ABSTRACT
Oxazolidinones are efficacious in treating mycobacterial infections, including tuberculosis (TB) caused by drug-resistant Mycobacterium tuberculosis. In this study, we compared the in vitro activities and MIC distributions of delpazolid, a novel oxazolidinone, and linezolid against multidrug-resistant TB (MDR-TB) and extensively drug-resistant TB (XDR-TB) in China. Additionally, genetic mutations in 23S rRNA, rplC, and rplD genes were analyzed to reveal potential mechanisms underlying the observed oxazolidinone resistance. A total of 240 M. tuberculosis isolates were included in this study, including 120 MDR-TB isolates and 120 XDR-TB isolates. Overall, linezolid and delpazolid MIC90 values for M. tuberculosis isolates were 0.25 mg/liter and 0.5 mg/liter, respectively. Based on visual inspection, we tentatively set epidemiological cutoff (ECOFF) values for MIC determinations for linezolid and delpazolid at 1.0 mg/liter and 2.0 mg/liter, respectively. Although no significant difference in resistance rates was observed between linezolid and delpazolid among XDR-TB isolates (P > 0.05), statistical analysis revealed a significantly greater proportion of linezolid-resistant isolates than delpazolid-resistant isolates within the MDR-TB group (P = 0.036). Seven (53.85%) of 13 linezolid-resistant isolates were found to harbor mutations within the three target genes. Additionally, 1 isolate exhibited an amino acid substitution (Arg126His) within the protein encoded by rplD that contributed to high-level resistance to linezolid (MIC of >16 mg/liter), compared to a delpazolid MIC of 0.25. In conclusion, in vitro susceptibility testing revealed that delpazolid antibacterial activity was comparable to that of linezolid. A novel mutation within rplD that endowed M. tuberculosis with linezolid, but not delpazolid, resistance was identified.
KEYWORDS: delpazolid, linezolid, multidrug-resistant tuberculosis, MIC, minimal inhibitory concentration
INTRODUCTION
Tuberculosis (TB), caused by Mycobacterium tuberculosis complex, is one of humankind's deadliest diseases (1). According to estimates by the World Health Organization, there were 10.4 million new incident cases and 1.67 million deaths due to TB in 2016 (1). Despite the decreases in incidence and mortality rates during the past decade (2), the emergence of drug-resistant TB, especially multidrug-resistant TB (MDR-TB) (defined as TB with in vitro resistance to rifampin and isoniazid) and extensively drug-resistant TB (XDR-TB) (defined as MDR-TB with in vitro resistance to any fluoroquinolone and at least one of the second-line injectable drugs [kanamycin, amikacin, or capreomycin]), is creating major obstacles that will hinder disease control efforts worldwide (3). Due to emerging resistance to the two most potent first-line drugs, the treatment of MDR-TB requires more toxic, more costly, and less effective second-line treatment regimens, with poorer clinical outcomes than those achieved for drug-susceptible cases (3, 4). To make matters worse, additional resistance to fluoroquinolones and second-line injectable drugs leads to clinically almost incurable results for treatment of XDR-TB infections using current second-line regimens (4). Therefore, the epidemic of MDR- and XDR-TB highlights an urgent need for new antibiotics with improved safety, tolerability, and efficacy (5).
The oxazolidinones, a new class of synthetic antibiotics, exhibit good activity against Gram-positive pathogenic bacteria, including those resistant to other agents. Delpazolid (research code LCB01-0371), a novel oxazolidinone containing a cyclic amidrazone group, was evaluated for safety, tolerability, and pharmacokinetics in a recently completed phase I clinical trial (6). Preliminary studies have demonstrated that oxazolidinones inhibit the biosynthesis of bacterial proteins at an early stage of translation by binding to domain V of 23S rRNA (7). As a consequence, mutations in 23S rRNA and two ribosomal proteins, i.e., L3 (rplC) and L4 (rplD), are involved in the major mechanism employed by various pathogenic organisms conferring resistance to oxazolidinones (8, 9). Linezolid is efficacious in treating mycobacterial infections, including drug-resistant TB (10–13). A recent meta-analysis revealed that more than 90% of MDR-TB cases achieved culture conversion after treatment with individualized regimens containing linezolid, underscoring the excellent in vivo efficacy of the drug against MDR-TB (5). Recently, the novel oxazolidinone delpazolid was developed to produce improved antibacterial activity and safety (14, 15). In vitro studies and pharmacological evidence have indicated that this new agent is more active than linezolid against various Gram-positive bacteria (14); however, data regarding the in vitro activity of delpazolid against MDR- and XDR-TB are limited. In this study, we compared the in vitro activity and MIC distribution of the novel oxazolidinone delpazolid with those of linezolid against MDR- and XDR-TB in China. In addition, genetic mutations in 23S rRNA, rplC, and rplD were analyzed to explore potential mechanisms underlying M. tuberculosis oxazolidinone resistance.
RESULTS
Linezolid and delpazolid MICs for MDR- and XDR-TB isolates.
The MICs of linezolid and delpazolid against M. tuberculosis isolates and the percentages of resistant strains are summarized in Table 1. Overall, the linezolid and delpazolid MIC90 values for M. tuberculosis isolates were 0.25 mg/liter and 0.5 mg/liter, respectively. Against MDR-TB, the MIC90 of delpazolid (MIC90, 0.5 mg/liter) was lower than that of linezolid (MIC90, 1.0 mg/liter). In contrast, the MIC90 of delpazolid (MIC90, 1.0 mg/liter) against XDR-TB was 4-fold higher than the MIC90 of linezolid (MIC90, 0.25 mg/liter).
TABLE 1.
Distribution of M. tuberculosis isolates with different linezolid and delpazolid MIC values
| Classification and druga | No. (%) of strains with MIC of: |
MIC50 (mg/liter) | MIC90 (mg/liter) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <0.016 mg/liter | 0.016 mg/liter | 0.032 mg/liter | 0.064 mg/liter | 0.13 mg/liter | 0.25 mg/liter | 0.5 mg/liter | 1 mg/liter | 2 mg/liter | 4 mg/liter | 8 mg/liter | 16 mg/liter | >16 mg/liter | Total | |||
| MDR-TB | ||||||||||||||||
| Linezolid | 4 (3.3) | 6 (5.0) | 24 (20.0) | 49 (40.8) | 11 (9.2) | 9 (7.5) | 6 (5.0) | 3 (2.5) | 7 (5.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.8) | 120 | 0.064 | 1 |
| Delpazolid | 2 (1.7) | 3 (2.5) | 3 (2.5) | 6 (5.0) | 13 (10.8) | 32 (26.7) | 53 (44.2) | 6 (5.0) | 1 (0.8) | 1 (0.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 120 | 0.5 | 0.5 |
| XDR-TB | ||||||||||||||||
| Linezolid | 3 (2.5) | 4 (3.3) | 12 (10.0) | 21 (17.5) | 49 (40.8) | 24 (20.0) | 1 (0.8) | 1 (0.8) | 1 (0.8) | 1 (0.8) | 0 (0.0) | 2 (1.7) | 1 (0.8) | 120 | 0.13 | 0.25 |
| Delpazolid | 1 (0.8) | 2 (1.7) | 2 (1.7) | 3 (2.5) | 5 (4.2) | 35 (29.2) | 57 (47.5) | 9 (7.5) | 1 (0.8) | 2 (1.7) | 1 (0.8) | 0 (0.0) | 2 (1.7) | 120 | 0.5 | 1 |
| Total | ||||||||||||||||
| Linezolid | 7 (2.9) | 10 (4.2) | 36 (15.0) | 70 (29.2) | 60 (25.0) | 33 (13.8) | 7 (2.9) | 4 (1.7) | 8 (3.3) | 1 (0.4) | 0 (0.0) | 2 (0.8) | 2 (0.8) | 240 | 0.064 | 0.25 |
| Delpazolid | 3 (1.3) | 5 (2.1) | 5 (2.1) | 9 (3.8) | 18 (7.5) | 67 (27.9) | 110 (45.8) | 15 (6.3) | 2 (0.8) | 3 (1.3) | 1 (0.4) | 0 (0.0) | 2 (0.8) | 240 | 0.5 | 0.5 |
XDR-TB, extensively drug-resistant tuberculosis; MDR-TB, multidrug-resistant tuberculosis.
We further analyzed the tentative epidemiological cutoff (ECOFF) values for linezolid and delpazolid. As shown in Fig. 1, the MIC distributions for linezolid and delpazolid were bimodal. Therefore, on the basis of visual inspection, we set tentative ECOFFs for MIC determinations at 1.0 mg/liter and 2.0 mg/liter for linezolid and delpazolid, respectively. Notably, the ECOFFs of linezolid were consistent with the breakpoints used for the determination of in vitro linezolid resistance in previous studies. When 1.0 mg/liter was used as the cutoff value, 8 (6.67% [8/120 isolates]) and 5 (4.17% [5/120 isolates]) MDR- and XDR-TB isolates, respectively, were resistant to linezolid. For delpazolid, resistance was noted for 1 (0.83% [1/120 isolates]) and 5 (4.2% [5/120 isolates]) MDR- and XDR-TB isolates, respectively. Although there was no significant difference in the resistance rates for linezolid and delpazolid among the XDR-TB isolates tested (P > 0.05), statistical analysis revealed that the proportion of linezolid-resistant isolates was significantly greater than the proportion of delpazolid-resistant isolates within the MDR group (P = 0.036). Of the 13 linezolid-resistant isolates, 6 (46.2%) were resistant to delpazolid, whereas the other 7 isolates were susceptible to delpazolid, including 1 isolate with a MIC of 0.25 mg/liter, 3 with MICs of 0.5 mg/liter, 1 with a MIC of 1.0 mg/liter, and 2 with MICs of 2 mg/liter. In addition, 3 of 4 isolates with high-level resistance to linezolid (MICs of ≥8 mg/liter) belonged to the XDR-TB group, while 77.78% of isolates (7/9 isolates) with low-level resistance to linezolid belonged to the MDR-TB group.
FIG 1.
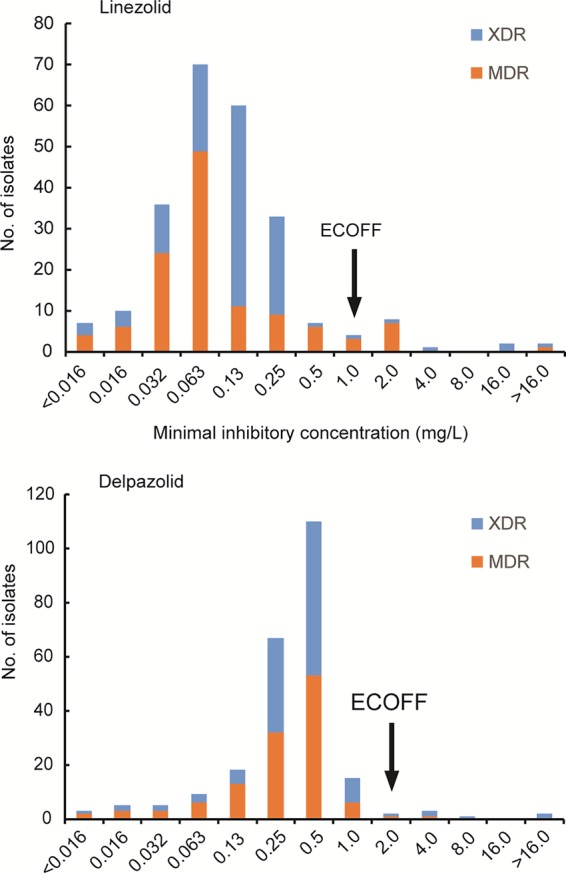
MIC distributions for MDR- and XDR-TB strains. The arrows indicate the proposed linezolid and delpazolid ECOFF values for M. tuberculosis isolates.
Mutations conferring linezolid and delpazolid resistance.
The entire 23S rRNA, rplC, and rplD genes were sequenced for all resistant isolates, to identify potential mutations associated with linezolid and delpazolid resistance. As shown in Table 2, the DNA sequence chromatograms demonstrated that 7 (53.85%) of 13 linezolid-resistant isolates harbored mutations within the three target genes. The remaining 6 linezolid-resistant isolates (46.15%) exhibited wild-type sequences at all loci. The most frequently observed mutation (T460C; n = 3) was observed in the rplC gene and coded for a nonconservative amino acid substitution, Cys154Arg. In addition, 2 linezolid-resistant isolates exhibited a mutation at position 2061 of 23S rRNA, resulting in high-level resistance to both linezolid and delpazolid (MICs of ≥16 mg/liter). Interestingly, 1 isolate contained an amino acid substitution from Arg to His at codon 126 of the rplD gene, which contributed to high-level linezolid resistance (MIC, >16 mg/liter) but not delpazolid resistance (MIC, 0.25 mg/liter). Among the linezolid-susceptible isolates, we identified several synonymous single-nucleotide polymorphisms (SNPs) within rplC, including 1 isolate with Arg93Arg (AGG→AGA) and 1 isolate with Gly153Gly (GGA→GGG). In addition, two types of synonymous SNPs within the coding region of rplD were found among 4 isolates, including Gln89Gln (CAG→CAA; n = 3) and Gln47Gln (CAG→CAA; n = 1).
TABLE 2.
MICs and 23S rRNA, rplC, and rplD mutations for the 13 oxazolidinone-resistant clinical strains
| Strain | MIC (mg/liter) |
Resistance genotype |
|||
|---|---|---|---|---|---|
| Linezolid | Delpazolid | 23S rRNAa | rplC | rplD | |
| XDR014 | >16 | >16 | G2061T | WT | WT |
| MDR052 | 16 | >16 | G2061T | WT | WT |
| XDR042 | >16 | 0.25 | WT | WT | CGC377CAC (Arg126His) |
| XDR037 | 16 | 4 | WT | TGC460CGC (Cys154Arg) | WT |
| XDR021 | 4 | 8 | WT | TGC460CGC (Cys154Arg) | WT |
| MDR055 | 2 | 4 | WT | TGC460CGC (Cys154Arg) | WT |
| MDR087 | 2 | 4 | WT | CAC463GAC (His155Asp) | WT |
| XDR075 | 2 | 2 | WT | WT | WT |
| MDR046 | 2 | 2 | WT | WT | WT |
| MDR077 | 2 | 1 | WT | WT | WT |
| MDR085 | 2 | 0.5 | WT | WT | WT |
| MDR098 | 2 | 0.5 | WT | WT | WT |
| MDR112 | 2 | 0.5 | WT | WT | WT |
The nucleotide positions of the mutations are listed according to Escherichia coli numbering. WT, wild type.
DISCUSSION
In this study, we first compared the in vitro activities of linezolid and delpazolid against MDR- and XDR-TB isolates. Our data demonstrated a delpazolid MIC90 of 0.5 mg/liter against severe forms of drug-resistant TB, similar to MIC90 values obtained for Staphylococcus aureus (MIC90, 0.5 mg/liter) and Streptococcus pneumoniae (MIC90, 1 mg/liter) (14). The most important finding of this study is that an ECOFF value of 2.0 mg/liter is suggested for delpazolid. On the basis of this ECOFF value, delpazolid showed antibacterial activity comparable to that of linezolid, while only 2.9% of drug-resistant TB strains exhibited resistance to this novel antimicrobial agent. Notably, the proportion of delpazolid-resistant isolates was significantly smaller than that of linezolid-resistant isolates within the MDR-TB group. Although linezolid has been reported to be one of the most potent antibiotics against infections caused by drug-resistant TB, the long-term use of linezolid produces high rates of adverse events, such as myelosuppression and peripheral neuropathy (5, 16). Previous experimental evidence showed that, compared with linezolid, delpazolid exhibited superior pharmacokinetic parameters and good safety profiles (14). In a recent clinical trial, Choi and colleagues demonstrated that LCB01-0371 was well tolerated in healthy male subjects after administration of multiple doses of up to 1,200 mg twice daily for 21 days (17). Therefore, the impressive in vitro effectiveness and favorable tolerability of delpazolid make it a promising candidate for use in combination treatment with other anti-TB drugs against MDR- and XDR-TB. In view of the prolonged administration needed for treatment of drug-resistant TB, additional trials are urgently needed to evaluate the efficacy and safety of delpazolid for the management of patients with drug-resistant TB.
Resistance of clinical M. tuberculosis isolates to oxazolidinones has primarily been shown to be due to mutations in 23S rRNA and rplC, with G2061T and G2576T mutations in 23S rRNA having been shown to cause high-level resistance to linezolid (8). Consistent with previous reports (7), 2 strains with a G2061T mutation in 23S rRNA studied here demonstrated high-level resistance to both linezolid and delpazolid. In addition, we observed that mutations in rplC led to great diversity in linezolid susceptibility, such that the MIC values of strains ranged from 2 to 16 mg/liter. The rplD gene encodes ribosomal protein L4, the main portion of which is positioned close to the ribosomal peptidyl transferase center (PTC) (8). Many studies have associated rplD mutations with linezolid resistance in several bacterial species, such as Staphylococcus epidermidis and Enterococcus faecium (18, 19), while no study has reported the role of rplD in any M. tuberculosis linezolid resistance mechanism. In the present study, we first identified a novel mutation within the rplD gene that potentially confers linezolid resistance to M. tuberculosis. Compared with the greater frequency of the rplC mutation, the rarity of the rplD mutation in linezolid-resistant bacterial isolates is likely due to the fact that L3 residues are in close proximity to the PTC (8). Interestingly, the mutation in the L4 ribosomal protein causes decreased susceptibility only to linezolid and not to delpazolid, indicating that these two oxazolidinones may exhibit different binding sites within the PTC. Further structural data on the PTC-oxazolidinone complex will extend our knowledge of the molecular mechanisms of oxazolidinone resistance.
Although multiple mutations conferring linezolid resistance have been identified, nearly one-half of linezolid-resistant strains in this study lacked target gene mutations. On one hand, in addition to the targets sequenced in our study, several other modifications of 23S rRNA may play important roles in the occurrence of linezolid resistance (20). In addition, 23S RNA mutations apparently confer high-level resistance, while efflux pumps and other mechanisms usually result in low-level resistance (7). Of note, all M. tuberculosis isolates in this study without detected genetic mutations exhibited low-level resistance (MICs of <4 mg/liter). Therefore, we hypothesize that the efflux-mediated mechanism may play an important role in these linezolid-resistant isolates. On the other hand, the poor correlation between genetic mutations and the linezolid resistance phenotype suggests that the current set of target genes is not suitable for prediction of linezolid resistance among MDR-TB isolates. Given the projected widespread future use of linezolid in the treatment of drug-resistant TB, there is an urgent need to broaden our knowledge of the mechanisms of linezolid resistance in M. tuberculosis, to provide a critical component for the development of favorable molecular diagnostic approaches.
We also acknowledge several obvious limitations of this study. First, the determination of critical concentrations for delpazolid should be based not only on the ECOFF value but also on pharmacokinetic/pharmacodynamic and clinical outcome data, as evaluated in prospective studies (21). Second, due to the small number of oxazolidinone-resistant M. tuberculosis isolates, the second MIC distribution peak was relatively unobvious, compared with the first peak, within the bimodal distribution of MIC values, which may undermine the reliability of tentative ECOFFs. Third, the novel rplD mutation that is potentially associated with resistance to linezolid but not delpazolid was not confirmed by further experimental evidence. Sequence analysis of rplD genes from a larger number of linezolid-resistant isolates should confirm our hypothesis. In addition, directed mutagenesis and heterologous expression studies will help us to conclusively link this mutation to linezolid resistance.
In conclusion, we first established an ECOFF value of 2.0 mg/liter for delpazolid. In vitro susceptibility tests revealed that delpazolid shows antibacterial activity comparable to that of linezolid, with only 2.9% of drug-resistant TB strains exhibiting resistance to this novel antimicrobial agent. In addition, nucleotide mutations of 23S rRNA yielded high-level resistance to both linezolid and delpazolid, while mutations of rplC led to great diversity in linezolid susceptibility. A novel mutation within rplD that conferred resistance to linezolid but not delpazolid was identified. Further studies are urgently needed to elucidate the role of rplD in the decreased susceptibility to linezolid observed for M. tuberculosis strains in this study.
MATERIALS AND METHODS
Ethics statement.
The protocols applied in this study were approved by the Ethics Committee of Beijing Chest Hospital, Capital Medical University. All of the patients provided signed informed consent forms prior to their enrollment in this study.
Bacterial strains.
A total of 120 MDR-TB strains and 120 XDR-TB strains were randomly selected from the Tuberculosis BioBank maintained at the National Clinical Laboratory on Tuberculosis. These strains were obtained from consecutive patients who sought health care in Beijing Chest Hospital between January 2017 and October 2017. Each M. tuberculosis strain was isolated from a unique patient. The drug susceptibility profiles were retrospectively reviewed using in vitro drug susceptibility testing (DST) results determined in the National Clinical Laboratory on Tuberculosis. Tests for first- and second-line antituberculosis drug susceptibilities were performed using the absolute concentration method with Löwenstein-Jensen (L-J) medium containing the corresponding anti-TB drugs, as reported previously (22).
MIC determinations.
The microplate alamarBlue assay (MABA), which employs alamarBlue reagent for the determination of growth, was performed to determine the MICs of MDR- and XDR-TB against linezolid and delpazolid (7). Prior to in vitro susceptibility testing, the strains were recovered on L-J medium after incubation for 4 weeks at 37°C. Briefly, fresh bacterial clones were harvested from the surface of L-J slants. After vigorous mixing for 1 min on a vortex mixer, a suspension of each M. tuberculosis strain was prepared in sterile saline solution and adjusted to a density of 1.0 McFarland standard. The inoculum was further diluted 1:20 with Middlebrook 7H9 broth containing 10% Middlebrook oleic-albumin-dextrose-catalase (OADC) enrichment supplement (containing oleic acid and bovine serum albumin along with sodium chloride, dextrose, and catalase). Next, 100 μl of this inoculum was added to wells of 96-well plates containing 100 μl of antimicrobial serial dilutions in broth per well, to yield a highest final concentration of 16 mg/liter. After 7 days of incubation at 37°C, 70 μl of alamarBlue solution was added to each well, plates were further incubated for 24 h at 37°C, and then color changes were read by visual inspection. The results were interpreted by two independent individuals and inconsistent MIC values were read again by a third individual, to avoid potential bias. The MIC was defined as the lowest concentration of antimicrobial agent that prevented a color change from blue to pink. The standard strain H37Rv served as a control in the MABA assay. The final concentrations of linezolid and delpazolid in the test panel ranged from 0.016 mg/liter to 16 mg/liter. The MIC breakpoint for linezolid was defined as 1.0 mg/liter on the basis of a previous report (7). For delpazolid, we set tentative ECOFFs for MIC determination using the MABA method (23).
DNA sequencing.
Extraction of genomic DNA from M. tuberculosis strains was performed with freshly cultured bacteria, as reported previously (7). Crude DNA served as the template for PCR amplification to generate gene fragments from isolates exhibiting oxazolidinone resistance. Sequencing of PCR products was performed using the Sanger method, with primers designed to be specific for 23S rRNA, rplC, or rplD. The primers used in this study are listed in Table S1 in the supplemental material and were synthesized by Tsingke Biotech Co. (Beijing, China). The 50-μl reaction mixtures were prepared as follows: 25 μl of 2× PCR mixture (Genstar, Beijing, China), 0.2 μM each primer, and 4 μl of template DNA. PCR cycling consisted of 94°C for 5 min and then 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 5 min. PCR products were submitted to Tsingke Biotech Co. for DNA sequencing. The sequencing results were analyzed by alignment against corresponding sequences of the reference M. tuberculosis strain H37Rv (ATCC 27294).
Data analysis.
Comparisons of the rates of resistance of M. tuberculosis isolates to linezolid and delpazolid were performed using Pearson's chi-square test, with P values of <0.05 being considered significant. All statistical analyses were carried out using SPSS software (version 17.0; SPSS Inc., Chicago, IL, USA).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Beijing Municipal Administration of Hospitals' Youth Programme (grant QML20171601), Capital Health Research and Development of Special (grant 2016-2-1041), and the Natural Science Fund of China (grant 81672065).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00165-18.
REFERENCES
- 1.World Health Organization. 2017. Global tuberculosis report 2017. WHO/HTM/TB/2017.23 World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/handle/10665/259366/9789241565516-eng.pdf;jsessionid=2CDE1255E5813851E3CBC49470A41260?sequence=1. [Google Scholar]
- 2.Gunther G. 2014. Multidrug-resistant and extensively drug-resistant tuberculosis: a review of current concepts and future challenges. Clin Med (Lond) 14:279–285. doi: 10.7861/clinmedicine.14-3-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falzon D, Mirzayev F, Wares F, Baena IG, Zignol M, Linh N, Weyer K, Jaramillo E, Floyd K, Raviglione M. 2015. Multidrug-resistant tuberculosis around the world: what progress has been made? Eur Respir J 45:150–160. doi: 10.1183/09031936.00101814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caminero JA, Sotgiu G, Zumla A, Migliori GB. 2010. Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Lancet Infect Dis 10:621–629. doi: 10.1016/S1473-3099(10)70139-0. [DOI] [PubMed] [Google Scholar]
- 5.Sotgiu G, Centis R, D'Ambrosio L, Alffenaar JW, Anger HA, Caminero JA, Castiglia P, De Lorenzo S, Ferrara G, Koh WJ, Schecter GF, Shim TS, Singla R, Skrahina A, Spanevello A, Udwadia ZF, Villar M, Zampogna E, Zellweger JP, Zumla A, Migliori GB. 2012. Efficacy, safety and tolerability of linezolid containing regimens in treating MDR-TB and XDR-TB: systematic review and meta-analysis. Eur Respir J 40:1430–1442. doi: 10.1183/09031936.00022912. [DOI] [PubMed] [Google Scholar]
- 6.Jeong JW, Jung SJ, Lee HH, Kim YZ, Park TK, Cho YL, Chae SE, Baek SY, Woo SH, Lee HS, Kwak JH. 2010. In vitro and in vivo activities of LCB01-0371, a new oxazolidinone. Antimicrob Agents Chemother 12:5359–5362. doi: 10.1128/AAC.00723-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z, Pang Y, Wang Y, Liu C, Zhao Y. 2014. Beijing genotype of Mycobacterium tuberculosis is significantly associated with linezolid resistance in multidrug-resistant and extensively drug-resistant tuberculosis in China. Int J Antimicrob Agents 43:231–235. doi: 10.1016/j.ijantimicag.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Long KS, Vester B. 2012. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob Agents Chemother 56:603–612. doi: 10.1128/AAC.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beckert P, Hillemann D, Kohl TA, Kalinowski J, Richter E, Niemann S, Feuerriegel S. 2012. rplC T460C identified as a dominant mutation in linezolid-resistant Mycobacterium tuberculosis strains. Antimicrob Agents Chemother 56:2743–2745. doi: 10.1128/AAC.06227-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee M, Lee J, Carroll MW, Choi H, Min S, Song T, Via LE, Goldfeder LC, Kang E, Jin B, Park H, Kwak H, Kim H, Jeon HS, Jeong I, Joh JS, Chen RY, Olivier KN, Shaw PA, Follmann D, Song SD, Lee JK, Lee D, Kim CT, Dartois V, Park SK, Cho SN, Barry CE III. 2012. Linezolid for treatment of chronic extensively drug-resistant tuberculosis. N Engl J Med 367:1508–1518. doi: 10.1056/NEJMoa1201964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray S, Mendel C, Spigelman M. 2016. TB Alliance regimen development for multidrug-resistant tuberculosis. Int J Tuberc Lung Dis 20:38–41. doi: 10.5588/ijtld.16.0069. [DOI] [PubMed] [Google Scholar]
- 12.Conradie F, Diacon AH, Everitt D, Mendel C, Niekerk C, Howell P, Comins K, Spigelman M. 2017. The NIX-TB trial of pretomanid, bedaquiline and linezolid to treat XDR-TB. Abstr Conf Retroviruses Opportunistic Infect, Seattle, Washington, 13 to 16 February 2017. [Google Scholar]
- 13.Cox H, Ford N. 2012. Linezolid for the treatment of complicated drug-resistant tuberculosis: a systematic review and meta-analysis. Int J Tuberc Lung Dis 16:447–454. doi: 10.5588/ijtld.11.0451. [DOI] [PubMed] [Google Scholar]
- 14.Jeong JW, Jung SJ, Lee HH, Kim YZ, Park TK, Cho YL, Chae SE, Baek SY, Woo SH, Lee HS, Kwak JH. 2010. In vitro and in vivo activities of LCB01-0371, a new oxazolidinone. Antimicrob Agents Chemother 54:5359–5362. doi: 10.1128/AAC.00723-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim TS, Choe JH, Kim YJ, Yang CS, Kwon HJ, Jeong J, Kim G, Park DE, Jo EK, Cho YL, Jang J. 2017. Activity of LCB01-0371, a novel oxazolidinone, against Mycobacterium abscessus. Antimicrob Agents Chemother 61:e02752-16. doi: 10.1128/AAC.02752-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolhuis MS, Tiberi S, Sotgiu G, De Lorenzo S, Kosterink JG, van der Werf TS, Migliori GB, Alffenaar JW. 2015. Linezolid tolerability in multidrug-resistant tuberculosis: a retrospective study. Eur Respir J 46:1205–1207. doi: 10.1183/13993003.00606-2015. [DOI] [PubMed] [Google Scholar]
- 17.Choi Y, Lee SW, Kim A, Jang K, Nam H, Cho YL, Yu KS, Jang IJ, Chung JY. 2018. Safety, tolerability and pharmacokinetics of 21 day multiple oral administration of a new oxazolidinone antibiotic, LCB01-0371, in healthy male subjects. J Antimicrob Chemother 73:183–190. doi: 10.1093/jac/dkx367. [DOI] [PubMed] [Google Scholar]
- 18.Wong A, Reddy SP, Smyth DS, Aguero-Rosenfeld ME, Sakoulas G, Robinson DA. 2010. Polyphyletic emergence of linezolid-resistant staphylococci in the United States. Antimicrob Agents Chemother 54:742–748. doi: 10.1128/AAC.00621-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendes RE, Deshpande LM, Farrell DJ, Spanu T, Fadda G, Jones RN. 2010. Assessment of linezolid resistance mechanisms among Staphylococcus epidermidis causing bacteraemia in Rome, Italy. J Antimicrob Chemother 65:2329–2335. doi: 10.1093/jac/dkq331. [DOI] [PubMed] [Google Scholar]
- 20.Mendes RE, Deshpande LM, Jones RN. 2014. Linezolid update: stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist Updat 17:1–12. doi: 10.1016/j.drup.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Wang G, Chen S, Wei G, Shang Y, Dong L, Schon T, Moradigaravand D, Parkhill J, Peacock SJ, Koser CU, Huang H. 2016. Wild-type and non-wild-type Mycobacterium tuberculosis MIC distributions for the novel fluoroquinolone antofloxacin compared with those for ofloxacin, levofloxacin, and moxifloxacin. Antimicrob Agents Chemother 60:5232–5237. doi: 10.1128/AAC.00393-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pang Y, Zong Z, Huo F, Jing W, Ma Y, Dong L, Li Y, Zhao L, Fu Y, Huang H. 2017. In vitro drug susceptibility of bedaquiline, delamanid, linezolid, clofazimine, moxifloxacin, and gatifloxacin against extensively drug-resistant tuberculosis in Beijing, China. Antimicrob Agents Chemother 61:e00900-17. doi: 10.1128/AAC.00900-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pang Y, Zheng H, Tan Y, Song Y, Zhao Y. 2017. In vitro activity of bedaquiline against nontuberculous mycobacteria in China. Antimicrob Agents Chemother 61:e02627-16. doi: 10.1128/AAC.02627-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


