ABSTRACT
A putative fosA gene in Kluyvera georgiana 14751 showed 99% nucleotide identity with plasmid-encoded fosA4. Due to a single-nucleotide insertion translating to a truncated protein, K. georgiana 14751 fosA does not confer fosfomycin resistance. However, analysis of another genome deposit (Kluyvera ascorbata WCH1410) that could be recategorized as K. georgiana after phylogenetic analysis revealed a fosA gene 100% identical to the plasmid-borne fosA4 gene. We suggest that Kluyvera georgiana represents the most probable origin of fosA4.
KEYWORDS: FosA, Kluyvera ascorbata, Kluyvera georgiana, whole-genome sequencing, fosfomycin resistance
TEXT
Fosfomycin is an old, broad-spectrum antibiotic that inhibits cell wall biosynthesis by inactivating UDP-N-acetylglucosamine-3-enolpyruvyltransferase (MurA), acting as a phosphoenolpyruvate analogue (1). It has regained attention in the past few years due to its activity against multidrug-resistant and extremely drug-resistant microorganisms, typically recovered from hospital-acquired infections.
Among several fosfomycin-modifying enzymes described, the FosA enzymes (FosA, FosA2, FosA3, FosA4, FosA5, and FosA6) are the most prevalent enzymes among Gram-negative organisms. They are found mainly in plasmids from Enterobacteriaceae but are also observed in Pseudomonas aeruginosa and Acinetobacter baumannii (1, 2). Recently, the origins of some plasmid-mediated fosA genes have been proposed; fosA2 in Tn2961 originated from the chromosome of Enterobacter cloacae (3), fosA4 occurred in an Escherichia coli isolate (4), fosA5 and fosA6 originated from the chromosome of Klebsiella pneumoniae (5, 6), and, more recently, Ito et al. proposed a chromosome-encoded fosA gene from Kluyvera georgiana YDC799 as the origin of plasmid-encoded fosA3, with 99% amino acid identity (7).
Upon whole-genome sequencing (WGS) with the Illumina platform, we searched for the putative chromosome-encoded fosA gene(s) in Kluyvera georgiana 14751, which was isolated from a bloodstream infection (SENTRY Antimicrobial Surveillance Program) in Louisville, Kentucky, USA, in 2002. De novo assembly of reads was achieved using the Velvet package (velveth and velvetg programs) (https://www.ebi.ac.uk/~zerbino/velvet/), resulting in a contigs.fa file with 1,589 nodes, an N50 value of 20,178, a longest contig of 105,293 bp, and a total assembly of 4,993,089 bp, using 1,264,850/1,278,330 reads. The fosA genes were screened in the contigs.fa file using NCBI BLAST.
Node 845 (8,626 bp; coverage of 23.09×) contained a fosA gene (fosAK14751) displaying 99% nucleotide identity (409/412 bp, including a single cytosine nucleotide insertion at position 339) with plasmid-encoded fosA4 (GenBank accession numbers CP023167.1 and CP016184.1, among others) and ∼93% nucleotide identity with fosA3 from K. georgiana YDC799 (GenBank accession number CP022114.1), which was previously proposed as the chromosomal origin of fosA3 (7). Interestingly, we also detected a sequence containing a chromosome-encoded fosA4 gene deposited in the nonredundant nucleotide databases as Kluyvera ascorbata WCH1410 (GenBank accession number NZ_LSME00000000.1), which we included in the analysis to compare both fosA4 genes and the surrounding sequences.
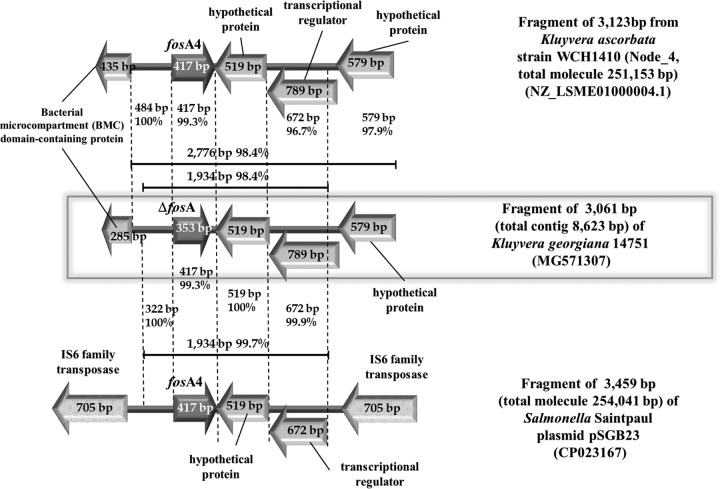
A 1,933-bp sequence is almost identical (99.7% nucleotide identity) between K. georgiana 14751 and plasmid pSGB23 from Salmonella enterica subsp. enterica serovar Saintpaul, including a 322-bp region upstream of fosA (100% nucleotide identity), the fosA gene (99.3% nucleotide identity [409/412 bp]), and a 1,195-bp region downstream of fosA (99.9% identity). The last fragment includes a 519-bp open reading frame (ORF) (100% nucleotide identity) and a 789-bp ORF in which a 672-bp segment has 99.9% nucleotide identity with a transcriptional regulator in plasmid pSGB23. In K. ascorbata strain WCH1410, the corresponding 322-bp and 519-bp regions demonstrate 100% nucleotide identity, and fosA has 99.3% nucleotide identity with K. georgiana 14751 fosA; however, the gene is 100% identical to its plasmidic counterpart. There is also a 792-bp transcriptional regulator with ∼96% nucleotide identity with that from K. georgiana 14751 (764/792 bp, including 3 gaps) (Fig. 1). Remarkably, chromosomal fosA from K. georgiana 14751 and the fosA genetic environment display greater identity with plasmid-borne fosA4 and the neighboring sequences (99.7% nucleotide identity [1,928/1,933 bp]) than with the equivalent chromosomal segment from K. ascorbata WCH1410 (98.7% nucleotide identity [1,901/1,933 bp]).
FIG 1.
Schematic representation of the partial sequence of node 845 of the Kluyvera georgiana 14751 genome assembly (middle) and comparison with the partial sequence of node 4 of the genome assembly from Kluyvera ascorbata WCH1410 (top) and the partial sequence of plasmid pSGB23 from Salmonella enterica subsp. enterica serovar Saintpaul strain SGB23 (bottom).
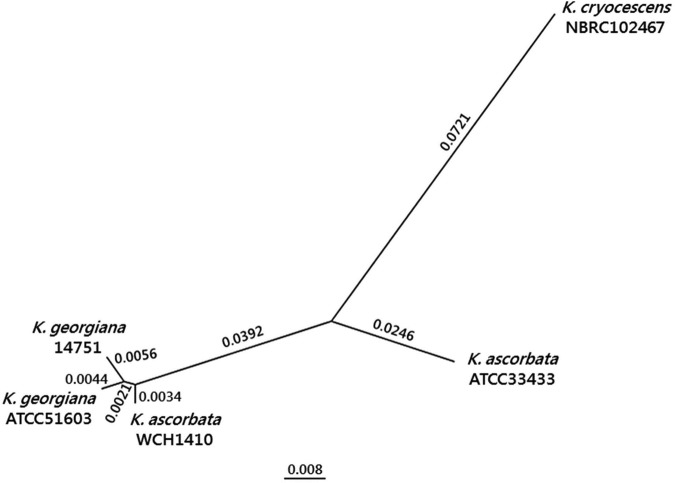
Additionally, we performed a phylogenetic analysis by concatenation of several housekeeping genes (16S rRNA, adk, gyrA, gyrB, recA, infB, and rpoB genes) from K. ascorbata WCH1410, K. ascorbata ATCC 33433, Kluyvera cryocrescens NBRC102167, K. georgiana ATCC 51603, and K. georgiana 14751. The analysis was conducted by using ClustalX (http://clustalx.software.informer.com/2.1) to align all sequences, and the molecular evolution model was estimated with jModelTest2 (http://github.com//ddarriba/jmodeltest2/releases). The resulting phylogenetic tree was obtained with PhyML (http://www.atgc-montpellier.fr/phyml/versions.php), using the Bayesian information criterion (BIC) parameters suggested by the jModelTest software, with 1,000 bootstraps. The phylogenetic tree was visualized and edited using FigTree (http://tree.bio.ed.ac.uk/software/figtree).
Housekeeping genes from K. ascorbata WCH1410 showed greater identity with the homologous genes from K. georgiana ATCC 51603 (99.1% nucleotide identity) and K. georgiana 14751 (99.0% nucleotide identity) than with the corresponding genes from K. ascorbata ATCC 33433 (95.0% nucleotide identity), as shown in Fig. 2. We suggest that K. ascorbata WCH1410 might be, in fact, a K. georgiana isolate. Therefore, a taxonomic reevaluation of the entire genus is currently necessary (data not shown; M. M. Rodriguez, B. Ghiglione, M. Almuzara, P. Power, T. Naas, and G. Gutkind, unpublished data).
FIG 2.
Phylogenetic tree (1,000 bootstraps) of housekeeping genes from K. ascorbata WCH1410, K. ascorbata ATCC 33433, K. cryocrescens NBRC102147, K. georgiana ATCC 51603, and K. georgiana 14751. Phylogenetic relationships are expressed in each branch as the substitutions per site (also expressed in the scale bar).
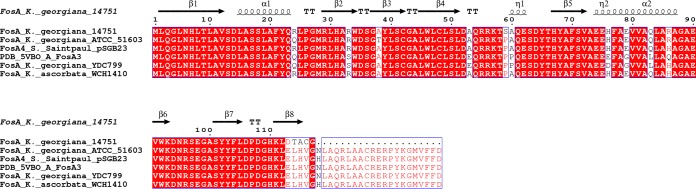
As a result of the previously mentioned single-nucleotide insertion generated at the 3′ end of the fosAK14751 gene, a shorter peptide seems to be translated due to the occurrence of a premature stop codon in the mRNA; this generates a deduced FosAK14751 enzyme with 95% amino acid identity with the main core of FosA4 (111/117 amino acids) from several species (GenBank accession numbers BAP18892.1, KXT28349.1, OJQ09299.1, OYF76970.1, OYI75904.1, ASZ39831.1, and PAY66171.1); the protein seems to conserve all proposed active site residues except for the last α-helix (Fig. 3).
FIG 3.
Amino acid sequence alignment of FosA proteins from Kluyvera georgiana 14751 and K. georgiana ATCC 51603, FosA4 from Salmonella enterica subsp. enterica serovar Saintpaul plasmid pSGB23 (GenBank accession number ASZ39831.1), FosA4 from Kluyvera ascorbata WCH1410 (100% amino acid identity), FosA3 from Escherichia coli (PDB accession number 5VB0), and FosA3 from K. georgiana YDC799 (GenBank accession number ASG63672.1). Putative secondary domains are shown above the sequences.
To test whether the expressed FosA protein has activity toward fosfomycin, we cloned the fosA gene from K. georgiana 14751 in a pK19 vector in frame with the vector's promoter, using the primers fosA4_HindIII_F (5′-AAGCTTCATGCTGCAGGGATTGAA-3′) and fosA4_EcoRI_R (5′-CGGCAGTAAGCTGAACGAATTCGTCA-3′), and transformed the recombinant plasmid in E. coli TOP10 cells. The sequence was confirmed by DNA sequencing at Macrogen (Korea). Fosfomycin susceptibility tests were performed using fosfomycin disks (200 μg) with glucose-6-phosphate (50 μg), according to CLSI guidelines (8). E. coli clones producing FosA were susceptible to fosfomycin and showed the same inhibition zones as the control strains, suggesting that the C-terminal deletion in the FosA protein (including the conserved Arg122 residue) (Fig. 3) indeed has a deleterious impact on fosfomycin resistance.
While our own sequence does not provide resistance (due to the single-nucleotide insertion and frameshift, resulting in premature termination of the protein), we still consider, based on the 100% identity of K. ascorbata WCH1410 fosA with fosA and the analysis of genetic contexts described above, that the origin of the plasmid-borne fosA4 gene, as well as other resistance genes, can be traced back to K. georgiana (9–11). The role of Kluyvera members as donors of chromosomal genes to be recruited by plasmid platforms is noteworthy. Therefore, compartmentalized evolution (as expected for microorganisms in soil, water, or sewage environments, with no epidemiological link) through which microevolution within different, originally chromosomal genes may occur, either (most probably) before or after recruitment, is proposed.
Accession number(s).
The genomic sequence of Kluyvera georgiana 14751 was deposited in GenBank under accession number MG571307.
ACKNOWLEDGMENTS
This work was supported by grants from the University of Buenos Aires (grant UBACyT 2014-2017 to P.P. and grant 2013-2015 to G.G.), the Agencia Nacional de Promoción Científica y Tecnológica (grant PICT 2014-0457 to P.P. and grant BID PICT 2015-1925 to G.G.), the Assistance Publique-Hôpitaux de Paris, Université Paris Sud (grant EA 7361), and LabEx LERMIT (supported by grant ANR-10-LABX-33 from the French National Research Agency). This work was also funded in part by a grant from the Joint Programme Initiative on Antimicrobial Resistance (grant ANR-14-JAMR-0002).
M.M.R., B.G., P.P., and G.G. are members of Carrera del Investigador Cientifico, CONICET, Argentina.
We thank H. Sader and R. N. Jones (JMI Laboratories, North Liberty, IA) for kindly providing the bacterial strain for the study.
REFERENCES
- 1.Thompson MK, Keithly ME, Sulikowski GA, Armstrong RN. 2015. Diversity in fosfomycin resistance proteins. Perspect Sci 4:17–23. doi: 10.1016/j.pisc.2014.12.004. [DOI] [Google Scholar]
- 2.Silver LL. 2017. Fosfomycin: mechanism and resistance. Cold Spring Harb Perspect Med 7:a025262. doi: 10.1101/cshperspect.a025262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu H, Miao V, Kwong W, Xia R, Davies J. 2011. Identification of a novel fosfomycin resistance gene (fosA2) in Enterobacter cloacae from the Salmon River, Canada. Lett Appl Microbiol 52:427–429. doi: 10.1111/j.1472-765X.2011.03016.x. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura G, Wachino J, Sato N, Kimura K, Yamada K, Jin W, Shibayama K, Yagi T, Kawamura K, Arakawa Y. 2014. Practical agar-based disk potentiation test for detection of fosfomycin-nonsusceptible Escherichia coli clinical isolates producing glutathione S-transferases. J Clin Microbiol 52:3175–3179. doi: 10.1128/JCM.01094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Y, Xu X, Guo Q, Wang P, Wang W, Wang M. 2015. Characterization of fosA5, a new plasmid-mediated fosfomycin resistance gene in Escherichia coli. Lett Appl Microbiol 60:259–264. doi: 10.1111/lam.12366. [DOI] [PubMed] [Google Scholar]
- 6.Guo Q, Tomich AD, McElheny CL, Cooper VS, Stoesser N, Wang M, Sluis-Cremer N, Doi Y. 2016. Glutathione-S-transferase FosA6 of Klebsiella pneumoniae origin conferring fosfomycin resistance in ESBL-producing Escherichia coli. J Antimicrob Chemother 71:2460–2465. doi: 10.1093/jac/dkw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito R, Pacey MP, Mettus RT, Sluis-Cremer N, Doi Y. 2018. Origin of the plasmid-mediated fosfomycin resistance gene fosA3. J Antimicrob Chemother 73:373–376. doi: 10.1093/jac/dkx389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing; 27th informational supplement. M100-S27. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 9.Olson AB, Silverman M, Boyd DA, McGeer A, Willey BM, Pong-Porter V, Daneman N, Mulvey MR. 2005. Identification of a progenitor of the CTX-M-9 group of extended-spectrum β-lactamases from Kluyvera georgiana isolated in Guyana. Antimicrob Agents Chemother 49:2112–2115. doi: 10.1128/AAC.49.5.2112-2115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poirel L, Kampfer P, Nordmann P. 2002. Chromosome-encoded Ambler class A β-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum β-lactamases. Antimicrob Agents Chemother 46:4038–4040. doi: 10.1128/AAC.46.12.4038-4040.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez MM, Power P, Sader H, Galleni M, Gutkind G. 2010. Novel chromosome-encoded CTX-M-78 β-lactamase from a Kluyvera georgiana clinical isolate as a putative origin of CTX-M-25 subgroup. Antimicrob Agents Chemother 54:3070–3071. doi: 10.1128/AAC.01615-09. [DOI] [PMC free article] [PubMed] [Google Scholar]