Scedosporium spp. cause infections (scedosporiosis) in both immunocompetent and immunocompromised individuals and may persistently colonize the respiratory tract in patients with cystic fibrosis (CF).
KEYWORDS: azole resistance, Cyp51, antifungal agents
ABSTRACT
Scedosporium spp. cause infections (scedosporiosis) in both immunocompetent and immunocompromised individuals and may persistently colonize the respiratory tract in patients with cystic fibrosis (CF). They are less susceptible against azoles than are other molds, such as Aspergillus spp., suggesting the presence of resistance mechanisms. It can be hypothesized that the decreased susceptibility of Scedosporium spp. to azoles is also CYP51 dependent. Analysis of the Scedosporium apiospermum and Scedosporium aurantiacum genomes revealed one CYP51 gene encoding the 14-α-lanosterol demethylase. This gene from 159 clinical or environmental Scedosporium isolates and three Lomentospora prolificans isolates has been sequenced and analyzed. The Scedosporium CYP51 protein clustered with the group of known CYP51B orthologues and showed species-specific polymorphisms. A tandem repeat in the 5′ upstream region of Scedosporium CYP51 like that in Aspergillus fumigatus could not be detected. Species-specific amino acid alterations in CYP51 of Scedosporium boydii, Scedosporium ellipsoideum, Scedosporium dehoogii, and Scedosporium minutisporum isolates were located at positions that have not been described as having an impact on azole susceptibility. In contrast, two of the three S. apiospermum-specific amino acid changes (Y136F and G464S) corresponded to respective mutations in A. fumigatus CYP51A at amino acid positions 121 and 448 (Y121F and G448S, respectively) that had been linked to azole resistance.
INTRODUCTION
At present, the genus Scedosporium contains the following 10 opportunistic pathogenic species: S. aurantiacum, S. minutisporum, S. desertorum, S. cereisporum, and S. dehoogii, as well as the S. apiospermum complex that comprises S. angustum, S. apiospermum, S. boydii, S. ellipsoideum, and S. fusoideum (1). This complex is distantly related to Lomentospora prolificans and Petriellopsis africana (2). Scedosporium species cause infections in immunocompetent and immunocompromised patients and may colonize the lungs, especially in patients with cystic fibrosis (CF) (3–6). Multilocus sequence typing (MLST) studies for S. apiospermum and S. boydii isolates revealed that nearly every patient hosts a strain with an individual sequence type (7, 8). Invasive scedosporiosis is difficult to treat and is associated with a high mortality rate (5, 9). Of the already-small set of clinically relevant antifungal drugs (polyene, azoles, echinocandins, pyrimidine analogues, and allylamines), Scedosporium species show in vitro resistance to amphotericin B and 5-flucytosine (10, 11). The therapeutic efficacy to echinocandins is under discussion (12).
Scedosporium species are considered to be susceptible to azoles, including voriconazole (VCZ), but the MICs are higher than those of other molds, such as Aspergillus fumigatus (12–15). Although VCZ is the first-line treatment for scedosporiosis (12), a synergistic effect has been found for a combination of VCZ and an echinocandin (16, 17). L. prolificans is resistant in vitro against all currently marketed antimycotic drugs.
The mechanism of action of azoles (e.g., itraconazole [ICZ], posaconazole [PCZ], and VCZ) is based on the inhibition of the 14-α-lanosterol demethylase (ERG11/CYP51). The inhibition of ERG11/CYP51 results in cell membrane stress, which is caused by the decrease in ergosterol, an accumulation of lanosterol, and other toxic sterol intermediates (18, 19). The CYP51 protein family is present in nearly all taxonomic kingdoms (20). These proteins are characterized by six highly conserved substrate recognition sites (SRSs) and the heme-binding region (HBR) (21).
For molds, like A. fumigatus or Penicillium digitatum, the azole resistance mechanisms involving CYP51 have been analyzed. Target alterations in CYP51 (so-called “hot spot” mutations) are found as one resistance mechanism, resulting in structural changes to the protein which appear to block the binding of azoles (22). For A. fumigatus, CYP51A amino acid changes at positions G54, L98, G138, M220, Y431, and G448 have been linked to azole resistance (19, 23, 24). Also, upregulation of CYP51 gene expression has been documented to lower azole susceptibility. A. fumigatus isolates were described with insertions of tandem-repeat sequences (34, 46, or 53 bp long) in the promoter region of CYP51A as assigned in this species without or in combination with point mutations leading to amino acid substitutions in the CYP51A protein (TR46/Y121/T289), and respective isolates showed increased azole resistance (23, 25). In azole-resistant Penicillium digitatum isolates, CYP51 expression was increased, and this correlated with a presence of a 126-bp sequence in the promoter region of CYP51 that was repeated five times (26). This 126-bp sequence acts as a transcriptional enhancer, and its repetition increased the level of expression of P. digitatum CYP51.
The molecular mechanisms of the increased azole MICs in Scedosporium spp. have not yet been elucidated. Analogous to other molds, it could be hypothesized that the decreased susceptibility of Scedosporium spp. to azoles is also CYP51 dependent and is caused by CYP51 proteins with sequences comparable to those of resistance-associated CYP51 isoforms as known from A. fumigatus and/or caused by a higher abundance of the enzyme.
The aim of this study was to identify and analyze CYP51 sequences from a collection of clinical Scedosporium species isolates, for which the MICs for azoles had been determined by a microdilution reference standard.
RESULTS
Unlike the situation in A. fumigatus and Fusarium oxysporum, only one CYP51 orthologous protein was identified by the tblastx analysis in the genomes of S. apiospermum (GenBank accession no. JOWA01000088) and S. aurantiacum (GenBank accession no. JUDQ01000082). The identities of the respective blast hits at the protein level were up to 54% for AfumCYP51A/FoxyCYP51A and up to 76% for AfumCYP51B/FoxyCYP51B, respectively (see Table S1 in the supplemental material). Further blast hits identified hypothetical proteins or distantly related proteins of the cytochrome P450 group.
The CYP51 gene in the genus Scedosporium was between 1,750 and 1,760 bp long (Table 1). In the published genome of S. apiospermum (GenBank accession no. JOWA01000088), the CYP51 gene was already annotated with three exons separated by two introns. A comparison of the genomic DNA and respective cDNA sequence of CYP51 (strain CBS 117407T) could confirm the annotation of the introns. The functionality of the gene was proven by heterologous gene expression of an S. apiospermum CYP51 in S. cerevisiae BY4741 by replacing the endogenous S. cerevisiae orthologue, ERG11, with a yeast codon use-adapted S. apiospermum CYP51 version, which resulted in fully viable transgenic yeasts (data not shown).
TABLE 1.
Variability between the Scedosporium CYP51 gene and the CYP51 proteina
CYP51, 14-α-lanosterol demethylase.
bC, clinical isolate; E, environmental isolate; V, veterinary isolate.
cCF, cystic fibrosis.
dnt, nucleotides; SNPs, single nucleotide polymorphisms.
e5′ UTR, 5′ untranslated region.
fFor isolate RKI 11-0901, it was not possible to analyze the 5′ UTR.
gFor isolate RKI 11-0093, it was not possible to analyze the 5′ UTR.
hPartial protein.
iPolymorphic residues detected in isolates as single amino acid changes compared to the respective species' consensus sequence; amino acids in parentheses were always observed en bloc in isolates exhibiting that Cyp51 type.
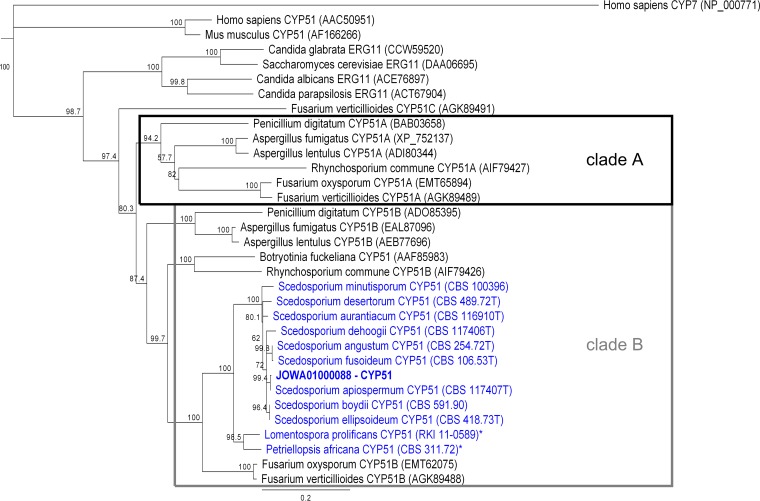
The CYP51 cDNA encodes a 528-amino acid (aa) protein. The predicted molecular mass is about 59.8 kDa, and the isoelectric point is between 8.40 and 8.65. The transmembrane region was predicted between aa 22 and aa 41 at the N terminus of Scedosporium CYP51 (data not shown). The predicted protein identity within the genus Scedosporium ranged from 94.7% to 100%. The CYP51 protein sequences for S. boydii and S. ellipsoideum were identical (Table S2). The identity between Scedosporium spp. and L. prolificans CYP51 (LproCYP51) and P. africana CYP51 (PafrCYP51) was 87%. (Table S2). All derived CYP51 proteins of Scedosporium spp., L. prolificans, and P. africana detected here clustered with the group of known CYP51B orthologues (Fig. 1).
FIG 1.
Phylogenetic tree of protein CYP51/ERG11 orthologues. Scedosporium/L. prolificans/P. africana CYP51 clustered in the group of CYP51B orthologues. The Homo sapiens cholesterol 7-alpha-monooxygenase (CYP7) was used as the outgroup. The best result from the blast analysis of the S. apiospermum genome with Afum/FoyxCYP51A/B is shown in bold (JOWA01000088–CYP51). CBS, Centraalbureau voor Schimmelcultures; RKI, Robert Koch Institute. *, partial protein. Accession numbers or strain identifiers are shown in parentheses.
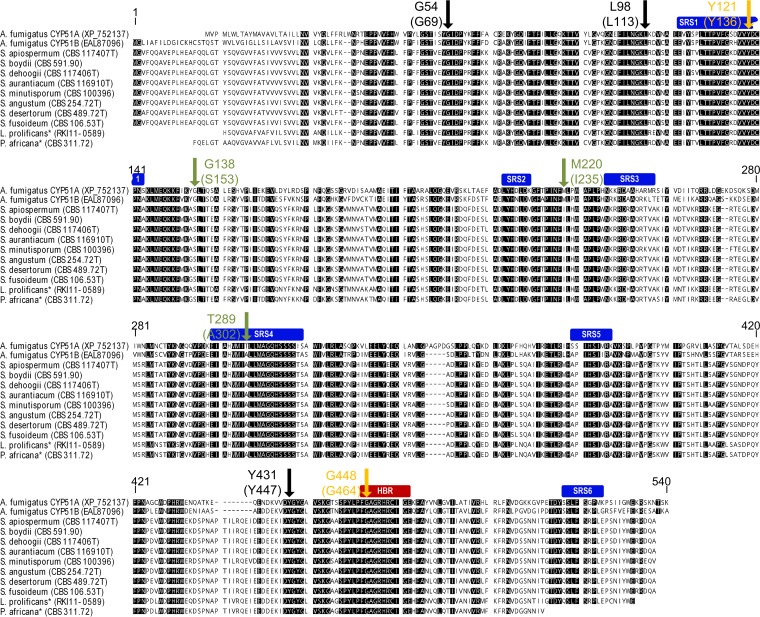
Alignment of Scedosporium spp. CYP51, LproCYP51, and PafrCYP51 protein sequences with those of A. fumigatus CYP51 (AfumCYP51A and AfumCYP51B) indicated that SRSs and the HBR were mostly conserved (Fig. 2; see also File S1 in the supplemental material). Six selected amino acids of AfumCYP51A (G54, L98, G138, M220, Y431, and G448) which are described as being subject to hot spot mutations linked to azole resistance were compared with the corresponding amino acids in proteins of Scedosporium species, L. prolificans, and P. africana. Three of these six hot spot mutation sites were identical in all shown CYP51 sequences to those in azole-susceptible A. fumigatus isolates (AfumCYP51 G54, L98, or Y431; Fig. 2, black arrow). In contrast, all studied Scedosporium species, L. prolificans, and P. africana isolates showed an amino acid change (Scedosporium CYP51 S153 and I235, respectively; Fig. 2, green arrow) corresponding to positions AfumCYP51A G138 and AfumCYP51A M220 compared to the protein sequence of nonresistant strain Af293. In AfumCYP51B, Scedosporium CYP51, LproCYP51, and PafrCYP51, a uniform change (Scedosporium CYP51 A302; Fig. 2, green arrow) affected the amino acid corresponding to position AfumCYP51A T289. T289 is one of 2 aa which are characteristic of a tandem-repeat-dependent azole-resistant (TR46/Y121/T289) AfumCYP51A allele. Changes corresponding to amino acid AfumCYP51A Y121 and the hot spot mutation site AfumCYP51A G448 were detected only in three S. apiospermum isolates, altering amino acids in positions 136 and 464 of S. apiospermum CYP51 (SapioCYP51), respectively (Fig. 2, orange arrow; see below).
FIG 2.
Amino acid alignment of CYP51 from A. fumigatus and Scedosporium species. One hundred percent conserved residues are in black. The six P450 substrate recognition sites (SRSs) are marked with blue boxes, and the heme-binding region (HBR) is marked with a red box. The arrow indicates eight selected amino acids of A. fumigatus CYP51A (AfumCYP51A G54, L98, Y121, G138, M220, T289, Y431, and G448; Stensvold et al. [19]). An alteration in these amino acids is associated with azole-resistant A. fumigatus isolates. The AfumCYP51 protein sequences shown here originated from the nonresistant strain Af293. The corresponding amino acids from Scedosporium spp. are shown in parentheses. The black arrows mark the conserved amino acids in all Scedosporium species isolates. The green arrows indicate the amino acids which in all Scedosporium species isolates changed in comparison to AfumCYP51A from strain Af293. The orange arrows indicate amino acid alterations in three S. apiospermum isolates (with SapioCYP51 Y136 and SapioCYP51 G464). Because of the protein identity of S. boydii (CBS 591.90) and S. ellipsoideum (CBS 418.73T), the CYP51 of S. ellipsoideum is not shown. *, partial sequences. Accession numbers and strain identifiers are shown in parentheses.
Scedosporium CYP51 sequences showed species-specific polymorphisms. More than one allele type with single nucleotide polymorphisms (SNPs) of CYP51 was found in S. apiospermum, S. boydii, S. ellipsoideum, S. dehoogii, S. aurantiacum, and S. minutisporum isolates (Table 1). In 79/81 S. apiospermum isolates, 15 different allele types were found to be represented by 33 SNPs. Only three of these 33 SNPs in the coding regions of SapioCYP51 resulted in amino acid changes (Y136F, D446A, or G464S; Table 1). These were found in five isolates which had all been isolated from patients with cystic fibrosis (CF; patients 19, 81, and 87, and strain CBS 117430). In 25/26 S. boydii isolates, 25 SNPs defined seven allele types of the S. boydii CYP51 gene (SboyCYP51). Again, two SNPs resulted in amino acid changes for SboyCYP51 (E9Q or K77N). These amino acid changes were found in isolates from two patients, one with CF and one with a eumycetoma (CBS 101.22T, isolated in 1921). In 23/24 S. ellipsoideum isolates, three SNPs defined four allele types of the S. ellipsoideum CYP51 gene (SellCYP51). One SNP resulted in amino acid changes for SellCYP51 in the last amino acid before the stop codon (A528V). This protein was found in isolates from two patients, one patient with CF and one patient with another underlying disease. Remarkably, for S. boydii and S. ellipsoideum, the CYP51 protein sequence without the amino acid changes in CYP51 E9Q, K77N, or A528V was identical. In 15/16 S. dehoogii isolates, up to six allele types were found in the S. dehoogii CYP51 gene (SdehCYP51; 32 SNPs). Five SNPs in the coding region of SdehCYP51 resulted in amino acid changes (Y173F, A255S, R267K, K425R, and V496I). Remarkably, all changes (SdehCYP51 Y173F, A255S, R267K, K425R, and V496I) occurred together in three different environmental isolates. A single change, SdehCYP51 A255S, was detected in eight isolates (from three patients and two environmental isolates). For the two S. minutisporum isolates studied, 11 SNPs were found in the coding region of the S. minutisporum CYP51 gene (SminCYP51), and three SNPs resulted in amino acid changes (SminCYP51 P345L, S498G, and V528A). For the S. aurantiacum isolates, seven SNPs in CYP51 were detected, but these SNPs were synonymous. Of note, no correlation was found between the MLST type and the allele type of CYP51 of S. apiospermum, S. boydii, S. ellipsoideum, and S. dehoogii (all data in Table S3). Finally, the CYP51 gene of the three L. prolificans isolates showed no polymorphism.
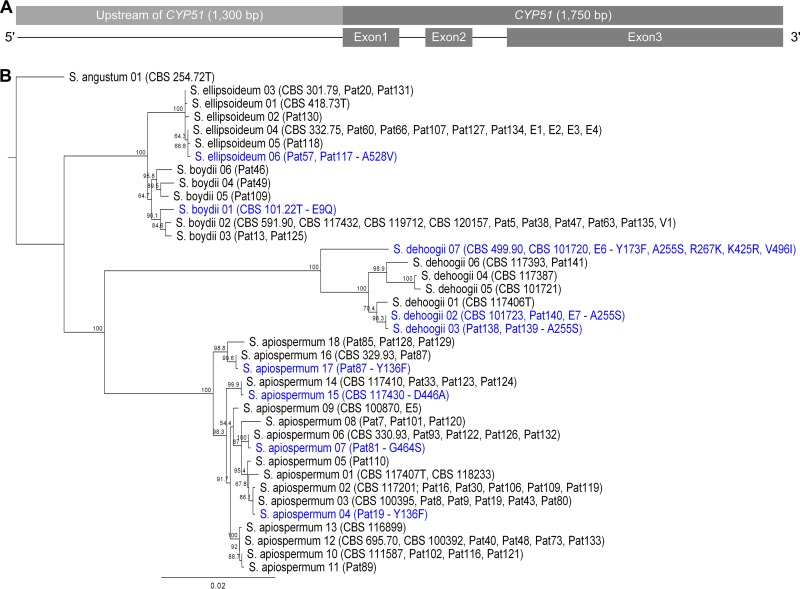
Additionally, the 5′ upstream region (up to 2,000 bp) of CYP51 in 146 isolates of S. apiospermum, S. boydii, S. ellipsoideum, S. dehoogii, and S. angustum was sequenced, and around 1,300 nucleotides upstream of the start codon were analyzed for sequence polymorphisms in this predicted regulatory region. The combination of 5′ upstream and coding region of CYP51 increased the number of distinguishable sequence types for S. apiospermum, S. boydii, S. ellipsoideum, and S. dehoogii to 18, six, six, and seven, respectively (Table 1 and Fig. 3). However, an analysis of the 5′ upstream region to identify tandem-repeat sequences of a range from 30 to 150 bp with a 10-bp step size between failed to reveal repeats like those described for A. fumigatus (34 bp, 46 bp, and 53 bp) or P. digitatum (126 bp) CYP51A.
FIG 3.
(A) Schematic representation of CYP51 sequence (3,050 bp). (B) Phylogenetic tree of 5′ upstream region CYP51 together with CYP51 gene sequences of S. apiospermum, S. boydii, S. ellipsoideum, S. dehoogii, and S. angustum (n = 146; around 3,050 bp). The tree was rooted to the S. angustum sequences. Branch labels are the percent consensus support. Isolates with amino acid changes are marked in blue. CBS, Centraalbureau voor Schimmelcultures; Pat, patient; E, environmental isolate; V, veterinary isolate.
The results of the in vitro azole susceptibility testing of S. apiospermum revealed that the MICs for ICZ, PCZ, and VCZ ranged from 0.5 to >16 μg/ml, 0.25 to >16 μg/ml, and 0.25 to 4 μg/ml, respectively. These values for S. boydii were from 0.5 to >16 μg/ml, 0.25 to >16 μg/ml, and 0.125 to 4 μg/ml and for S. ellipsoideum were 0.25 to >16 μg/ml, 0.5 to >16 μg/ml, and 0.25 to 4 μg/ml. The MICs of S. dehoogii isolates for ICZ, PCZ, and VCZ corresponded to ≥16 μg/ml, 2 to >16 μg/ml, and 0.5 to >16 μg/ml. In contrast, Lomentospora isolates had MICs of ≥16 μg/ml for all azoles (Table 2). The complete resistogram of the isolates is shown in Table S3.
TABLE 2.
In vitro antifungal susceptibility patterns of Scedosporium spp. and L. prolificans against azolesa
| Species | Total no. of isolates | ICZ |
PCZ |
VCZ |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Range | MIC50 | MIC90 | GM | n | Range | MIC50 | MIC90 | GM | n | Range | MIC50 | MIC90 | GM | ||
| S. apiospermum | 81 | 80 | 0.5 to >16 | >16 | >16 | 14.5 | 80 | 0.25 to >16 | 4 | >16 | 5.8 | 81 | 0.25 to 4 | 2 | 4 | 1.5 |
| S. boydii | 26 | 22 | 0.5 to >16 | >16 | >16 | 11.3 | 24 | 0.25 to >16 | 2 | >16 | 2.8 | 26 | 0.125 to 4 | 1 | 4 | 0.8 |
| S. ellipsoideum | 24 | 21 | 0.25 to >16 | >16 | >16 | 14.0 | 22 | 0.5 to >16 | 4 | >16 | 6.2 | 24 | 0.25 to 4 | 1 | 2 | 1.2 |
| S. dehoogii | 16 | 13 | >16 | >16 | >16 | 32.0 | 13 | 2 to >16 | 2 | >16 | 6.1 | 13 | 0.5 to >16 | 1 | >16 | 2.4 |
| S. aurantiacum | 7 | 3 | >16 | 32.0 | 4 | 1 to >16 | 3.4 | 4 | 0.25 to >16 | 1.4 | ||||||
| L. prolificans | 3 | 1 | >16 | 32.0 | 2 | >16 | 32.0 | 2 | >16 | 32.0 | ||||||
GM, geometric mean; ICZ, itraconazole; PCZ, posaconazole; VCZ, voriconazole.
DISCUSSION
Based on a lower susceptibility or even resistance of Scedosporium spp. to azoles and other drugs, treatment is considered more challenging than that for candidosis or aspergillosis. Over the last years, many studies have been analyzing the development of azole resistance in different yeast species or molds (like Candia spp. or A. fumigatus) under prolonged therapy or associated with the large-scale use of azoles in agriculture (27, 28). These studies identified mutations in the drug target, 14-α-lanosterol demethylase (CYP51), as one reason for resistance. The CYP51 protein family is present in almost all taxonomic kingdoms (20). A number of plants and fungi contain more than one CYP51 gene, and some species express additional pseudogenes of CYP51 (20, 29). The study presented here identified a single CYP51 gene in Scedosporium spp., and the same was found for the orthologue genes in the two phylogenetically distinct species L. prolificans and P. africana. CYP51 proteins are characterized by six highly conserved SRSs and the HBR, with a characteristic iron-binding cysteine (21). These conserved sites were found in all newly identified CYP51 proteins. Phylogenetic analysis places the CYP51 proteins of Scedosporium spp., LproCYP51 and PafrCYP51, in a monophyletic clade with CYP51B orthologues. This result is consistent with the finding of phylogenetic analyses of CYP51s in other fungal organisms, with only one isoform also clustering with the CYP51B group (30, 31). Hawkins et al. (31) found that Rhynchosporium commune, a filamentous ascomycete causing scald or leaf blotch on barley, encodes CYP51B. However, they also observed isolates with an additional CYP51A, and these were less sensitive to azoles than those without a CYP51A orthologue (31). In the genome data, we did not find any indication that the presence of additional paralogues accounted for resistance in Scedosporium where only one CYP51 was detectable.
It has been hypothesized that the possibility to detect any alteration in CYP51 should be increased in isolates with a high in vitro MIC to VCZ. As a consequence of our selective criteria, the geometric means (GM; Table 2) of in vitro MICs against azoles in the selected S. apiospermum, S. boydii, S. ellipsoideum, and S. dehoogii isolates were higher in this study than what was found in other published data (13, 14) and are not representative of the genus Scedosporium. Nevertheless, the published isolates and the ones presented here display an inherent low susceptibility for Scedosporium isolates against azoles compared to susceptible A. fumigatus isolates (32, 33).
For A. fumigatus, a single amino acid change in CYP51A or a tandem repeat in combination with amino acid substitutions in CYP51A were associated with increased azole resistance (19, 34). Amino acid changes are thought to affect drug binding and tandem repeats in 5′ upstream region to affect CYP51A expression levels. A tandem repeat, like that in A. fumigatus or P. digitatum, could not be detected in the 5′ upstream region of the Scedosporium isolates studied here, but three of the amino acid alterations that were thought to reduce azole binding to target in AfumCYP51A were observed in all studied Scedosporium, L. prolificans, and P. africana isolates (amino acid position in Scedosporium CYP51 S153, I235, and A302). Thus, we propose that this combination of changes in Scedosporium CYP51 explains the inherent low susceptibility against azoles of Scedosporium species. Formal proof that this was indeed the case would require homologous replacement of the endogenous gene in Scedosporium spp., with a version designed so that these alterations would be changed to mimic a CYP51A sequence known from susceptible Aspergillus isolates. However, a respective genetic tool box for Scedosporium is not yet available.
Furthermore, species-specific polymorphisms were identified. However, all additionally detected species-specific amino acid changes in CYP51 of S. boydii, S. ellipsoideum, S. dehoogii, and S. minutisporum isolates were located outside the SRSs, HBR, or on positions that have not been described as having an impact on azole susceptibility. Of note, these domains determined for CYP51A in A. fumigatus are presently only predicted based on homology in Scedosporium spp., and although there is strong conservation of these domains in other fungi, experimental proof of the correct assignment is currently lacking. In contrast, two of three additional species-specific amino acid alterations found in S. apiospermum (SapioCYP51 Y136F and SapioCYP51 G464S) corresponded to other A. fumigatus CYP51A mutations linked to azole resistance, namely, a hot spot mutation and a substitution which was described in combination with a tandem repeat in mutant aspergilli (19, 34). All S. apiospermum isolates with such additional amino acid changes were collected from patients with CF. All previous or follow-up isolates from these patients belonged to individual MLSTs. The allele type of CYP51 did not correlate with a certain MLST, indicating independent evolutionary pressures acting upon the respective traits. With respect to azole susceptibility and resistance, this adds important information to previous inconclusive attempts to correlate clinical and MLST data. Interestingly, the isolate with SapioCYP51 Y136F (from patient 87) and the two isolates from patient 81 (with SapioCYP51 G464S) showed lower VCZ susceptibilities than the other S. apiospermum isolates. Patient 87 was obviously colonized by isolates with different CYP51 proteins, but this particular isolate with SapioCYP51 Y136F may have an additional property that distinguishes it from the one from patient 19 that also possessed the amino acid mutation SapioCYP51 Y136F. In analogy to the situation in A. fumigatus, a probable explanation would be that this phenotypic difference is linked to the differences in the 5′ upstream region of CYP51 between these isolates which entail 48 SNPs. SapioCYP51 D446A found in CBS 117430 did not result in a decreased susceptibility for VCZ in vitro. It is tempting to speculate that the differences noted in these isolates may emerge as a result of selective drug pressure. Indeed, monotherapy with ICZ or VCZ for up to 2 months before the isolates were collected has been documented in three of the four cases (data not shown). S. apiospermum is the most frequently isolated Scedosporium species in patients with CF in Germany (7, 13). Thus, azole-based monotherapy may select or induce resistance mutations in these patients, which may also hold true for other Scedosporium species colonizing patients with CF. Longitudinal sampling data that are correlated with clinical data need to be gathered to substantiate this alarming finding.
The detected CYP51 polymorphisms may not be the only determinants of reduced azole susceptibility in Scedosporium species. Nevertheless, the presented data provide the first evidence that CYP51 is likely to play a significant role in the respective mechanisms at work in Scedosporium.
MATERIALS AND METHODS
Fungal isolates and identification.
The study was performed with isolates from the culture collection of fungi at the Robert Koch Institute. All isolates had been identified by micromorphology and molecular methods, sequencing of the internal transcribed spacer (ITS) region of the rDNA gene cluster, and/or hybridization to specific gene probes targeting the ITS2 region of the rDNA gene cluster (8). MLST for S. apiospermum, S. boydii, S. ellipsoideum, and S. dehoogii isolates was performed, as described by Bernhardt et al. (8). MLST analysis for S. aurantiacum was performed according to http://mlst.mycologylab.org. New sequence types were submitted to the MLST database for Scedosporium spp. (http://mlst.mycologylab.org). The isolates had been cryopreserved at −70°C.
The study is based on 159 Scedosporium species isolates (clinical, n = 134; veterinary, n = 2; environmental, n = 23, including 37 CBS strains), as follows: S. apiospermum, n = 81; S. boydii, n = 26; S. ellipsoideum, n = 24; S. dehoogii, n = 16; S. aurantiacum, n = 7; S. minutisporum, n = 2; and S. angustum, S. desertorum, and S. fusoideum, n = 1 (Table 1). The S. apiospermum, S. boydii, and S. ellipsoideum isolates were selected, if possible, based on a low (≤1 μg/ml) or high (≥4 μg/ml) in vitro MIC of VCZ, and preference was given to isolates from patients for whom isolates obtained at different times were available (Table 2). The decision to analyze isolates with different in vitro susceptibilities was based on the hypothesis that the detection of any alteration in CYP51 should be increased in isolates with a high in vitro MIC to VCZ. Additionally, two phylogenetically distinct species, Lomentospora prolificans (n = 3) and Petriellopsis africana (n = 1), were included (Table 1). The clinical isolates had been isolated from 85 patients, including 44 patients with cystic fibrosis (mean age, 22.0 years) and 41 patients with other or no underlying diseases (mean age, 47.8 years). The isolates from patients with CF (n = 83/94 isolates) had been cultured from the respiratory samples, and one isolate was from soft tissue. The origins of isolates from patients with other or no underlying disease (n = 43) were the skin/soft tissue (n = 13), respiratory tract (n = 12), eye (n = 4), ear (n = 3), blood (n = 3), and brain (n = 2). The sources of 16 isolates was unknown (patients with CF, n = 10; other patients, n = 6). The origins of the isolates are listed in Table S3.
In vitro susceptibility against azoles.
In vitro susceptibility testing of the Scedosporium isolates, if possible, had been performed for azoles (ICZ, PCZ, and VCZ; Sigma-Aldrich, Munich, Germany) in a microdilution assay, according to Clinical and Laboratory Standards Institute M38-A2 guidelines (35). The drug concentration ranged in 2-fold steps from 0.03 to 16 μg/ml. Candida krusei (ATCC 6258) was used as a quality control strain to confirm the correct testing conditions. The incubation time was 72 h. GMs and MICs were calculated as described by Lackner et al. (14).
Genomic DNA extraction.
The genomic DNA extraction of filamentous fungi was performed with the formalin-fixed paraffin-embedded (FFPE) tissue LEV DNA purification kit designed for the Maxwell 16 instrument (Promega, Mannheim, Germany), with minor modifications of the manufacturer's protocol, as follows: (i) DNA was purified directly from fungal cultures incubated on potato dextrose agar for 7 days at 37°C; (ii) an inoculating loop of mycelium was suspended in 300 μl of lysis buffer and incubated with shaking at 70°C for 10 min; and (iii) the extraction volume was 120 μl of bidest. water.
RNA extraction and cDNA synthesis.
Six-day-old seedlings (germinated in liquid RPMI 1640 medium with final 2% glucose) from S. apiospermum CBS 117407T were blot dried, frozen with liquid nitrogen, and ground to powder using a pestle and mortar. The RNA was extracted from the seedling powder the with NucleoSpin RNA plant kit (Macherey-Nagel, Düren, Germany), following the manufacturer's protocol. The first-strand cDNA synthesis was performed on 350 ng of RNA by SuperScript II reverse transcriptase (Invitrogen, Darmstadt, Germany), with an oligo(dT)12–18 primer (Invitrogen), following the manufacturer's protocol. The partial CYP51 cDNA was amplified with primers rtCYPSabp166F and Pb-CYP51_1214r (Table S4); the PCR product was purified and sequenced (see below). To identify the location of the introns, the obtained cDNA sequence and the sequence of genomic CYP51 were compared.
Identification of Scedosporium CYP51 orthologues through alignment and phylogenetic analysis.
Protein sequences of CYP51A and CYP51B in Aspergillus fumigatus (AfumCYP51A/AfumCYP51B, RefSeq accession numbers XP_752137/EAL87096) and in Fusarium oxysporum (FoxyCYP51A/FoxyCYP51B, RefSeq accession numbers EMT65894/EMT62075) were used to analyze the available genome sequences of S. apiospermum (CBS 117430; GenBank accession no. JOWA01000000) and S. aurantiacum (WM09-24; GenBank accession no. JUDQ01000000) by the tblastx algorithm from Geneious (36, 37). Aspergillus as well as Fusarium protein sequences were derived from the respective RefSeq sequences for comparison. Fusarium proteins have a closer phylogenetic relationship to Scedosporium proteins than to Aspergillus proteins.
CYP51 sequencing and analysis.
Based on the S. aurantiacum and S. apiospermum genome sequences for the CYP51 orthologue, CYP51 primers were developed with the tool “design new primer” as part of the program Geneious. For all Scedosporium isolates, L. prolificans and P. africana CYP51 genes were amplified and sequenced. For five Scedosporium species (S. apiospermum, S. boydii, S. ellipsoideum, S. dehoogii, and S. angustum), the 5′ upstream region of CYP51 was amplified and sequenced (up to 2,000 bp). The primer sequences of the 5′ upstream region were based on the sequence of S. apiospermum (CBS 117430; GenBank accession no. JOWA01000000). The amplification and sequencing primers are listed in Table S4.
The PCR assay (25 μl) included 1 to 5 μl of fungal DNA, 1 μM each gene-specific primer (Table S4) (TIB Molbiol, Berlin, Germany), 0.2 mM each deoxynucleoside triphosphate (Roche, Mannheim, Germany), 1× DreamTaq buffer with 20 mM MgCl2 (Thermo Fisher Scientific, Darmstadt, Germany) and 0.625 U DreamTaq DNA polymerase (Thermo Fisher Scientific). Amplification of the genes was performed in a T1 thermocycler (Biometra, Göttingen, Germany), as follows: 5 min of initial denaturation at 95°C, followed by 35 cycles at 95°C for 30 s, gene-specific annealing temperature for 30 s (Table S4), and 72°C for 1 min per 1 kb PCR product length. The final extension step was 7 min at 72°C.
The PCR products were purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany). Sequencing was performed using the BigDye Terminator version 3.1 cycle sequencing kit, and the reactions were run on the ABI Prism 3130 genetic analyzer (Applied Biosystems, Darmstadt, Germany). Sequencing analysis was performed using the program Geneious.
All CYP51 allele types were numbered, starting with the type strain of the species, if possible.
The phylogenetic trees were generated by ClustalW alignment using a Geneious tree (neighbor-joining consensus tree) with 1,000 bootstrap replications. The amino acid alignment was performed using the ClustalW alignment tool from Geneious.
Functional complementation study.
The deletion of the ERG11 gene in S. cerevisiae is lethal, and the growth of recombinant strains with SaipoCYP51 provides evidence for the functionality of the gene. The recombinant strains were constructed according to Parker et al. (38). The cDNA sequences of S. apiospermum CYP51 (SapioCYP51; allele type 1, CBS 11740T) and S. cerevisiae ERG11 (ScereERG11; strain S288c) were optimized for expression in S. cerevisiae by GeneArt gene synthesis and synthesized by Invitrogen. The genes were cloned into the EcoRI and HindIII (New England BioLabs GmbH, Frankfurt am Main, Germany) sites in pUC19 (T4 ligase; Thermo Fisher Scientific). The URA3-loxP marker was excised from pUG72 (Swiss-Prot accession no. P30117; Euroscarf, Frankfurt am Main, Germany), using SpeI and PstI, and was ligated in all pUC19(CYP51/ERG11) constructs. For transformation experiments, the S. cerevisiae strain BY4741 (MATa his3-D1 leu2-D0 met15-D0 ura3-D0; strain obtained from Euroscarf) was used. BY4741 was grown on synthetic dextrose (SD) minimal medium (pH 5.7) containing 1.7 g/liter yeast nitrogen base without amino acid and ammonium sulfate (Sigma-Aldrich), 5 g/liter ammonium sulfate (Merck, Germany) and 20 g/liter glucose, with 50 μg/ml histidine (Sigma-Aldrich), and leucine (Sigma-Aldrich), methionine (Carl Roth, Karlsruhe, Germany), and uracil (pH 8.5; Sigma-Aldrich) added at 100 μg/ml each. S. cerevisiae strain BY4741 was transformed by electroporation (39), with modification of the protocol as follows: (i) the cell pellets were collected by centrifugation (1,100 × g, 4°C, 5 min) and suspended in 20 ml of sterilized ice-cold bidest. water. (ii) After centrifugation, the cells were treated with 20 ml of sorbitol-TE-LiAc (16 ml of 1 M sorbitol, 2 ml of 10× TE buffer [100 mM Tris-HCl, 10 mM EDTA {pH 7.5}], 2 ml of 1 M lithium acetate [pH 7.5]) at 30°C for 30 min, followed by adding 0.2 ml of 1 M dithiothreitol (DTT) and incubating at 30°C for 15 min. (iii) The cells were washed twice in 20 ml of ice-cold 1 M sorbitol and suspended in 200 μl of 1 M sorbitol. (iv) Fifty microliters of cell suspension was used for each transformation. (v) After the electroporation, the cells were incubated in 1 ml of 1 M sorbitol for 90 min at 30°C, centrifuged (3,000 × g, 1 min), suspended in 150 μl of 1 M sorbitol, and then spread on selection plates. The transformed yeast strains grew on SD medium without the addition of uracil. All yeast strains were incubated at 35°C for up to 10 days. Insertion of the constructs was confirmed by PCR (primers in Table S4) and the replacement of the endogenous ScereERG11 by the codon-optimized ERG11/CYP51 by quantitative PCR (qPCR). The genomic DNA extraction of yeast was performed with MasterPure yeast DNA purification kit (Biozym Scientific GmbH, Hessisch Oldendorf, Germany) (40). For the qPCR, the yeast RNA was extracted from 1.5 ml of a 2-day-old cell culture using the NucleoSpin RNA plant kit (Macherey-Nagel), following the manufacturer's protocol. Cell grinding was performed with glass beads and the MP Biomedicals FastPrep-24 5G instrument. The first-strand cDNA synthesis was performed on 350 ng of RNA by SuperScript II reverse transcriptase (Invitrogen) with an oligo(dT)12–18 primer (Invitrogen), following the manufacturer's protocol. The qPCR assay (25 μl) included 1 μl of yeast cDNA, 0.6 μM each gene-specific primer (endogenous ScereERG11, codon-optimized ERG11/CYP51, and S. cerevisiae actin gene; TIB Molbiol) (Table S4), 1 mM GeneAMP dNTP blend with dUTP (Thermo Fisher Scientific), 0.65 × EvaGreen dye (Biotium, Fremont, CA, USA), 1× ROX reference dye (Invitrogen), 3 mM MgCl2 (Invitrogen), 1× Platinum Taq buffer without MgCl2, and 2 U Platinum Taq DNA polymerase (Invitrogen). Amplification was performed in an ABI 7500 cycler (Applied Biosystems), as follows: 2 min at 50°C and 10 min at 95°C, followed by 45 cycles at 95°C for 15 s, 55°C for 30 s, and 72°C for 40 s. The data collection was done during the extension step. Gel electrophoresis confirmed the results.
Accession number(s).
The accession numbers (MH120874 to MH121036) are listed in Table S3.
Supplementary Material
ACKNOWLEDGMENTS
We declare no conflicts of interest.
We thank Elisabeth Antweiler and Birgid Raddatz for identification of the clinical isolates, Chang-Ok Han and Michael Seibold for assistance with antifungal susceptibility testing, Julia Tesch, Julia Hinzmann, and Silvia Muschter for sequencing, and Ursula Erikli for copyediting.
The research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02599-17.
REFERENCES
- 1.Ramirez-Garcia A, Pellon A, Rementeria A, Buldain I, Barreto-Bergter E, Rollin-Pinheiro R, de Meirelles JV, Xisto M, Ranque S, Havlicek V, Vandeputte P, Govic YL, Bouchara JP, Giraud S, Chen S, Rainer J, Alastruey-Izquierdo A, Martin-Gomez MT, Lopez-Soria LM, Peman J, Schwarz C, Bernhardt A, Tintelnot K, Capilla J, Martin-Vicente A, Cano-Lira J, Nagl M, Lackner M, Irinyi L, Meyer W, de Hoog S, Hernando FL. 2018. Scedosporium and Lomentospora: an updated overview of underrated opportunists. Med Mycol 56:102–125. doi: 10.1093/mmy/myx113. [DOI] [PubMed] [Google Scholar]
- 2.Lackner M, de Hoog GS, Yang LY, Moreno LF, Ahmed SA, Andreas F, Kaltseis J, Nagl M, Lass-Florl C, Risslegger B, Rambach G, Speth C, Robert V, Buzina W, Chen S, Bouchara JP, Cano-Lira JF, Guarro J, Gene J, Silva FF, Haido R, Haase G, Havlicek V, Garcia-Hermoso D, Meis JF, Hagen F, Kirchmair M, Rainer J, Schwabenbauer K, Zoderer M, Meyer W, Gilgado F, Schwabenbauer K, Vicente VA, Pieckova E, Regenermel M, Rath PM, Steinmann J, de Alencar XW, Symoens F, Tintelnot K, Ulfig K, Velegraki A, Tortorano AM, Giraud S, Mina S, Rigler-Hohenwarter K, Hernando FL, Ramirez-Garcia A, Pellon A, et al. . 2014. Proposed nomenclature for Pseudallescheria, Scedosporium and related genera. Fungal Divers 67:1–10. doi: 10.1007/s13225-014-0295-4. [DOI] [Google Scholar]
- 3.Tintelnot K, Wagner N, Seibold M, de Hoog GS, Horre R. 2008. Re-identification of clinical isolates of the Pseudallescheria boydii-complex involved in near-drowning. Mycoses 51(Suppl 3):S11–S16. doi: 10.1111/j.1439-0507.2008.01579.x. [DOI] [PubMed] [Google Scholar]
- 4.Cortez KJ, Roilides E, Quiroz-Telles F, Meletiadis J, Antachopoulos C, Knudsen T, Buchanan W, Milanovich J, Sutton DA, Fothergill A, Rinaldi MG, Shea YR, Zaoutis T, Kottilil S, Walsh TJ. 2008. Infections caused by Scedosporium spp. Clin Microbiol Rev 21:157–197. doi: 10.1128/CMR.00039-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tammer I, Tintelnot K, Braun-Dullaeus RC, Mawrin C, Scherlach C, Schluter D, Konig W. 2011. Infections due to Pseudallescheria/Scedosporium species in patients with advanced HIV disease–a diagnostic and therapeutic challenge. Int J Infect Dis 15:e422–429. doi: 10.1016/j.ijid.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Guarro J, Kantarcioglu AS, Horre R, Rodriguez-Tudela JL, Cuenca Estrella M, Berenguer J, de Hoog GS. 2006. Scedosporium apiospermum: changing clinical spectrum of a therapy-refractory opportunist. Med Mycol 44:295–327. doi: 10.1080/13693780600752507. [DOI] [PubMed] [Google Scholar]
- 7.Bernhardt A, Sedlacek L, Wagner S, Schwarz C, Wurstl B, Tintelnot K. 2013. Multilocus sequence typing of Scedosporium apiospermum and Pseudallescheria boydii isolates from cystic fibrosis patients. J Cyst Fibros 12:592–598. doi: 10.1016/j.jcf.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Bernhardt A, Seibold M, Rickerts V, Tintelnot K. 2015. Cluster analysis of Scedosporium boydii infections in a single hospital. Int J Med Microbiol 305:724–728. doi: 10.1016/j.ijmm.2015.08.024. [DOI] [PubMed] [Google Scholar]
- 9.Castiglioni B, Sutton DA, Rinaldi MG, Fung J, Kusne S. 2002. Pseudallescheria boydii (Anamorph Scedosporium apiospermum). infection in solid organ transplant recipients in a tertiary medical center and review of the literature. Medicine (Baltimore) 81:333–348. [DOI] [PubMed] [Google Scholar]
- 10.Capilla J, Mayayo E, Serena C, Pastor FJ, Guarro J. 2004. A novel murine model of cerebral scedosporiosis: lack of efficacy of amphotericin B. J Antimicrob Chemother 54:1092–1095. doi: 10.1093/jac/dkh468. [DOI] [PubMed] [Google Scholar]
- 11.Zeng J, Kamei K, Zheng Y, Nishimura K. 2004. Susceptibility of Pseudallescheria boydii and Scedosporium apiospermum to new antifungal agents. Nihon Ishinkin Gakkai Zasshi 45:101–104. doi: 10.3314/jjmm.45.101. [DOI] [PubMed] [Google Scholar]
- 12.Tortorano AM, Richardson M, Roilides E, van Diepeningen A, Caira M, Munoz P, Johnson E, Meletiadis J, Pana ZD, Lackner M, Verweij P, Freiberger T, Cornely OA, Arikan-Akdagli S, Dannaoui E, Groll AH, Lagrou K, Chakrabarti A, Lanternier F, Pagano L, Skiada A, Akova M, Arendrup MC, Boekhout T, Chowdhary A, Cuenca-Estrella M, Guinea J, Guarro J, de Hoog S, Hope W, Kathuria S, Lortholary O, Meis JF, Ullmann AJ, Petrikkos G, Lass-Florl C, European Society of Clinical Microbiology and Infectious Diseases Fungal Infection Study Group, European Confederation of Medical Mycology . 2014. ESCMID and ECMM joint guidelines on diagnosis and management of hyalohyphomycosis: Fusarium spp., Scedosporium spp. and others. Clin Microbiol Infect 20(Suppl 3):S27–S46. doi: 10.1111/1469-0691.12465. [DOI] [PubMed] [Google Scholar]
- 13.Sedlacek L, Graf B, Schwarz C, Albert F, Peter S, Wurstl B, Wagner S, Klotz M, Becker A, Haase G, Laniado G, Kahl B, Suerbaum S, Seibold M, Tintelnot K. 2015. Prevalence of Scedosporium species and Lomentospora prolificans in patients with cystic fibrosis in a multicenter trial by use of a selective medium. J Cyst Fibros 14:237–241. doi: 10.1016/j.jcf.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 14.Lackner M, de Hoog GS, Verweij PE, Najafzadeh MJ, Curfs-Breuker I, Klaassen CH, Meis JF. 2012. Species-specific antifungal susceptibility patterns of Scedosporium and Pseudallescheria species. Antimicrob Agents Chemother 56:2635–2642. doi: 10.1128/AAC.05910-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castanheira M, Messer SA, Rhomberg PR, Pfaller MA. 2016. Antifungal susceptibility patterns of a global collection of fungal isolates: results of the SENTRY Antifungal Surveillance Program (2013). Diagn Microbiol Infect Dis 85:200–204. doi: 10.1016/j.diagmicrobio.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Cuenca-Estrella M, Alastruey-Izquierdo A, Alcazar-Fuoli L, Bernal-Martinez L, Gomez-Lopez A, Buitrago MJ, Mellado E, Rodriguez-Tudela JL. 2008. In vitro activities of 35 double combinations of antifungal agents against Scedosporium apiospermum and Scedosporium prolificans. Antimicrob Agents Chemother 52:1136–1139. doi: 10.1128/AAC.01160-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen SC, Playford EG, Sorrell TC. 2010. Antifungal therapy in invasive fungal infections. Curr Opin Pharmacol 10:522–530. doi: 10.1016/j.coph.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, Kelly DE, Kelly SL. 2010. A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14alpha-demethylase) and ERG5 (encoding C22 desaturase) is cross resistant to azoles and amphotericin B. Antimicrob Agents Chemother 54:3578–3583. doi: 10.1128/AAC.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stensvold CR, Jørgensen LN, Arendrup MC. 2012. Azole-resistant invasive aspergillosis: relationship to agriculture. Curr Fungal Infect Rep 6:178–191. doi: 10.1007/s12281-012-0097-7. [DOI] [Google Scholar]
- 20.Lepesheva GI, Waterman MR. 2011. Structural basis for conservation in the CYP51 family. Biochim Biophys Acta 1814:88–93. doi: 10.1016/j.bbapap.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Debeljak N, Fink M, Rozman D. 2003. Many facets of mammalian lanosterol 14alpha-demethylase from the evolutionarily conserved cytochrome P450 family CYP51. Arch Biochem Biophys 409:159–171. doi: 10.1016/S0003-9861(02)00418-6. [DOI] [PubMed] [Google Scholar]
- 22.Denning DW, Perlin DS. 2011. Azole resistance in Aspergillus: a growing public health menace. Future Microbiol 6:1229–1232. doi: 10.2217/fmb.11.118. [DOI] [PubMed] [Google Scholar]
- 23.Verweij PE, Chowdhary A, Melchers WJ, Meis JF. 2016. Azole resistance in Aspergillus fumigatus: can we retain the clinical use of mold-active antifungal azoles? Clin Infect Dis 62:362–368. doi: 10.1093/cid/civ885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albarrag AM, Anderson MJ, Howard SJ, Robson GD, Warn PA, Sanglard D, Denning DW. 2011. Interrogation of related clinical pan-azole-resistant Aspergillus fumigatus strains: G138C, Y431C, and G434C single nucleotide polymorphisms in cyp51A, upregulation of cyp51A, and integration and activation of transposon Atf1 in the cyp51A promoter. Antimicrob Agents Chemother 55:5113–5121. doi: 10.1128/AAC.00517-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bader O, Tunnermann J, Dudakova A, Tangwattanachuleeporn M, Weig M, Gross U. 2015. Environmental isolates of azole-resistant Aspergillus fumigatus in Germany. Antimicrob Agents Chemother 59:4356–4359. doi: 10.1128/AAC.00100-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamamoto H, Hasegawa K, Nakaune R, Lee YJ, Makizumi Y, Akutsu K, Hibi T. 2000. Tandem repeat of a transcriptional enhancer upstream of the sterol 14alpha-demethylase gene (CYP51) in Penicillium digitatum. Appl Environ Microbiol 66:3421–3426. doi: 10.1128/AEM.66.8.3421-3426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verweij PE, Snelders E, Kema GH, Mellado E, Melchers WJ. 2009. Azole resistance in Aspergillus fumigatus: a side-effect of environmental fungicide use? Lancet Infect Dis 9:789–795. doi: 10.1016/S1473-3099(09)70265-8. [DOI] [PubMed] [Google Scholar]
- 28.Tumbarello M, Caldarola G, Tacconelli E, Morace G, Posteraro B, Cauda R, Ortona L. 1996. Analysis of the risk factors associated with the emergence of azole resistant oral candidosis in the course of HIV infection. J Antimicrob Chemother 38:691–699. doi: 10.1093/jac/38.4.691. [DOI] [PubMed] [Google Scholar]
- 29.Lepesheva GI, Waterman MR. 2007. Sterol 14alpha-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim Biophys Acta 1770:467–477. doi: 10.1016/j.bbagen.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becher R, Weihmann F, Deising HB, Wirsel SG. 2011. Development of a novel multiplex DNA microarray for Fusarium graminearum and analysis of azole fungicide responses. BMC Genomics 12:52. doi: 10.1186/1471-2164-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawkins NJ, Cools HJ, Sierotzki H, Shaw MW, Knogge W, Kelly SL, Kelly DE, Fraaije BA. 2014. Paralog re-emergence: a novel, historically contingent mechanism in the evolution of antimicrobial resistance. Mol Biol Evol 31:1793–1802. doi: 10.1093/molbev/msu134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraczek MG, Bromley M, Buied A, Moore CB, Rajendran R, Rautemaa R, Ramage G, Denning DW, Bowyer P. 2013. The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J Antimicrob Chemother 68:1486–1496. doi: 10.1093/jac/dkt075. [DOI] [PubMed] [Google Scholar]
- 33.ATCC. 2017. ATCC multidrug-resistant & antimicrobial testing reference strains. American Type Culture Collection, Manassas, VA: https://www.atcc.org/~/media/PDFs/Multidrug%20Resistant%20%20Antimicrobial%20Strains.ashx. [Google Scholar]
- 34.Wiederhold NP, Gil VG, Gutierrez F, Lindner JR, Albataineh MT, McCarthy DI, Sanders C, Fan H, Fothergill AW, Sutton DA. 2016. First detection of TR34 L98H and TR46 Y121F T289A Cyp51 mutations in Aspergillus fumigatus isolates in the United States. J Clin Microbiol 54:168–171. doi: 10.1128/JCM.02478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.CLSI. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard, 2nd ed CLSI document M38-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 36.Pérez-Bercoff A, Papanicolaou A, Ramsperger M, Kaur J, Patel HR, Harun A, Duan SY, Elbourne L, Bouchara JP, Paulsen IT, Nevalainen H, Meyer W, Huttley GA. 2015. Draft genome of Australian environmental strain WM 09.24 of the opportunistic human pathogen Scedosporium aurantiacum. Genome Announc 3:e01526-. doi: 10.1128/genomeA.01526-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandeputte P, Ghamrawi S, Rechenmann M, Iltis A, Giraud S, Fleury M, Thornton C, Delhaes L, Meyer W, Papon N, Bouchara JP. 2014. Draft genome sequence of the pathogenic fungus Scedosporium apiospermum. Genome Announc 2:e00988-14. doi: 10.1128/genomeA.00988-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker JE, Merkamm M, Manning NJ, Pompon D, Kelly SL, Kelly DE. 2008. Differential azole antifungal efficacies contrasted using a Saccharomyces cerevisiae strain humanized for sterol 14 alpha-demethylase at the homologous locus. Antimicrob Agents Chemother 52:3597–3603. doi: 10.1128/AAC.00517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson JR, Register E, Curotto J, Kurtz M, Kelly R. 1998. An improved protocol for the preparation of yeast cells for transformation by electroporation. Yeast 14:565–571. doi:. [DOI] [PubMed] [Google Scholar]
- 40.Sanchini A, Smith IM, Sedlacek L, Schwarz R, Tintelnot K, Rickerts V. 2014. Molecular typing of clinical Cryptococcus neoformans isolates collected in Germany from 2004 to 2010. Med Microbiol Immunol 203:333–340. doi: 10.1007/s00430-014-0341-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.