ABSTRACT
The linkage of the protease-chaperon system, SmeYZ pump, and aminoglycoside resistance was assessed in Stenotrophomonas maltophilia. The clpA, clpS, clpP, and htpX genes were upregulated in response to kanamycin exposure. Of these, clpA and htpX were the primary determinants responsible for intrinsic aminoglycoside (AG) resistance. Inactivation of clpA and htpX compromised protease-mediated intrinsic aminoglycoside resistance and weakened SmeYZ pump-mediated aminoglycoside resistance, signifying HtpX and ClpA as potential AG adjuvant targets for treatment of S. maltophilia infections.
KEYWORDS: Stenotrophomonas maltophilia, aminoglycoside resistance, chaperone, protease
TEXT
Defective proteins, either mistranslated or misfolded, are unavoidably generated during bacterial growth. Two different systems, protease and chaperone, have evolved to maintain protein homeostasis (1). Protease systems are ubiquitous in bacterial cells to degrade defective proteins. A Gram-negative model organism, Escherichia coli, harbors seven main protease systems, ClpXP, ClpAP, ClpS, Lon, HslUV, FtsH, and HtpX (2, 3). Among the chaperone systems, DnaK/DnaJ/GrpE, GroES/GroEL, and ClpB are highly conserved in several bacteria, and they assist protein folding and prevent protein aggregation (4). Furthermore, the concerted action of the DnaK system and ClpB chaperon can rescue the proteins from aggregated states by resolubilizing and refolding aggregated proteins.
Aminoglycoside (AG) antibiotics target the 30S subunit of the ribosome and interfere with translational fidelity (5). Bacterial resistance to AGs typically results from aminoglycoside-modifying enzymes (AMEs), efflux pumps, and 16S rRNA modification (6). In addition, inactivation or overexpression of the protease and chaperone genes has been linked to a decrease or increase in AG resistance (7, 8). Therefore, some proteases and chaperones contribute to intrinsic AG resistance.
Stenotrophomonas maltophilia is an important nosocomial pathogen (9). The main problem in the treatment of infections caused by S. maltophilia is inherent resistance to a variety of antimicrobials, particularly β-lactams and aminoglycosides. The mechanism of AG resistance in S. maltophilia is the expression of AMEs and efflux pumps (SmeIJK, SmeOP, and SmeYZ) (10). The 16S rRNA modification-mediated AG resistance is rarely seen in S. maltophilia. However, the linkage between protein homeostasis and intrinsic AG resistance in S. maltophilia remains poorly understood.
clpA, clpS, clpP, and htpX were upregulated in response to kanamycin exposure.
To find out the possible candidates which are linked to intrinsic AG resistance in S. maltophilia strain KJ, the expression of a subset of protease and chaperone genes in response to 2-h kanamycin exposure was assessed by reverse transcription-quantitative PCR (qRT-PCR). RNA extracts, cDNA preparation, and qRT-PCR were carried out as described previously (11). The primers used for qRT-PCR are listed in Table 1. Normalized expression levels of the target gene transcripts were calculated relative to those of 16S rRNA genes using the ΔΔCT method (12). Compared to the untreated counterpart, four protease genes of strain KJ cells, clpP, clpA, clpS, and htpX, were upregulated (Fig. 1). However, no significant upregulations were observed in the other protease (clpX, lon, hslU, hslV, and ftsH) and chaperone (grpE, dnaK, dnaJ, groEL, groES, and clpB) genes (Fig. 1).
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Species, strain, plasmid, or primer | Genotype or properties |
|---|---|
| S. maltophilia | |
| KJ | Wild type, a clinical isolate from Taiwan |
| KJΔClpA | S. maltophilia KJ mutant of clpA; ΔclpA |
| KJΔClpP | S. maltophilia KJ mutant of clpP; ΔclpP |
| KJΔClpS | S. maltophilia KJ mutant of clpS; ΔclpS |
| KJΔHtpX | S. maltophilia KJ mutant of htpX; ΔhtpX |
| KJΔYZ | S. maltophilia KJ mutant of smeYZ operon; ΔsmeYZ |
| KJΔClpAΔHtpX | S. maltophilia KJ mutant of clpA and htpX; ΔclpA ΔhtpX |
| KJΔClpPΔHtpX | S. maltophilia KJ mutant of clpP and htpX; ΔclpP ΔhtpX |
| KJΔClpSΔHtpX | S. maltophilia KJ mutant of clpS and htpX; ΔclpS ΔhtpX |
| KJΔYZΔClpAΔHtpX | S. maltophilia KJ mutant of smeYZ operon, clpS, and htpX; ΔsmeYZ ΔclpS ΔhtpX |
| E. coli | |
| DH5a | F− ϕ80d/acZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rk− mk+) phoA supE44λ thi-1 gyrA96 relA1 |
| S17-1 | λ pir+ mating strain |
| Plasmids | |
| pEX18Tc | sacB oriT, Tcr |
| pRK415 | Mobilizable broad-host-range plasmid cloning vector, RK2 origin; Tcr |
| pΔClpA | pEX18Tc with an internal-deletion clpA gene; Tcr |
| pΔClpP | pEX18Tc with an internal-deletion clpP gene; Tcr |
| pΔClpS | pEX18Tc with an internal-deletion clpS gene; Tcr |
| pΔHtpX | pEX18Tc with an internal-deletion htpX gene; Tcr |
| pΔYZ | pEX18Tc with an internal-deletion smeYZ operon; Tcr |
| pClpA | pRK415 with an intact clpA gene; Tcr |
| pClpP | pRK415 with an intact clpP gene; Tcr |
| pClpS | pRK415 with an intact clpS gene; Tcr |
| pHtpX | pRK415 with an intact htpX gene; Tcr |
| Primers | |
| ClpAN-F | 5′-TCGGTACCGCTGCTGATCTTC-3′ |
| ClpAN-R | 5′-CCGAGCTCTGCACGTGGTACA-3′ |
| ClpAC-F | 5′-GATCTAGATGGAGATGCTGC-3′ |
| ClpAC-R | 5′-GCGTCGACCAGCTATGTCTT-3′ |
| ClpPN-F | 5′-CTGAGCTCTTCGACCTACGA-3′ |
| ClpPN-R | 5′-CGGGATCCGGCGAGTTGATGT-3′ |
| ClpPC-F | 5′-AGCGGATCCACTTCAAGAGCG-3′ |
| ClpPC-R | 5′-ACGGATCCGTCGAGCACCTC-3′ |
| ClpSN-F2 | 5′-GCGAGCTCGAACTGCTTGGT-3′ |
| ClpSN-R2 | 5′-GCTCTAGATGGGAATCGGGG-3′ |
| ClpSC-F | 5′-CGATCTAGAAGGCCTGACCGG-3′ |
| ClpSC-R | 5′-GGAGCATGCGTTCGATTACAC-3′ |
| HtpXN-F | 5′-GGGAGCTCTGGCAGACAAAG-3′ |
| HtpXN-R | 5′-CGGGTACCTGGCTTGGATTG-3′ |
| HtpXC-F | 5′-GTGGTACCGCCTATTACATC-3′ |
| HtpXC-R | 5′-GCGGATCCTTTCTTTGGTTA-3′ |
| SmeYZ-F | 5′-CGCAAGCTTGACCTGCGCTATGCC-3′ |
| SmeYZ-R | 5′-TGCGAATTCCAGCAGCATCGCCTCC-3′ |
| ClpA-F2 | 5′-GCTCTAGAAGGGTTCGCTCAT-3′ |
| ClpA-R | 5′-CCGAGCTCGTCATTTCCAGTCT-3′ |
| ClpP-F | 5′-CCTCTAGAACCGTGTGATGG-3′ |
| ClpP-R | 5′-CCGGATCCAGAGCAGTAGAG-3′ |
| ClpS-F | 5′-GGTCTAGACCAGCTTGTCCA-3′ |
| ClpS-R | 5′-GCGGTACCGTTCAGGGATGA-3′ |
| HtpX-F | 5′-CCTCTAGACGCCGTTATGTC-3′ |
| HtpX-R | 5′-AAGAGGTGCGGGTGTCTGAG-3′ |
| ClpXQ-F | 5′-GAACTGGCGAAGTCGAACAT-3′ |
| ClpXQ-R | 5′-CGACGTCGTAGTCGCACTT-3′ |
| ClpPQ-F | 5′-CAAGCCGAATGTGAGCACTA-3′ |
| ClpPQ-R | 5′-GAACGCAGGGTCAGGATCT-3′ |
| ClpAQ-F | 5′-AGAACAACCCGCTGTACGTC-3′ |
| ClpAQ-R | 5′-GTTGGGCACCTTCTTCAGTG-3′ |
| ClpSQ-F | 5′-TACCAGGTGATGCTGCTCAA-3′ |
| ClpSQ-R | 5′-ACCTTGGTCTCGGCCACT-3′ |
| LonQ-F | 5′-CGCAACGAACTGAACAAGC-3′ |
| LonQ-R | 5′-ACCGCCAGGTATTCAAGGAT-3′ |
| HslUQ-F | 5′-CCAGCAGCTGAAGTCGATGT-3′ |
| HslUQ-R | 5′-GCGACCTTGTCGATCTCGT-3′ |
| HslVQ-F2 | 5′-CGATGCCTTCACCCTGTTC-3′ |
| HslVQ-R2 | 5′-CCTCCGGTTCGATCACATC-3′ |
| FtsHQ-F | 5′-AGCACATCCTCAAGGTCCAC-3′ |
| FtsHQ-R | 5′-GCACCCATCAGGATCTTGTC-3′ |
| HtpXQ-F | 5′-GCTTCGGTGGCTCCTTCAT-3′ |
| HtpXQ-R | 5′-CCAGTGGCGAAGGCGTTGAT-3′ |
| GrpEQ-F | 5′-CTGGAGAACCAGCGCAAG-3′ |
| GrpEQ-R | 5′-AGCAGGACCAGTCCGTTGT-3′ |
| DnaKQ-F | 5′-GCGTCATCGAGTACCTGGTT-3′ |
| DnaKQ-R | 5′-GGGTCAGCTTGATGTTCAGG-3′ |
| DnaJQ-F | 5′-CGATATCGGCTACGTGATGG-3′ |
| DnaJQ-R | 5′-CTGCTGCATGGCGAAGAT-3′ |
| GroELQ-F | 5′-TCGCCGTACTTCATCAACAA-3′ |
| GroELQ-R | 5′-GATGCCACGGATGGTGTT-3′ |
| GroESQ-F | 5′-GATCGAAGCCGACGAAATC-3′ |
| GroESQ-R | 5′-CGCAGGACCTTGTATTCGAC-3′ |
| ClpBQ-F | 5′-TTCATCGACGAACTGCACAC-3′ |
| ClpBQ-R | 5′-GCCCACGAACACCTTCTG-3′ |
| SmeZQ-F | 5′-TGTCCAGCGTCAAGCACC-3′ |
| SmeZQ-R | 5′-GCCGACCAGCATCAGGAAG-3′ |
| rDNA-F | 5′-GACCTTGCGCGATTGAATG-3′ |
| rDNA-R | 5′-CGGATCGTCGCCTTGGT-3′ |
FIG 1.
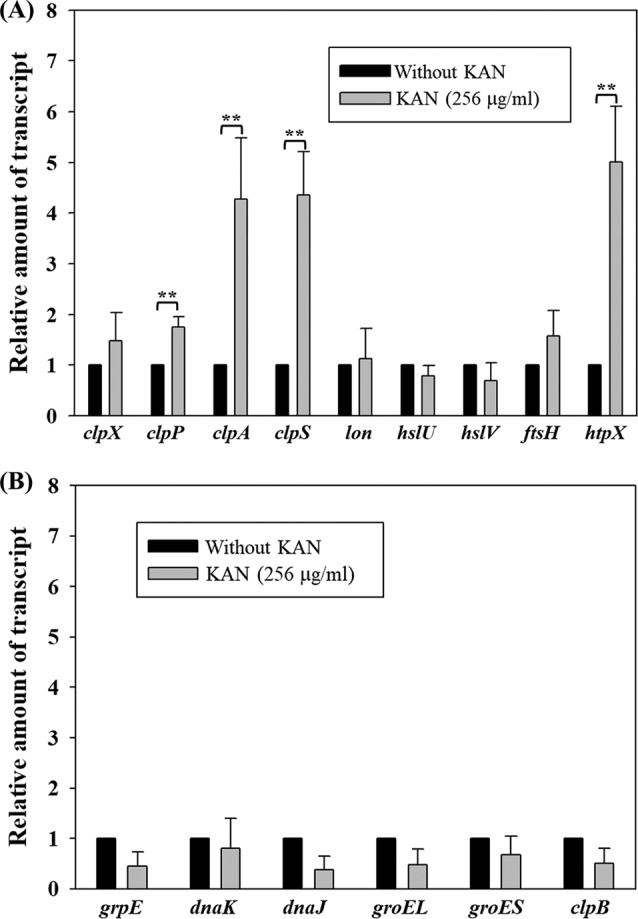
qRT-PCR analysis of protease and chaperone genes in response to kanamycin exposure. RNAs from the logarithmic phase of S. maltophilia KJ cells, with or without 2-h kanamycin exposure, were prepared, and the transcript amounts of genes were comparatively quantified by qRT-PCR. The relative amount of transcript assayed was calculated using the KJ cells without kanamycin treatment as 1. The data presented are representatives from three independent experiments, and standard deviations are shown as vertical bars. *, P < 0.5; **, P < 0.05; significance calculated by Student's t test. (A) qRT-PCR analysis of the protease genes. (B) qRT-PCR analysis of the chaperone genes.
The phenomenon that protease and/or chaperone genes are upregulated in response to AG challenge has been reported in several microorganisms, including for clpP and clpX of Bacillus subtilis, dnaK and groEL of Acinetobacter baumannii, and dnaK-dnaJ-grpE, groEL-groES, and clpB of E. coli (13–15); however, this is the first report of this phenomenon in S. maltophilia. Distinct from other microorganisms, the protease system, rather than the chaperone system, appears to play a critical role in alleviating AG-mediated stress in S. maltophilia.
clpP, clpA, clpS, and htpX contributed to intrinsic AG resistance.
Next, we considered the contribution of the clpP, clpA, clpS, and htpX genes to the intrinsic AG resistance of S. maltophilia. Individual or combined in-frame deletions of each gene were introduced into the chromosome of S. maltophilia KJ, and the impact of gene deletion on AG resistance was investigated by susceptibility test. Plasmids pΔClpP, pΔClpS, pΔClpA, and pΔHtpX (Table 1) were prepared for the construction of mutants. Two PCR amplicons, corresponding to the upstream and downstream regions of the target gene, were amplified and subsequently cloned into pEX18Tc to yield the recombinant plasmid, in which the cloned target gene was partially deleted. The primer sets used were ClpPN-F/ClpPN-R and ClpPC-F/ClpPC-R for plasmid pΔClpP; ClpSN-F2/ClpSN-R2 and ClpSC-F/ClpSC-R for plasmid pΔClpS; ClpAN-F/ClpAN-R and ClpAC-F/ClpAC-R for plasmid pΔClpA; and HtpXN-F/HtpXN-R and HtpXC-F/HtpXC-R for plasmid pΔHtpX (Table 1). Plasmid mobilization, transconjugant selection, and deletion mutant confirmation were performed as described previously (11). For complementary test, the intact clpP, clpS, clpA, and htpX genes were amplified by PCR using the primer sets of ClpP-F/ClpP-R, ClpS-F/ClpS-R, ClpA-F2/ClpA-R, and HtpX-F/HtpX-R and then cloned into pRK415, yielding plasmids pClpP, pClpS, pClpA, and pHtpX, respectively (Table 1). The MICs of AG for the strains were determined by a standard 2-fold serial agar dilution method, according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (16). All of the resultant single mutants (KJΔClpP, KJΔClpA, KJΔClpS, and KJΔHtpX) were more susceptible to AG (2- to 16-fold reduction in MICs), and this phenotype was reversed by cloned wild-type copies of the corresponding genes, although the complementation by clpA was less complete than expected (Table 2). None of the deletion mutants showed any change in susceptibility to spectinomycin (SPT), a bacteriostatic antibiotic that causes translational blockage without misreading (Table 2). Notably, of the three double mutants examined, KJΔClpAΔHtpX exhibited the most substantial decrease in AG resistance (Table 2). Thus, HtpX and ClpA are the primary determinants responsible for intrinsic AG resistance, and both are potential aminoglycoside adjuvant targets.
TABLE 2.
Antibiotic susceptibility of S. maltophilia KJ and its derived mutants
| Strain | MIC (μg/ml) fora: |
||||
|---|---|---|---|---|---|
| AMK | KAN | GEM | TOB | SPT | |
| KJ | 1,024 | 256 | 1,024 | 256 | 4,096 |
| KJΔHtpX | 512 | 64 | 256 | 128 | 4,096 |
| KJΔHtpX(pHtpX) | 1,024 | 512 | 1,024 | 256 | 4,096 |
| KJΔClpP | 256 | 64 | 128 | 64 | 4,096 |
| KJΔClpP(pClpP) | 512 | 128 | 512 | 256 | 4,096 |
| KJΔClpA | 64 | 32 | 64 | 32 | 4,096 |
| KJΔClpA(pClpA) | 128 | 32 | 128 | 128 | 4,096 |
| KJΔClpS | 512 | 64 | 128 | 128 | 4,096 |
| KJΔClpS(pClpS) | 1,024 | 512 | 512 | 256 | 4,096 |
| KJΔClpSΔHtpX | 256 | 32 | 128 | 64 | 4,096 |
| KJΔClpPΔHtpX | 512 | 128 | 256 | 128 | 4,096 |
| KJΔClpAΔHtpX | 16 | 4 | 4 | 8 | 4,096 |
| KJΔYZ | 8 | 8 | 8 | 16 | 4,096 |
| KJΔYZΔClpAΔHtpX | 8 | 2 | 2 | 8 | 4,096 |
AMK, amikacin; GEN, gentamicin; KAN, kanamycin; TOB, tobramycin; SPT, spectinomycin.
ClpP is structured as a self-compartmentalized protease, and it requires association to AAA-positive (AAA+) ATPases (ClpA or ClpX) to efficiently process the defective cytoplasmic proteins. The type of AAA+ ATPase determines the substrate specificity of the barrel-like proteolytic complex. The upregulation of clpA was more apparent than that of clpX in response to kanamycin exposure (Fig. 1), and the AG resistance compromise in the clpA mutant was notable (Table 2), suggesting that the ClpA protein, rather than the ClpX protein, may specify the AG-mediated defective proteins. In addition, ClpA proteins can function as independent molecular chaperones even in the absence of ClpP (17), which provides a reasonable explanation why the clpA mutant (KJΔClpA) was more susceptible to AG than was the clpP mutant (KJΔClpP) (Table 2). Distinct from the role of ClpA in the degradation of cytoplasmic defective proteins, HtpX is a membrane-localized proteolytic system. Mutations in membrane proteases leading to increased susceptibility to AG have been reported, such as the FtsH of Pseudomonas aeruginosa (7, 14). Of all the protease mutants surveyed, KJΔClpAΔHtpX displayed the most significant compromise in AG resistance, indicating that ClpA and HtpX are critical cytoplasmic and membrane-bound proteases, respectively, for alleviating AG-mediated stress in S. maltophilia.
The linkage between protease-mediated and SmeYZ efflux pump-mediated AG resistance.
S. maltophilia KJ is a clinical isolate of high resistance to AG (18) (Table 2). The SmeYZ efflux pump has been identified as the main determinant of AG resistance in S. maltophilia KJ (18). To determine whether the decreased AG resistance of KJΔClpAΔHtpX is related to the SmeYZ pump, its expression, with or without kanamycin treatment, was comparatively determined in wild-type KJ and KJΔClpAΔHtpX strains by qRT-PCR. The smeZ transcripts were comparable in both strains, regardless of the presence of kanamycin. This observation suggested that protease system-mediated and SmeYZ pump-mediated AG resistance could have an additive effect. As a further test, a KJΔYZΔClpAΔHtpX mutant was constructed. To our surprise, the KJΔClpAΔHtpX, and KJΔYZΔClpAΔHtpX strains had comparable AG susceptibilities. The differences in AG susceptibility among them were not obvious enough to support the occurrence of an additive effect between protease system-mediated and SmeYZ-mediated AG resistance (Table 2).
The SmeYZ pump is known to be a key determinant for AG resistance in S. maltophilia KJ, since inactivation of smeYZ causes a 16- to 128-fold reduction in the MICs of AG (18) (Table 2). Two interesting results obtained in this study attracted our attention, namely, (i) the expression level of the smeYZ operon in KJΔClpAΔHtpX was similar to that in the wild-type KJ strain and (ii) KJΔClpAΔHtpX and KJΔYZΔClpAΔHtpX displayed comparable AG susceptibility (Table 2). These observations indicate that smeYZ expression in KJΔClpAΔHtpX contributes little to AG resistance, which further supports the applicability of ClpA and HtpX as the aminoglycoside adjuvant targets.
ACKNOWLEDGMENTS
This study was supported by grant MOST 104-2320-B-010-023-MY3 from the Ministry of Science and Technology of Taiwan and grant 40419001 from the Professor Tsuei-Chu Mong Merit Scholarship.
REFERENCES
- 1.Mogk A, Huber D, Bukau B. 2011. Integrating protein homeostasis strategies in prokaryotes. Cold Spring Harb Perspect Biol 3:a004366. doi: 10.1101/cshperspect.a004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bittner LM, Arends J, Narberhaus F. 2017. When, how and why? Regulated proteolysis by the essential FtsH protease in Escherichia coli. Biol Chem 398:625–635. [DOI] [PubMed] [Google Scholar]
- 3.Sakoh M, Ito K, Akiyama Y. 2005. Proteolytic activity of HtpX, a membrane-bound and stress-controlled protease from Escherichia coli. J Biol Chem 280:33305–33310. doi: 10.1074/jbc.M506180200. [DOI] [PubMed] [Google Scholar]
- 4.Thomas JG, Baneyx F. 2000. ClpB and HtpG facilitate de novo protein folding in stressed Escherichia coli cells. Mol Microbiol 36:1360–1370. doi: 10.1046/j.1365-2958.2000.01951.x. [DOI] [PubMed] [Google Scholar]
- 5.Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH. 2007. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol 14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 6.Garneau-Tsodikova S, Labby KJ. 2016. Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. Medchemcomm 7:11–27. doi: 10.1039/C5MD00344J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinz A, Lee S, Jacoby K, Manoil C. 2011. Membrane proteases and aminoglycoside antibiotic resistance. J Bacteriol 193:4790–4797. doi: 10.1128/JB.05133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.George J, Halami PM. 2017. Sub-inhibitory concentrations of gentamicin triggers the expression of acc(6′)le-aph(2′′)la, chaperons and biofilm related genes in Lactobacillus plantarum MCC 3011. Res Microbiol 168:722–731. doi: 10.1016/j.resmic.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Palleroni NJ, Bradbury JF. 1993. Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 1983. Int J Syst Bacteriol 43:606–609. [DOI] [PubMed] [Google Scholar]
- 10.Sánchez MB. 2015. Antibiotic resistance in the opportunistic pathogen Stenotrophomonas maltophilia. Front Microbiol 6:658. doi: 10.3389/fmicb.2015.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang TC, Huang YW, Hu RM, Huang SC, Lin YT. 2009. AmpDI is involved in expression of the chromosomal L1 and L2 beta-lactamases of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 53:2902–2907. doi: 10.1128/AAC.01513-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCTmethod. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Cardoso K, Gandra RF, Wisniewski ES, Osaku CA, Kadowaki MK, Felipach-Neto V, Haus LF, Simão Rde C. 2010. DnaK and GroEL are induced in response to antibiotic and heat shock in Acinetobacter baumannii. J Med Microbiol 59:1061–1068. doi: 10.1099/jmm.0.020339-0. [DOI] [PubMed] [Google Scholar]
- 14.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. 2008. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin JT, Connelly MB, Amolo C, Otani S, Yaver DS. 2005. Global transcriptional response of Bacillus subtilis to treatment with subinhibitory concentrations of antibiotics that inhibit protein synthesis. Antimicrob Agents Chemother 49:1915–1926. doi: 10.1128/AAC.49.5.1915-1926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. 2017. performance standards for antimicrobial susceptibility testing, 27th ed CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 17.Frees D, Brøndsted L, Ingmer H. 2013. Bacterial proteases and virulence. Subcell Biochem 66:161–192. doi: 10.1007/978-94-007-5940-4_7. [DOI] [PubMed] [Google Scholar]
- 18.Lin YT, Huang YW, Chen SJ, Chang CW, Yang TC. 2015. The SmeYZ efflux pump of Stenotrophomonas maltophilia contributes to drug resistance, virulence-related characteristics, and virulence in mice. Antimicrob Agents Chemother 59:4067–4073. doi: 10.1128/AAC.00372-15. [DOI] [PMC free article] [PubMed] [Google Scholar]


