The gonococcal NorM efflux pump exports substrates with a cationic moiety, including quaternary ammonium compounds such as berberine (BE) and ethidium bromide (EB) as well as antibiotics such as ciprofloxacin and solithromycin. The norM gene is part of a four-gene operon that is transcribed from a promoter containing a polynucleotide tract of 6 or 7 thymidines (T's) between the −10 and −35 hexamers; the majority of gonococcal strains analyzed in this study contained a T-6 sequence.
KEYWORDS: NorM, efflux pumps, gonorrhea, resistance
ABSTRACT
The gonococcal NorM efflux pump exports substrates with a cationic moiety, including quaternary ammonium compounds such as berberine (BE) and ethidium bromide (EB) as well as antibiotics such as ciprofloxacin and solithromycin. The norM gene is part of a four-gene operon that is transcribed from a promoter containing a polynucleotide tract of 6 or 7 thymidines (T's) between the −10 and −35 hexamers; the majority of gonococcal strains analyzed in this study contained a T-6 sequence. Primer extension analysis showed that regardless of the length of the poly(T) tract, the same transcriptional start site (TSS) was used for expression of norM. Interestingly, the T-6 tract correlated with a higher level of both norM expression and gonococcal resistance to NorM substrates BE and EB. Analysis of expression of genes downstream of norM showed that the product of the tetR-like gene has the capacity to activate expression of norM as well as murB, which encodes an acetylenolpyroylglucosamine reductase predicted to be involved in the early steps of peptidoglycan synthesis. Moreover, loss of the TetR-like transcriptional regulator modestly increased gonococcal susceptibility to NorM substrates EB and BE. We conclude that both cis- and trans-acting regulatory systems can regulate expression of the norM operon and influence levels of gonococcal susceptibility to antimicrobials exported by NorM.
INTRODUCTION
Neisseria gonorrhoeae is a strict human pathogen and is the etiologic agent of the sexually transmitted infection (STI) termed gonorrhea, which is the second most prevalent bacterial STI in the United States and had a worldwide incidence in 2012 of an estimated 78 million infections (1). The gonococcus has adapted numerous strategies to survive attack by antimicrobials, including classical antibiotics used in treatment of infection and those of host origin that participate in innate host defense. In this respect, gonococci use efflux pumps to resist the antimicrobial action of beta-lactam and macrolide antibiotics as well as cationic antimicrobial peptides and long-chain fatty acids (2–4). The capacity of gonococci to utilize efflux pumps to resist clinically useful antibiotics is of interest given the emergence of strains resistant to current and past front-line antibiotics (2, 5–7). The contribution of efflux pumps in aiding bacterial evasion of antimicrobials can be enhanced by mutations that derepress expression of efflux pump-encoding genes (8). With respect to gonococci, previous work revealed that overexpression of the mtrCDE efflux pump operon due to cis- or trans-acting mutations can contribute to clinically relevant levels of antibiotic resistance (9, 10) and increase bacterial fitness during experimental infection of the lower genital tract of female mice, presumably due to enhanced resistance to host antimicrobials (11, 12).
In this study, we investigated the regulation of the gonococcal norM gene. NorM belongs to the multidrug and toxic compound extrusion (MATE) family of efflux proteins, which are Na+- or H+-coupled transporters and are present in all living organisms (13). Gonococcal NorM is highly similar (56%) to NorM of Vibrio parahaemolyticus (14). We previously reported that NorM can export substrates with a cationic moiety, including berberine (BE), ciprofloxacin (CIP), and ethidium bromide (EB) (15). Additionally, the loss of the NorM efflux pump in multidrug-resistant strain H041 was found by Golparian et al. (6) to increase gonococcal susceptibility to solithromycin. In this study, we investigated cis- and trans-acting regulatory mechanisms that influence norM expression and the consequence of such on antimicrobial resistance. Importantly, we identified a heretofore undescribed TetR-like regulator that activated the norM gene as well as a single-base-pair deletion that resulted in a stronger norM promoter.
RESULTS AND DISCUSSION
cis-Acting transcriptional regulation of norM in N. gonorrhoeae and influence on antimicrobial resistance.
Bioinformatics analysis (http://www.ncbi.nlm.nih.gov/gene) indicated that norM (NGO0395) is the first gene of an operon that also contains three downstream genes annotated as murB (NGO0394), which encodes a putative UDP-N-acetylenolpyruvoylglucosamine reductase involved in the initial steps of the peptidoglycan synthesis (Fig. 1A) (24), NGO0393, which encodes a TetR-like family transcriptional regulator homolog, and NGO0392, which encodes a hypothetical protein. Using total RNA prepared from strain FA19StrR in reverse transcription-PCR (RT-PCR) experiments, we confirmed transcriptional linkage of norM and murB as well as murB and tetR (see Fig. S1 in the supplemental material), which supports the hypothesis that the genes form an operon. Primer extension analysis of this RNA indicated the presence of 2 distinct transcriptional start sites (TSSs). One TSS was located upstream of norM that corresponded to that described previously by our group (15) as well as another TSS located upstream of tetR. This result suggests the presence of two distinct promoters that express genes within the operon, one capable of directing transcription of the entire operon and the second driving the transcription of tetR and possibly NGO0392 (Fig. 1B).
FIG 1.
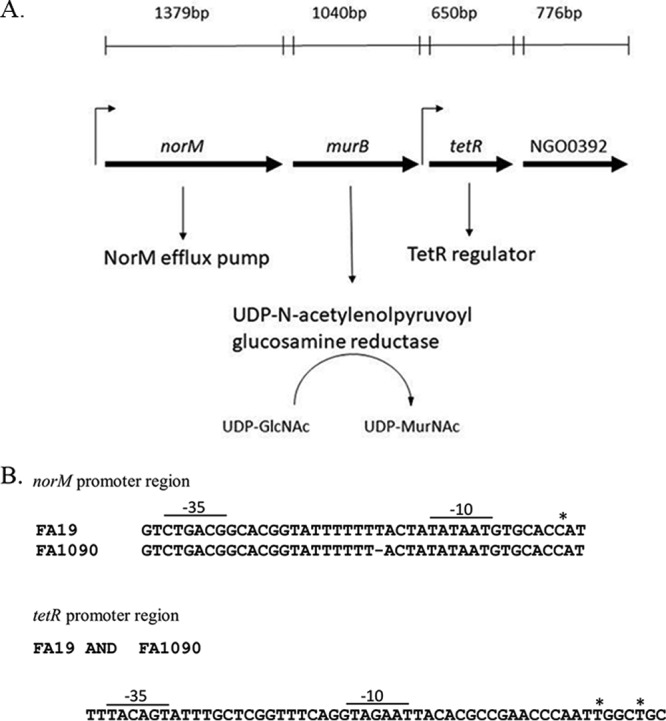
(A) The organization of the norM operon is depicted. The length and transcriptional direction (arrows) of the genes are shown. (B) Sequences of the norM and tetR promoter regions from strains FA19 and FA1090. The −10 and −35 hexamers are indicated. The asterisk represents the TSS.
DNA sequencing of the norM promoter region of strain FA19StrR revealed the presence of a stretch of 7 T's between the −10 and −35 hexamers (Fig. 1B). In order to learn if this poly(T) stretch is common among gonococci, we performed a bioinformatics analysis of a 200-bp region upstream of the norM translational start codon using 31 gonococcal whole-genome sequences that are available online (http://blast.ncbi.nlm.nih.gov). This analysis revealed that the majority (77.4%) of gonococcal strains had a stretch of 6 T's (including strains FA1090 and certain WHO reference strains), while the rest (22.6%) had 7 T's (including strains FA19 and MS11). Using a PCR-generated product, we also sequenced this norM upstream region from 10 clinical isolates and found that 9 strains had the T-6 sequence (Table S1). Thus, we conclude that the T-6 sequence predominates in gonococci. In contrast, our analysis of whole-genome sequences of 86 Neisseria meningitidis strains that are publicly available (http://blast.ncbi.nlm.nih.gov) revealed that 85 (99%) have a norM promoter with a T-7 repeat sequence (data not presented).
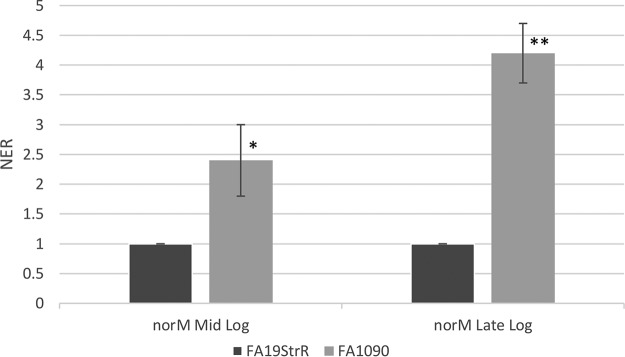
Despite the difference in T repeat length, primer extension analysis revealed that the TSS positioned upstream of the norM promoter was possessed by strains FA19 (T-7) and FA1090 (T-6), which was identified as a C residue located 6 bp downstream of the −10 hexamer (data not presented; summarized in Fig. 1B). The level of the norM transcript in strains FA19StrR and FA1090 was determined by quantitative RT-PCR (qRT-PCR) analysis using total RNA prepared from mid-logarithmic and late logarithmic cultures, which showed that the norM transcript was 2.4- and 4.2-fold higher in strain FA1090 than in FA19StrR at the mid-logarithmic and late logarithmic phases, respectively (Fig. 2). Previous studies on the regulation of the mtrCDE efflux pump-encoding operon revealed that the distance between the −10 and −35 promoter hexamers can significantly influence transcription and levels of gonococcal resistance to antimicrobials exported by MtrCDE (9, 16). Guided by this work, we constructed norM mutants of FA1090 and FA19StrR by insertional inactivation with the nonpolar aphA-3 cassette and found that while the loss of norM influenced gonococcal susceptibility to NorM substrates (BE and EB), the impact was greater in strain FA1090 (T-6 promoter) than in strain FA19StrR (T-7 promoter) (Table 1). In order to determine if inactivation of norM would increase susceptibility of a more recent gonococcal clinical isolate displaying resistance to multiple antibiotics (6), we constructed a norM::kan transformant of strain H041 (T-6 promoter) (Table S1). We found that the loss of the NorM efflux pump decreased resistance of H041 to both BE and EB as well as solithromycin (4-fold decrease in MIC [Table S2]).
FIG 2.
Quantitative RT-PCR results with norM in strains FA19StrR and strain FA1090 at the mid-logarithmic and late logarithmic phases of growth. Error bars represent SDs from the means of 3 independent experiments. Normalized expression ratios (NER) were calculated using 16S rRNA expression. *, P = 0.018; **, P = 0.004 (for comparison of values of FA1090 versus FA19StrR). The statistical significance was determined by Student's t test.
TABLE 1.
Susceptibility of gonococcal strains to NorM substrates
| Strain | MIC (μg/ml)a |
|
|---|---|---|
| BE | EB | |
| FA19StrR | 5 | 1.25 |
| FA19StrR norM::kan | 1.25 | 0.625 |
| FA19StrR norM::kanC3 | 5 | 1.25 |
| FA19StrR tetR::kan | 2.5 | 0.625 |
| FA19StrR tetR::kanC4 | 5 | 1.25 |
| FA19StrRPnorMFA1090 | 10 | 2.5 |
| FA 1090 | 20 | 5 |
| FA 1090 norM::kan | 1.25 | 0.625 |
| FA 1090 tetR::kan | 20 | 5 |
All results are representative of those from from 3 or more independent determinations. BE, berberine; EB, ethidium bromide.
In order to determine the influence of the norM promoter T repeat sequence on gonococcal expression of the norM operon and resistance to NorM substrates, we exchanged the norM promoter region of FA19StrR with that of FA1090 by transformation. DNA sequencing of a PCR fragment from a representative transformant strain (FA19StrRPnorMFA1090) confirmed the presence of the T-6 instead of T-7 repeat element (data not presented). Importantly, FA19StrRPnorMFA1090 displayed a 5-fold increase in expression of norM as assessed by qRT-PCR (Fig. 3) and displayed a 2-fold increase in resistance to EB and BE compared to parental strain FA19StrR (Table 1). These combined results indicated that the length of the T tract can influence levels of gonococcal expression of norM and resistance to NorM substrates.
FIG 3.
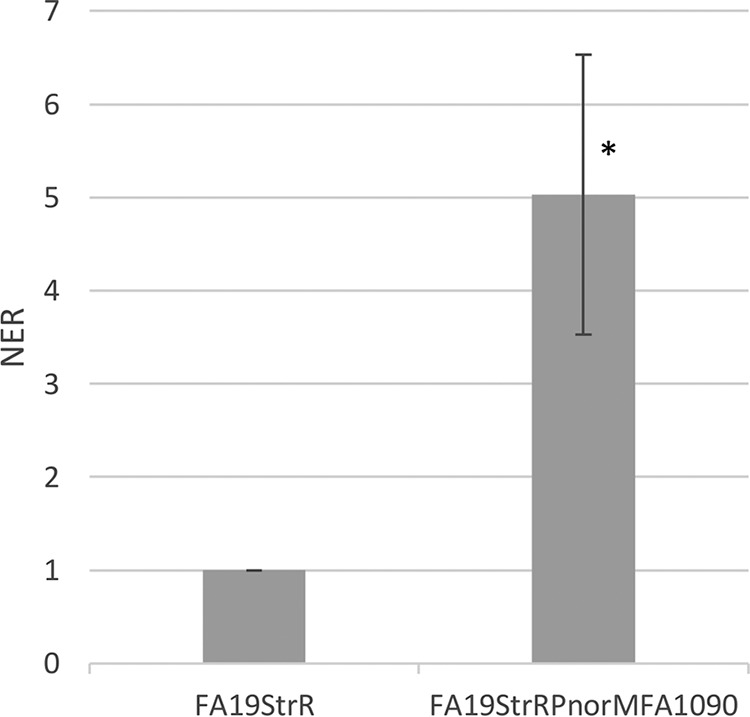
Quantitative RT-PCR results with norM in strains FA19StrR and strain FA19StrRPnorMFA1090 at mid-logarithmic phase of growth. Error bars represent SDs from the means of 3 independent experiments. NER were calculated using 16S rRNA expression. *, P = 0.01. The statistical significance of the results was determined by Student's t test.
trans-Acting transcriptional regulation of norM and influence on antimicrobial resistance.
Bioinformatics analysis revealed that the putative TetR-like protein (216 amino acids) encoded by a gene within the norM operon is highly conserved in gonococci. This finding is exemplified by 100% amino acid identity of the protein that would be produced by strains FA19 and FA1090; the protein is also highly similar (97% identical) to a counterpart encoded by meningococci (data not presented). That this TetR-like protein can act as a transcriptional regulator was suggested by the presence of a helix-turn-helix DNA-binding domain at the N terminus (amino acids 15 to 61). Further, the position of tetR downstream of norM suggested that the TetR-like protein might exert transcriptional control of norM and other genes (e.g., murB) in the operon. In order to test this possibility, tetR::kan mutants of strains FA19StrR and FA1090 were constructed and analyzed for changes in susceptibility to NorM substrates and levels of gene transcripts within the operon. We noted that with strain FA19StrR, but not FA1090, insertional inactivation of tetR reproducibly resulted in 2-fold decrease in gonococcal resistance to BE and EB (Table 1). Although the impact of loss of the TetR-like protein was modest, complementation of the FA19StrR tetR::kan strain with a pGCC4 construct bearing the wild-type tetR gene expressed at the aspC-lctP locus from an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lac promoter restored wild-type levels of antimicrobial susceptibility (Table 1).
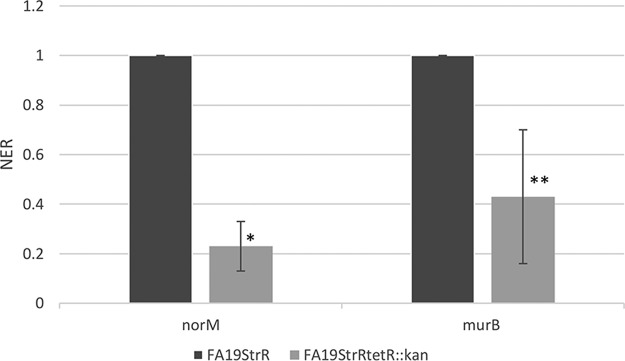
Based on the above-described results, we used strain FA19StrR to ascertain if the TetR-like protein could regulate the norM operon. Results from qRT-PCR experiments indicated that loss of the TetR-like protein decreased expression of both norM and murB (Fig. 4), which is consistent with the transcriptional linkage of these genes by a promoter upstream of norM. To investigate if the TetR homolog could directly activate transcription of the norM operon, a recombinant His-tagged TetR protein was purified and employed in competitive electrophoretic mobility shift assays (EMSAs) that used a radiolabeled PCR probe containing 344 bp of DNA upstream of norM. The results from DNA binding experiments showed that TetR could bind to the probe in a specific manner, as binding could be inhibited by the unlabeled norM PCR product, but not by a nonspecific PCR probe (Fig. 5). Thus, this gene regulator serves as a transcriptional activator of the norM operon.
FIG 4.
Quantitative RT-PCR results with norM and murB in strain FA19StrR and strain FA19StrRtetR::kan at the mid-logarithmic phase of growth. Error bars represent SDs from the means of 3 independent experiments. NER were calculated using 16S rRNA expression. *, P = 0.0004; **, P = 0.022 (for comparison of values of FA19StrR tetR::kan versus FA19StrR). The statistical significance of the results was determined by Student's t test.
FIG 5.
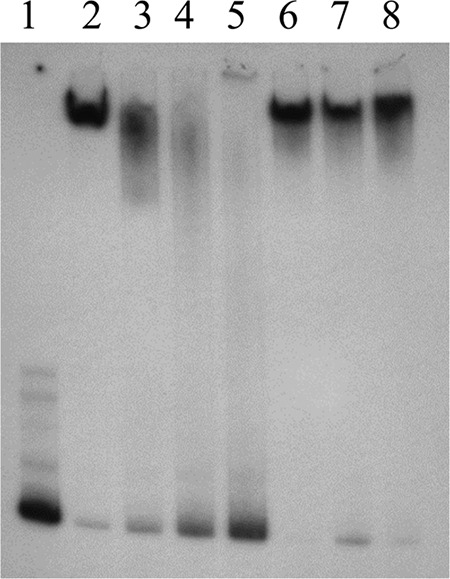
Competitive EMSA. The purified TetR-His protein binds to the norM promoter from strain FA19 in a specific manner. Lane 1, hot probe N11/N14* alone; lane 2, hot probe N11/N14* plus 8 μg of TetR-His; lanes 3 to 5, hot probe N11/N14* plus 8 μg of TetR-His plus 100×, 200×, and 400×, respectively, unlabeled N11/N14; lanes 6 to 8, hot probe N11/N14* plus 8 μg of TetR-His plus 100×, 200×, and 400×, respectively, unlabeled rnpB.
As a member of the MATE family of efflux pumps, the gonococcal NorM efflux pump has the capacity to export antimicrobial quaternary ammonium compounds (reference 15 and Table 1). The conservation of norM among gonococci suggests a role for NorM in the survival of gonococci. Thus, we hypothesized that NorM might also export host-derived antimicrobials and promote survival of gonococci during infection. However, using the established female mouse model of lower genital tract infection previously employed to determine the biological significance of the gonococcal MtrCDE efflux pump and cognate regulatory systems (11, 17), we did not detect a fitness or survival defect of gonococci (FA19StrR and FA1090) bearing a null mutation in norM when competed with the wild-type parent strains (Fig. S2). It is important to note that this model may not fully recapitulate the repertoire of antimicrobials present at human female or male mucosal surfaces. Moreover, the infection model employed is limited to the lower genital tract of female mice, and distinct antimicrobials in the upper tract that might serve as NorM substrates could exist. For instance, differences in the presence and level of antimicrobial peptides have been reported at mucosal surfaces of the human ectocervix and endocervix (18). Hence, the possibility that NorM promotes survival of gonococci during human infection by promoting resistance to a host antimicrobial cannot be discounted.
As with other bacterial efflux pump-encoding genes (8, 9, 16), we conclude that the gonococcal norM gene is subject to transcriptional control that influences its expression and levels of bacterial resistance to antimicrobials that can be exported by NorM. It is of interest that both cis- and trans-acting regulatory processes identified in this study can modulate expression of norM and that these regulatory schemes seem dependent on the length a poly(T) tract in the norM promoter. The majority of gonococci contain a T-6 tract in the promoter that seems to enhance transcription of norM. In contrast, strain FA19, which we have used extensively in our work on gonococcal efflux pumps (3, 4, 15–19), is representative of the minority of gonococcal strains harboring a T-7 sequence. Since mutations that increase or decrease spacing between the −10 and −35 hexamers can influence the fidelity of gene expression due to impacting interactions of RNA polymerase, as has been observed with nucleotide deletions or insertions within the mtrR promoter (16, 19), it is likely that the single T difference can impact norM expression in gonococcal strains with a T-6 or T-7 sequence by influencing promoter recognition by RNA polymerase.
In addition to this cis-acting regulatory mechanism, the TetR-like protein encoded by a gene within the norM operon can influence expression of norM in strain FA19. Importantly, the TetR DNA-binding protein also activates expression of murB, which is consistent with its transcriptional linkage with norM. It is of interest that a gene (murB) encoding an enzyme involved in the earliest steps of peptidoglycan biosynthesis (19) is coregulated with norM by both cis- and trans-acting regulatory schemes. Thus, the fidelity of early stages of peptidoglycan biosynthesis may be modulated by transcriptional control systems that also influence expression of norM and levels of gonococcal resistance to antimicrobials exported by NorM.
The chemical characteristics of known substrates of the gonococcal NorM efflux pump suggest that the clinical efficacy of future antimicrobials having similar properties may be influenced by constitutive or inducible changes in norM expression. We hypothesize that increased expression of norM coupled with other mutations could result in clinical resistance to antibiotics used in the future for treatment of gonorrhea. Based on earlier work with multidrug-resistant strain H041 by Golparian et al. (6) and our findings with this clinical isolate, this possibility should be considered for solithromycin and its future use in treatment of gonorrhea. In a broader sense, derepression of bacterial efflux pump genes due to constitutive mutations as well as inducible activation systems should be a contributing factor by which gonococci (or other bacteria) might develop clinical resistance to antibacterials under development.
MATERIALS AND METHODS
Gonococcal strains, growth conditions, and determination of susceptibility to antimicrobial agents.
Strains FA19, FA19StrR and FA1090 were the primary gonococcal strains used in this study. These strains and their genetic derivatives as well as their susceptibilities to antimicrobials are presented in Table 1. We also sequenced the norM promoter regions from 10 clinical isolates (Table S1; see below). Gonococcal strains were grown overnight at 37°C under 5% (vol/vol) CO2 on GC agar containing defined supplements I and II (9). The susceptibilities of test strains to antibiotics were determined by the agar dilution method and reported as the MIC (20). IPTG was added at a final concentration of 1 mM to MIC plates to allow complementation by the pGCC4 vector (21). Antibiotics were purchased from Sigma Chemical Co. (St. Louis, MO). Solithromycin was obtained from Med Chem Express (Monmouth, NJ). E. coli strains were grown overnight at 37°C on LB agar.
Sequencing of the norM promoter.
The norM promoter region was PCR amplified from gonococci using primers norMPac1 (5′-GATCTTAATTAACAATGCCGTCAAGTCGTTAAA-3′) and N10 (5′-CATCACGGTATCGACGAAACGATGCCC-3′). The resulting PCR product was sequenced using norMPac1.
Construction of the norM and tetR-negative mutants and their complemented strains.
The pBADnorM::kan construct (15) was transformed into FA19StrR, and transformants were selected on GC agar supplemented with 50 μg/ml of kanamycin (Kan). FA19StrR norM::kan transformants were verified by PCR and sequencing. The pGCC3 vector (21) was used to complement FA19StrR norM::kan. This complementation system allows the integration of a wild-type copy of norM under its own promoter at the transcriptionally silent intergenic region between lctP and aspC. norMPac1 and norMpme1 (5′-GATCGTTTAAACTATCGGATGGGTTGCATGGT-3′) were used to amplify the norM gene and its own promoter. The resulting PCR product was cloned into the pGCC3 vector. The pGCC3norM construct was verified by sequencing and then transformed into FA19StrR norM::kan. FA19StrR norM::kanC3 transformants were selected on GC agar plates supplemented with 1 μg/ml of erythromycin (Ery) and verified by colony PCR and sequencing. The norM gene from FA1090 was amplified using primers N6 (5′-TCGGTATCGGATGGGTTGC-3′) and N4 (5′-ATGCTGCTCGACCTCGACC-3′), and the resulting PCR product was cloned into pBAD. pBADnorM was then digested by NaeI, and a nonpolar Kan resistance cassette from pUC18K (22) was inserted. The resulting construct was transformed into FA1090, and transformants were selected on GC agar plates supplemented with 50 μg/ml of Kan. FA1090 norM::kan transformants were verified by colony PCR and sequencing. To construct the tetR-negative mutant, pUC19 vector was digested by BamHI and EcoRI and PCR was performed on FA19 genomic DNA with primers E1tetR (5′-GGAATTCCTGTATGGGCAGGTTGATGTC-3′) and Sma1R (5′-TCCCCCGGGGGATCGCCCAACAATTCGGCAC-3′) and B1tetR (5′-CGCGGATCCGCGCTGAAGGGCTTCCAAATCGG-3′) and Sma1F (5′-TCCCCCGGGGGAACACAATACCTTTACCCAAGC-3′). The resulting PCR products were ligated into pUC9 digested with BamHI and EcoRI to create a Sma1 site 356 bp downstream the ATG of the tetR gene. The resulting construct was verified by PCR and then digested by SmaI. The Kan resistance cassette was PCR amplified with Pfu using primers AphF (5′-GTGACTAACTAGGAGGAATAAAT-3′) and AphR (5′-GGTCATTATTCCCTCCAGGTA-3′) and pUC18K (21) as a template. The Kan resistance cassette was then cloned into the SmaI-digested pUC19tetR. The ligation was transformed into E. coli DH5α, and transformants were selected on LB agar plates supplemented with 50 μg/ml of Kan. The resulting construct was then verified by sequencing and used to transform strains FA19StrR and FA1090 for resistance to Kan (50 μg/ml). The pGCC4 vector was used to complement FA19StrR tetR::kan. This complementation system allows the integration of a wild-type copy of tetR under the control of an IPTG-inducible promoter at the transcriptionally silent intergenic region between lctP and aspC. tetRpac1 (5′-GATCTTAATTAAAGCCTGTAAATCCAAGGAGTA-3′) and tetRpme1 (5′-GATCCGTTTAAACCGTCTGAAGGCTGATTCGG-3′) were used to amplify the tetR gene. The resulting PCR product was cloned into the pGCC4 vector. The pGCC4tetR construct was verified by sequencing and then transformed into FA19StrR tetR::kan. Transformant FA19StrR tetR::kanC4 was obtained by selection with 1 μg/ml of chloramphenicol (CMP), and the genotype was verified by colony PCR.
Construction of FA19StrRPnorMFA1090.
Primers N5 (5′-GGATGAACATCGGCACCTTG-3′) and norMPac1 (5′-GATCTTAATTAACAATGCCGTCAAGTCGTTAAA-3′) were used to amplify the norM promoter region of strain FA1090. The resulting 1,385-bp PCR product was then transformed into strain FA19StrR, and transformants were selected on GC agar containing defined supplements I and II supplemented with EB (1 μg/ml). Transformants were then verified by DNA sequencing of a PCR product generated using primers N5 and norMPacI.
Mapping transcriptional start sites by primer extension analysis.
Total RNA from strains FA19 and FA1090 was prepared at the late logarithmic phase of growth in GC broth as described above by the TRIzol method as directed by the manufacturer (Thermo Fisher Scientific, Waltham, MA). Primer extension experiments were performed as described previously (9, 16) on 6 μg of total RNA with primer N11 (5′-CGGTCAGCAGGCGGATTTCTTTCAGG-3′) for norM and primer tetRPE (5′-TGGCGTCGATGATGCGGG-3′) for tetR. Primer extension transcription start sites (TSSs) were determined by electrophoresis of the extension products on a 6% (wt/vol) DNA sequencing acrylamide gel adjacent to reference sequencing reactions.
Qualitative and quantitative RT-PCR.
For RT-PCR and qRT-PCR analyses of transcript levels, RNA was extracted from strains FA19StrR, FA1090, their respective norM-negative and tetR-negative mutants, and FA19StrRPnorMFA1090 grown in GC broth plus supplements to mid-logarithmic and late logarithmic phases by the TRIzol method as directed by the manufacturer (Thermo Fisher Scientific, Waltham, MA). Genomic DNA (gDNA) was removed by RNase-free DNase treatment and gDNA Wipeout (Qiagen, Germantown, MD). The resulting RNA was then reverse transcribed to cDNA using the QuantiTect reverse transcriptase kit (Qiagen). Quantitative real-time RT-PCR was performed using the generated cDNA, and results were normalized to 16S rRNA expression for each strain. Primers 16Smai_qRTF (5′-CCATCGGTATTCCTCCACATCTCT-3′) and 16Smai_qRTR (5′-CGTAGGGTGCGAGCGTTAATC-3′) were used for the 16S rRNA, while primers tetR_qRTR (5′-TTCCACATCAGAGGGCAACA-3′) and tetR_qRTF (5′-GCAACATCAGCACCAACCAT-3′) were used for the tetR gene. Primers N4 and N10 (5′-CATCACGGTATCGACGAAACCGATGCCC-3′) were used for the norM gene. Primers murB_qRTF (5′-TAAACACGCCGACGAATTGC-3′) and murB_qRTR (5′-TCTCGCGTATGCCCTTGTTT-3′) were used for the murB gene. All qRT-PCRs were performed in experimental duplicates and biological triplicates. For RT-PCR, random hexamers were used for the reverse transcription, while murB_qRTF and tetRSma1R (5′-TCCCCCGGGGGATCGCCCAACAATTCGGCAC-3′), N8 (5′-CCGTTCGGACTGACAGCG-3′), and murB_qRTR were used for PCRs on the cDNA, for the controls without reverse transcriptase, and for the genomic DNA.
Purification of the TetR protein.
Construction of pET15btetR was done by amplifying the tetR open reading frame using the primers pETtetR_F (5′-TCGATCCATATGCCCGTGACCCGCATTG-3′) and pETtetR_R (5′-GATCGGATCCTTACGGGTTGCCGTTGCCG-3′). The resulting PCR product along with the pET15b vector was digested with NdeI and BamHI, ligated overnight, and transformed into E. coli DH5α. The pET15btetR construct was confirmed by sequencing with vector-specific primers T7F (5′-TTAATACGACTCACTATAGG-3′) and T7R (5′-GCTAGTTATTGCTCAGCGG-3′). For protein expression, pET15btetR was transformed into E. coli BL21(DE3) cells. Cultures (5 ml) of BL21(DE3)-pET15btetR were grown overnight at 30°C and added to 500 ml of LB broth the next morning. The culture was grown at 30°C until mid-log phase, then induced with 0.3 mM IPTG, and grown overnight at 30°C. Cells were harvested and resuspended in 20 ml of 10 mM Tris (pH 7.5)–200 mM NaCl, and EDTA-free protease inhibitor was added to the bacterial suspension. The cells were lysed by use of a French press cell as described previously (23), membranes and unbroken cells were removed by centrifugation at 100,000 × g, and the supernatant was collected and filtered. TetR-His was purified over a 2-ml nickel-nitrilotriacetic acid (Ni2+-NTA) column. After the supernatant was allowed to flow over the Ni2+-NTA column, the resin was washed successively with buffer containing 20 mM and 50 mM imidazole to remove contaminants and weakly bound proteins, and TetR-His was eluted successively with buffer containing 100 and 200 mM imidazole. The fractions containing TetR-His were concentrated, and the imidazole-containing buffer was removed by dialysis into storage buffer (10 mM Tris-HCl [pH 7.5], 200 mM NaCl, and 1 mM EDTA). Dithiothreitol and glycerol were added to final concentrations of 1 mM and 10% (wt/vol), respectively.
EMSA.
A DNA probe encompassing the norM promoter region was amplified by PCR from FA19 genomic DNA using either of the upstream primers N11 (5′-CGGTCAGCAGGCGGATTTCTTTCAGG-3′) and N14 (5′-TCTGCCTTCTGTTTTATCCTG-3′). When making radioactive probes, the desired PCR products were labeled with [γ-32P]dATP using T4 polynucleotide kinase (New England BioLabs). The labeled DNA fragments were incubated with 8 μg of TetR-His in 30 μl of reaction buffer at room temperature. For the competition assays, the same nonlabeled probe or a nonlabeled PCR product using primers rnpBF1 (5′-CGGGACGGGCAGACAGTCGC-3′) and rnpBR1 (5′-GGACAGGCGGTAAGCCGGGTTC-3′) were added in the reaction. Samples were subjected to electrophoresis in a 6% native polyacrylamide gel at 4°C, followed by autoradiography as described previously (23).
Competitive infection of female mice to measure gonococcal fitness.
The female mouse model of lower genital tract infection was used to assess whether loss of NorM imposed an in vivo fitness cost or benefit. Mice were inoculated vaginally with equal numbers of CFU of parent strains FA19StrR and FA1090 with their respective norM::kan transformants, and the relative numbers of mutant and wild-type bacteria recovered were compared. The details of the experimental procedures have been described previously (11, 12, 23). Animal experiments were conducted in the laboratory animal facility at USUHS, which is fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care, under a protocol approved by the USUHS Institutional Animal Care and Use Committee.
Supplementary Material
ACKNOWLEDGMENTS
The contents of this article are solely the responsibility of the authors and do not necessarily reflect the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, or the U.S. government.
We have no competing interest to declare.
We thank C. del Rio, J. Dillon, R. Nicholas, and M. Unemo for providing clinical isolates.
This work was supported by NIH grants R37AI21150-33 (W.M.S.) and U19 AI113170-04 (A.E.J.) and in part by VA Merit Award 510 1BX000112-09 (W.M.S.) from the Biomedical Laboratory Research and Development Service of the U.S. Department of Veterans Affairs. W.M.S. is the recipient of a Senior Research Career Scientist Award from the Biomedical Laboratory Research and Development Service of the U.S. Department of Veterans Affairs.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00821-18.
REFERENCES
- 1.Newman L, Rowley J, Vander Hoorn S, Saman Wijesooriya N, Unemo M, Low N, Stevens G, Gottlieb S, Kiarie J, Temmerman M. 2015. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One 10(12):e0143304. doi: 10.1371/journal.pone.0143304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Unemo M, Shafer WM. 2014. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century—past, evolution, and future. Clin Microbiol Rev 27:587–613. doi: 10.1128/CMR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shafer WM, Qu X-D, Waring AJ, Lehrer RI. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc Natl Acad Sci U S A 95:1829–1833. doi: 10.1073/pnas.95.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee EH, Shafer WM. 1999. The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol Microbiol 33:839–845. doi: 10.1046/j.1365-2958.1999.01530.x. [DOI] [PubMed] [Google Scholar]
- 5.Fifer H, Natarajan U, Alexander S, Hughes G, Golparian D, Unemo M. 2016. Failure of dual antimicrobial therapy in treatment of gonorrhea. N Engl J Med 374:25. doi: 10.1056/NEJMc1512757. [DOI] [PubMed] [Google Scholar]
- 6.Golparian D, Shafer WM, Ohnishi M, Unemo M. 2014. Importance of multi-drug efflux pumps in the antimicrobial resistance property of clinical multi-drug resistant isolates of Neisseria gonorrhoeae: rationale for targeting efflux systems for drug development. Antimicrob Agents Chemother 58:3556–3559. doi: 10.1128/AAC.00038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Unemo M, Del Rio C, Shafer WM. 2016. Antimicrobial resistance expressed by Neisseria gonorrhoeae: a major global public health problem in the 21st century. Microbiol Spectr 4:EI10-0009-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weston N, Sharma P, Ricci V, Piddock LJV. 8 November 2017. Regulation of the AcrAB-TolC efflux system in Enterobacteriaceae. Res Microbiol doi: 10.1016/j.resmic.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Hagman KE, Pan W, Spratt BG, Balthazar JT, Judd RC, Shafer WM. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- 10.Veal WL, Nicholas RA, Shafer WM. 2002. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J Bacteriol 184:5619–5624. doi: 10.1128/JB.184.20.5619-5624.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warner DM, Folster JP, Shafer WM, Jerse AE. 2007. Regulation of the MtrC-MtrD-MtrE efflux-pump system modulates the in vivo fitness of Neisseria gonorrhoeae. J Infect Dis 196:1804–1812. doi: 10.1086/522964. [DOI] [PubMed] [Google Scholar]
- 12.Warner DM, Shafer WM, Jerse AE. 2008. Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol Microbiol 70:462–478. doi: 10.1111/j.1365-2958.2008.06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuroda T, Tsuchiya T. 2009. Multidrug efflux transporters in the MATE family. Biochim Biophys Acta 1794:763–768. doi: 10.1016/j.bbapap.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 14.Morita Y, Kodama K, Shiota S, Mine ST, Kataoka A, Mizushima T, Tsuchiya T. 1998. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother 42:1778–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouquette-Loughlin C, Dunham SA, Kuhn M, Balthazar JT, Shafer WM. 2003. The NorM efflux pump of Neisseria gonorrhoeae and Neisseria meningitidis recognizes antimicrobial cationic compounds. J Bacteriol 185:1101–1106. doi: 10.1128/JB.185.3.1101-1106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagman KE, Shafer WM. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J Bacteriol 177:4162–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jerse AE, Sharma ND, Simms AN, Crow ET, Snyder LA, Shafer WM. 2003. A gonococcal efflux pump system enhances bacterial survival in a female mouse model of genital tract infection. Infect Immun 71:5576–5582. doi: 10.1128/IAI.71.10.5576-5582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burgener A, Tjerlund A, Kaldensjo T, Abou M, McCorrister S, Westmacott GR, Mogk K, Ambrose E, Broliden K, Ball B. 2013. A systems biology evaluation of the human female genital tract shows compartmentalization of immune factor expression. J Virol 87:5141–5150. doi: 10.1128/JVI.03347-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zarantonelli L, Borthagaray G, Lee EH, Veal W, Shafer WM. 2001. Decreased susceptibility to azithromycin and erythromycin mediated by a novel mtrR promoter mutation in Neisseria gonorrhoeae. J Antimicrob Chemother 47:651–654. doi: 10.1093/jac/47.5.651. [DOI] [PubMed] [Google Scholar]
- 20.Sarubbi FA Jr, Blackman E, Sparling PF. 1974. Genetic mapping of linked antibiotic resistance loci in Neisseria gonorrhoeae. J Bacteriol 120(3):1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skaar EP, Lecuyer B, Lenich AG, Lazio MP, Perkins-Balding D, Seifert HS, Karls AC. 2005. Analysis of the Piv recombinase-related gene family of Neisseria gonorrhoeae. J Bacteriol 187:1276–1286. doi: 10.1128/JB.187.4.1276-1286.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ménard R, Sansonetti PJ, Parsot C. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol 175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rouquette-Loughlin CE, Zalucki YM, Dhulipala VL, Balthazar JT, Doyle RG, Nicholas RA, Begum AA, Raterman EL, Jerse AE, Shafer WM. 2017. Control of gdhR expression in Neisseria gonorrhoeae by autoregulation and a master repressor (MtrR) of a drug efflux pump operon. mBio 8(2):e00449-17. doi: 10.1128/mBio.00449-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizyed S, Oddone A, Byczynski B, Hugues DW, Berti PJ. 2005. UDP-N-acetylmuramic acid (UDP-MurNAc) is a potent inhibitor of MurA (enolpyruvyl-UDP-GlcNAc synthase). Biochemistry 44:4011–4017. doi: 10.1021/bi047704w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.