A multidrug-resistant Klebsiella pneumoniae 1210 isolate with reduced carbapenem susceptibility revealed the presence of a novel plasmid-encoded blaOXA-48-like gene, named blaOXA-519. The 60.7-kb plasmid (pOXA-519) was similar to the IncL-OXA-48 prototypical plasmid except for a ca.
KEYWORDS: carbapenemase, mutant, steady-state kinetics, OXA-48 like, detection
ABSTRACT
A multidrug-resistant Klebsiella pneumoniae 1210 isolate with reduced carbapenem susceptibility revealed the presence of a novel plasmid-encoded blaOXA-48-like gene, named blaOXA-519. The 60.7-kb plasmid (pOXA-519) was similar to the IncL-OXA-48 prototypical plasmid except for a ca. 2-kb deletion due to an IS1R insertion. OXA-519 differed from OXA-48 by a Val120Leu substitution, which resulted in an overall reduced β-lactam-hydrolysis profile, except those for ertapenem and meropenem, which were increased. Thus, detection of OXA-519 producers using biochemical tests that monitor imipenem hydrolysis will be difficult.
TEXT
Class D β-lactamases (DBLs), or OXA-type β-lactamases (OXAs), form a very diverse family of enzymes (1–3), the diversity of which is reflected at both genetic and biochemical levels. OXA-48, the main carbapenem-hydrolyzing class D β-lactamase (CHDL) encountered in Enterobacteriaceae in many countries around the Mediterranean area, was initially identified from a carbapenem-resistant Klebsiella pneumoniae isolate from Turkey in 2001 (4, 5). Although OXA-48 hydrolyzes penicillins at a high level and carbapenems at a low level, it shows very weak or no activity against expanded-spectrum cephalosporins (6). Along with the spread of OXA-48, several variants have been reported that differ from OXA-48 by up to 5 amino acid substitutions or deletions (http://bldb.eu/BLDB.php?class=D#OXA) (7). The aim of this study was to characterize OXA-519, a novel OXA-48-like β-lactamase detected in a clinical K. pneumoniae isolate recovered in Belgium in 2015.
K. pneumoniae 1210 was recovered from a urine sample of an 87-year-old patient who had not been hospitalized in the last 12 months. The urine displayed hyperleukocyturia, and the culture grew >105 CFU/ml of K. pneumoniae, which was identified using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MALDI Biotyper; Bruker Daltonics, Illkirch, France). Disk diffusion antibiotic susceptibility testing was done and interpreted according to the EUCAST guidelines (http://www.eucast.org). The isolate K. pneumoniae 1210 was resistant to penicillins and expanded-spectrum cephalosporins, was intermediate to ertapenem, and remained susceptible to meropenem and imipenem (Table 1). The Carba NP test, β-Carba test (Bio-Rad, Marnes-La-Coquette, France), Bogaerts-Yunus-Glupczynski (BYG) Carba test, and Maldi-TOF Star-BL assay (Brucker Daltonics) were performed as previously described (9–12). Carba NP and BYG Carba tests gave negative test results while β-Carba and Star BL gave weak but reproducible positive results. OXA-48 K-SeT (Coris BioConcept, Gembloux, Belgium) and the NG Carba 5 (NG Biotech, Rennes, France) immunochromatographic assays revealed a positive OXA-48 band (13, 14). PCR experiments and subsequent sequencing, as previously described (15), revealed the presence of a novel blaOXA-48-like gene, designated the blaOXA-519 gene, which showed a single nucleotide change resulting in a single amino acid Val to Leu substitution at position 120, according to DBL numbering (3, 16, 17).
TABLE 1.
MICs of β-lactams for K. pneumoniae and E. coli strainsa
| Antibiotic | MIC (mg/liter) against: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| K. pneumoniae 1210 |
E. coli TOP10 |
E. coli TOP10 |
E. coli HB4 |
E. coli HB4 | |||||||
| pNOXA-519 | pR-OXA-48 | pR-OXA-519 | pR-OXA-163 | pR-OXA-48 | pR-OXA-519 | pR-OXA-163c | pN-OXA-519 | ||||
| Amoxicillin | >256 | >256 | >256 | >256 | >256 | 2 | >256 | >256 | >256 | >256 | 16 |
| Amoxicillin + CLAb | 48 | 32 | 192 | 32 | 96 | 2 | >256 | 32 | >256 | 8 | |
| Cefotaxime | >32 | 0.047 | 0.094 | 0.047 | 3 | 0.06 | 8 | 0.75 | 128 | 1.5 | 1 |
| Ceftazidime | 48 | 0.125 | 0.19 | 0.125 | 16 | 0.12 | 1 | 0.5 | 256 | 1 | 0.75 |
| Cefepime | 16 | 0.023 | 0.047 | 0.023 | 0.5 | 0.023 | 12 | 0.75 | 1 | 0.75 | |
| Imipenem | 0.25 | 0.25 | 0.38 | 0.25 | 0.25 | 0.25 | >32 | 1.5 | 0.5 | >32 | 0.125 |
| Meropenem | 0.75 | 0.19 | 0.047 | 0.19 | 0.023 | 0.016 | >32 | >32 | 4 | >32 | 0.38 |
| Ertapenem | 2 | 0.125 | 0.047 | 0.125 | 0.032 | 0.003 | >32 | >32 | 32 | >32 | 0.75 |
| Temocillin | >1,024 | >1,024 | >1,024 | >1,024 | 32 | 4 | >1,024 | >1,024 | 64 | >1,024 | 24 |
| Aztreonam | >256 | 0.047 | 0.047 | 0.047 | 2 | 0.047 | 0.5 | 0.38 | 0.5 | 0.38 | |
K. pneumoniae 1210, E.coli TOP10 pTOPO OXA-48, E. coli TOP10 pTOPO-519, E. coli TOP 10 pTOPO-163, E. coli TOP10, E. coli HB4 pOXA-48, E. coli HB4 pOXA-519, and E. coli HB4.
CLA, clavulanic acid (2 mg/liter).
Values from Oueslati et al. (15).
The blaOXA-48, blaOXA-163, and blaOXA-519 genes were amplified using preOXA-48A (5′-TATATTGCATTAAGCAAGGG-3′) and cloning-OXA-48B (5′-AAAAGGATCCCTAGGGAATAATTTTTTCCTGTTTGAGCA-3′) primers and were subsequently cloned into the pCR-Blunt II-TOPO plasmid (Invitrogen, Illkirch, France), resulting in recombinant pR-OXA-48, pR-OXA-163, and pR-OXA-519 plasmids, respectively. OXA-519 conferred similar, but reduced resistance profiles on Escherichia coli TOP10 compared to those conferred by OXA-48, consisting of susceptibility to expanded-spectrum cephalosporins, resistance to temocillin and piperacillin-tazobactam, and decreased susceptibility to imipenem (MIC, 0.25 μg/μl). OXA-519 presented slightly higher MICs for meropenem and ertapenem compared to those presented by OXA-48 (∼4-fold and ∼3-fold, respectively) (Table 1), and it was more inhibited by clavulanic acid compared to OXA-48 and OXA-163 (Table 1). In order to see whether these enzymes may lead to carbapenem resistance in bacterial hosts with impaired outer-membrane permeability, pR-OXA-48, pR-OXA-163, and pR-OXA-519 plasmids were transformed into E. coli HB4 lacking the major porins OmpF and OmpC (15). In E. coli HB4, all three enzymes presented increased MICs for all of the β-lactams tested, including the carbapenems. For OXA-519, MIC values increased by more than ∼256-fold for ertapenem and meropenem, thus resulting in resistance to these two molecules, but only by 6-fold for imipenem (Table 1), resulting in a MIC of 1.5 μg/μl, which is still in the susceptibility range.
The blaOXA-519 gene was cloned into the expression vector pET41b (+) (Novagen, VWR International, Fontenay-sous-Bois, France) using the PCR-generated fragment with primers INF-OXA-48Fw (5′-AAGGAGATATACATATGGTAGCAAAGGAATGGCAAG-3′) and INF-OXA-48Rv (5′-GGTGGTGGTGCTCGAAGGGAATAATTTTTTCCTGTTTGAG-3′), and the NEBuilder HiFi DNA assembly cloning kit (New England BioLabs Inc., United Kingdom), following the manufacturer's instructions. Recombinant plasmid pET41-OXA-519 was electroporated into E. coli strain BL21(DE3) and expressed at 25°C with 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) as inducer. OXA-519 was purified by one-step pseudo-affinity chromatography, using a nitrilotriacetic acid (NTA)-nickel column (GE Healthcare, Freiburg, Germany). Kinetic parameters were determined as previously reported (15). The lowest catalytic efficiencies were observed for cephalosporins. Interestingly, and unlike for OXA-48, hydrolysis of ceftazidime could be detected for OXA-519, although at lower level than that for OXA-163. With respect to carbapenems, OXA-519 had an increased catalytic efficiency for ertapenem (10-fold) and meropenem (2-fold), but the kcat/Km value for imipenem was 176-fold smaller than that of OXA-48 (Table 1). Unlike OXA-48, which is not inhibited by clavulanic acid and is well inhibited by NaCl (50% inhibitory concentration [IC50], 7 mM) (4), OXA-519 was significantly inhibited by clavulanic acid (IC50, 83 μM) and weakly inhibited by NaCl (IC50, 200 mM). This inhibition profile is similar to that of OXA-163, which has IC50 values of 13.4 μM and 270 mM for clavulanic acid and NaCl, respectively (4, 15).
An analysis of all OXA-48-like sequences available in the Beta-Lactamase DataBase (7) showed that position 120 is very conserved within this family. The mutated residue in position 120 is situated at the bottom of the binding site (data not shown), in the close vicinity of the active Ser70 and the β5-β6 loop, and thus it is likely involved in the β-lactamase activity of OXA-48 (6, 18, 19). In OXA-51, the chromosomal CHDL of Acinetobacter baumannii that hydrolyzes carbapenems less efficiently than other CHDLs, position 120 is occupied by Ile instead of Val (20). The carbon δ of this Ile would cause a steric clash with the hydroxyethyl group of carbapenems, leading to an increase in the Km values of OXA-51 (8, 21). Likewise, the bulkier side chain of Leu120 in OXA-519, compared to that of Val120 in OXA-48, hampers the approach of β-lactam substrate, resulting in a decrease of the substrate affinity. This is in agreement with higher Km values determined for OXA-519 compared with those for OXA-48 (Table 2).
TABLE 2.
Steady-state kinetic parameters of β-lactamases OXA-48, OXA-519, and OXA-163
| Substrate | Kinetic parametera |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
Km (μM) |
kcat (s−1) |
kcat/Km (mM−1/s−1) |
|||||||
| OXA-48 | OXA-519 | OXA-163 | OXA-48 | OXA-519 | OXA-163 | OXA-48 | OXA-519 | OXA-163 | |
| Ampicillin | 395 | 776 | 315 | 955 | 131 | 23 | 2,418 | 169 | 70 |
| Piperacillin | 410 | 109 | ND | 75 | 36 | ND | 180 | 328 | ND |
| Oxacillin | 95 | 338 | 90 | 130 | 8 | 34 | 1368 | 23 | 370 |
| Temocillin | 45 | >1,000 | NH | 0.30 | >12 | NH | 6.6 | 8.8 | ND |
| Cephalothin | 195 | >1,000 | 10 | 44 | >13 | 3.0 | 226 | 3.5 | 300 |
| Cefoxitin | >200 | >1,000 | ND | >0.05 | >4 | ND | 0.26 | 1.7 | ND |
| Ceftazidime | NH | 373 | >1,000 | NH | 0.02 | 8 | ND | 0.04 | 3 |
| Cefotaxime | >900 | >1,000 | 45 | >9 | >1.3 | 10 | 10 | 0.39 | 230 |
| Cefepime | >550 | >1,000 | ND | >0.60 | >1.3 | ND | 1.1 | 0.31 | ND |
| Imipenem | 13 | 982 | 520 | 4.8 | 2.1 | 0.03 | 369 | 2.1 | 0.06 |
| Meropenem | 11 | 358 | >2,000 | 0.07 | 3.4 | 0.10 | 6.2 | 9.5 | 0.03 |
| Ertapenem | 100 | 83 | 130 | 0.13 | 1.1 | 0.05 | 1.3 | 13 | 0.30 |
NH, hydrolysis could not be detected with concentrations of substrate and enzymes up to 1,000 mM and 400 nM, respectively; ND, not determined.
The whole genome of K. pneumoniae 1210 was sequenced using Illumina technology, as previously reported (22). The genome was 5,549,801 bp in size, with a mean coverage of 57×. Multilocus sequence typing (MLST) of K. pneumoniae 1210, determined using MLST 1.8 software (23), revealed a novel ST, which was assigned the novel allelic profile ST2728 (1-1-211-1-1-1-1) by the K. pneumoniae MLST database (http://bigsdb.web.pasteur.fr). This sequence type is a single-locus variant (SLV) of ST15 (1-1-1-1-1-1-1-1), a pandemic clone widely described in association with carbapenemases or ESBLs and sometimes involved in outbreaks (24–26). The acquired resistance genes were identified using ResFinder server v2.1 (http://cge.cbs.dtu.dk/services/ResFinder-2.1) (27). K. pneumoniae 1210 presented four β-lactamases genes, including the naturally occurring blaSHV-28, the acquired blaOXA-1, the blaCTX-M-15 ESBL, and the blaOXA-519 carbapenemase genes. The blaOXA-1 gene, the aac(6′)-Ib-cr gene conferring resistance to kanamycin, tobramycin, and amikacin and decreased susceptibility to fluoroquinolones (28), and the catB4 gene conferring chloramphenicol resistance were part of a class 1 integron, as previously described (29). A fosA-like gene involved in the decreased susceptibility to fosfomycin in K. pneumoniae was also evidenced. In addition, GyrA topoisomerase exhibited several substitutions (S83F and D87A) that are known to confer high-level resistance to fluoroquinolones in Gram-negative rods (30). However, the gyrB, parC, and parE genes did not display any mutation in the quinolone resistance-determining region (QRDR) (data not shown). Finally, the ompK35 gene coding for the OMPK35 porin was disrupted by the insertion sequence belonging to the IS1 family, while the ompK36 gene was not altered.
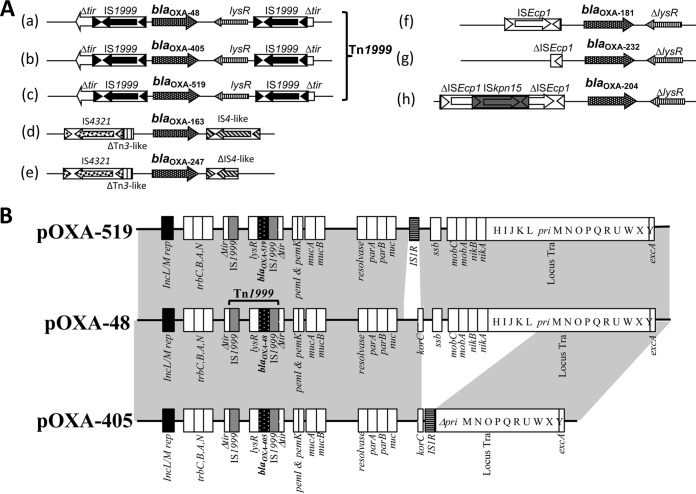
The blaOXA-519 gene was located on a Tn1999 composite transposon, unlike the blaOXA-163 or blaOXA-247 genes, which are associated with IS4361 and IS4 elements, and the blaOXA-181, blaOXA-204, and blaOXA-232 genes, which are associated with the ISEcp1 element (Fig. 1A). Three different plasmid-replication origins belonging to the incompatibility groups, IncFIB, IncL, and IncFII(K) were identified using PlasmidFinder 1.3 (https://cge.cbs.dtu.dk/services/). The blaOXA-519 gene was carried by an IncL-type plasmid of ca. 60 kb, similar to the prototypical pOXA-48 plasmid (GenBank accession number JN626286) but differing by a 1,986-bp deletion encompassing the korC gene (31–33) (Fig. 1B). Conjugation assays, performed as previously described (34), revealed a high conjugation frequency of pN-OXA-519 (1.05 × 10−1 ± 0.003), which was similar to that of pOXA-48 (1.13 × 10−1 ± 0.002), suggesting that the deletion has no impact on the conjugation efficiency.
FIG 1.
(A) Schematic representation of the genetic environment of blaOXA-48-like β-lactamases. BlaOXA-48 (a), blaOXA-405 (b), blaOXA-519 (c), blaOXA-163(d), blaOXA-247 (e), blaOXA-181 (f), blaOXA-232 (g), and blaOXA-204 (h) genes. The Tn1999 composite is formed by two copies of insertion sequence IS1999 bracketing a fragment containing the blaOXA-48, blaOXA-405, and blaOXA-519 genes. (B) Major structural features of the plasmids pNOXA-519 from K. pneumoniae 1210 with the prototypical IncL blaOXA-48 plasmid (pOXA-48) (GenBank accession number JN626286) and pOXA-405. Common structures are highlighted in gray.
We report here a novel OXA-48-like β-lactamase, OXA-519, which presents reduced activity toward imipenem compared to that of OXA-48. It is important to stress that, even though OXA-519 has reduced imipenem-hydrolytic activity compared to that of OXA-48, it has increased meropenem and ertapenem catalytic efficiency, and when expressed in a porin-deficient strain, the MICs rose significantly in the resistance range. Of note, OXA-519 may spread silently, since conventional biochemical tests based on carbapenem hydrolysis failed to detect this variant. The mutation Val120Leu is located at the bottom of the active site cavity of the protein. The bulkier side chain of Leu in OXA-519 compared with that of Val in OXA-48 induces a decrease in substrate affinity. Considering the high transfer frequency of pOXA-519, which is similar to that of pOXA-48, the risk of dispersion of this gene in the gut of patients may result in high-level carbapenem resistance, even though the initial isolate may be susceptible.
Accession number(s).
The nucleotide sequences of the blaOXA-519 gene and of its natural plasmid pOXA-519 have been submitted to the EMBL/GenBank nucleotide sequence database under the accession numbers KX349732 and KY215945, respectively. This whole-genome shotgun sequence of strain K. pneumoniae 1210 has been deposited at DDBJ/ENA/GenBank under the accession number PCGD00000000. The version described in this paper is PCGD01000000.
ACKNOWLEDGMENTS
This work was supported by the Assistance Publique—Hôpitaux de Paris (AP-HP), the University Paris-Sud, the Laboratory of Excellence in Research on Medication and Innovative Therapeutics (LERMIT) supported by a grant from the French National Research Agency (grant ANR-10-LABX-33) and by the Joint Programming Initiative on Antimicrobial Resistance (JPIAMR) DesInMBL (grant ANR-14-JAMR-002), and by DIM Malinf, Ile de France, for L.D.'s PhD fellowship.
We declare no competing interests.
REFERENCES
- 1.Ambler RP. 1980. The structure of β-lactamases. Philos Trans R Soc London B Biol Sci 289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 2.Lamotte-Brasseur J, Knox J, Kelly A, Charlier P, Fonzé E, Dideberg O, Frére JM. 1994. The structures and catalytic mechanisms of active-site serine β-lactamases. Biotechnol Genet Eng. Rev 12:189–230. doi: 10.1080/02648725.1994.10647912. [DOI] [PubMed] [Google Scholar]
- 3.Naas T, Nordmann P. 1999. OXA-type β-lactamases. Curr Pharm Des 5:865–879. [PubMed] [Google Scholar]
- 4.Poirel L, Heritier C, Tolun V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aubert D, Naas T, Héritier C, Poirel L, Nordmann P. 2006. Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of beta-lactam resistance genes. J Bacteriol 188:6506–6514. doi: 10.1128/JB.00375-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Docquier JD, Calderone V, De Luca F, Benvenuti M, Giuliani F, Bellucci L, Tafi A, Nordmann P, Botta M, Rossolini GM, Mangani S. 2009. Crystal structure of the OXA-48 β-lactamase reveals mechanistic diversity among class D carbapenemases. Chem Biol 16:540–547. doi: 10.1016/j.chembiol.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, Retailleau P, Iorga BI. 2017. Beta-Lactamase DataBase (BLDB) – structure and function. J Enz Inh Med Chem 32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith CA, Antunes NT, Stewart NK, Frase H, Toth M, Kantardjieff KA, Vakulenko S. 2015. Structural basis for enhancement of carbapenemase activity in the OXA-51 family of class D β-lactamases. ACS Chem Biol 10:1791–1796. doi: 10.1021/acschembio.5b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriacea. Emerg Infect Dis 18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernabeu S, Dortet L, Naas T. 2017. Evaluation of the β-CARBA™ test, a colorimetric test for the rapid detection of carbapenemase activity in Gram-negative bacilli. J Antimicrob Chemother 72:1646–1658. doi: 10.1093/jac/dkx061. [DOI] [PubMed] [Google Scholar]
- 11.Bogaerts P, Yunus S, Massart M, Huang TD, Glupczynski Y. 2016. Evaluation of the BYG carba test, a new electrochemical assay for rapid laboratory detection of carbapenemase-producing Enterobacteriaceae. J Clin Microbiol 54:349–358. doi: 10.1128/JCM.02404-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogaerts P, Oueslati S, Meunier D, Nonhoff C, Yunus S, Massart M, Denis O, Woodford N, Hopkins KL, Naas T, Dortet L, Huang TD, Glupczynski Y. 2017. Multicentre evaluation of the BYG Carba v2.0 test, a simplified electrochemical assay for the rapid laboratory detection of carbapenemase-producing Enterobacteriaceae. Sci Rep 7(1):9937. doi: 10.1038/s41598-017-09820-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boutal H, Vogel A, Bernabeu S, Devilliers K, Creton E, Cotellon G, Plaisance M, Oueslati S, Dortet L, Jousset A, Simon S, Naas T, Volland H. 2018. A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 73:909–915. doi: 10.1093/jac/dkx521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dortet L, Jousset A, Sainte-Rose V, Cuzon G, Naas T. 2016. Prospective evaluation of the OXA-48 K-SeT assay, an immunochromatographic test for the rapid detection of OXA-48-type carbapenemases. J Antimicrob Chemother 71:1834–1840. doi: 10.1093/jac/dkw058. [DOI] [PubMed] [Google Scholar]
- 15.Oueslati S, Nordmann P, Poirel L. 2015. Heterogeneous hydrolytic features for OXA-48-like β-lactamase. J Antimicrob Chemother 70:1059–1063. doi: 10.1093/jac/dku524. [DOI] [PubMed] [Google Scholar]
- 16.Poirel P, Naas T, Nordmann P. 2010. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob Agents Chemother 54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couture F, Lachapelle J, Levesque RC. 1992. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol Microbiol 6:1693–1705. doi: 10.1111/j.1365-2958.1992.tb00894.x. [DOI] [PubMed] [Google Scholar]
- 18.De Luca F, Benvenuti M, Carboni F, Pozzi C, Rossolini GM, Mangani S, Docquier JD. 2011. Evolution to carbapenem-hydrolyzing activity in noncarbapenemase class D β-lactamase OXA-10 by rational protein design. Proc Natl Acad Sci U S A 108:18424–18429. doi: 10.1073/pnas.1110530108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stojanoski V, Chow DC, Fryszczyn B, Hu L, Nordmann P, Poirel L, Sankaran B, Prasad BV, Palzkill T. 2015. Structural basis for different substrate profiles of two closely related class D β-lactamases and their inhibition by halogens. Biochemistry 54:3370–3380. doi: 10.1021/acs.biochem.5b00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell JM, Leonard DA. 2014. Common clinical substitutions enhance the carbapenemase activity of OXA-51-like class D β-lactamases from Acinetobacter spp. Antimicrob Agents Chemother 58:7015–7016. doi: 10.1128/AAC.03651-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.June CM, Muckenthaler TJ, Schroder EC, Klamer ZL, Wawrzak Z, Powers RA, Szarecka A, Leonard DA. 2016. The structure of a doripenem-bound OXA-51 class D β-lactamase variant with enhanced carbapenemase activity. Protein Sci 25:2152–2163. doi: 10.1002/pro.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jousset AB, Dabos L, Bonnin RA, Girlich D, Potron A, Cabanel N, Dortet L, Glaser P, Naas T. 2018. CTX-M-15-producing Shewanella species clinical isolate expressing OXA-535, a chromosome-encoded OXA-48 variant, putative progenitor of the plasmid-encoded OXA-436. Antimicrob Agents Chemother 62:e01879-17. doi: 10.1128/AAC.01879-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodrigues C, Bavlovič J, Machado E, Machado E, Amorim J, Peixe L, Novais Â. 2016. KPC-3-producing Klebsiella pneumoniae in Portugal linked to previously circulating non-CG258 lineages and uncommon genetic platforms (Tn4401d-IncFIA and Tn4401d-IncN). Front Microbiol 7:1000. doi: 10.3389/fmicb.2016.01000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou K, Lokate M, Deurenberg RH, Tepper M, Arends JP, Raangs EG, Lo-Ten-Foe J, Grundmann H, Rossen JW, Friedrich AW. 2016. Use of whole-genome sequencing to trace, control and characterize the regional expansion of extended-spectrum β-lactamase producing ST15 Klebsiella pneumoniae. Sci Rep 6:20840. doi: 10.1038/srep20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewers C, Stamm I, Pfeifer Y, Wieler LH, Kopp PA, Schønning K, Prenger-Berninghoff E, Scheufen S, Stolle I, Günther S, Bethe A. 2014. Clonal spread of highly successful ST15-CTX-M-15 Klebsiella pneumoniae in companion animals and horses. J Antimicrob Chemother 69:2676–2680. doi: 10.1093/jac/dku217. [DOI] [PubMed] [Google Scholar]
- 27.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raherison S, Jove T, Gaschet M, Pinault E, Tabesse A, Torres C, Ploy MC. 2017. Expression of the aac(6′)-Ib-cr gene in class 1 integrons. Antimicrob Agents Chemother 61:e02704-16. doi: 10.1128/AAC.02704-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic support and diversity of acquired extended-spectrum β-lactamases in Gram-negative rods. Infect Genet Evol 12:883–893. doi: 10.1016/j.meegid.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Fabrega A, Madurga S, Giralt E, Vila J. 2009. Mechanism of action of and resistance to quinolones. Microb Biotechnol 2:40–61. doi: 10.1111/j.1751-7915.2008.00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, Poirel L. 2011. Characterization of OXA-181, a carbapenem-hydrolyzing class D β-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother 55:4896–4899. doi: 10.1128/AAC.00481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potron A, Rondinaud E, Poirel L, Belmonte O, Boyer S, Camiade S, Nordmann P. 2013. Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D β-lactamase from Enterobacteriaceae. Int J Antimicrob Agents 41:325–329. doi: 10.1016/j.ijantimicag.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother 56:559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potron A, Poirel L, Nordmann P. 2014. Derepressed transfer properties leading to the efficient spread of the plasmid encoding carbapenemase OXA-48. Antimicrob Agents Chemother 58:467–471. doi: 10.1128/AAC.01344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]