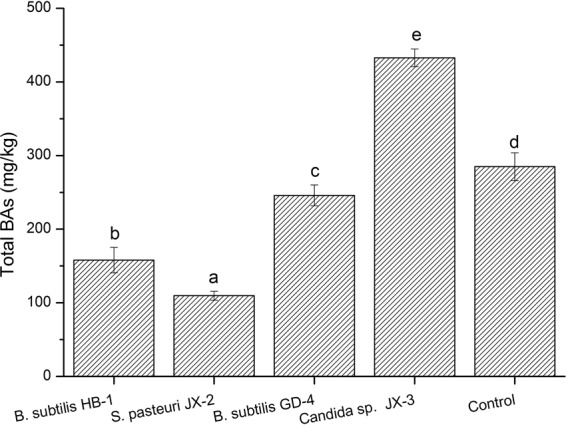
BAs contents in fermented soybean product model inoculated with different strians. Different letters (a, b, c, etc.) indicate significantly different means at P < 0.05.
Conclusion
In summary, the BAs profiles of 15 representative Douchi samples were analyzed and the correlation between microbes and BAs were also investigated. The total BAs contents of all the Douchi samples were within the acceptable dose level, while histamine and β-phenethylamine showed above the toxic level in some samples, indicating that evaluation and control of specific amine are important for Douchi safety. High-throughput sequencing showed that Bacillus and Candida were the dominant genera. Nineteen strains were isolated from Douchi samples to investigate microbial contribution to BAs accumulation. B. subtilis HB-1 and S. pasteuri JX-2 showed no BAs formation ability, and B. subtilis GD-4 and Candida sp. JX-3 revealed high BAs degradation abilities. Moreover, fermented soybean model further confirmed that B. subtilis HB-1 and S. pasteuri JX-2 would be the potential candidates for BAs control. This study not only explained the microbial contribution to BAs in Douchi, but also provided potential strains to reduce the BAs in fermented soybean products.
Electronic supplementary material
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 31501468), and the Fundamental Research Funds for the Central Universities (No. 2662016PY121).
Author Contributions
X. Wei designed the study. L. Li, L. Ruan and A. Ji executed the experimental work. L. Li analyzed the data. X. Wei contributed reagents and materials. L. Li, X. Wei, Z. Wen, S. Chen and L. Wang wrote and revised the manuscript. All authors read and approved the final manuscript.
Data availability
All relevant data are within the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-30456-z.
References
- 1.Halász A, Baráth Á, Simon-Sarkadi L, Holzapfel W. Biogenic amines and their production by microorganisms in food. Trends. Food Sci. Tech. 1994;5:42–49. doi: 10.1016/0924-2244(94)90070-1. [DOI] [Google Scholar]
- 2.Lee S, Yoo M, Shin D. The identification and quantification of biogenic amines in Korean turbid rice wine, Makgeolli by HPLC with mass spectrometry detection. LWT-Food Sci. Technol. 2015;62:350–356. doi: 10.1016/j.lwt.2015.01.016. [DOI] [Google Scholar]
- 3.Yang J, Ding X, Qin Y, Zeng Y. Safety assessment of the biogenic amines in fermented soya beans and fermented bean curd. J. Agric. Food. Chem. 2014;62:7947–7954. doi: 10.1021/jf501772s. [DOI] [PubMed] [Google Scholar]
- 4.Kim B, Byun BY, Mah JH. Biogenic amine formation and bacterial contribution in Natto products. Food Chem. 2012;135:2005–2011. doi: 10.1016/j.foodchem.2012.06.091. [DOI] [PubMed] [Google Scholar]
- 5.Cheng W, Sun D-W, Cheng J-H. Pork biogenic amine index (BAI) determination based on chemometric analysis of hyperspectral imaging data. LWT-Food Sci. Technol. 2016;73:13–19. doi: 10.1016/j.lwt.2016.05.031. [DOI] [Google Scholar]
- 6.Tsai Y-H, et al. Occurrence of histamine and histamine-forming bacteria in salted mackerel in Taiwan. Food Microbiol. 2005;22:461–467. doi: 10.1016/j.fm.2004.11.003. [DOI] [Google Scholar]
- 7.Toro-Funes N, Bosch-Fuste J, Latorre-Moratalla ML, Veciana-Nogués MT, Vidal-Carou MC. Biologically active amines in fermented and non-fermented commercial soybean products from the Spanish market. Food Chem. 2015;173:1119–1124. doi: 10.1016/j.foodchem.2014.10.118. [DOI] [PubMed] [Google Scholar]
- 8.Jung WY, Jung JY, Lee HJ, Jeon CO. Functional Characterization of Bacterial Communities Responsible for Fermentation of Doenjang: A Traditional Korean Fermented Soybean Paste. Front Microbiol. 2016;7:827. doi: 10.3389/fmicb.2016.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzzi G, Torriani S. Editorial: Biogenic amines in foods. Front Microbiol. 2015;6:472. doi: 10.3389/fmicb.2015.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai X, Byun BY, Mah J-H. Formation and destruction of biogenic amines in Chunjang (a black soybean paste) and Jajang (a black soybean sauce) Food Chem. 2013;141:1026–1031. doi: 10.1016/j.foodchem.2013.03.054. [DOI] [PubMed] [Google Scholar]
- 11.Chen T, Wang M, Li S, Wu Q, Wei H. Molecular identification of microbial community in surface and undersurface douchi during postfermentation. J. Food Sci. 2014;79:M653–658. doi: 10.1111/1750-3841.12417. [DOI] [PubMed] [Google Scholar]
- 12.del Rio B, et al. The dietary biogenic amines tyramine and histamine show synergistic toxicity towards intestinal cells in culture. Food Chem. 2017;218:249–255. doi: 10.1016/j.foodchem.2016.09.046. [DOI] [PubMed] [Google Scholar]
- 13.Shalaby AR. Significance of biogenic amines to food safety and human health. Food Res. Int. 1996;29:675–690. doi: 10.1016/S0963-9969(96)00066-X. [DOI] [Google Scholar]
- 14.Chen T, et al. Investigation of the microbial changes during koji-making process of Douchi by culture-dependent techniques and PCR-DGGE. Int. J. Food Sci. Tech. 2011;46:1878–1883. doi: 10.1111/j.1365-2621.2011.02696.x. [DOI] [Google Scholar]
- 15.Chen T, et al. Molecular identification of microbial community in Chinese douchi during post-fermentation process. Food Sci. BioTechnol. 2011;20:1633–1638. doi: 10.1007/s10068-011-0225-0. [DOI] [Google Scholar]
- 16.Yang L, Yang HL, Tu ZC, Wang XL. High-Throughput Sequencing of Microbial Community Diversity and Dynamics during Douchi Fermentation. PLoS One. 2016;11:e0168166. doi: 10.1371/journal.pone.0168166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai Y-H, Kung H-F, Chang S-C, Lee T-M, Wei C-I. Histamine formation by histamine-forming bacteria in douchi, a Chinese traditional fermented soybean product. Food Chem. 2007;103:1305–1311. doi: 10.1016/j.foodchem.2006.10.036. [DOI] [Google Scholar]
- 18.Kasankala LM, Xiong YL, Chen J. The influence of douchi starter cultures on the composition of extractive components, microbiological activity, and sensory properties of fermented fish pastes. J. Food Sci. 2011;76:C154–161. doi: 10.1111/j.1750-3841.2010.01971.x. [DOI] [PubMed] [Google Scholar]
- 19.Gong X, et al. Determination of biogenic amines in traditional Chinese fermented foods by reversed-phase high-performance liquid chromatography (RP-HPLC) Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2014;31:1431–1437. doi: 10.1080/19440049.2014.926402. [DOI] [PubMed] [Google Scholar]
- 20.Li L, et al. Evaluation of the Biogenic Amines Formation and Degradation Abilities of Lactobacillus curvatus From Chinese Bacon. Front Microbiol. 2018;9:1015. doi: 10.3389/fmicb.2018.01015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladero V, et al. Genetic and functional analysis of biogenic amine production capacity among starter and non-starter lactic acid bacteria isolated from artisanal cheeses. Eur. Food Res. Technol. 2015;241:377–383. doi: 10.1007/s00217-015-2469-z. [DOI] [Google Scholar]
- 22.Landete JM, de las Rivas B, Marcobal A, Muñoz R. Molecular methods for the detection of biogenic amine-producing bacteria on foods. Int. J. Food Microbiol. 2007;117:258–269. doi: 10.1016/j.ijfoodmicro.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Takebe Y, et al. Evaluation of the Biogenic Amine-production Ability of Lactic Acid Bacteria Isolated from Tofu-misozuke. Food Sci. Technol Res. 2016;22:673–678. doi: 10.3136/fstr.22.673. [DOI] [Google Scholar]
- 24.Xia X, Zhang Q, Zhang B, Zhang W, Wang W. Insights into the Biogenic Amine Metabolic Landscape during Industrial Semidry Chinese Rice Wine Fermentation. J. Agric. Food. Chem. 2016;64:7385–7393. doi: 10.1021/acs.jafc.6b01523. [DOI] [PubMed] [Google Scholar]
- 25.Liu SP, et al. Sequencing-based screening of functional microorganism to decrease the formation of biogenic amines in Chinese rice wine. Food Control. 2016;64:98–104. doi: 10.1016/j.foodcont.2015.12.013. [DOI] [Google Scholar]
- 26.Chen T, et al. Application of denaturing gradient gel electrophoresis to microbial diversity analysis in Chinese Douchi. J. Sci. Food Agric. 2012;92:2171–2176. doi: 10.1002/jsfa.5604. [DOI] [PubMed] [Google Scholar]
- 27.Chen C, et al. Identification of key micro-organisms involved in Douchi fermentation by statistical analysis and their use in an experimental fermentation. J. Appl. Microbiol. 2015;119:1324–1334. doi: 10.1111/jam.12917. [DOI] [PubMed] [Google Scholar]
- 28.Liu C-j, et al. Natural populations of lactic acid bacteria in douchi from Yunnan Province, China. J. Zhejiang Univ Sci B. 2012;13:298–306. doi: 10.1631/jzus.B1100221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Meth. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 34.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.ten Brink B, Damink C, Joosten HMLJ, Huis in ‘t Veld JHJ. Occurrence and formation of biologically active amines in foods. Int. J. Food Microbiol. 1990;11:73–84. doi: 10.1016/0168-1605(90)90040-C. [DOI] [PubMed] [Google Scholar]
- 36.Santos MHS. Biogenic amines: their importance in foods. Int. J. Food Microbiol. 1996;29:213–231. doi: 10.1016/0168-1605(95)00032-1. [DOI] [PubMed] [Google Scholar]
- 37.Tang T, et al. Monitoring the contents of biogenic amines in sufu by HPLC with SPE and pre-column derivatization. Food Control. 2011;22:1203–1208. doi: 10.1016/j.foodcont.2011.01.018. [DOI] [Google Scholar]
- 38.Rauscher-Gabernig E, et al. Dietary exposure assessment of putrescine and cadaverine and derivation of tolerable levels in selected foods consumed in Austria. Eur. Food Res. Technol. 2012;235:209–220. doi: 10.1007/s00217-012-1748-1. [DOI] [Google Scholar]
- 39.Quigley L, et al. High-Throughput Sequencing for Detection of Subpopulations of Bacteria Not Previously Associated with Artisanal Cheeses. Appl. Environ. Microbiol. 2012;78:5717–5723. doi: 10.1128/AEM.00918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chettri R, Tamang JP. Bacillus species isolated from tungrymbai and bekang, naturally fermented soybean foods of India. Int. J. Food Microbiol. 2015;197:72–76. doi: 10.1016/j.ijfoodmicro.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 41.Nam Y-D, Lee S-Y, Lim S-I. Microbial community analysis of Korean soybean pastes by next-generation sequencing. Int. J. Food Microbiol. 2012;155:36–42. doi: 10.1016/j.ijfoodmicro.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 42.Nam Y-D, Yi S-H, Lim S-I. Bacterial diversity of cheonggukjang, a traditional Korean fermented food, analyzed by barcoded pyrosequencing. Food Control. 2012;28:135–142. doi: 10.1016/j.foodcont.2012.04.028. [DOI] [Google Scholar]
- 43.Cobo Molinos A, Abriouel H, Ben Omar N, López RL, Gálvez A. Microbial diversity changes in soybean sprouts treated with enterocin AS-48. Food Microbiol. 2009;26:922–926. doi: 10.1016/j.fm.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 44.Zhang W, et al. Microbial diversity in two traditional bacterial douchi from Gansu province in northwest China using Illumina sequencing. PLoS One. 2018;13:e0194876. doi: 10.1371/journal.pone.0194876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeyaram K, et al. Molecular identification of dominant microflora associated with ‘Hawaijar’ — A traditional fermented soybean (Glycine max (L.)) food of Manipur, India. Int. J. Food Microbiol. 2008;122:259–268. doi: 10.1016/j.ijfoodmicro.2007.12.026. [DOI] [PubMed] [Google Scholar]
- 46.Yan YZ, Qian YL, Ji FD, Chen JY, Han BZ. Microbial composition during Chinese soy sauce koji-making based on culture dependent and independent methods. Food Microbiol. 2013;34:189–195. doi: 10.1016/j.fm.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 47.Garcia NFL, et al. Production of β-glucosidase on solid-state fermentation by Lichtheimia ramosa in agroindustrial residues: Characterization and catalytic properties of the enzymatic extract. Electron. J. Biotechnol. 2015;18:314–319. doi: 10.1016/j.ejbt.2015.05.007. [DOI] [Google Scholar]
- 48.Mi YY, Kim YJ, Hwang HJ. Properties and safety aspects of Enterococcus faecium strains isolated from Chungkukjang, a fermented soy product. LWT-Food Sci. Technol. 2008;41:925–933. doi: 10.1016/j.lwt.2007.05.024. [DOI] [Google Scholar]
- 49.Zhen F, Wei G, Dan R, Xi C, Juan‐juan L. Evaluation of bacterial flora during the ripening of Kedong sufu, a typical Chinese traditional bacteria-fermented soybean product. J. Sci. Food Agric. 2013;93:1471–1478. doi: 10.1002/jsfa.5918. [DOI] [PubMed] [Google Scholar]
- 50.Eom JS, Seo BY, Choi HS. Biogenic Amine Degradation by Bacillus Species Isolated from Traditional Fermented Soybean Food and Detection of Decarboxylase-Related Genes. J. Microbiol. Biotechn. 2015;25:1519. doi: 10.4014/jmb.1506.06006. [DOI] [PubMed] [Google Scholar]
- 51.Kim S-J, Dunlap CA, Kwon S-W, Rooney AP. Bacillus glycinifermentans sp. nov., isolated from fermented soybean paste. Int. J. Syst. Evol. Micr. 2015;65:3586–3590. doi: 10.1099/ijsem.0.000462. [DOI] [PubMed] [Google Scholar]
- 52.Linjiang Z, Zihao F, Hui K, Qi L. Batch‐batch stable microbial community in the traditional fermentation process of huyumei broad bean pastes. Lett. Appl. Microbiol. 2017;65:226–233. doi: 10.1111/lam.12765. [DOI] [PubMed] [Google Scholar]
- 53.Marcobal A, De LRB, Muñoz R. First genetic characterization of a bacterial beta-phenylethylamine biosynthetic enzyme in Enterococcus faecium RM58. FEMS Microbiol. Lett. 2006;258:144–149. doi: 10.1111/j.1574-6968.2006.00206.x. [DOI] [PubMed] [Google Scholar]
- 54.Lee Y-C, Lin C-S, Liu F-L, Huang T-C, Tsai Y-H. Degradation of histamine by Bacillus polymyxa isolated from salted fish products. J. Food. Drug. Anal. 2015;23:836–844. doi: 10.1016/j.jfda.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mah JH, Hwang HJ. Inhibition of biogenic amine formation in a salted and fermented anchovy by Staphylococcus xylosus as a protective culture. Food Control. 2009;20:796–801. doi: 10.1016/j.foodcont.2008.10.005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the paper.