ABSTRACT
Unlike most antimicrobial peptides (AMPs), the main mode of action of the subclass of proline-rich antimicrobial peptides (PrAMPs) is not based on disruption of the bacterial membrane. Instead, PrAMPs exploit the inner membrane transporters SbmA and YjiL/MdtM to pass through the bacterial membrane and enter the cytosol of specific Gram-negative bacteria, where they exert an inhibitory effect on protein synthesis. Despite sharing a high proline and arginine content with other characterized PrAMPs, the PrAMP Bac5 has a low sequence identity with them. Here we investigated the mode of action of three N-terminal Bac5 fragments, Bac5(1-15), Bac5(1-25), and Bac5(1-31). We show that Bac5(1-25) and Bac5(1-31) retained excellent antimicrobial activity toward Escherichia coli and low toxicity toward eukaryotic cells, whereas Bac5(1-15) was inactive. Bac5(1-25) and Bac5(1-31) inhibited bacterial protein synthesis in vitro and in vivo. Competition assays suggested that the binding site of Bac5 is within the ribosomal tunnel, where it prevents the transition from the initiation to the elongation phase of translation, as reported for other PrAMPs, such as the bovine PrAMP Bac7. Surprisingly, unlike Bac7, Bac5(1-25) exhibited species-specific inhibition, being an excellent inhibitor of protein synthesis on E. coli ribosomes but a poor inhibitor on Thermus thermophilus ribosomes. This indicates that while Bac5 most likely has an overlapping binding site with Bac7, the mode of interaction is distinct, suggesting that Bac5 fragments may be interesting alternative lead compounds for the development of new antimicrobial agents.
KEYWORDS: proline-rich antimicrobial peptide, PrAMPs, Bac5, ribosome, translation, protein synthesis inhibitor, proline-rich antimicrobial agents
INTRODUCTION
The increasing spread of multidrug-resistant pathogens is highlighting the urgent need for new antimicrobial compounds that can be used for the treatment of human and livestock infections. Antimicrobial peptides (AMPs) are considered a good starting point for the development of future antibiotics (1), and among AMPs, proline-rich antimicrobial peptides (PrAMPs) are very promising candidates (2). As a result of convergent evolution, PrAMPs are present in some insects and mammals, where they act as important effectors of the innate immunity (3, 4). Indeed, PrAMPs have a potent antimicrobial effect on many different Gram-negative bacteria (3, 5). Unlike most other AMPs that have a lytic mode of action, the majority of the characterized PrAMPs pass through the membrane and access the bacterial cytosol, where they target intracellular processes (6–8). Generally, PrAMPs damage and permeabilize the bacterial membrane only at concentrations much above those sufficient to inhibit the growth of the bacteria; therefore, their lytic mode of action is limited to a secondary effect (7–9), with the exception of that observed in Pseudomonas aeruginosa strains (10). Moreover, because of their intracellular mode of action, PrAMPs display lower toxicity toward eukaryotic cells (8) than lytic AMPs.
PrAMPs take advantage of two inner membrane transport proteins to pass through the bacterial membranes, predominantly using the SbmA transporter, but also the YjiL-MdtM transport system at higher concentrations (7, 11). Once in the bacterial cytosol, PrAMPs prevent bacterial growth by binding to ribosomes and inhibiting the protein synthesis (4). So far, two distinct mechanisms of action have been identified for PrAMPs. The type I PrAMPs bind within the ribosomal exit tunnel and allow translation initiation but block the transition into the elongation phase, presumably by preventing aminoacyl-tRNA accommodation at the peptidyltransferase center of the ribosome (12, 13). Type I PrAMPs encompass insect PrAMPs, such as oncocin, pyrrhocoricin, and metalnikowin, as well as mammalian PrAMPs, such as the bovine Bac7 and its dolphin orthologue, Tur1A (12–14). In contrast, type II PrAMPs also bind within the ribosomal tunnel but do not interfere with translation initiation and elongation but, rather, inhibit the termination step by blocking the dissociation of release factors (RFs; RF1 and RF2) from the ribosome (15). Type II PrAMPs include the insect PrAMP apidaecin 1b and its synthetic derivative Api137 (15).
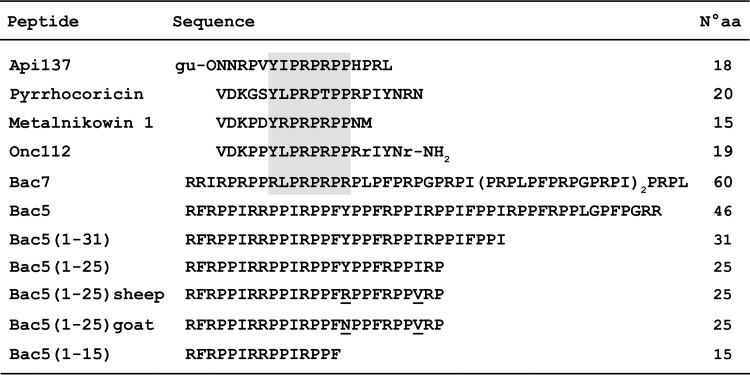
Because of the high prevalence of arginine and proline residues in the PrAMPs, it is not always obvious how to use sequence similarity to predict which mechanistic class that a particular PrAMP belongs to. For example, the PrAMP Bac5 is also highly rich in proline and arginine and intermediate in length (43 amino acids [aa]) between Bac7 (60 aa) and oncocin (19 aa). However, despite its high content of proline and arginine, the Bac5 sequence cannot be aligned easily with those of other PrAMPs (Fig. 1). Bac5 was first isolated from bovine neutrophils as a peptide with remarkable antimicrobial potency (16), and its orthologues were subsequently found in other mammals, including sheep and goats (17–19). An old pioneering study suggested that Bac5 (and Bac7) had a permeabilizing mode of action (20); however, a subsequent study demonstrated that a shortened version of the ovine homolog of Bac5 [Bac5(1-24)] did not strongly affect bacterial membrane integrity (21). Moreover, an Escherichia coli mutant lacking the SbmA transporter was less susceptible to the antimicrobial activity of the Bac5(1-31) fragment (7), suggesting that Bac5 could also exploit this transporter to access the bacterial cytosol and inhibit essential bacterial processes, such as protein synthesis.
FIG 1.
Alignment of the native Bac5 and Bac5 fragments with other PrAMPs. The gray box indicates a conserved region among ribosome-targeting PrAMPs. r, d-arginine; gu, N,N,N′,N′-tetramethylguanidino; O, ornithine. Underlined residues are those that differ among the Bac5(1-25) orthologues.
In this study, we characterized N-terminal fragments of Bac5, namely, Bac5(1-15), Bac5(1-25), and Bac5(1-31). We observed that the Bac5 fragments indeed inhibit bacterial growth using a nonlytic mode of action. We show that Bac5 binds to ribosomes and blocks translation using a mechanism of action analogous to that reported previously for oncocin and Bac7 but unlike that of apidaecin. Despite also binding within the ribosomal tunnel, Bac5 fragments exhibited a mode of interaction with the ribosome distinct from that exhibited by oncocin and Bac7. Specifically, we observed that the Bac5 fragments, like Bac7, are excellent inhibitors of translation on E. coli ribosomes; however, unlike Bac7, the Bac5 fragments were poor inhibitors of translation on Thermus thermophilus ribosomes. Moreover, our findings suggest that PrAMPs such as Bac5 can exert a bactericidal effect at concentrations where cell membrane integrity is not affected, thus indicating that the bactericidal effect results from interaction with the ribosome, as reported for the aminoglycoside and ketolide classes of antibiotics (22, 23).
RESULTS
Antimicrobial activity of Bac5 fragments.
To assess the antimicrobial activity of the Bac5(1-15), Bac5(1-25), and Bac5(1-31) fragments, the MIC and minimum bactericidal concentration (MBC) were determined using E. coli strain BW25113 (Table 1). Both the Bac5(1-25) and Bac5(1-31) fragments displayed good antimicrobial activity, with MICs of 1 μM and 4 μM, respectively. In contrast, the shortest fragment, Bac5(1-15), was inactive. A previous study reported that the native Bac5 had an MIC value (2 μM) for E. coli similar to the MICs observed here for Bac5(1-25) and Bac5(1–35), suggesting that these N-terminal Bac5 fragments retained full antimicrobial activity. However, we note that these comparisons could be affected by the difference in E. coli strains used, namely, ATCC 25922 (16) versus BW25113, used in this study.
TABLE 1.
MIC and MBC values of Bac5 fragments for E. coli BW25113 and E. coli BW25113 ΔsbmA
| E. coli strain | MIC (μM)a |
MBC (μM)a |
|||||
|---|---|---|---|---|---|---|---|
| Bac5(1-15) | Bac5(1-25) | sBac5(1-25) | gBac5(1-25) | Bac5(1-31) | Bac5(1-25) | Bac5(1-31) | |
| BW25113 | >32 | 1 | 1 | 1 | 4 | 4 | 16 |
| BW25113 ΔsbmA | >32 | 16 | 8 | >32 | 16–32 | >16 | >32 |
Results are the mode from three independent experiments performed in duplicate (n = 6). Results for sheep Bac5(1-25) [sBac5(1-25)] and goat Bac5(1-25) [gBac5(1-25)] orthologues are also shown.
Both of the longer fragments, Bac5(1-25) and Bac5(1-31), also displayed good bactericidal activity, with MBCs of 4 μM and 16 μM, respectively, i.e., MBCs 4-fold higher than the respective MIC values in both cases (Table 1). This is in agreement with the bactericidal activity reported for other PrAMPs. The fragments from aa 1 to 16 and aa 1 to 35 of the bovine peptide Bac7 displayed bactericidal properties when used on E. coli cells at 4 μM and 1 μM, respectively, i.e., concentrations 2-fold higher than the respective MICs (7, 9). A killing activity on E. coli cells at 4 μM was also reported for the porcine PrAMP PR-39 (24). However, a bactericidal effect of PrAMPs is not limited to peptides from mammals since lethality has also been reported under some conditions for insect AMPs, e.g., for pyrrhocoricin (25) and apidaecin (26).
To assess whether the Bac5 fragments display a mode of action involving their internalization in bacteria through the SbmA transporter, the MIC values of the Bac5 fragments were determined for the E. coli strain BW25113 lacking the sbmA gene (BW25113 ΔsbmA). Indeed, the absence of the SbmA transporter led to 16-fold and 4-fold increases in the MICs of Bac5(1-25) and Bac5(1-31), respectively, indicating that SbmA plays a major role in the uptake of Bac5 fragments into the bacterial cell. Additionally, the absence of SbmA also led to an increase in the MBC for Bac5(1-25) and Bac5(1-31) (Table 1). These observations support a nonlytic mode of action for Bac5 fragments.
We also determined the MIC of the Bac5(1-25) orthologues from sheep and goats, which have high sequence similarity with bovine Bac5 (Fig. 1). Both the sheep and goat Bac5(1-25) peptides displayed good antimicrobial activity against E. coli BW25113 with an MIC of 1 μM, i.e., an MIC identical to that determined for the bovine Bac5(1-25) fragment (Table 1). This was not unexpected, given that only two sequence differences were present, one of which was a conservative Ile23Val substitution (Fig. 1). Apparently, replacing Tyr16 in the bovine Bac5(1-25) with either Arg16 in the sheep Bac5 or Asn16 in the goat Bac5 has little effect on the antimicrobial activity of this PrAMP. Similarly, the absence of SbmA led to an increase in the MIC for sheep and goat Bac5(1-25), indicating that SbmA also plays a major role in the uptake of these Bac5 orthologues into the bacterial cell.
Membrane permeabilization activity of Bac5 fragments.
To directly assess whether the Bac5 fragments were capable of disrupting the bacterial membrane, a permeabilization assay using flow cytometry previously used for assessing the permeabilization activity of Bac7 fragments was employed (7, 27). The integrity of the bacterial membrane was assessed by exposing E. coli BW25113 cells to either 1 μM, 10 μM, or 50 μM the Bac5 fragments in the presence of propidium iodide (PI) for up to 1 h. Under these conditions, Bac5(1-15) did not permeabilize the bacterial membrane and Bac5(1-25) permeabilized less than 1% of the bacterial cells even at a concentration of 10 μM (data not shown), which was well above both the MIC and the MBC values (1 μM and 4 μM, respectively). This result is consistent with a nonlytic mechanism of action for Bac5(1-25). Similarly, Bac5(1-31) permeabilized less than 5% of the bacterial cells at a concentration of 10 μM, which was above the MIC value of 4 μM, indicating that Bac5(1-31), like Bac5(1-25), also utilizes a primarily nonlytic mechanism. Interestingly, Bac5(1-25) did not permeabilize the E. coli membrane even at 10 μM, a concentration more than 2-fold the MBC for Bac5(1-25), suggesting that not only the bacteriostatic effect but also the bactericidal effect of this fragment can be ascribed to the targeting of bacterial cytosolic structures. Nevertheless, the relatively high content of charged arginine residues (seven) in the longer Bac5 peptides, together with a larger amount of hydrophobic residues, likely explains why high concentrations (50 μM) of both Bac5(1-25) and Bac5(1-31) led to permeabilization of more than 90% of the bacterial cells (data not shown).
Bac5(1-25) inhibits protein synthesis in living bacteria.
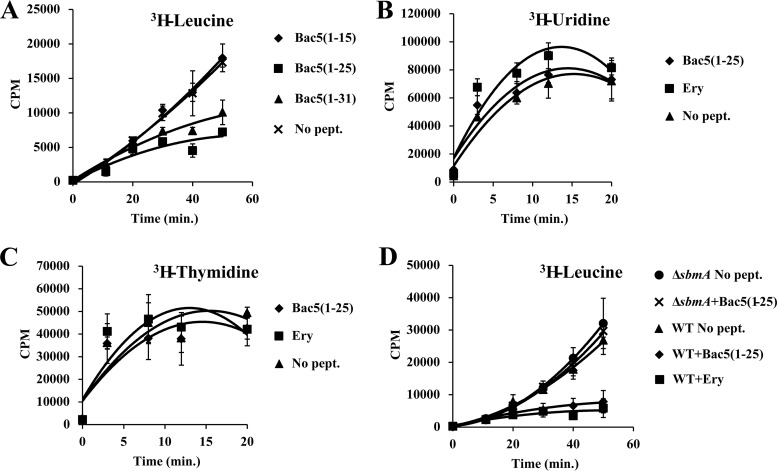
Because the high arginine and proline content of Bac5 is similar to that of other PrAMPs, such as Bac7, we hypothesized that Bac5 most likely targets the ribosome and inhibits protein synthesis in vivo. However, the inability to align the Bac5 sequence to the sequences of other PrAMPs (Fig. 1) raised the possibility that Bac5 may have a different mechanism of action. To assess the intracellular target of Bac5 in living bacteria, a culture of E. coli strain BW25113 was exposed to 1 μM each of the Bac5 fragments in the presence of radioactive leucine. The incorporation of the radioactive amino acid into newly synthesized proteins was then assessed to evaluate the effect of the Bac5 fragments on protein synthesis (Fig. 2A). As expected, in the absence of Bac5 fragments, there was a linear increase of radioactive leucine incorporation with increasing time. A similar trend was observed in the presence of the shortest fragment, Bac5(1-15), consistent with the lack of inhibitory activity of this peptide on the growth of E. coli cells (Table 1). In contrast, Bac5(1-31) and especially Bac5(1-25) inhibited the incorporation of leucine into newly synthesized proteins in vivo (Fig. 2A). This is also consistent with the good MICs of Bac5(1-31) and especially Bac5(1-25) observed in the antimicrobial activity assays (Table 1). The specificity of the Bac5-induced inhibition on protein synthesis was evaluated by also monitoring the effect of the Bac5(1-25) fragment on RNA (Fig. 2B) and DNA (Fig. 2C) synthesis, in addition to protein synthesis (Fig. 2A and D), in living bacteria under similar conditions. The experiments were also performed in the presence of the macrolide erythromycin, a well-known inhibitor of protein synthesis (28, 29). As expected, Bac5(1-25) and erythromycin had no effect on the incorporation of radioactive uridine into RNA or on incorporation of radioactive thymidine into DNA (Fig. 2B and C), indicating that Bac5(1-25) and erythromycin do not inhibit RNA or DNA synthesis. In contrast, the inhibitory activity of Bac5(1-25) on protein synthesis was comparable to that of erythromycin (Fig. 2D). Moreover, to validate the role of the SbmA transporter in the mode of uptake of Bac5 fragments, the incorporation of radioactive leucine was performed not only on the wild-type strain E. coli BW25113 but also on the mutant E. coli BW25113 ΔsbmA lacking SbmA (Fig. 2D). The inhibition of the protein synthesis by Bac5(1-25) was abolished in the absence of the SbmA transporter, indicating the critical role that SbmA has for internalization of Bac5 fragments. The specific action on protein synthesis and the lack of activity on DNA and RNA synthesis as well as the dependency on SbmA for inhibition of protein synthesis are all additional arguments in favor of an intracellular and nonlytic mode of action of the two Bac5 fragments.
FIG 2.
Incorporation of radioactive isotopes in the E. coli BW25113 and BW25113 ΔsbmA strains. (A) Incorporation of radioactive leucine in an E. coli BW25113 culture in the absence (no peptide [No pept.]) or the presence of 1 μM Bac5(1-15), Bac5(1-25), or Bac5(1-31). (B to D) Incorporation of radioactive uridine (B), thymidine (C), and leucine (D) in an E. coli BW25113 culture in the absence (no peptide) or the presence of 1 μM Bac5(1-25) or erythromycin (Ery). (D) The incorporation of radioactive leucine in an E. coli BW25113 ΔsbmA culture was also determined in the absence (no peptide) or the presence of 1 μM Bac5(1-25). Error bars represent the standard deviation from the mean from three independent experiments. WT, wild type.
Bac5 fragments specifically inhibit bacterial translation in vitro.
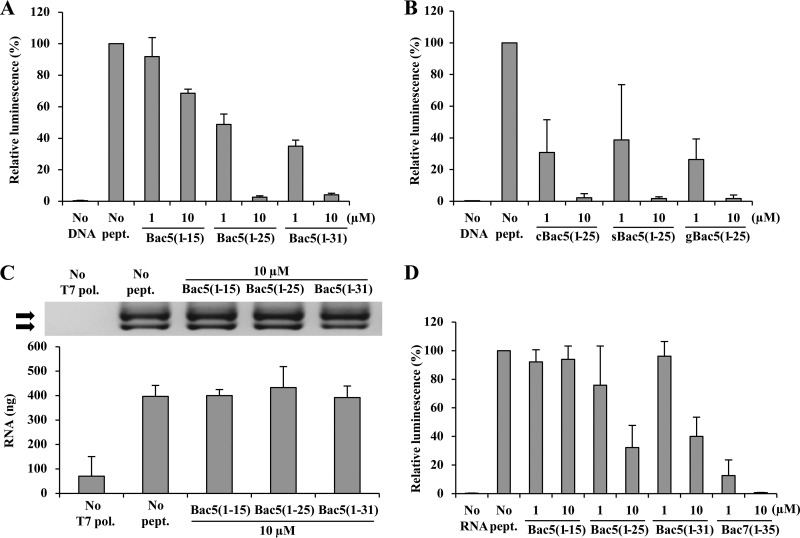
To directly assess the effect of the Bac5 fragments on transcription and translation, we employed in vitro transcription and translation assays. First, we monitored the effect of 1 μM and 10 μM each of the Bac5 fragments on inhibition of the synthesis of a luciferase reporter protein using an E. coli in vitro coupled transcription-translation system (Fig. 3A). As expected, no reporter protein was synthesized in the absence of the luciferase DNA template, whereas luciferase activity was observed in the absence of the Bac5 peptides, which was normalized to 100% (Fig. 3A). Bac5(1-15) was a poor inhibitor, displaying no significant inhibition at 1 μM and only a 30% reduction in luciferase production at 10 μM (Fig. 3A). This suggests that the lack of antimicrobial activity of Bac5(1-15) observed on living cells is due to the loss of activity of the peptide, rather than to the lack of uptake of the peptide or reduced peptide stability. In contrast, Bac5(1-25) and Bac5(1-31) were potent inhibitors, displaying an approximately 50% reduction of luciferase synthesis at 1 μM and nearly complete inhibition of luciferase synthesis at 10 μM (Fig. 3A). We note that similar results were also obtained for the sheep and goat Bac5(1-25) orthologues (Fig. 3B), consistent with the excellent MIC activities that these PrAMPs had in vivo (Table 1). To distinguish between an inhibitory effect on transcription and/or translation by the Bac5 peptides, we assessed the influence of the Bac5 fragments on a T7 RNA polymerase-based in vitro transcription assay. As seen in Fig. 3C, none of the Bac5 fragments displayed any inhibitory effect on transcription, even at 10 μM, indicating that Bac5(1-25) and Bac5(1-31) are potent inhibitors of translation and do not affect transcription. These findings are in agreement with the specific inhibitory effect of Bac5(1-25) and Bac5(1-31) on protein synthesis (Fig. 2).
FIG 3.
Effect of Bac5 fragments on in vitro transcription and translation assays. (A) Effect of Bac5 fragments in in vitro prokaryotic coupled transcription/translation assays (E. coli lysate). The presence of luciferase was checked and quantified using luminescence. As a negative control, a reaction was performed in the absence of DNA template (No DNA). (B) Effect of Bac5(1-25) homologues on E. coli in vitro transcription/translation reactions. The cow [cBac5(1-25)], sheep [sBac5(1-25)], and goat [gBac5(1-25)] orthologues were used. As a negative control, a reaction was performed in the absence of DNA template (No DNA). (C) In vitro mRNA synthesis in the presence of Bac5 fragments. The presence of two different RNA products (arrows) was checked and quantified by agarose gel electrophoresis (top) and spectrophotometric quantification (A260) (bottom). As a negative control, a reaction was performed in the absence of T7 RNA polymerase (No T7 pol.). (D) Effect of Bac5 fragments on in vitro prokaryotic translation assay (T. thermophilus lysate). The luciferase was quantified by luminescence. As a negative control, a reaction was performed in the absence of RNA template (No RNA). Bac7(1-35) was used for comparison. Error bars represent the standard deviation from the average from three independent experiments. Results are expressed as a percentage of the value for the positive controls, namely, reactions performed in the presence of water instead of peptides, for which the results were defined as 100%.
Species-specific inhibition by Bac5 fragments.
Previous studies have successfully utilized the T. thermophilus 70S ribosomes to determine the X-ray structures of PrAMPs in complex with the bacterial ribosome, including Bac7 (12, 30). However, all attempts to obtain a structure of Bac5(1-25) bound to the T. thermophilus 70S ribosome were unsuccessful (Axel Innis, personal communication), raising the question as to whether Bac5 can actually inhibit protein synthesis on T. thermophilus ribosomes. To assess this, we prepared a T. thermophilus lysate-based translation system, as described previously (14), and monitored the effect of the Bac5 fragments on the synthesis of firefly luciferase (Fig. 3D). Consistent with the results of the crystallization experiments, all Bac5 fragments were shown to be poor inhibitors of translation on T. thermophilus ribosomes. Partial inhibition of translation was observed for Bac5(1-25) and Bac5(1-31) at 10 μM, whereas at these concentrations, complete inhibition was observed in the E. coli lysate-based system (Fig. 3A). As a positive control, we could demonstrate that Bac7(1-35) is a potent inhibitor in the T. thermophilus translation system, displaying good inhibition at 1 μM and complete inhibition at 10 μM. These findings suggest that Bac5 has a lower affinity for T. thermophilus 70S ribosomes than for E. coli 70S ribosomes, which may explain the unsuccessful crystallization attempts. Therefore, it may be interesting in the future to investigate structurally the interaction of Bac5 fragments using E. coli ribosome complexes, as was successfully performed recently for the PrAMP apidaecin using cryo-electron microscopy (15).
Bac5 peptides inhibit the first translation elongation step.
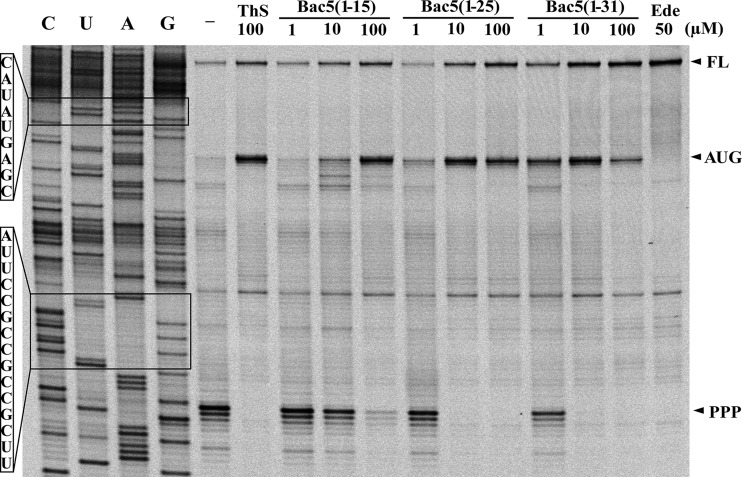
Given that Bac5 appears to interact with the ribosome differently than Bac7, we decided to investigate which step of the protein synthesis is targeted by the Bac5 fragments using toe-printing, an assay in which reverse transcription monitors the position of the bacterial ribosomes on an mRNA template (31). Translation reactions used an mRNA encoding the N terminus of the E. coli protein H-NS and were performed in the presence of Bac5(1-15), Bac5(1-25), and Bac5(1-31) (at 1 μM, 10 μM, and 100 μM) and the control antibiotics thiostrepton (ThS; 100 μM) and edeine (Ede; 50 μM) as well as in the absence of antibiotic (Fig. 4). In the absence of antibiotic, ribosomes initiate at the AUG start codon and elongate until becoming stalled at an in-frame proline triplet (PPP) because of the absence of EF-P in the translation system (32, 33). In the presence of thiostrepton, ribosomes initiate on the AUG start codon but cannot enter into the elongation phase, whereas edeine prevents initiation (22), leading to a signal for the full-length mRNA. In the presence of 1 μM and 10 μM Bac5(1-15), ribosomes were observed to stall at the PPP motif, indicating that translation was not efficiently inhibited, whereas ribosome accumulation at the AUG start codon occurred only at very high concentrations (100 μM). These findings indicate that Bac5(1-15) is a poor translation inhibitor, in agreement with the in vivo and in vitro data (Table 1; Fig. 2 and 3). In contrast, the presence of 10 μM either Bac5(1-25) or Bac5(1-31) was sufficient to completely inhibit the first translation elongation step, leading to an accumulation of ribosomes at the AUG start codon and loss of the stall signal at the PPP triplet (Fig. 4). Therefore, the mechanism of action of Bac5 appears to be similar to that reported previously for the PrAMPs Onc112 (13) and Bac7(1–35) (12, 30) and differs from that observed for apidaecin (15). However, whether Bac5 fragments influence A-site substrate binding or accommodation, peptide bond formation, or elongation factor G-dependent translocation remains to be elucidated.
FIG 4.
Toe-printing analysis to monitor the effect of Bac5 fragments on translation. Fluorescence scan of a polyacrylamide gel of toe-print samples obtained in the absence or presence of increasing concentrations of the Bac5(1-15), Bac5(1-25), and Bac5(1-31) peptides. Water instead of antibiotics or peptide was added to a reaction mixture as a control (lane −). Thiostrepton (ThS) was chosen as a control for the inhibition of elongation, and edeine (Ede) was chosen as a control for the inhibition of initiation. ThS stalls ribosomes at the AUG site, and Ede prevents the formation of the initiation complex. PPP, the position where ribosomes stall on the proline triplet motif; FL, the position of the reverse transcription band corresponding to the full-length mRNA; lanes C, U, A, and G, the DNA sequencing lanes of the template.
Bac5 fragments compete with erythromycin to bind to the ribosomal exit tunnel.
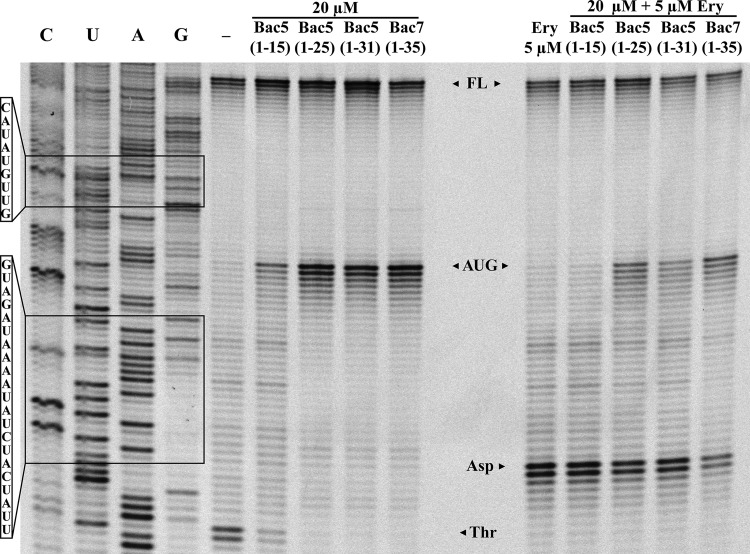
The structures reported for PrAMPs, such as for Onc112 (13) and Bac7(1-35) (12), as well as, more recently, for Api137 (15), show that all these PrAMPs bind within the ribosomal exit tunnel, overlapping the binding position of the macrolide antibiotic erythromycin. Moreover, Onc112 and Bac7 were shown to compete with erythromycin for ribosome binding (12). To assess whether the Bac5 binding site also overlaps the erythromycin binding site, we employed a competitive toe-printing assay (Fig. 5). In the absence of PrAMPs, ribosomes initiate and translate a portion of the ErmBL mRNA until they become stuck on a downstream threonine (Thr) codon, because the amino acid threonine (Thr) is lacking from the translation mix. As before, the inclusion of the Bac5 fragments led to the appearance of a toe-print signal corresponding to ribosomes stuck on the AUG start codon, as well as a reduction [Bac5(1-15)] or a complete loss [Bac5(1-25) and Bac5(1-31)] of ribosomes stalled at the downstream Thr codon. In the presence of erythromycin but in the absence of PrAMP, ribosomes translate the ErmBL mRNA but become stalled at the aspartate (Asp) codon because the ErmBL nascent chain and erythromycin interplay within the tunnel to induce translation arrest (34, 35). When the reaction mixtures were pretreated with the Bac5 peptides and then erythromycin was added to the reaction mixtures, stalling at the Asp codon was observed, indicating that erythromycin could displace some Bac5 molecules to allow translation until the Asp codon, where ErmBL-dependent arrest ensued (Fig. 5). Erythromycin was particularly efficient at displacing Bac5(1-15) but could only partially displace Bac5(1-25), Bac5(1-31), and Bac7(1-35). These findings support the suggestion that Bac5 binds within the ribosomal exit tunnel analogously to Bac7 and other PrAMPs.
FIG 5.
Toe-printing assay on Bac5 fragments and Bac7 alone and in the presence of erythromycin. A fluorescence scan of a polyacrylamide gel of toe-print samples obtained in the absence or presence of increasing concentrations of peptides. Water only instead of antibiotics or peptide was added to a reaction mixture as a control (lane −). The already characterized Bac7(1-35) peptide was used for comparison. All the peptides were used at the identical concentration of 20 μM. Samples in the right panel were pretreated with the peptides and then exposed to 5 μM erythromycin. The positions of the AUG, Asp, and Thr sites are indicated by arrowheads. Lanes C, U, A, and G, the DNA sequencing lanes of the template; FL, full-length RNA.
Influence of rRNA and ribosomal protein mutations on antimicrobial activity of Bac5 fragments.
Studies on E. coli have shown that specific mutations within the ribosomal exit tunnel, which can confer resistance to erythromycin, can also reduce the sensitivity of strains to the PrAMPs oncocin and apidaecin (15, 30) but not Bac7(1-35) (30) or Tur1A (14). We used these E. coli erythromycin-resistant strains to evaluate the MIC of Bac5(1-25) and Bac5(1-31), to assess whether there was any reduced sensitivity. In particular, we tested three strains bearing the 23S rRNA mutation A2503C, A2059G, or A2059C (15) as well as two strains with alterations in the ribosomal protein L4 (K63E) or L22 (Δ82MRK84) (36, 37) (Table 2). The MIC of erythromycin was also determined in parallel as a control, and, as expected, all the mutant strains displayed higher erythromycin MIC values than their respective wild-type parental strains (Table 2). In contrast, little to no change in the MIC against Bac5(1-25) and Bac5(1-31) was observed between the wild-type and most mutant strains, similar to the findings observed previously with Bac7(1-35) and Tur1A (14). A slight decrease (2- to 4-fold) in the sensitivity of E. coli to the Bac5 fragments was observed for strains bearing the A2503C mutation; however, structural analysis will be required to validate whether this nucleotide is directly involved in contacting the Bac5 peptide within the ribosomal tunnel.
TABLE 2.
MICs of Bac5(1-25), Bac5(1-31), and erythromycin for erythromycin-resistant E. coli strains
| Antimicrobial | MICa (μM) |
||||||
|---|---|---|---|---|---|---|---|
| AB301 (wt) | AB301 N281 (L22) | AB301 N282 (L4) | SQ110 ΔtolC (wt) | SQ110 ΔtolC A2059C | SQ110 ΔtolC A2059G | SQ110 ΔtolC A2503C | |
| Bac5(1-25) | 8 | 4–8 | 16 | 1 | 1–2 | 1 | 4 |
| Bac5(1-31) | 8 | 8 | 8 | 2–4 | 4–8 | 4 | 8 |
| Erythromycin | 128 | 1,024 | 1,024 | 2 | 4,096 | 2,048 | 2,048 |
Data are the mode from at least three independent experiments. wt, wild type.
Bac5 fragments are poor inhibitors of eukaryotic translation in vitro.
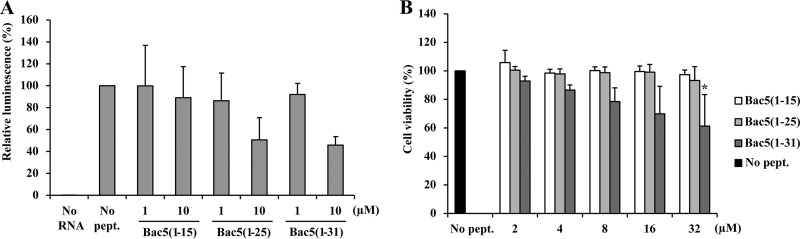
To understand if Bac5 fragments can also bind to eukaryotic ribosomes, we tested the effect of the Bac5 peptides in a cell-free translation system based on a rabbit reticulocyte lysate (Fig. 6A). Each Bac5 fragment (1 μM and 10 μM) was added to the translation reaction mixtures, and the effect on the production of luciferase was evaluated. As expected, Bac5(1-15) was completely inactive, as observed also in the bacterial translation assays (Fig. 3A). Similarly, Bac5(1-25) and Bac5(1-31) were also poor inhibitors of eukaryotic translation, reducing the translation activity by 50% at 10 μM (Fig. 6A), whereas in the E. coli system, the same peptides obtained a comparable effect already at 1 μM (Fig. 3A). The difference in the activity of the peptides on E. coli and eukaryotic translation was therefore assessed as being approximately 10-fold, suggesting a certain selectivity of the Bac5 fragments for the bacterial over the eukaryotic translation machinery, as previously reported for Bac7(1-35) (12).
FIG 6.
Effect of Bac5 fragments on eukaryotic translation and cell viability. (A) Effect of Bac5 fragments on an in vitro eukaryotic translation assay (rabbit reticulocyte lysate). The luciferase was quantified by luminescence. As a negative control, a reaction was performed in the absence of RNA template (No RNA). (B) NIH 3T3 murine fibroblast cells were exposed to Bac5 fragments for 24 h, and then cell viability was measured by the tetrazolium (MTT) assay by reading the optical density at 570 nm. As a negative control for toxicity, sterile water was added instead of the peptides (No pept.). The error bars represent the standard deviation from the mean from three independent experiments performed with internal duplicates (n ≥ 6). *, statistically significant compared with the value for the untreated control (P ≤ 0.05, the Student t test).
Cytotoxicity of Bac5 fragments.
Given the slight permeabilization activity of the Bac5 fragments at high peptide concentrations on E. coli cells, we also determined the cytotoxicity of the peptides on eukaryotic cells, specifically, a murine fibroblast cell line (NIH 3T3). The fibroblast cells were exposed to increasing concentrations of Bac5(1-15), Bac5(1-25), and Bac5(1-31) for 24 h, and cell viability was estimated using the tetrazolium assay, a colorimetric assay evaluating cell viability as a function of mitochondrial activity (38). Bac5(1-15) and Bac5(1-25) had no significant effect on cell viability at concentrations up to 32 μM (Fig. 6B). On the other hand, Bac5(1-31) displayed a concentration-dependent cytotoxicity, but this became statistically significant only at 32 μM, the highest concentration tested (Fig. 6B).
DISCUSSION
During the last few years, the molecular details of the mode of action of many PrAMPs has been unraveled. Insect-derived PrAMPs, such as metanikowin, oncocin, and pyrrhocoricin, but also the mammalian dolphin-derived Tur1A PrAMP and some fragments of the bovine-derived Bac7 PrAMP, have been shown to block the bacterial protein synthesis (4, 14, 39, 40). They act by binding to ribosomes (39, 40) and preventing them from entering the elongation phase (4, 12–14, 30). All these peptides share not only the molecular target but also a quite conserved region of their sequence, which is likely very important for their mechanism of action (4). Other well-characterized PrAMPs include the apidaecins, which have significant sequence similarity with ribosome-targeting PrAMPs (4) but a distinct mode of action (15). Rather than blocking the first translation elongation step, the apidaecins inhibit termination by trapping the release factors RF1 or RF2 on the ribosome, thereby depleting the pool of free RFs that bacteria need to efficiently carry on the protein synthesis (15). The Bac5 sequence cannot be satisfactorily aligned with any of the peptides reported above (4), yet the proline- and arginine-rich content of Bac5 suggested nevertheless that the mode of action is likely to be via inhibition of protein synthesis. Here we have confirmed this hypothesis, demonstrating both in vivo and in vitro that Bac5 targets protein synthesis. More specifically, we could demonstrate that Bac5 displays many characteristics similar to those of Bac7 since (i) Bac5 fragments enter the bacterial cell mainly via the SbmA transporter (Table 1), (ii) once they are in the cytosol, they most likely bind within the ribosomal exit tunnel with a binding site at least partially overlapping that of erythromycin (Fig. 5), and (iii) they inhibit protein synthesis by blocking the transition from the initiation to the elongation phase of translation (Fig. 4). Curiously, Bac5 also displays characteristics distinct from those of Bac7 since Bac5 fragments (i) could not be crystallized with the T. thermophilus 70S ribosomes, as other PrAMPs were (12, 13, 30, 41), (ii) did not inhibit the protein synthesis in a T. thermophilus lysate (Fig. 3D), and (iii) appeared to have a slightly reduced activity in strains bearing the 23S rRNA A2503C mutation (Table 2), unlike Bac7(1-35) and Tur1A (14). These findings are in agreement with the similar but distinct amino acid sequence and proline pattern that Bac5 has compared to other PrAMPs, suggesting that while Bac5 most likely has a binding site within the exit tunnel that overlaps that of erythromycin and other PrAMPs, the mode of interaction appears to be distinct. This is unexpected, given the high structural conservation between components that comprise the exit tunnel of T. thermophilus and E. coli ribosomes; however, species-specific interactions of ligands, such as antibiotics, have been observed before within the upper region of the tunnel (42). Structural studies using E. coli ribosomes should be able to elucidate the details of the interaction of Bac5 within its ribosomal binding site.
The inactivity of the Bac5(1-15) fragment and the good inhibitory activity of the Bac5(1-25) and Bac5(1-31) fragments suggest that residues 16 to 25 establish critical interactions with the ribosome. The best antimicrobial effect on E. coli cells was reported for Bac5(1-25) (MIC = 1 μM; Table 1), whereas the addition of six more residues slightly increased the MIC (4 μM; Table 1). This differs from the findings for other PrAMPs, such as Bac7 fragments, which showed an antimicrobial potency directly proportional to the length of the peptide (5). Despite having different MIC values, Bac5(1-25) and Bac5(1-31) exerted comparable inhibition toward protein synthesis in vitro (Fig. 3A), which may suggest that their different effects on bacterial growth could be due to different transport efficiencies across the inner membrane by the SbmA transporter, to different peptide stabilities, or to dissimilar peptide attachment to the bacterial membrane.
The increase in the MIC and MBC values of Bac5 fragments in the absence of the transporter SbmA and the lack of any lytic effect on E. coli membranes by Bac5(1-25) not only at its MIC but also at its MBC suggest that the interaction of this peptide with the ribosome is sufficient to explain the Bac5(1-25) bactericidal mechanism. The specific in vivo inhibition of protein synthesis by the Bac5(1-25) peptide, leaving DNA and RNA synthesis unaffected, as well as the dependency on the SbmA transporter for protein synthesis inhibition, growth inhibition, and killing, is in agreement with this proposition. This is not the first report of a nonlytic bactericidal PrAMP effect. Podda and colleagues (9) reported a lethal effect of Bac7(1-35) on Salmonella enterica serovar Typhimurium occurring above the MIC value without significant membrane permeabilization (i.e., approximately 3% permeabilization at 1 μM, 2-fold the MIC, and approximately 10% permeabilization at 10 μM, 20-fold the MIC). Recent studies have shown that while bactericidal antibiotics, such as the ketolide telithromycin, and bacteriostatic antibiotics, such as the macrolide erythromycin, have similar affinities for the ribosome, the rate of dissociation of ketolides is much lower than that of macrolides (23). This may suggest that the bactericidal effect of PrAMPs is also due to a low dissociation rate constant of the peptide fragments from the ribosome; however, this remains to be tested.
Because Bac5, like Bac7, is expressed as a prepropeptide (43), the active Bac5 is not likely present in the cytosol of eukaryotic cells. Nevertheless, we show that even if Bac5 can access the cytosol, the peptide appears to have a low affinity for the eukaryotic ribosome since the peptides inhibited protein synthesis in an E. coli lysate system approximately 10-fold more efficiently than in a rabbit reticulocyte system (Fig. 3A and 4A). Moreover, the bacteriostatic and even bactericidal concentrations of Bac5(1-25) and Bac5(1-31) are still not significantly toxic toward eukaryotic cells (Table 1 and Fig. 6).
Taken together, our results indicate that among the tested Bac5 fragments, Bac5(1-25) is the most promising molecule for future studies. It has the lowest MIC value toward E. coli cells (1 μM) and is not toxic for eukaryotic fibroblasts even at 32 μM. This peptide surely deserves further screening to evaluate its efficacy against other bacterial strains and clinically isolated pathogen strains. Additionally, further structure-activity relationship studies are desirable to improve the already promising Bac5(1-25) antimicrobial activity. A rational optimization of Bac5(1-25) would provide an excellent candidate for the development of a new antimicrobial agent.
MATERIALS AND METHODS
Peptide synthesis and quantification.
The peptides Bac5(1-15), Bac5(1-25), and Bac5(1-31) were synthesized using solid-phase 9-fluorenylmethoxy carbonyl chemistry and purified by high-performance liquid chromatography and their quality was assessed by mass spectrometry, as previously described (27). The ovine and caprine orthologues of Bac5(1-25), as well as Bac7(1-35), were purchased from NovoPro (China). All the peptides were lyophilized three times from 10 mM HCl solutions to remove the trifluoroacetate group present as the counterion of the arginine residues, and then the peptides were resuspended in sterile Milli-Q water. The concentration of the peptides was estimated spectrophotometrically. The absorbance of the peptides at 214 nm and at 280 nm was measured, and the concentration was calculated using the molar extinction coefficients of tyrosine (ε = 1,500 M−1 cm−1) and of the whole peptide. The latter value was calculated by the in-house-developed program ConCalc using the values proposed previously (44), i.e., ε = 93,800 for Bac5(1-31), ε = 74,500 for Bac5(1-25), and ε = 40,500 for Bac5(1-15).
Bacterial strains.
All the bacterial strains used in this study were grown at 37°C in Müller-Hinton broth (MHB) with shaking (140 rpm). The E. coli BW25113, N281, N282, and AB301 strains were grown without antibiotics, whereas E. coli BW25113 ΔsbmA (JW0368-1) (45) required a final concentration of 50 μg/ml kanamycin. The E. coli strains SQ110 ΔtolC, SQ110 ΔtolC A2059C, SQ110 ΔtolC A2059G, and SQ110 ΔtolC A2503C were grown in the presence of a final concentration of 25 μg/ml kanamycin and 50 μg/ml spectinomycin. The E. coli strains BW25113 and BW25113 ΔsbmA::Kmr are part of the KEIO Collection (45). E. coli strains AB301 with its mutants N281 (mutation on L22) and N282 (mutation on L4) and SQ110 ΔtolC::Kmr and its mutants SQ110 ΔtolC::Kmr Spr A2059C, SQ110 ΔtolC::Kmr Spr A2059G, and SQ110 ΔtolC::Kmr Spr A2503C were generously provided by Alexander Mankin and Nora Vasquez-Laslop, University of Illinois, Chicago, IL, USA.
MIC and MBC determination.
A bacterial overnight culture was diluted 1:50 in new MHB (with antibiotics, as required) and incubated at 37°C with agitation until an optical density at 600 nm (OD600) of ≈0.3 was reached. The culture was then diluted to 5 × 105 CFU/ml with fresh MHB. The peptide was serially diluted in 50 μl of MHB in a microtiter plate (round bottom), before adding 50 μl of the bacterial suspensions (5 × 105 CFU/ml) to each well. The plate was sealed using Parafilm and incubated at 37°C overnight. The MIC was calculated as the lowest peptide concentration that inhibited the visible growth of bacteria. Then, 25 μl of medium from each clear well was collected after pipetting and was spread on Müller-Hinton agar plates. The colonies were counted after incubation overnight at 37°C. The minimum bactericidal concentration (MBC) was calculated as the lowest peptide concentration killing 99.9% of the original bacterial inoculum.
Membrane integrity evaluation.
To evaluate the bacterial membrane integrity, propidium iodide (PI) uptake was evaluated by flow cytometry using a Cytomics FC 500 instrument (Beckman-Coulter, Inc., Fullerton, CA), as described previously (27, 46). A bacterial mid-log-phase culture diluted to 1 × 106 CFU/ml in MHB was incubated at 37°C for 15 min, 30 min, and 60 min in the presence of 1 μM, 10 μM, or 50 μM peptide and of a final concentration of 10 μg/ml PI. A total of 104 cells were used for each measurement. PI cannot cross an intact bacterial envelope; therefore, the percentage of PI-positive cells indicates the extent of membrane damage.
In vitro transcription/translation using an E. coli lysate.
Reaction mixtures were set up using an S30 T7 high-yield protein expression system (Promega) or an RTS 100 E. coli HY system (Biotech Rabbit), with 1 μl or 0.1 μl, respectively, of RNase inhibitor (RNasin; 20 to 40 U/μl; Promega) being added to each reaction mixture. In the positive controls, nuclease-free water was added instead of the peptides. In the negative control, nuclease-free water was added instead of the peptides and the DNA template. For the S30 T7 kit, 2 μl of peptide solution was added to each reaction mixture to get a final concentration of 1 μM or 10 μM in a final volume of 40 μl. Samples were incubated at 37°C for 1 h and subjected to 12 cycles of 1 min of vigorous mixing (1,200 rpm) and 4 min of resting. Subsequently, the presence of the reporter protein (Renilla reniformis luciferase) was assessed, and the reporter protein was quantified using a commercial kit (Renilla luciferase assay system; Promega), as indicated in the S30 T7 high-yield protein expression system kit, using black flat-bottom 96-well plates and a luminometer (Chameleon plate; Hidex) with MikroWin 2000 software. For experiments performed using the RTS 100 kit, 1 μl of peptide solution was added to each reaction mixture to reach a final concentration of 1 μM or 10 μM in a final volume of 6 μl. Samples were incubated at 30°C for 1 h with shaking (750 rpm). To stop the reaction, 2 µl of reaction mixture was added to 8 µl of kanamycin (50 mg/ml) and mixed, before being diluted with 40 μl of luciferase assay reagent (Promega) and placed into a 96-well white flat-bottom microtiter plate (Greiner). The presence of the reporter protein (Photinus pyralis luciferase) was assessed, and the reporter protein was quantified using a Tecan Infinite M1000 plate reader. In all the luminescence analyses, the relative values were calculated as a percentage of the value for the positive control.
In vitro translation using Thermus thermophilus lysate.
The lysate of T. thermophilus was prepared in-house using a protocol slightly modified from a previously reported one (47). Bacterial cells were grown in 1× YT (yeast extract and tryptone) medium at 70°C to an OD600 of ≈0.6, harvested by centrifugation at 5,000 × g at 4°C for 15 min, and then washed 3 times in buffer A (10 mM Tris-acetate buffer [pH 8.2], 14 mM magnesium acetate [MgOAc], 60 mM potassium acetate[KOAc], 1 mM dithiothreitol [DTT], 6 mM 2-mercaptoethanol). The pellet was flash frozen in liquid nitrogen. Cells were then thawed, resuspended in buffer B (10 mM Tris-acetate buffer [pH 8.2], 14 mM MgOAc, 60 mM KOAc, 1 mM DTT) and disrupted by three passages through a French press at >15,000 lb/in2. The lysate was cleared by centrifugation at 12,000 × g for 10 min at 4°C. The supernatant was aliquoted, flash frozen in liquid nitrogen, and stored at −80°C. The in vitro translation reactions were performed with a final volume of 25 μl (containing 240 mM HEPES [pH 8], 0.6 mM polyethylene glycol 8000, 60 mM glucose, 4.4 μg bulk tRNA from E. coli MRE600 [Sigma], 1.2 mM ATP, 1.2 mM GTP, 0.85 μg folinic acid, 1 mM DTT, 90 mM potassium glutamate. 80 mM ammonium acetate, 20 mM K2HPO4, 1.8 mM each amino acid, 12.6 mM magnesium acetate, and 200 ng of firefly luciferase mRNA synthesized in vitro as reported previously [48]) using 6.75 μl of bacterial lysate and 1 μl of peptide solution where required. The reaction mixtures were incubated at 30°C for 1 h under agitation (550 rpm). The reaction was stopped by the addition of 2 μl of 50 mg/ml kanamycin to 8 μl of each reaction mixture. All samples were diluted with 40 μl of luciferase assay substrate (Promega) and placed into a white 96-well chimney flat-bottom microtiter plate (Greiner). The luminescence was then measured using a Tecan Infinite M1000 plate reader. Relative values were determined by defining the luminescence value of the sample without inhibitor as 100%.
In vitro transcription.
Reactions were carried out using a commercial kit (Riboprobe System T7; Promega) in the absence or presence of 4 μl of peptide solution (final concentration, 1 μM or 10 μM). For the negative control, no RNA polymerase was added to the reaction mixture. The in vitro-transcribed RNA was subsequently purified using the TRIzol reagent (Life technologies), as suggested by the supplier for RNA isolation from suspension cell samples. The purified synthetic mRNA was resuspended in 20 μl of RNase-free water and quantified spectrophotometrically using a NanoDrop 2000 spectrophotometer, and then 2 μl of each sample was separated by electrophoresis on a 1% agarose gel containing 0.05% NaClO (49).
In vitro translation using rabbit reticulocyte lysate.
Reaction mixtures with or without Bac5 fragments were set up with a commercial rabbit reticulocyte lysate system kit (Promega), according to the manufacturer's description, but the total volume was scaled down to 6 μl and 0.1 μl of an RNase inhibitor (RNasin; 20 to 40 U/1 μl; Promega) was added to each reaction mixture. The reaction mixtures were incubated for 1 h at 30°C with shaking (400 rpm). The reaction was stopped by the addition of 7 μl kanamycin (50 mg/ml) to 3 μl of each reaction mixture. All samples were diluted with 40 μl of luciferase assay substrate (Promega), and the samples were placed into a white 96-well chimney flat-bottom microtiter plate (Greiner). Luminescence was then measured using a Tecan Infinite M1000 plate reader. Relative values were determined by defining the luminescence value of the sample without inhibitor as 100%.
Toe-printing assay.
For toe-printing assays, an in vitro transcription/translation kit (PURExpress in vitro protein synthesis kit; NEB) was used. Reaction mixtures were set up by mixing 2 μl of solution A, 1.5 μl of solution B, 1 μl (0.5 pmol) of DNA template, 0.1 μl of RNasin (20 to 40 U/μl; Promega), and 1 μl of peptide (to get a final concentration of 1 μM, 10 μM, or 100 μM) or 1 μl of antibiotic (to obtain a final concentration of 100 μM for thiostrepton and 50 μM for edeine) in PCR tubes. The DNA template codes for the first 36 N-terminal residues of the E. coli DNA-binding protein H-NS (UniProt accession number P0ACF8) were modified to replace the residues at positions 20 to 22 of the original protein with three proline residues (5′-ATTAATTACGACTCACTATAGGGATATAAGGAGGAAAACATATGAGCGAAGCACTTAAAATTCTGAACAACCTGCGTACTCTTCGTGCGCAGGCAATTCCGCCGCCGCTTGAAACGCTGGAAGAAATGCTGGAAAAATTAGAAGTTGTTGTTTAAGTGATAGAATTCTATCGTTAATAAGCAAAATTCATTATAACC-3′; the start, proline, and stop codons are highlighted in bold). The control had nuclease-free water instead of inhibitors. Samples were incubated for transcription/translation for 15 min at 37°C with agitation (550 rpm), cooled down on ice for 5 min, and equilibrated at room temperature for 2 min. Then, 1 μl (2 pmol) of Alexa Fluor 647 5′-labeled NV-1 toe-print primer (5′-GGTTATAATGAATTTTGCTTATTAAC-3′) was added to each reaction mixture, and the samples were incubated for 5 min at 37°C. For reverse transcription, 0.5 μl of avian myeloblastosis virus reverse transcriptase (NEB), 0.1 μl deoxynucleoside triphosphate mix (10 mM), and 0.4 μl Pure system buffer were added to each reaction mixture and the samples were incubated for 20 min at 37°C. To stop the reaction and degrade the RNA, 1 μl of 5 M NaOH was added to each reaction mixture and the samples were incubated for 15 min at 37°C. Samples were neutralized by adding 0.7 μl 25% (vol/vol) HCl and 20 μl of toe-print resuspension buffer. Samples were then purified by using a QIAquick nucleotide removal kit (Qiagen) and adding 200 μl of PN1 buffer following the supplier's instructions. DNA was eluted using 80 μl of RNase-free water, dried in a vacuum centrifuge, and resuspended in 4 μl of formamide loading dye (10 mM EDTA, 1 mg/ml bromophenol blue, 1 mg/ml xylene cyanol in formamide). Samples were heated for 5 min at 95°C and then separated by electrophoresis on a 6% polyacrylamide gel (19:1) containing 7 M urea at a setting of 2,000 V and blocking the run before the dye exited the gel. Results were acquired using a GE Typhoon FLA9500 imaging system.
For the competition toe-print assay with erythromycin, the PURExpress in vitro protein synthesis kit (without the amino acid mix and tRNA; NEB) was used. The reaction mixtures were set up by mixing 1 μl of solution A (without the amino acid mix and tRNA), 0.5 μl of 5 mM amino acid mix (without Thr), 0.5 μl of a tRNA mix, and 1.5 μl of solution B, 0.5 μl of peptide (to get a final concentration of 1 μM, 10 μM, or 100 μM) or 0.5 μl of antibiotic (to get a final concentration of 100 μM for thiostrepton and 50 μM for edeine). Samples were incubated for 5 min at 37°C at 550 rpm, and then 1 μl (0.9 pmol) of a DNA template encoding part of the slightly modified ermBL sequence (5′-ATTAATACGACTCACTATAGGGATATAAGGAGGAAAACATATGTTGGTATTCCAAATGCGTAATGTAGATAAAATATCTACTATTTTGAAATAAGTGATAGAATTCTATCGTTAATAAGCAAAATTCATTATAACC-3′; the start, aspartate, threonine, and stop codons are highlighted in bold) was added to each sample. After 5 min, some reaction mixtures also received 0.5 μl of erythromycin to obtain a final drug concentration of 5 μM. To the other samples, 0.5 μl of RNase-free water was added to equalize the volumes. Then, the toe-print assay was carried out as described above.
Sequencing of the toe-print DNA template.
The sequencing of the toe-print DNA template was carried out by mixing 1 μl (0.5 to 0.8 pmol) of DNA template, 5 μl of sequencing buffer, 9 μl of nuclease-free water, 1 μl (10 pmol) of Alexa Fluor 647-labeled NV-1 toe-print primer (5′-GGTTATAATGAATTTTGCTTATTAAC-3′), and 1 μl of Hemo Klen Taq polymerase. Then, 4 μl of this mix was added to 2 μl of ddATP, ddTTP, ddGTP, or ddCTP and incubated in a thermocycler (1 cycle of 2 s at 95°C; 30 cycles of 30 s at 95°C, 30 s at 42°C, and 1 min at 70°C; and 1 cycle of 1 min at 70°C with storage at 4°C). Samples were then heated and loaded on a gel as indicated above for the toe-print assay.
In vivo assays for radioactive isotope incorporation into bacterial macromolecules.
The incorporation of radioactive isotopes into bacterial macromolecules was performed as previously described (39). Briefly, mid-log-phase E. coli BW25113 cells grown in MHB medium were centrifuged (2,000 × g) and resuspended in M9 salts containing 5 mM glucose and 1% (vol/vol) MHB to a final concentration of 1.5 × 107 CFU/ml, and 650 μl of this culture was incubated at 37°C in 2-ml tubes in a dry-block bath for 15 min. Subsequently, the radioactive compounds and Bac5 peptides were added to the bacterial suspension. The assay was performed in the presence of 7.5 μCi [3H]leucine (1 mCi/ml; PerkinElmer), 4 μCi [3H]uridine (1 mCi/ml; PerkinElmer), or 4 μCi [3H]thymidine (1 mCi/ml; PerkinElmer) for protein, RNA, and DNA synthesis, respectively. All the Bac5 fragments and the erythromycin were used at a final concentration of 1 μM. After incubation at 37°C for the times indicated in Fig. 2, 100 μl of the bacterial culture was mixed with 5 ml of ice-cold 10% trichloroacetic acid followed immediately by the addition of bovine serum albumin to a final concentration of 200 μg/ml. The bacteria were lysed, and bacterial macromolecules were precipitated by incubation on ice and then filtered on 0.22-μm-pore-size filters (GSTF; Millipore). The filters were air dried and then incubated at room temperature in the dark in a scintillation tube with 4 ml of scintillation fluid (Ecolite[+] liquid scintillation cocktail; MP Biomedicals). The radioactivity was quantified using an LS-600-SC liquid scintillation analyzer (Beckman), with the radioactivity in each sample being measured for 10 min. Some reaction mixtures were set up using sterile Milli-Q water instead of peptides as controls. Results are the averages from three independent experiments, and error bars in the figures represent the standard deviation from the mean.
Cytotoxicity and cell viability assay.
The cytotoxicity of the Bac5 fragments for NIH 3T3 murine fibroblasts was assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (38). Cells were grown to subconfluence in 50 μl of Dulbecco's modified Eagle's medium (Sigma-Aldrich) containing 10% fetal bovine serum (EuroClone), 2.4 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin, using 96-well flat-bottom microtiter plates at 37°C in 5% CO2. Serial 2-fold dilutions of the Bac5 fragments were prepared in cell growth medium, and 50 μl of diluted peptides was added to the cells. After 20 h of incubation, 20 μl of MTT (Sigma-Aldrich) in phosphate-buffered saline was added to each well to a final concentration of 1 mg/ml. After 4 h of incubation at 37°C in 5% CO2, 100 μl of 10% Igepal (Sigma-Aldrich) in 10 mM HCl was added to each well and the plate was incubated overnight at 37°C. On the following day, the absorbance at 570 nm was measured using a plate reader (Tecan Infinite M1000) to evaluate cell viability.
ACKNOWLEDGMENTS
We acknowledge Axel Innis and his research group (University of Bordeaux) for the attempts to obtain a crystal structure for Bac5 on the T. thermophilus ribosome and Monica Benincasa (University of Trieste) for her assistance during flow cytometry assays.
Mario Mardirossian acknowledges the Talents3 fellowship program from the Operative Regional Programme of European Social Fund 2014-2020 of the Autonomous Region of Friuli Venezia Giulia, Italy. Quentin Barrière is the recipient of a Ph.D. fellowship from the Université Paris-Sud. The research in the Wilson group was supported by grants (FOR1805 and WI3285/6-1) from the Deutsche Forschungsgemeinschaft (DFG). The research in the Scocchi group was supported by the Finanziamento di Ateneo per la Ricerca Scientifica 2016 program (FRA 2016) of the University of Trieste. Peter Mergaert acknowledges the Agence National de la Recherche (ANR; grant no. ANR-17-CE20-0011-02).
REFERENCES
- 1.Kang SJ, Park SJ, Mishig-Ochir T, Lee BJ. 2014. Antimicrobial peptides: therapeutic potentials. Expert Rev Anti Infect Ther 12:1477–1486. doi: 10.1586/14787210.2014.976613. [DOI] [PubMed] [Google Scholar]
- 2.Li W, Tailhades J, O'Brien-Simpson NM, Separovic F, Otvos L Jr, Hossain MA, Wade JD. 2014. Proline-rich antimicrobial peptides: potential therapeutics against antibiotic-resistant bacteria. Amino Acids 46:2287–2294. doi: 10.1007/s00726-014-1820-1. [DOI] [PubMed] [Google Scholar]
- 3.Scocchi M, Tossi A, Gennaro R. 2011. Proline-rich antimicrobial peptides: converging to a non-lytic mechanism of action. Cell Mol Life Sci 68:2317–2330. doi: 10.1007/s00018-011-0721-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graf M, Mardirossian M, Nguyen F, Seefeldt AC, Guichard G, Scocchi M, Innis CA, Wilson DN. 2017. Proline-rich antimicrobial peptides targeting protein synthesis. Nat Prod Rep 34:702–711. doi: 10.1039/C7NP00020K. [DOI] [PubMed] [Google Scholar]
- 5.Benincasa M, Scocchi M, Podda E, Skerlavaj B, Dolzani L, Gennaro R. 2004. Antimicrobial activity of Bac7 fragments against drug-resistant clinical isolates. Peptides 25:2055–2061. doi: 10.1016/j.peptides.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Casteels P, Tempst P. 1994. Apidaecin-type peptide antibiotics function through a nonporeforming mechanism involving stereospecificity. Biochem Biophys Res Commun 199:339–345. doi: 10.1006/bbrc.1994.1234. [DOI] [PubMed] [Google Scholar]
- 7.Mattiuzzo M, Bandiera A, Gennaro R, Benincasa M, Pacor S, Antcheva N, Scocchi M. 2007. Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Mol Microbiol 66:151–163. doi: 10.1111/j.1365-2958.2007.05903.x. [DOI] [PubMed] [Google Scholar]
- 8.Scocchi M, Mardirossian M, Runti G, Benincasa M. 2016. Non-membrane permeabilizing modes of action of antimicrobial peptides on bacteria. Curr Top Med Chem 16:76–88. doi: 10.2174/1568026615666150703121009. [DOI] [PubMed] [Google Scholar]
- 9.Podda E, Benincasa M, Pacor S, Micali F, Mattiuzzo M, Gennaro R, Scocchi M. 2006. Dual mode of action of Bac7, a proline-rich antibacterial peptide. Biochim Biophys Acta 1760:1732–1740. doi: 10.1016/j.bbagen.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Runti G, Benincasa M, Giuffrida G, Devescovi G, Venturi V, Gennaro R, Scocchi M. 2017. The mechanism of killing by the proline-rich peptide Bac7(1-35) against clinical strains of Pseudomonas aeruginosa differs from that against other Gram-negative bacteria. Antimicrob Agents Chemother 61:e01660-16. doi: 10.1128/AAC.01660-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krizsan A, Knappe D, Hoffmann R. 2015. Influence of the yjiL-mdtM gene cluster on the antibacterial activity of proline-rich antimicrobial peptides overcoming Escherichia coli resistance induced by the missing SbmA transporter system. Antimicrob Agents Chemother 59:5992–5998. doi: 10.1128/AAC.01307-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seefeldt AC, Graf M, Perebaskine N, Nguyen F, Arenz S, Mardirossian M, Scocchi M, Wilson DN, Innis CA. 2016. Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res 44:2429–2438. doi: 10.1093/nar/gkv1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seefeldt AC, Nguyen F, Antunes S, Perebaskine N, Graf M, Arenz S, Inampudi KK, Douat C, Guichard G, Wilson DN, Innis CA. 2015. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat Struct Mol Biol 22:470–475. doi: 10.1038/nsmb.3034. [DOI] [PubMed] [Google Scholar]
- 14.Mardirossian M, Perebaskine N, Benincasa M, Gambato S, Hofmann S, Huter P, Muller C, Hilpert K, Innis CA, Tossi A, Wilson DN. 2018. The dolphin proline-rich antimicrobial peptide Tur1A inhibits protein synthesis by targeting the bacterial ribosome. Cell Chem Biol 25:530–539.e7. doi: 10.1016/j.chembiol.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florin T, Maracci C, Graf M, Karki P, Klepacki D, Berninghausen O, Beckmann R, Vazquez-Laslop N, Wilson DN, Rodnina MV, Mankin AS. 2017. An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome. Nat Struct Mol Biol 24:752–757. doi: 10.1038/nsmb.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gennaro R, Skerlavaj B, Romeo D. 1989. Purification, composition, and activity of two bactenecins, antibacterial peptides of bovine neutrophils. Infect Immun 57:3142–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shamova O, Brogden KA, Zhao C, Nguyen T, Kokryakov VN, Lehrer RI. 1999. Purification and properties of proline-rich antimicrobial peptides from sheep and goat leukocytes. Infect Immun 67:4106–4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huttner KM, Lambeth MR, Burkin HR, Burkin DJ, Broad TE. 1998. Localization and genomic organization of sheep antimicrobial peptide genes. Gene 206:85–91. doi: 10.1016/S0378-1119(97)00569-6. [DOI] [PubMed] [Google Scholar]
- 19.Treffers C, Chen L, Anderson RC, Yu PL. 2005. Isolation and characterisation of antimicrobial peptides from deer neutrophils. Int J Antimicrob Agents 26:165–169. doi: 10.1016/j.ijantimicag.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Skerlavaj B, Romeo D, Gennaro R. 1990. Rapid membrane permeabilization and inhibition of vital functions of gram-negative bacteria by bactenecins. Infect Immun 58:3724–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson RC, Hancock RE, Yu PL. 2004. Antimicrobial activity and bacterial-membrane interaction of ovine-derived cathelicidins. Antimicrob Agents Chemother 48:673–676. doi: 10.1128/AAC.48.2.673-676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson DN. 2009. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol 44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 23.Svetlov MS, Vazquez-Laslop N, Mankin AS. 2017. Kinetics of drug-ribosome interactions defines the cidality of macrolide antibiotics. Proc Natl Acad Sci U S A 114:13673–13678. doi: 10.1073/pnas.1717168115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boman HG, Agerberth B, Boman A. 1993. Mechanisms of action on Escherichia coli of cecropin P1 and PR-39, two antibacterial peptides from pig intestine. Infect Immun 61:2978–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cociancich S, Dupont A, Hegy G, Lanot R, Holder F, Hetru C, Hoffmann JA, Bulet P. 1994. Novel inducible antibacterial peptides from a hemipteran insect, the sap-sucking bug Pyrrhocoris apterus. Biochem J 300(Pt 2):567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castle M, Nazarian A, Yi SS, Tempst P. 1999. Lethal effects of apidaecin on Escherichia coli involve sequential molecular interactions with diverse targets. J Biol Chem 274:32555–32564. doi: 10.1074/jbc.274.46.32555. [DOI] [PubMed] [Google Scholar]
- 27.Guida F, Benincasa M, Zahariev S, Scocchi M, Berti F, Gennaro R, Tossi A. 2015. Effect of size and N-terminal residue characteristics on bacterial cell penetration and antibacterial activity of the proline-rich peptide Bac7. J Med Chem 58:1195–1204. doi: 10.1021/jm501367p. [DOI] [PubMed] [Google Scholar]
- 28.Kannan K, Kanabar P, Schryer D, Florin T, Oh E, Bahroos N, Tenson T, Weissman JS, Mankin AS. 2014. The general mode of translation inhibition by macrolide antibiotics. Proc Natl Acad Sci U S A 111:15958–15963. doi: 10.1073/pnas.1417334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson DN. 2014. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 30.Gagnon MG, Roy RN, Lomakin IB, Florin T, Mankin AS, Steitz TA. 2016. Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition. Nucleic Acids Res 44:2439–2450. doi: 10.1093/nar/gkw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartz D, McPheeters DS, Traut R, Gold L. 1988. Extension inhibition analysis of translation initiation complexes. Methods Enzymol 164:419–425. doi: 10.1016/S0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 32.Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. 2013. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 33.Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. 2013. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 34.Arenz S, Ramu H, Gupta P, Berninghausen O, Beckmann R, Vazquez-Laslop N, Mankin AS, Wilson DN. 2014. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat Commun 5:3501. doi: 10.1038/ncomms4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arenz S, Bock LV, Graf M, Innis CA, Beckmann R, Grubmuller H, Vaiana AC, Wilson DN. 2016. A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest. Nat Commun 7:12026. doi: 10.1038/ncomms12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouck N, Adelberg EA. 1970. Mechanism of action of nalidixic acid on conjugating bacteria. J Bacteriol 102:688–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wittmann HG, Stoffler G, Apirion D, Rosen L, Tanaka K, Tamaki M, Takata R, Dekio S, Otaka E. 1973. Biochemical and genetic studies on two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal proteins. Mol Gen Genet 127:175–189. doi: 10.1007/BF00333665. [DOI] [PubMed] [Google Scholar]
- 38.Tada H, Shiho O, Kuroshima K, Koyama M, Tsukamoto K. 1986. An improved colorimetric assay for interleukin 2. J Immunol Methods 93:157–165. doi: 10.1016/0022-1759(86)90183-3. [DOI] [PubMed] [Google Scholar]
- 39.Mardirossian M, Grzela R, Giglione C, Meinnel T, Gennaro R, Mergaert P, Scocchi M. 2014. The host antimicrobial peptide Bac71-35 binds to bacterial ribosomal proteins and inhibits protein synthesis. Chem Biol 21:1639–1647. doi: 10.1016/j.chembiol.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 40.Krizsan A, Volke D, Weinert S, Strater N, Knappe D, Hoffmann R. 2014. Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70S ribosome. Angew Chem Int Ed Engl 53:12236–12239. doi: 10.1002/anie.201407145. [DOI] [PubMed] [Google Scholar]
- 41.Roy RN, Lomakin IB, Gagnon MG, Steitz TA. 2015. The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nat Struct Mol Biol 22:466–469. doi: 10.1038/nsmb.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson DN. 2011. On the specificity of antibiotics targeting the large ribosomal subunit. Ann N Y Acad Sci 1241:1–16. doi: 10.1111/j.1749-6632.2011.06192.x. [DOI] [PubMed] [Google Scholar]
- 43.Zanetti M, Litteri L, Gennaro R, Horstmann H, Romeo D. 1990. Bactenecins, defense polypeptides of bovine neutrophils, are generated from precursor molecules stored in the large granules. J Cell Biol 111:1363–1371. doi: 10.1083/jcb.111.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuipers BJH, Gruppen H. 2007. Prediction of molar extinction coefficients of proteins and peptides using UV absorption of the constituent amino acids at 214 nm to enable quantitative reverse phase high-performance liquid chromatography-mass spectrometry analysis. J Agric Food Chem 55:5445–5451. doi: 10.1021/jf070337l. [DOI] [PubMed] [Google Scholar]
- 45.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio Collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benincasa M, Pacor S, Gennaro R, Scocchi M. 2009. Rapid and reliable detection of antimicrobial peptide penetration into gram-negative bacteria based on fluorescence quenching. Antimicrob Agents Chemother 53:3501–3504. doi: 10.1128/AAC.01620-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim TW, Keum JW, Oh IS, Choi CY, Park CG, Kim DM. 2006. Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system. J Biotechnol 126:554–561. doi: 10.1016/j.jbiotec.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 48.Leoni G, De Poli A, Mardirossian M, Gambato S, Florian F, Venier P, Wilson DN, Tossi A, Pallavicini A, Gerdol M. 2017. Myticalins: a novel multigenic family of linear, cationic antimicrobial peptides from marine mussels (Mytilus spp.). Mar Drugs 15:E261. doi: 10.3390/md15080261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aranda PS, LaJoie DM, Jorcyk CL. 2012. Bleach gel: a simple agarose gel for analyzing RNA quality. Electrophoresis 33:366–369. doi: 10.1002/elps.201100335. [DOI] [PMC free article] [PubMed] [Google Scholar]