ABSTRACT
The opportunistic Gram-negative pathogen Pseudomonas aeruginosa, known for its intrinsic and acquired antibiotic resistance, has a notorious ability to form biofilms, which often facilitate chronic infections. The evolutionary paths to antibiotic resistance have mainly been investigated in planktonic cultures and are less studied in biofilms. We experimentally evolved P. aeruginosa PAO1 colony biofilms and stationary-phase planktonic cultures for seven passages in the presence of subinhibitory levels (0.1 mg/liter) of ciprofloxacin (CIP) and performed a genotypic (whole-bacterial population sequencing) and phenotypic assessment of the populations. We observed a higher proportion of CIP resistance in the CIP-evolved biofilm populations than in planktonic populations exposed to the same drug concentrations. However, the MICs of ciprofloxacin were lower in CIP-resistant isolates selected from the biofilm population than the MICs of CIP-resistant isolates from the planktonic cultures. We found common evolutionary trajectories between the different lineages, with mutations in known CIP resistance determinants as well as growth condition-dependent adaptations. We observed a general trend toward a reduction in type IV-pilus-dependent motility (twitching) in CIP-evolved populations and a loss of virulence-associated traits in the populations evolved in the absence of antibiotic. In conclusion, our data indicate that biofilms facilitate the development of low-level mutational resistance, probably due to the lower effective drug exposure than in planktonic cultures. These results provide a framework for the selection process of resistant variants and the evolutionary mechanisms involved under the two different growth conditions.
KEYWORDS: Pseudomonas aeruginosa, biofilm, drug resistance evolution
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic Gram-negative pathogen causing acute and persistent infections. Biofilm infections represent a major medical challenge and are implicated in up to 60% of bacterial infections, which complicates diagnosis and treatment (1). Persistent infections caused by biofilm-growing P. aeruginosa strains can either be device or tissue associated (2). Infections of catheters, intratracheal tubes in artificially ventilated patients, and implants are examples of device-associated biofilms. Infections of chronic wounds, bone infections, and lung infections in patients with cystic fibrosis (CF) are examples of tissue-associated biofilms (1, 2). The main reason for the persistence of biofilms implicated in infection is their multifactorial intrinsic tolerance toward antimicrobial agents and the immune system, which has been intensively studied (1). Such persistence of P. aeruginosa in biofilms causes a reoccurrence of the infection and requires repeated courses of antibiotics that may encourage the development of antibiotic resistance.
Biofilms are spatially structured heterogeneous environments due to the impaired diffusion of nutrients and due to the consumption of oxygen by the metabolically active superficial bacterial layers (3). The colony biofilm model chosen in this study was shown to be spatially heterogeneous and able to diversify due to the concentration gradients established within the colony (4). Zhang et al. (5) has convincingly demonstrated that concentration gradients can strongly promote antibiotic resistance development. Therefore, we hypothesize that the biofilm environment might be supportive of resistance development, and there is already some body of evidence suggesting that biofilm growth provides a specific niche for mutational events (6, 7). For instance, endogenous oxidative stress in the biofilm of P. aeruginosa has been shown to promote mutagenesis via mutagenic repair of double-strand breaks in DNA (8).
The major mechanisms of intrinsic and acquired mutational resistance of P. aeruginosa have been elucidated, but the evolutionary trajectories of resistance development have been studied mainly in planktonically grown cultures (9) or in continuous cultures where both planktonic and biofilm populations were maintained (10). One of the purposes of evolution studies is to predict resistance development, which may aid in the design of superior treatment regimes. However, the predictive value of such experiments depends largely on the degree to which they represent relevant in vivo conditions (11). In particular, planktonic cultures of exponentially grown bacterial populations experiencing homogenous environments may not represent an appropriate laboratory model for chronic infections involving bacterial biofilms.
P. aeruginosa infection in the cystic fibrosis lung starts as a focal infection (12) and spreads to new areas of the lung during the progression of the disease We established in this study a biofilm evolution model trying to reproduce this dynamic in which bacteria with a history of biofilm growth and exposure to antibiotics form new biofilms.
In the colony biofilm model used in this study, the bacteria are fed from the surface on which they are growing and therefore can be a model for biofilms associated with soft tissue infections (13).
The fluoroquinolone ciprofloxacin (CIP), the only antipseudomonal drug available for oral administration, is extensively used to treat biofilm infections caused by P. aeruginosa (14). It is used together with colistin or tobramycin for the prevention of chronic lung infections in CF patients with intermittent P. aeruginosa colonization of their lungs and as maintenance therapy in chronically infected patients (15, 16). However, resistance to this antibiotic is easily acquired through mutations (17). In addition to selecting resistant mutants, fluoroquinolones have been shown to promote evolution in planktonic cells by means of different pathways involving oxidative stress, SOS response, or direct DNA interactions (18).
Experimental evolution during exposure to inhibitory concentrations of the antibiotic has repeatedly shown the development of resistance to ciprofloxacin in P. aeruginosa (19, 20). In clinical settings, however, compartments where ciprofloxacin does not reach bacterial inhibitory concentrations are created, such as inside the sputum in the conductive zones of the airways (21). In addition, ciprofloxacin is excreted in sweat and present on the skin at low concentrations for long periods of time (22), with rapid development of resistance in skin bacteria. Therefore, it is relevant to address the role of subinhibitory concentrations of ciprofloxacin in the development of resistance, and we have already published a study on the fast development of resistance in planktonic experimental evolution (23) and in flow cell biofilms using a monitor for mutations in nfxB, the negative regulator of the MexCD-OprJ efflux pump (24); similar findings were reported by Macià et al. (25).
In the present study, we tested the hypothesis that the dynamics of resistance development differs between biofilms and planktonic cultures. To do this, we conducted experimental evolution in both biofilm and planktonic cultures exposed to a constant subinhibitory concentration of CIP (Fig. 1). We demonstrate that a larger CIP-resistant population evolved in biofilms than in planktonic cultures. Further, we sequenced entire bacterial populations evolved as biofilms or as batch cultures to identify common evolutionary pathways among different lineages of the evolved populations. While mutations in genes with similar functions were found across setups, culture-specific variants were also identified, indicating that the mode of growth is an important aspect of resistance development.
FIG 1.
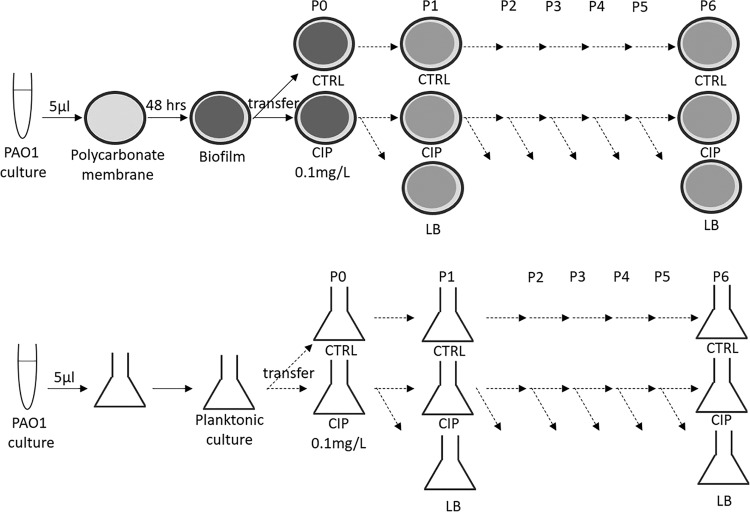
Setup of the experimental evolution in colony biofilms and stationary-phase planktonic cultures. Top, a colony biofilm was formed on polycarbonate membrane from an overnight culture of PAO1. In the step called passage 0 (P0), membranes containing 48-h colony biofilms were transferred to fresh LB plates with either CIP or without CIP for 48 h (CTRL). Every 48 h, the colony biofilms were dispersed, and the bacterial suspension was used to start new biofilms. The bacterial suspensions of CIP biofilms were used to start new biofilms on plates with either CIP or without CIP (LB). The bacterial suspensions of CTRL biofilms were used to start new biofilms on LB plates. This was repeated for 6 passages (P1 to P6) with four replicates per population (A, B, C, and D). Bottom, the same setup was used for planktonic batch cultures. A 48-h stationary culture in LB was started from an overnight culture. From the 48-h culture, two flasks were inoculated: one with 0.1 mg/liter CIP and one in LB (CTRL) (P0). From CIP stationary cultures, every 48 h, new planktonic cultures were established in flasks with CIP or without CIP (LB). From CTRL stationary cultures, every 48 h, new planktonic cultures were established in flasks with LB. This was repeated for 6 passages (P1 to P6), with four replicates per each population (A, B, C, and D).
RESULTS
Evolution of low-level resistance occurred at a higher rate in biofilms than in planktonic cultures.
Analysis of CIP resistance development in CIP-evolved P. aeruginosa PAO1 populations showed that, at the last passage, the size of the resistant subpopulations recovered from the different lineages plated on LB containing 1 mg/liter and 2 mg/liter CIP was significantly larger for biofilm-evolved populations than for planktonic populations (Fig. 2A). As expected, the size of these resistant subpopulations was also significantly larger in CIP-evolved populations than in control populations (Fig. 2A).
FIG 2.
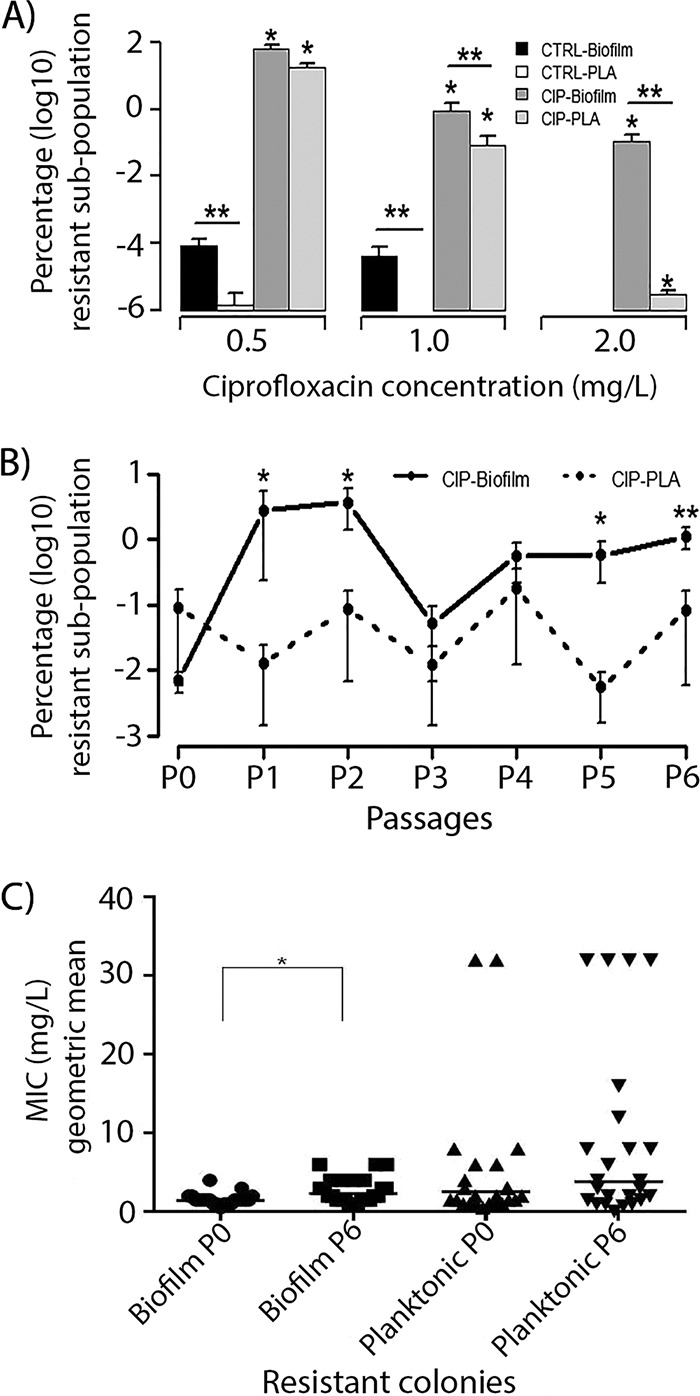
(A) Size of the PAO1 biofilm (biofilm) and planktonic (PLA) populations recovered from 0.5, 1, and 2 mg/liter CIP after evolution in the presence of CIP (0.1 mg/liter) (CIP) or in the absence of antibiotic (CTRL). The values represent the mean (SEM) of eight lineages for each growth condition. *, the size of the resistant population is significantly different in CIP-evolved population compared to the CTRL (P ≤ 0.0001); **, the size of the resistant population was significantly higher in biofilm than in planktonic-evolved populations (P = 0.008 for populations recovered from 1 mg/liter CIP and P = 0.0013 for populations recovered from 2 mg/liter CIP). (B) The development of the resistant population recovered from 1 mg/liter CIP in CIP-evolved biofilm and planktonic cultures during passages (P0 to P6). The values represent the mean (SEM) of eight lineages for each growth condition. *, the sizes of the resistant population in CIP-biofilm populations was significantly higher than that in CIP-planktonic populations at passages 1, 2, 5, and 6 (P = 0.0329, 0.0401, 0.0283, and 0.0084, respectively); **, the size of the resistant population in the last passage was significantly different (P = 0.004) from the first passage (P0). (C) The MIC levels of ciprofloxacin (geometric means) for isolated colonies recovered from CIP-biofilm and planktonic cultures at P0 and P6. The MIC values of 24 independent clones for each growth condition and passage are presented. *, the MIC level was significantly different between the colonies isolated at P0 and P6 in biofilm (P = 0.005).
The dynamic of resistance development under the two growth conditions during the evolution experiment showed that at passages P1, P2, P5, and P6, the size of the resistant subpopulations recovered from 1 mg/liter was larger in biofilms than in planktonic cultures (P = 0.03, 0.04, 0.03, and 0.008, respectively; Fig. 2B). A significant increase in the resistant subpopulation occurred from first (P0) to the last passage (P6) in CIP-evolved biofilm and planktonic populations recovered from 0.5 mg/liter CIP (P < 0.001 and 0.002, respectively). For biofilms, unlike planktonic growth, the increase in resistant population from P6 compared to P0 was also significant for populations recovered from 1 mg/liter CIP (P = 0.004). (The percentages of resistant subpopulations recovered on 0.5 mg/liter, 1 mg/liter, and 2 mg/liter CIP during the seven passages are shown in Table S1A to C in the supplemental material, respectively.) The fluctuations in the sizes of the resistant subpopulations during the experimental evolution, observed especially in the planktonic cultures, might reflect oscillations of resistant mutants in the bacterial population caused by competition between clones with a different fitness cost.
Interestingly, in biofilms, even during evolution in the absence of CIP (control), the size of the resistant subpopulations recovered from 0.5 mg/liter CIP increased significantly (22-fold, P = 0.01) in the last passage compared to the first passage (Table S2). A resistant subpopulation significantly larger than the initial population was observed already after passage 3. The initial inoculum (single colonies used to start the evolution experiments) was tested by population analysis for the presence of preexisting resistant subpopulations, and no growth was detected on 0.5, 1, and 2 mg/liter CIP.
The level of resistance (MIC) of ciprofloxacin for selected resistant colonies from CIP-evolved biofilm populations increased significantly during the experimental evolution from passage 0 to passage 6 (P = 0.005) but not significantly for colonies from planktonic cultures (Fig. 2C). However, clones with higher MICs of ciprofloxacin were isolated at the last passage (P6) in selected resistant colonies from CIP-evolved planktonic cultures compared to isolates from CIP-evolved biofilm cultures (Fig. 2C). The MIC levels of ciprofloxacin for individual colonies from CIP-evolved populations of the different passages are presented in Table S3.
Enrichment of biofilms with resistant subpopulations during passages in the presence of CIP.
Cells dispersed by sonication from a CIP-exposed biofilm were used to initiate new biofilms in the presence of CIP or on LB (see Fig. 1 for experimental setup). After each passage, the sizes of the new biofilm populations established by plating on CIP and LB plates were estimated and used for calculation of an enrichment level (see Materials and Methods).
At the first passage, the population size of new biofilms formed in the presence of 0.1 mg/liter CIP (half the biofilm preventive concentration) was reduced by 1 log10 compared to the biofilms formed on LB. The inhibitory effect of CIP on formation of new biofilms diminished gradually during evolution and disappeared after 4 passages. From passage 4 to passage 6, there was no significant difference between the sizes of the biofilm populations established on CIP and LB growth media.
These results show a gradual enrichment of the CIP-evolved biofilm population with CIP-resistant bacteria during the first three passages (Fig. 3). During batch cultures (see Fig. 1 for experimental setup), the 0.1 mg/liter CIP concentration did not affect the size of the 48-h stationary-phase cultures. However, this CIP concentration decreased the size of a 24-h culture by 1 log10 due to its inhibitory effect on the growth rate (PAO1 doubling time increased from 34.5 ± 0.5 min to 71.8 ± 4.5 min).
FIG 3.
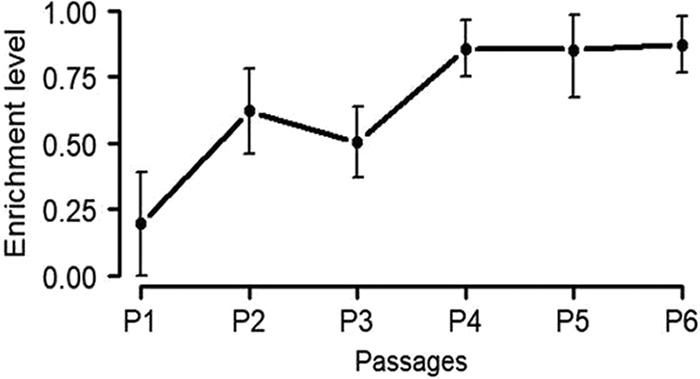
Enrichment of CIP-resistant subpopulation in CIP-evolved biofilm cultures during several passages (passages 1 to 6) in the presence of subinhibitory levels (0.1 mg/liter) of CIP. Enrichment level = 1 − (log10 CFU of CIP population grown on LB plates − log10 CFU of CIP population grown on CIP plates).
Different genetic responses in evolved biofilms and planktonic cultures.
To understand the genetic mechanisms underlying the evolution of antibiotic resistance during the two modes of growth, we sequenced the biofilm or planktonic populations after evolution in the presence or absence of CIP.
The distribution of mutated genes based on the differences in the frequency (%) of each mutation within both biofilm and planktonic CIP and control (CTRL)-evolved populations were clustered according to their function, as mentioned in the Pseudomonas Genome Database (26), and these are shown in Fig. 4. The sequence coverage for each lineage is shown in Table S4, and the list of mutations identified in the populations compared to the reference genome of PAO1 is shown in Table S5.
FIG 4.
Heatmap showing mutations in in CIP and CTRL-evolved populations in both biofilm and planktonic cultures. Genes are clustered according to their function reported in the Pseudomonas Genome Database (26). The intensity of color reflects the frequency of mutations in each replicate population.
We identified a significantly (P = 0.05) higher number of nonsynonymous mutations in the CIP-evolved biofilm and planktonic populations than in the CTRL biofilm and planktonic populations. A total of 53 and 37 mutations were identified in the CIP-evolved biofilm and planktonic populations, respectively. In the control populations evolved without CIP, the numbers of nonsynonymous mutations were 21 in biofilms and 30 in planktonic evolved populations. The mutational spectrum was different in biofilms from that in planktonic cultures. The numbers of indels in CIP-evolved biofilms and planktonic cultures were 25 and 5, the numbers of transitions were 3 and 21, and the numbers of transversions were 25 and 11, respectively. For control biofilm and planktonic populations, the numbers of indels were 8 and 7, the numbers of transitions were 1 and 11, and the numbers of transversions were 12 and 12, respectively. Statistical analysis showed that transitions were significantly more frequent in planktonic populations, while transversion and indels were significantly more frequent in biofilm populations (chi-square, 19 and 27, respectively; P < 0.001).
The CIP-evolved populations were preselected on 1 mg/liter CIP (see Materials and Methods) to ensure that only the resistant subpopulation was assessed.
Mutations in either mexR, nfxB, or mexS, which are regulators of the MexAB-OprM, MexCD-OprJ, and MexEF-OprN efflux pumps, respectively, were frequent in lineages evolved in CIP biofilms, whereas mutations in nalC and nalD (regulators of MexAB-OprM) and CIP target genes gyrA and gyrB were exclusive to the CIP-evolved planktonic populations (Fig. 4 and Table S5). Interestingly, although found in a small proportion of reads in lineages C and D of the CIP-evolved biofilm populations only, mutations in PA1252 encoding a malate dehydrogenase involved in the tricarboxylic acid (TCA) cycle were observed (Table S5). In addition, mutations in a gene encoding NADH dehydrogenase that is also involved in the TCA cycle were only detected in two lineages, A and B, of CIP-evolved biofilm populations, with a frequency of approximately 10% (Fig. 4 and Table S5).
In both biofilm and planktonic CIP-evolved populations, common evolutionary effects of type IV pili were identified as mutations in pil genes (Fig. 4 and Table S5). Whereas different pil genes and different types of mutations were identified under the two growth conditions (Table S5), these genetic changes correlated with a decreased type IV pilus-dependent twitching motility of isolates from the CIP-evolved populations compared to that in the control populations (Table S6).
Among genes mutated in the untreated (control) biofilm populations were lasR and prmC (Table S5). LasR is a regulator of the N-acyl-homoserine lactone quorum-sensing system, which controls different virulence factors. The mutations in lasR in lineages B and C (Table S5) correlated with a decreased amount of protease in isolates from these lineages (Table S7). In lineage D, mutation in prmC encoding a posttranscriptional regulator was observed, a deficiency of which has been shown to lead to a quorum-sensing-independent reduction in virulence factors (27). In untreated planktonic populations, various genes encoding proteins involved in flagellum-dependent motility (swimming) of P. aeruginosa were found to contain mutations (Table S5). In accordance, a decreased swimming motility of colonies from the CTRL planktonic populations was observed (data not shown).
DISCUSSION
Short-term evolution of P. aeruginosa PAO1 biofilms and stationary batch cultures in the presence of subinhibitory concentrations of CIP led to an increase of CIP-resistant subpopulations under both growth conditions. By the end of the experiment, the size of the resistant subpopulation was significantly higher in biofilms than in batch cultures. This might be due to the better adaptive environment offered by the spatially structured and heterogeneous biofilms than the relatively more homogenous environment of a planktonic culture where all cells are subjected to the same drug concentration (28).
Although the relative proportion of resistant cells was higher in CIP-evolved biofilms than in batch cultures, the MIC of ciprofloxacin for isolates from biofilm populations was lower than the MIC for isolates from planktonic cultures. In accordance, the genetic analysis revealed the presence of mutations in CIP target genes gyrA and gyrB conferring high levels of resistance only in CIP-evolved planktonic but not in biofilm populations. For example, in lineage B of CIP-evolved planktonic populations, the gyrA C248T mutation leading to Thr83Ile, one of the most common mutations encountered in CIP-resistant isolates (29), was found with a frequency of 90%, and the CIP-resistant isolates collected from this lineage had a ciprofloxacin MIC of 32 mg/liter (Table S3). A possible explanation might be the higher selective pressure imposed by 0.1 mg/liter CIP on stationary batch cultures than on biofilm-embedded cells. However, it is important to notice that we have previously shown that even at a lower selective pressure of 0.05 mg/liter CIP, gyrA mutations causing high-level ciprofloxacin resistance were selected during planktonic experimental evolution of PAO1 (23), suggesting that the selection of different CIP resistance mechanisms depends on the bacterial mode of growth. This is in agreement with the clinically observed high prevalence of mutations in CIP target genes in P. aeruginosa isolates from acute infections (urinary tract infections and wounds), which are less common in isolates from chronic biofilm infections (20). In addition, our own previous observations of antibiotic resistance levels in P. aeruginosa CF isolates with mucoid (typical biofilm phenotype) and nonmucoid phenotypes (typical planktonic phenotype) showed that the mucoid isolates were more sensitive than the nonmucoid counterparts (30), supporting the results obtained in this experimental study.
The different mutational spectrum observed in biofilms from that of the planktonically evolved population is in accordance with recent studies showing the role played by the environmental conditions for the mutational landscape (31, 32). The association of transversions and indels with the biofilm mode of growth might be due to the biofilm-related stress-induced mutagenesis.
Common adaptations toward mutations in genes involved in the regulation of multidrug efflux pumps mexR (MexAB-OprM), nfxB (MexCD-OprJ), and mexS (MexEF-OprN), with all of them accommodating CIP as the substrate, were observed in CIP-evolved biofilm populations. Mutations in these genes were found in several lineages of the CIP-evolved biofilm populations and conferred a lower level of CIP resistance than the gyrase mutations. For example, in lineage A of the CIP-evolved biofilm populations, the mexS G141T leading to Trp47Cys had reached fixation, and the CIP-resistant isolates collected from this lineage had a CIP MIC of 2 to 4 mg/liter (Table S3). This is in accordance with previously published resistance mechanisms that were found to be selected in PAO1 flow cell biofilms exposed to CIP in pharmacokinetic/pharmacodynamic (PK/PD)-adjusted doses (25). It has previously been shown for P. aeruginosa planktonic cultures that resistant subpopulations selected at low CIP concentrations are associated with the overexpression of efflux pumps, while high CIP concentrations lead to mutations in gyrase and topoisomerase IV (33). Mutations in genes involved in the regulation of MexAB-OprM and MexCD-OprD were also present in some lineages of the planktonic populations, though different types of mutations were identified under the two growth conditions (Table S5). Mutations in the known regulators nalD and nalC of the MexAB-OprM efflux pump, as well as in PA3228, a recently described transcriptional regulator of antibiotic resistance in P. aeruginosa (34), were present only in the CIP-evolved planktonic populations. Interestingly, many of these regulators of multidrug efflux pumps were identified as pathoadaptive genes for P. aeruginosa evolution in the CF lungs (35), suggesting that antibiotic stress is one of the main drivers of evolution in biofilms in vivo.
A common selection against type IV pili, involved in surface attachment/adhesion, cell-cell aggregation, biofilm formation, virulence, and twitching motility of P. aeruginosa (36), was also observed in lineages of CIP-evolved planktonic and biofilm populations as mutations in different pil genes were identified. Type IV pili are among the P. aeruginosa surface molecules that also function as phage receptors for Pf4 prophages of PAO1. These prophages were shown to be activated during biofilm growth and CIP treatment (37). The presence of the filamentous bacteriophage Pf4 in the matrix of the colony biofilm model has been described previously (38). In addition, phage-driven mutations in pil genes have also been described in PAO1 flow cell biofilm populations after 7 days of evolution (39). Although we did not investigate the presence of active phages in this study, we suggest that protection against infection with Pf4 is a possible explanation for the differential mutation pattern of these genes in the CIP-treated samples.
In the control planktonic populations, mutations in swimming motility genes (fli, flagellar assembly) were observed. In P. aeruginosa, swimming motility is driven by a single unipolar flagellum which in PAO1 is dependent on 41 genes encoding structural/assembly and regulatory components of the flagellar organelle (40). In accordance, phenotypic characterization of P. aeruginosa populations during planktonic experimental evolution showed a decrease in the swimming motility of the populations (41), and this was also confirmed in the present study (data not shown). This suggests that during evolution under conditions of good oxygenation and sufficient nutrition, swimming motility is probably an energy-expensive property that is easily lost.
In biofilm, but not planktonic populations, a small resistant subpopulation (85 × 10−4%) developed during evolution in the absence of CIP-selective pressure that might be explained by stress responses related to the biofilm growth (42). In accordance with this finding, the development of antibiotic resistance in biofilms in the absence of antibiotics has been previously reported (43). As the control populations were not enriched for CIP-resistant phenotypes before sequencing, there might have been genetic alterations that we did not detect at the applied sequencing depths.
In the nontreated biofilm population, a common evolutionary path toward the loss of virulence factors was observed. Mutations in the regulatory gene of the N-acyl-homoserine lactone quorum-sensing system (lasR) were identified in a high percentage of reads (more than 90% of reads) in two different lineages and correlated with a reduced production of proteases in isolates from these two lineages (Table S6). A loss of virulence traits in in vitro PAO1 evolution has been reported previously in planktonic cultures (41), showing that the occurrence of this phenotype in evolutionary studies is independent of in vivo factors, e.g., immune selection. Intriguingly, mutations in the quorum-sensing system were not present in the CIP-evolved biofilm populations, indicating a selection for the maintenance of quorum-sensing-regulated factors under evolution at subinhibitory concentrations of CIP. In accordance, we have previously observed increased levels of quorum-sensing molecules in planktonic evolved populations in the presence of 0.05 mg/liter CIP compared to the controls (41). One explanation for these observations could be the protection conferred by QS-regulated catalases and superoxide-dismutases against antibiotic-related oxidative stress (44), as we have shown increased production of reactive oxygen species in P. aeruginosa biofilms exposed to ciprofloxacin (45). Kohanski et al. showed that hydroxyl radicals resulted from the activation of the bacterial TCA cycle are involved in the bactericidal effect of quinolones, and mutants in enzymes involved in the TCA cycle were protected against antibiotic killing (46). In accordance with the metabolic changes caused by exposure to CIP, we found mutations in PA1252 encoding l-malate dehydrogenase, an enzyme of the TCA cycle, in two independent CIP-evolved biofilm populations, probably as a protective mechanism against hydroxyl radical formation and antibiotic killing. Additionally, two mutations in NADH dehydrogenase were only observed in CIP-evolved biofilm populations. A mutation in NADH dehydrogenase is involved in the inhibition of malate dehydrogenase in the TCA cycle (47). It has been shown previously that P. aeruginosa mutants in NADH dehydrogenase are more tolerant to tobramycin (48).
Although we may claim that the evolution of antibiotic resistance studied in an in vitro biofilm model is more representative of the development of resistance during chronic infections, there are several limitations of our study. Among them, the use of a laboratory growth medium and the exposure to constant subinhibitory concentration of CIP are clear simplifications not reflected in the nutrient and pharmacokinetic fluctuations encountered by the biofilms in vivo.
The survival of resistant mutants in a bacterial population depends also on the fitness cost of various mutations (49). An accumulation of compensatory mutations improving the adaptive capacity of the resistant mutants to a particular environment has been observed during evolution in CF sputum medium (19). Our findings suggest that isolates with a history of biofilm growth and environmental exposure to subinhibitory concentrations of antibiotics are more likely to contain mutations causing low-level resistance, which might accelerate the in vivo stepwise development of resistance. From a clinical perspective, this underlines the importance of using combinations of antibiotics to treat biofilm infections in order to prevent mutational resistance and the benefit of achieving high antibiotic concentrations at the infection site in order to prevent the seeding of new biofilms by resistant mutants detached from existing biofilms. For future studies, temporal sequencing and competition studies of the CIP-resistant isolates selected during experimental evolution would deepen our understanding of the quantitative dynamics of resistance development under different conditions.
MATERIALS AND METHODS
Bacterial strains, media, and antibiotics.
P. aeruginosa strain PAO1 (planktonic MIC ciprofloxacin, 0.094 mg/liter) was used to test the development of antibiotic resistance during experimental evolution in the colony biofilm model (4) and in planktonic batch cultures. Both biofilms and planktonic cultures were grown in Luria-Bertani (LB) medium and exposed to CIP (ciprofloxacin hydrochloride; Bayer; Germany), as described below. The MIC for a 48-h stationary PAO1 culture was 0.2 mg/liter, the minimal biofilm inhibitory concentration was 0.5 mg/liter, and the biofilm preventive concentration was 0.2 mg/liter (methods are described in the supplemental material).
Experimental evolution of colony biofilm cultures.
A single colony of P. aeruginosa PAO1 was used to inoculate LB for an overnight culture, incubated aerobically at 37°C. Five microliters of the diluted overnight culture containing approximately 106 cells (optical density at 600 nm [OD600] adjusted to 0.005) was spot-inoculated on the top of polycarbonate membrane filters (Whatman Nuclepore track-etched membranes, 25-mm diameter, 0.2-μm pore size) and incubated at 37°C to form 48-h colony biofilms on LB plates. Membranes containing 48-h colony biofilms were transferred to fresh LB plates with either 0.1 mg/liter CIP (1/5 minimal biofilm inhibitory concentrations) (CIP) or without CIP for 48 h (CTRL) (passage 0 consisted of a 4-day-old biofilm, 2 days on LB, followed by 2 days on LB with CIP). Every 48 h, the CIP and CTRL membranes were transferred to 10-ml tubes with saline, and biofilms were dispersed by vortexing (2 min) and sonication (10 min). After adjusting the OD600 to 0.005, 5 μl of the bacterial suspension of CTRL biofilms was used to start new biofilms on LB plates without antibiotics, and 5 μl of the bacterial suspensions of CIP biofilms was used to start new biofilms on plates with either 0.1 mg/liter CIP or on LB (Fig. 1).
A total of seven exposures (6 passages) to CIP with 4 independent lineages per experiment were performed in duplicate (a total of 8 lineages). Different single colonies were used to start each experiment. After each 48 h, the CFU counts for each biofilm populations were measured, and the disrupted biofilm populations were kept in 20% glycerol at −80°C until further analysis. The enrichment level of bacterial populations with CIP resistance variants able to form biofilm in the presence of CIP was calculated by using the following equation: enrichment level = 1 − (log10 CFU of CIP population grown on LB plates − log10 CFU of CIP population grown on CIP plates).
Experimental evolution of planktonic cultures.
Planktonic experiments were conducted with the same experimental design as implemented with biofilm cultures, achieving similar numbers of bacterial generations in the two modes of growth. Briefly, 5 μl of an overnight culture (OD600 adjusted to 0.005) started from a single colony of PAO1 (the same single colony used to start the evolution experiment in biofilms) was used to inoculate 10 ml LB medium and incubated for 48 h with shaking on an orbital shaker (180 rpm) at 37°C. From the 48-h culture, two flasks were inoculated, one with 0.1 mg/liter CIP (CIP) and one in LB (CTRL) and this was called passage 0 (P0). From CIP stationary cultures, every 48 h, new planktonic cultures were established by passing a volume of 5 μl in flasks with CIP or without CIP (LB). From CTRL stationary cultures, every 48 h, new planktonic cultures were established in flasks with LB. This was repeated for 6 passages (P1 to P6) (Fig. 1). The effect of CIP 0.1 mg/liter on the growth rate of PAO1 was determined by OD600 measurements during 8 h of culture in LB in the presence or absence of 0.1 mg/liter CIP, in triplicate.
Population analysis.
Bacterial populations (100 μl of different dilutions) obtained after sonication and vortexing of membranes containing colony biofilms or of planktonic cultures from each passage and treatment group were plated on LB plates to estimate the size of the bacterial population and on LB plates containing 0.5, 1, and 2 mg/liter CIP and incubated for 48 h to estimate the size of the resistant subpopulations (growing on CIP concentrations higher than the MIC of the strain PAO1). The size of the resistant population was expressed as a percentage of the total bacterial population and was calculated by dividing the CFU per milliliter on CIP by the CFU per milliliter on LB and multiplying by 100.
Three colonies were selected from the plates with the highest CIP concentrations allowing growth. These were passed twice in antibiotic-free medium, and the MIC of CIP was determined by performing an Etest (bioMérieux), according to the manufacturer's instructions.
Genome sequencing of evolved bacterial populations.
The CIP-evolved populations (after 7 passages in the presence of 0.1 mg/liter CIP) from four biofilm lineages and four planktonic lineages were grown on LB plates containing 1 mg/liter CIP for 48 h (four different lineages/condition). The CTRL-evolved populations (after seven passages in the absence of CIP) from biofilm and planktonic cultures were grown on LB plates. All colonies for each population were collected in 3 ml saline (0.9% NaCl) for genomic DNA extraction using the Gentra Puregene yeast/bacteria DNA purification kit. The DNA was prepared for sequencing using the Illumina TruSeq DNA Nano kit and sequenced on an Illumina MiSeq platform, yielding approximately 5 million reads per sample corresponding to a coverage depth of approximately 120× (Table S3). Sequencing reads were mapped to the reference genome of P. aeruginosa PAO1 (GenBank accession no. NC_002516), and single- and multiple-nucleotide variants (SNVs and MNVs, respectively) were called using CLC Genomics Workbench (Qiagen). R (version 3.2.5) was used for further statistical analysis of the mutations detected in each population. Briefly, all mutations occurring in >10% of the reads at any given position were included in the analysis and filtered based on their presence in the CTRL populations (those evolved without CIP). Only mutations in annotated coding regions were included in the analysis.
Statistical analysis.
Graphs and statistical analysis were done using GraphPad Prism 7 software and R (version 3.2.5). We conducted a D'Agostino-Pearson omnibus test to check for normal distribution of the data. Student's t test was used for comparisons among populations with normal distribution and Mann-Whitney test when the data failed to pass the normality test. A chi-square test was conducted to test the distribution of the different types of mutations between bacterial populations. The differences were considered significant when the P value was ≤0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tina Wassermann for excellent technical assistance.
We thank Mostafa M. H. Ellabaan for assistance with detecting large genomic events. M.O.A.S. further acknowledges financial support from the Novo Nordisk Foundation, the Lundbeck Foundation, and the Danish Council for Independent Research.
The financial support of M.N.A. by The Egyptian Ministry of Higher Education is acknowledged.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00320-18.
REFERENCES
- 1.Hoiby N, Bjarnsholt T, Moser C, Bassi GL, Coenye T, Donelli G, Hall-Stoodley L, Hola V, Imbert C, Kirketerp-Moller K, Lebeaux D, Oliver A, Ullmann AJ, Williams C, ESCMID Study Group for Biofilms and Consulting External Expert Werner Zimmerli . 2015. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect 21(Suppl 1):S1–S25. doi: 10.1016/j.cmi.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 2.Lebeaux D, Ghigo JM, Beloin C. 2014. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart PS. 2015. Antimicrobial tolerance in biofilms. Microbiol Spectr 3:. doi: 10.1128/microbiolspec.MB-0010-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walters MC III, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother 47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q, Lambert G, Liao D, Kim H, Robin K, Tung CK, Pourmand N, Austin RH. 2011. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science 333:1764–1767. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 6.Driffield K, Miller K, Bostock JM, O'Neill AJ, Chopra I. 2008. Increased mutability of Pseudomonas aeruginosa in biofilms. J Antimicrob Chemother 61:1053–1056. doi: 10.1093/jac/dkn044. [DOI] [PubMed] [Google Scholar]
- 7.Luján AM, Macia MD, Yang L, Molin S, Oliver A, Smania AM. 2011. Evolution and adaptation in Pseudomonas aeruginosa biofilms driven by mismatch repair system-deficient mutators. PLoS One 6:e27842. doi: 10.1371/journal.pone.0027842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boles BR, Singh PK. 2008. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci U S A 105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacLean RC, Perron GG, Gardner A. 2010. Diminishing returns from beneficial mutations and pervasive epistasis shape the fitness landscape for rifampicin resistance in Pseudomonas aeruginosa. Genetics 186:1345–1354. doi: 10.1534/genetics.110.123083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller C, Kong J, Tran TT, Arias CA, Saxer G, Shamoo Y. 2013. Adaptation of Enterococcus faecalis to daptomycin reveals an ordered progression to resistance. Antimicrob Agents Chemother 57:5373–5383. doi: 10.1128/AAC.01473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sommer MOA, Munck C, Toft-Kehler RV, Andersson DI. 2017. Prediction of antibiotic resistance: time for a new preclinical paradigm? Nat Rev Microbiol 15:689–696. doi: 10.1038/nrmicro.2017.75. [DOI] [PubMed] [Google Scholar]
- 12.de Jong PA, Nakano Y, Lequin MH, Mayo JR, Woods R, Pare PD, Tiddens HA. 2004. Progressive damage on high resolution computed tomography despite stable lung function in cystic fibrosis. Eur Respir J 23:93–97. doi: 10.1183/09031936.03.00006603. [DOI] [PubMed] [Google Scholar]
- 13.McBain AJ. 2009. Chapter 4: in vitro biofilm models: an overview. Adv Appl Microbiol 69:99–132. doi: 10.1016/S0065-2164(09)69004-3. [DOI] [PubMed] [Google Scholar]
- 14.Høiby N. 2011. Recent advances in the treatment of Pseudomonas aeruginosa infections in cystic fibrosis. BMC Med 9:32. doi: 10.1186/1741-7015-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen CR, Pressler T, Høiby N. 2008. Early aggressive eradication therapy for intermittent Pseudomonas aeruginosa airway colonization in cystic fibrosis patients: 15 years experience. J Cyst Fibros 7:523–530. doi: 10.1016/j.jcf.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Frederiksen B, Koch C, Høiby N. 1997. Antibiotic treatment of initial colonization with Pseudomonas aeruginosa postpones chronic infection and prevents deterioration of pulmonary function in cystic fibrosis. Pediatr Pulmonol 23:330–335. doi:. [DOI] [PubMed] [Google Scholar]
- 17.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blázquez J, Couce A, Rodriguez-Beltran J, Rodriguez-Rojas A. 2012. Antimicrobials as promoters of genetic variation. Curr Opin Microbiol 15:561–569. doi: 10.1016/j.mib.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Wong A, Rodrigue N, Kassen R. 2012. Genomics of adaptation during experimental evolution of the opportunistic pathogen Pseudomonas aeruginosa. PLoS Genet 8:e1002928. doi: 10.1371/journal.pgen.1002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong A, Kassen R. 2011. Parallel evolution and local differentiation in quinolone resistance in Pseudomonas aeruginosa. Microbiology 157:937–944. doi: 10.1099/mic.0.046870-0. [DOI] [PubMed] [Google Scholar]
- 21.Bos AC, Passe KM, Mouton JW, Janssens HM, Tiddens HA. 2017. The fate of inhaled antibiotics after deposition in cystic fibrosis: how to get drug to the bug? J Cyst Fibros 16:13–23. doi: 10.1016/j.jcf.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Høiby N, Jarlov JO, Kemp M, Tvede M, Bangsborg JM, Kjerulf A, Pers C, Hansen H. 1997. Excretion of ciprofloxacin in sweat and multiresistant Staphylococcus epidermidis. Lancet 349:167–169. doi: 10.1016/S0140-6736(96)09229-X. [DOI] [PubMed] [Google Scholar]
- 23.Jørgensen KM, Wassermann T, Jensen PO, Hengzuang W, Molin S, Høiby N, Ciofu O. 2013. Sublethal ciprofloxacin treatment leads to rapid development of high-level ciprofloxacin resistance during long-term experimental evolution of Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:4215–4221. doi: 10.1128/AAC.00493-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaborskyte G, Andersen JB, Kragh KN, Ciofu O. 2016. Real-time monitoring of nfxB mutant occurrence and dynamics in P. aeruginosa biofilm exposed to sub-inhibitory concentrations of ciprofloxacin. Antimicrob Agents Chemother 61:e02292–16. doi: 10.1128/AAC.02292-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macià MD, Perez JL, Molin S, Oliver A. 2011. Dynamics of mutator and antibiotic-resistant populations in a pharmacokinetic/pharmacodynamic model of Pseudomonas aeruginosa biofilm treatment. Antimicrob Agents Chemother 55:5230–5237. doi: 10.1128/AAC.00617-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pustelny C, Brouwer S, Musken M, Bielecka A, Dotsch A, Nimtz M, Haussler S. 2013. The peptide chain release factor methyltransferase PrmC is essential for pathogenicity and environmental adaptation of Pseudomonas aeruginosa PA14. Environ Microbiol 15:597–609. doi: 10.1111/1462-2920.12040. [DOI] [PubMed] [Google Scholar]
- 28.Melnyk AH, Wong A, Kassen R. 2015. The fitness costs of antibiotic resistance mutations. Evol Appl 8:273–283. doi: 10.1111/eva.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole K. 2012. Bacterial stress responses as determinants of antimicrobial resistance. J Antimicrob Chemother 67:2069–2089. doi: 10.1093/jac/dks196. [DOI] [PubMed] [Google Scholar]
- 30.Ciofu O, Fussing V, Bagge N, Koch C, Høiby N. 2001. Characterization of paired mucoid/non-mucoid Pseudomonas aeruginosa isolates from Danish cystic fibrosis patients: antibiotic resistance, beta-lactamase activity and RiboPrinting. J Antimicrob Chemother 48:391–396. doi: 10.1093/jac/48.3.391. [DOI] [PubMed] [Google Scholar]
- 31.Maharjan RP, Ferenci T. 2017. A shifting mutational landscape in 6 nutritional states: stress-induced mutagenesis as a series of distinct stress input-mutation output relationships. PLoS Biol 15:e2001477. doi: 10.1371/journal.pbio.2001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shewaramani S, Finn TJ, Leahy SC, Kassen R, Rainey PB, Moon CD. 2017. Anaerobically grown Escherichia coli has an enhanced mutation rate and distinct mutational spectra. PLoS Genet 13:e1006570. doi: 10.1371/journal.pgen.1006570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morero NR, Monti MR, Argarana CE. 2011. Effect of ciprofloxacin concentration on the frequency and nature of resistant mutants selected from Pseudomonas aeruginosa mutS and mutT hypermutators. Antimicrob Agents Chemother 55:3668–3676. doi: 10.1128/AAC.01826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall CW, Zhang L, Mah TF. 2017. PA3225 is a transcriptional repressor of antibiotic resistance mechanisms in Pseudomonas aeruginosa. Antimicrob Agents Chemother 61:e02114-16. doi: 10.1128/AAC.02114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marvig RL, Sommer LM, Jelsbak L, Molin S, Johansen HK. 2015. Evolutionary insight from whole-genome sequencing of Pseudomonas aeruginosa from cystic fibrosis patients. Future Microbiol 10:599–611. doi: 10.2217/fmb.15.3. [DOI] [PubMed] [Google Scholar]
- 36.Leighton TL, Buensuceso RN, Howell PL, Burrows LL. 2015. Biogenesis of Pseudomonas aeruginosa type IV pili and regulation of their function. Environ Microbiol 17:4148–4163. doi: 10.1111/1462-2920.12849. [DOI] [PubMed] [Google Scholar]
- 37.Hui JGK, Mai-Prochnow A, Kjelleberg S, McDougald D, Rice SA. 2014. Environmental cues and genes involved in establishment of the superinfective Pf4 phage of Pseudomonas aeruginosa. Front Microbiol 5:654. doi: 10.3389/fmicb.2014.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Secor PR, Sweere JM, Michaels LA, Malkovskiy AV, Lazzareschi D, Katznelson E, Rajadas J, Birnbaum ME, Arrigoni A, Braun KR, Evanko SP, Stevens DA, Kaminsky W, Singh PK, Parks WC, Bollyky PL. 2015. Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe 18:549–559. doi: 10.1016/j.chom.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McElroy KE, Hui JG, Woo JK, Luk AW, Webb JS, Kjelleberg S, Rice SA, Thomas T. 2014. Strain-specific parallel evolution drives short-term diversification during Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci U S A 111:E1419–E1427. doi: 10.1073/pnas.1314340111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jyet J, Ramphal R. 2008. Flagella and pili of Pseudomonas aeruginosa, p 85–108. In Rehm BHA. (ed), Pseudomonas: model organism, pathogen, cell factory. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 41.Wassermann T, Meinike JK, Ivanyshyn K, Bjarnsholt T, Khademi SM, Jelsbak L, Høiby N, Ciofu O. 2016. The phenotypic evolution of Pseudomonas aeruginosa populations changes in the presence of subinhibitory concentrations of ciprofloxacin. Microbiology 162:865–875. doi: 10.1099/mic.0.000273. [DOI] [PubMed] [Google Scholar]
- 42.Torres-Barceló C, Kojadinovic M, Moxon R, MacLean RC. 2015. The SOS response increases bacterial fitness, but not evolvability, under a sublethal dose of antibiotic. Proc Biol Sci 282:20150885. doi: 10.1098/rspb.2015.0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyerman JG, Ponciano JM, Joyce P, Forney LJ, Harmon LJ. 2013. The evolution of antibiotic susceptibility and resistance during the formation of Escherichia coli biofilms in the absence of antibiotics. BMC Evol Biol 13:22. doi: 10.1186/1471-2148-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dwyer DJ, Kohanski MA, Collins JJ. 2009. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol 12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jensen PØ, Briales A, Brochmann RP, Wang H, Kragh KN, Kolpen M, Hempel C, Bjarnsholt T, Høiby N, Ciofu O. 2014. Formation of hydroxyl radicals contributes to the bactericidal activity of ciprofloxacin against Pseudomonas aeruginosa biofilms. Pathog Dis 70:440–443. doi: 10.1111/2049-632X.12120. [DOI] [PubMed] [Google Scholar]
- 46.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prüss BM, Nelms JM, Park C, Wolfe AJ. 1994. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J Bacteriol 176:2143–2150. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amini S, Hottes AK, Smith LE, Tavazoie S. 2011. Fitness landscape of antibiotic tolerance in Pseudomonas aeruginosa biofilms. PLoS Pathog 7:e1002298. doi: 10.1371/journal.ppat.1002298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andersson DI, Hughes D. 2014. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12:465–478. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.