Abstract
Objectives
There are limited data regarding efavirenz pharmacogenetics in admixed populations. The Brazilian population is highly admixed. In a Brazilian cohort, we sought to characterize associations between efavirenz adverse effects (all-cause and CNS) and polymorphisms in seven genes known or suspected to affect efavirenz metabolism and transport.
Methods
We studied 225 HIV-positive individuals who had been prescribed efavirenz-containing regimens at a hospital in Rio de Janeiro, Brazil. Eighty-nine cases had efavirenz adverse effects, including 43 with CNS adverse effects, while 136 controls had no adverse effect of any antiretroviral after treatment for at least 6 months. A total of 67 candidate polymorphisms in ABCB1, CYP2A6, CYP2B6, CYP3A4, CYP3A5, NR1I2 and NR1I3 genes were selected for association analysis. Admixture was assessed using 28 ancestry-informative polymorphisms previously validated for the Brazilian population. Associations were evaluated with logistic regression models adjusted for sex and genetic ancestry.
Results
There was extensive African, European and Native American admixture in the cohort. Increased all-cause adverse effects were associated with the CYP2B6 genotype combination 15582CC-516TT-983TT (OR = 7.26, P = 0.003) and with the CYP2B6 slow metabolizer group 516TT or 516GT-983CT (OR = 3.10, P = 0.04). CNS adverse effects were nominally associated with CYP3A4 rs4646437 (OR = 4.63, P = 0.014), but not after adjusting for multiple comparisons.
Conclusions
In a highly admixed Brazilian cohort, the CYP2B6 slow metabolizer genotype was associated with an increased risk of efavirenz adverse effects.
Introduction
Worldwide, ∼37 million people are living with HIV-1, of whom nearly 50% are receiving ART.1 The most recent WHO guidelines recommend efavirenz in combination with tenofovir and lamivudine as first-line ART.2 Intolerance and adverse effects are primary reasons for first regimen discontinuation. Nearly one-half of patients prescribed efavirenz experience adverse effects, particularly CNS disturbances (e.g. insomnia, dizziness, depression, psychosis and suicidal ideation), which has been related to higher plasma efavirenz concentrations.3–6 In a Brazilian cohort, efavirenz CNS adverse effects were the third most frequent reason for treatment discontinuation.4
Cytochrome P450 (CYP2) B6 is primarily responsible for efavirenz metabolism, with accessory pathways involving CYP2A6 and possibly CYP3A4/5.7,8 The CYP genes are transcriptionally regulated by nuclear receptor genes PXR, CAR and others.9 Efavirenz is also directly glucuronidated by uridine 5′-diphospho-glucuronosyltransferase (UGT) 2B7.10,11 SNPs in genes that encode these enzymes, especially CYP2B6, predict higher plasma efavirenz concentrations.12–15
In CYP2B6, 516G→T (rs3745274) has been most extensively studied for associations with increased plasma efavirenz exposure.12,16–19 In addition, CYP2B6 983T→C (rs28399499) and 15582C→T (rs4803419) have been associated with increased plasma efavirenz exposure.20–22 Associations with plasma efavirenz concentrations have also been reported in different populations with SNPs in ABCB1, which encodes the efflux transporter P-glycoprotein.23–27 The CYP2B6 SNPs that predict increased plasma efavirenz concentrations have also been associated with efavirenz CNS adverse effects.12,17,28
Polymorphism frequencies and effects on gene expression vary among populations depending on genomic structure. Consequently, associations in one population may not translate to others. In Brazil there is marked ancestry admixture, defined as what occurs when previously isolated populations interbreed. This raises the possibility that genetic associations reported in other less admixed populations may not translate to Brazil.29 In addition, there are limited studies of efavirenz pharmacogenetics in Brazilian populations. A previous study from our group did not find associations between SNPs in absorption, distribution, metabolism and excretion (ADME) genes and adverse effects of ART regimens containing either efavirenz or nevirapine, but that study only considered all-cause rather than specific adverse effects.30
The aim of the present study was to characterize, among Brazilians, associations between SNPs in genes that are known or suspected to affect efavirenz disposition and risk of efavirenz all-cause and CNS adverse effects.
Methods
Ethics statement
This study was approved by the Research Ethics Committee of Hospital Universitário Gaffreé e Guinle (HUGG), number 94/2011. All procedures were performed in accordance with the guidelines of the Helsinki Declaration. Written informed consent was obtained from all subjects.
Study subjects
The present study was based on retrospective review of medical records and included a total of 225 HIV-1-positive individuals who received routine clinic follow-up at the Clinical Immunology Service of HUGG. Individuals at least 18 years of age were eligible regardless of sex. Additional eligibility criteria included complete documentation of ART regimens in the medical record, previous or current use of an efavirenz-containing regimen, documentation of reason for ART change and no treatment for TB or viral hepatitis. Individuals were excluded for adverse drug effects that occurred during pregnancy and for adverse effects that were attributed to antiretrovirals other than efavirenz. Analyses were performed with all eligible participants (n = 225). Those who developed any adverse effects due to efavirenz were defined as cases (n = 89) while those with no documented adverse effect of any antiretroviral for at least 6 months were defined as controls (n = 136). Additional analyses considered only the subset of cases with CNS adverse effects (n = 43). Adverse effects were based on self-report by individuals during clinical follow-up. Severity of adverse effects was not reliably documented in medical records. Among controls, the median time on an efavirenz-containing regimen was 63 months. Among cases, the median time on an efavirenz-containing regimen was 5.5 months. Data were obtained by retrospective review of medical records.
Considering a minimum allele frequency of 1% and a 50% incidence of adverse effects due to efavirenz, the minimum OR value to achieve 80% power in our sample size (n = 225) would be OR = 3. For analysis including only CNS adverse effects (n = 179), with a medium incidence of 30%, the minimum OR value to reach 80% power would be OR = 4.5.
Selection of polymorphisms for analysis
Candidate genes were ABCB1, CYP2A6, CYP2B6, CYP3A4, CYP3A5, NR1I2 and NR1I3, in which 67 SNPs were selected for genotyping largely based on literature review (Table S1, available as Supplementary data at JAC Online).7–9,11,14 The SNPper tool was used to search for potentially informative SNPs in coding and non-coding regulatory regions. We also used the HapMap data bank to identify tagging SNPs and increase gene coverage. For HapMap, we used a minor allele frequency (MAF) of 0.05 in CEU (Utah residents with Northern and Western European ancestry) or YRI (Yoruba in Ibadan, Nigeria) populations and an r2 cut-off of 0.8 as parameters. To adjust for genetic ancestry, we used a panel of 28 ancestry-informative SNPs previously validated for the Brazilian population.31
DNA extraction and genotyping
Buffy coat samples were obtained from whole blood by centrifugation at 3000 g for 10 min and genomic DNA was extracted with a salting-out method. The CYP2A6, CYP3A4, CYP3A5, NR1I2 and NR1I3 SNPs and ancestry-informative markers were genotyped with TaqMan® Open Array® technology, using the QuantStudio™ 12K Flex Real-Time PCR System (Thermo Fisher Scientific, MA, USA), according to the manufacturer’s instructions. The CYP2B6 SNPs (except rs3745274, rs4803419, rs4803420 and rs7260525) were genotyped with SNaPshot® (Thermo Fisher Scientific), according to the manufacturer’s instructions. Purified products underwent capillary electrophoresis on an ABI3130 Genetic Analyzer (Thermo Fisher Scientific) using the standard fragment analysis protocol. GeneMapper software (version 4.0 Thermo Fisher Scientific) was used for genotyping. CYP2B6 rs3745274 and rs4803419 were genotyped with the TaqMan® Drug Metabolism SNP Genotyping Assay (Thermo Fisher Scientific), following the manufacturer’s instructions. CYP2B6 rs4803420 and rs7260525, and ABCB1 markers were genotyped with a Sequenom MassARRAY® iPLEX platform (Agena Bioscience™, CA, USA) at Vanderbilt Technologies for Advanced Genomics (VANTAGE) in Nashville, TN, USA. Data were analysed using Typer software (Agena Bioscience™, CA, USA). Other genotyping was done at Laboratório de Virologia Molecular, Departamento de Genética, Instituto de Biologia, Universidade Federal do Rio de Janeiro.
Estimates of genetic ancestry
Proportions of African, European and Native American genetic ancestries were estimated using Structure software, version 2.3.1.32–35 Proportions of each ancestry were estimated under an admixture model using data from European and African populations of the 1000 Genomes Project as a reference. Native American ancestry was estimated using an admixed Amazonian population from Santa Isabel do Rio Negro as a reference, all of whom reported recent indigenous ancestry.36
Statistical analyses
Statistical analyses were performed using R software (version 2.13.0) and the genetics, gap, SNPassoc, haplo.stats, LDheatmap, grid and coin packages.37 Deviations from Hardy–Weinberg equilibrium (HWE) were assessed by the χ2 test. Pairwise linkage disequilibrium (LD) patterns were determined using r2 statistics (cut-off of r2 ≥ 0.8). Stepwise logistic regression analysis was performed to select covariates to include in the model as possible confounders, including age at the start of efavirenz-containing regimen, sex and genetic ancestry. The variables sex, African ancestry and Native American ancestry were considered to be informative and were included in the model. Sex proportions between cases and controls were compared using the χ2 test. Age and genetic ancestry were compared using the Wilcoxon rank sum test.
Logistic regression models were performed to identify associations between SNPs and adverse effect outcomes. We primarily considered additive genetic models and secondarily considered dominant and recessive models. Bonferroni adjustment was applied to minimize type I error (P value cut-off for 46 SNPs = 1.1 × 10−3). Haplotype analyses were performed for SNPs with nominal P value <0.05. Haplotype frequencies were estimated through maximum likelihood and compared between cases and controls using logistic regression models, as described for SNP analyses.
CYP2B6 genotype levels were defined as described elsewhere, with each increasing plasma efavirenz concentration stratum predicted by specific combinations of rs4803419 (15582C→T), rs3745274 (516G→T) and rs28399499 (983T→C) alleles.20 We designated the 10 strata defined by these genotypes as level 1 to level 10, with level 1 being the composite genotype associated with the lowest concentrations and level 10 being associated with the highest concentrations. Levels 1, 2 and 3 are defined by 15582CC, CT and TT, respectively, together with 516GG and 983TT homozygosity. Levels 4 and 5 are defined by 516GT and 983CT heterozygosity, respectively, together with 15582CC homozygosity. Levels 6 and 7 are defined by 516GT and 983CT heterozygosity, respectively, together with 15582CT heterozygosity. Level 8 is defined by 516TT with 983TT, level 9 is defined by 516GT with 983CT and level 10 is defined by 516GG with 983CC, all together with 15582CC homozygosity.
We also assigned CYP2B6 genotypes into three metabolizer groups: extensive, intermediate and slow. CYP2B6 levels 1 and 2 were defined as extensive metabolizers, CYP2B6 levels 3, 4, 5, 6 and 7 were defined as intermediate metabolizers and CYP2B6 levels 8, 9 and 10 were defined as slow metabolizers. CIs were determined using the Wald modified method. Associations between CYP2B6 genotype levels and CNS adverse effects were determined by logistic regression models considering CYP2B6 genotype level both as an ordinal variable and comparing each level separately to the normal metabolizer genotype. CYP2B6 genotype level was also included as a covariate in analyses for associations between other candidate SNPs and efavirenz adverse effects.
Results
Patient characteristics
The distribution of sex, age at the start of efavirenz-containing regimen and genetic ancestry in all-cause cases, CNS cases and controls is presented in Table 1. The cohort was highly admixed, with the average individual having ∼50% European, ∼45% African and ∼13% Native American genetic ancestry. Covariates did not differ significantly between cases and controls, although there were somewhat fewer males among all-cause cases versus controls.
Table 1.
Characteristics of study participants
| All-cause cases (N = 89) | CNS cases (N = 43) | Controls (N = 136) | Pc | Pd | |
|---|---|---|---|---|---|
| Sexa, n (frequency) | |||||
| male | 43 (0.48) | 26 (0.60) | 84 (0.62) | 0.064 | 0.978 |
| female | 46 (0.52) | 17 (0.40) | 52 (0.38) | ||
| Age (years), mean ± SDb | 41.1 ± 11.4 | 40.9 ± 11.9 | 40.5 ± 10.3 | 0.840 | 0.940 |
| Genetic ancestry (%), mean ± SDb | |||||
| European | 48 ± 28 | 41 ± 27 | 51 ± 29 | 0.540 | 0.071 |
| African | 40 ± 26 | 45 ± 28 | 36 ± 28 | 0.400 | 0.110 |
| Native American | 12 ± 14 | 14 ± 16 | 13 ± 14 | 0.640 | 0.950 |
Comparisons were performed using the χ2 test.
Comparisons were performed using the Wilcoxon rank sum test.
P value based on all-cause cases versus controls.
P value based on CNS cases versus controls.
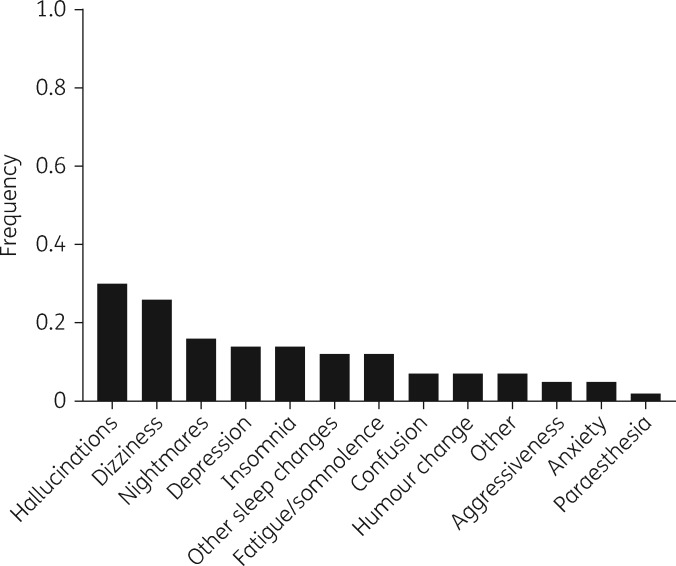
CNS adverse effects represented 48% of all adverse effect cases, with the most common symptoms being hallucinations (30%), dizziness (26%) and nightmares (16%) (Figure 1). Among the non-CNS cases (n = 46), 19% developed a rash, 11% were simply described as intolerant, 6% had malaise and 10% were unique cases of diabetes mellitus, dyslipidaemia, epigastralgia or hypertriglyceridaemia. Specific symptoms were not described for 24 individuals.
Figure 1.
Distribution of reported efavirenz CNS adverse effects among 43 cases. Some cases had more than one type of efavirenz CNS adverse effect documented. Each type of CNS adverse effect was counted separately for such individuals.
Genotypes including HWE and LD
We attempted to genotype 67 SNPs in seven drug metabolism and transport genes. Of these, 5 that deviated from HWE were excluded from analysis, as were an additional 13 monomorphic loci. Of the remaining SNPs, three pairs were in LD at r2 ≥ 0.8 in NR1I2 (rs1523127 and rs1523130) and ABCB1 (rs4148740 and rs10225473; rs3789244 and rs1128503). Only the most frequent SNP of each pair was maintained. The remaining 46 SNPs were included in association analyses (Table S1).
Association between CYP2B6 genotype levels and efavirenz adverse effects
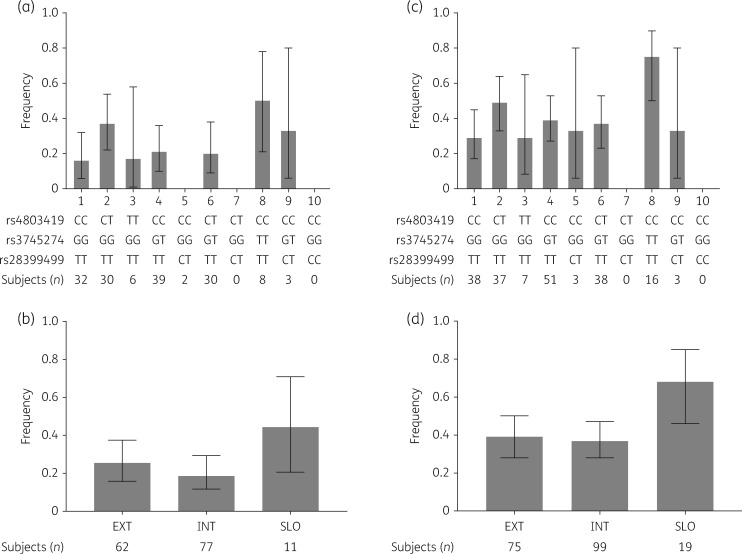
We assessed whether CYP2B6 genotype levels previously reported to predict plasma efavirenz concentrations were associated with efavirenz adverse effects.20 We found no significant association between CYP2B6 genotype level, considered as an ordinal variable, and CNS adverse effects by an unadjusted logistic regression model (OR = 1.06, P = 0.50) or with adjustment for sex, African ancestry and Native American ancestry (OR = 1.04, P = 0.67). Likewise, we found no significant association between CYP2B6 genotype level and all-cause efavirenz adverse effects in an unadjusted model (OR = 1.11, P = 0.14) or after adjustment (OR = 1.10, P = 0.13). Proportions of adverse effect cases within each genotype level for CNS adverse effects and for all-cause adverse effects are presented in Figure 2(a and c).
Figure 2.
Distribution of CYP2B6 level frequencies in cases. CIs were calculated using the modified Wald method. (a) Frequencies of CYP2B6 metabolizer genotype levels in CNS cases based on 10 possible levels (n = 43). (b) Frequencies of CYP2B6 metabolizer genotype levels in CNS cases collapsed into extensive, intermediate and slow metabolizer groups. (c) Frequencies of CYP2B6 metabolizer genotype levels in all-cause cases based on 10 possible levels (n = 89). (d) Frequencies of CYP2B6 metabolizer genotype levels in all-cause cases collapsed into extensive, intermediate and slow metabolizer groups. EXT, extensive metabolizer genotypes; INT, intermediate metabolizer genotypes; SLO, slow metabolizer genotypes. CYP2B6 genotype levels were collapsed as follows: extensive (levels 1 and 2), intermediate (levels 3, 4, 5, 6 and 7) and slow (levels 8, 9 and 10).20
In association analyses that compared each CYP2B6 genotype level pairwise to genotype level 1, which predicts the lowest plasma efavirenz concentrations, there was an association between the diplotype rs4803419CC-rs3745274TT-rs28399499TT (level 8) and increased risk of CNS adverse effects in an unadjusted model (OR = 8.67, P = 0.021). This association was marginally significant after adjustment for sex, African ancestry and Native American ancestry (OR = 5.46, P = 0.05). We also found an association between level 8 and all-cause efavirenz adverse effects in both an unadjusted model (OR = 7.26, P = 0.003) and after adjustment (OR = 6.83, P = 0.005).
We have also performed data analysis after collapsing CYP2B6 genotype levels into extensive, intermediate and slow metabolizer groups. Individuals with slow metabolizer genotypes were more likely to be among CNS adverse effect cases, although this difference was not statistically significant (Figure 2b). Using extensive metabolizers as reference, we found no significant association between the metabolizer group and CNS adverse effects in an unadjusted model and after adjusting for sex, African ancestry and Native American ancestry. The intermediate group had an OR of 0.68 (P = 0.34) and the slow group an OR of 2.29 (P = 0.23) in the adjusted model. However, when all-cause adverse effect cases were considered, we found a significant association between slow metabolizer status and increased risk for adverse effects both in unadjusted (OR = 3.44, P = 0.024) and adjusted (OR = 3.10, P = 0.04) analyses (Figure 2d).
Adverse effects and polymorphisms beyond CYP2B6
In logistic regression models that considered only CNS adverse effect cases, two SNPs in CYP3A4, rs4646437 (OR = 4.63, P = 0.014, additive model) and rs2740574 (OR = 2.86, P = 0.116, additive model; OR = 2.62, P = 0.043, recessive model), were nominally associated with increased risk. There was also a nominal association with ABCB1 rs1882478 (OR = 3.81, P = 0.040, TT genotype, additive model). After adjusting for sex, African ancestry and Native American ancestry, only CYP3A4 rs4646437 remained associated with efavirenz CNS adverse effects (OR = 4.95, P = 0.059, additive model; OR = 3.36, P = 0.029, dominant model) (Table 2). This association was not statistically significant after adjusting for multiple comparisons.
Table 2.
Significant associations between SNPs and efavirenz CNS adverse effects
| Genotype | Casesa | Controlsa | Unadjusted model OR (95% CI) | Adjusted model OR (95% CI)c |
|---|---|---|---|---|
| SNP rs1882478 (ABCB1) | ||||
| CC | 7 (0.17) | 41 (0.31) | reference | reference |
| CT | 22 (0.52) | 72 (0.54) | 1.79 (0.70–4.55) | 1.50 (0.54–4.17) |
| TT | 13 (0.31) | 20 (0.15) | 3.81 (1.32–11.02) | 2.62 (0.80–8.65) |
| 42 | 133 | P = 0.040b | P = 0.267b | |
| CC-CT | 29 (0.69) | 113 (0.85) | reference | reference |
| TT | 13 (0.31) | 20 (0.15) | 2.53 (1.13–5.69) | 1.94 (0.79–4.80) |
| P = 0.027d | P = 0.156d | |||
| SNP rs4646437 (CYP3A4) | ||||
| GG | 6 (0.17) | 47 (0.41) | reference | reference |
| GA | 16 (0.46) | 46 (0.40) | 2.72 (0.98–7.58) | 3.09 (0.96–9.95) |
| AA | 13 (0.37) | 22 (0.19) | 4.63 (1.55–13.79) | 4.95 (1.22–20.05) |
| 35 | 115 | P = 0.014b | P = 0.059b | |
| GG | 6 (0.17) | 47 (0.41) | reference | reference |
| GA-AA | 29 (0.83) | 68 (0.59) | 3.34 (1.29–8.68) | 3.36 (1.07–10.59) |
| P = 0.007d | P = 0.029d | |||
| SNP rs2740574 (CYP3A4) | ||||
| TT | 14 (0.39) | 60 (0.51) | reference | reference |
| TC | 12 (0.33) | 42 (0.36) | 1.22 (0.52–2.91) | 1.03 (0.39–2.70) |
| CC | 10 (0.28) | 15 (0.13) | 2.86 (1.06–7.68) | 2.02 (0.60–6.79) |
| 36 | 117 | P = 0.116b | P = 0.435b | |
| TT-TC | 26 (0.72) | 102 (0.87) | reference | reference |
| CC | 10 (0.28) | 15 (0.13) | 2.62 (1.05–6.49) | 1.98 (0.71–5.52) |
| P = 0.043d | P = 0.198d |
Results are shown as n (frequency) for SNP genotypes.
Overall P value for additive model (2 degrees of freedom).
Results adjusted for sex, African ancestry and Native American ancestry.
Overall P value for dominant or recessive model (1 degree of freedom).
Logistic regression analyses were also adjusted for CYP2B6 genotype level to see whether this affected the apparent associations with CYP3A4 rs4646437, CYP3A4 rs2740574 and/or ABCB1 rs1882478. Such adjustment did not substantially change ORs or P values of these associations (data not shown).
No associations were observed when all-cause efavirenz adverse effect cases were considered either before or after adjustment. The lowest P value was for CYP2A6 rs28399433 after adjustment for sex, African ancestry and Native American ancestry (P = 0.301).
Haplotype analysis
We tested for associations between CYP3A4 rs4646437-rs2740574 haplotypes noted above and efavirenz CNS adverse effects, as these two SNPs are in linkage equilibrium and were separately associated with the same risk effect. The haplotype containing both minor alleles, rs4646437A and rs2740574C, was associated with CNS adverse effects in both unadjusted (OR = 2.04, P = 0.012) and adjusted (OR = 2.08, P = 0.045) models. This haplotype did not have a stronger association with CNS adverse effects than each SNP considered separately (Table 3).
Table 3.
Association between CYP3A4 rs4646437 and rs2740574 and CNS adverse effects due to efavirenz
| Haplotype rs4646437/rs2740574 | Controlsa | Casesa | Unadjusted model OR (95% CI) | Adjusted model OR (95% CI)b |
|---|---|---|---|---|
| G/T | 0.6 | 0.39 | reference | reference |
| A/C | 0.29 | 0.43 | 2.04 (1.17–3.56; P = 0.012) | 2.08 (1.02–4.26; P = 0.045) |
| A/T | 0.1 | 0.16 | 2.70 (1.09–6.70; P = 0.033) | 2.94 (1.06–8.15; P = 0.04) |
| G/C | 0.01 | 0.02 | 1.99 (0.18–21.42; P = 0.571) | 1.83 (0.15–22.02; P = 0.634) |
Haplotype frequencies were estimated by maximum likelihood.
Results adjusted for sex, African ancestry and Native American ancestry.
Discussion
Research into the pharmacogenetics of efavirenz among Brazilians is limited. The present study characterized, in a highly admixed Brazilian population, associations between efavirenz adverse effects (both all-cause and CNS adverse effects) and SNPs in genes that are known or suspected to affect efavirenz disposition. In unadjusted analyses, CYP2B6 genotype rs4803419CC-rs3745274TT-rs28399499TT (genotype level 8) was significantly associated with efavirenz all-cause adverse effects. The CYP2B6 slow metabolizer group (rs3745274TT or rs3745274GT-rs28399499CT) was also significantly associated with all-cause efavirenz adverse effects. In addition, CYP3A4 rs4646437 was nominally associated with efavirenz CNS adverse effects, but not after adjusting for multiple comparisons.
The CYP2B6 516G→T (rs3745274) SNP is widely reported to be associated with efavirenz phenotypes, including higher efavirenz plasma concentrations and adverse effects.12,16–19,28 However, previous reports suggested that this effect may not occur in Brazilian populations.30,38,39 In a study of 50 individuals from the south of Brazil, Müller et al.38 found no association between rs3745274 and CNS adverse effects due to efavirenz. A limitation of that study was the small sample size. In a previous study by our group, rs3745274 was not associated with intolerance to regimens containing either efavirenz or nevirapine in 395 individuals from Rio de Janeiro, Brazil.30 Lack of focus on a specific intolerance could have limited our power to find an association. Similarly, Coelho et al.39 found no association between rs3745274 and treatment regimen failure.
In contrast to previous reports from Brazil, our analyses considered CYP2B6 genotype levels based on combinations of rs3745274, rs28399499 and rs4803419. We found that CYP2B6 level 8 (rs4803419CC-rs3745274TT-rs28399499TT) was associated with increased risk of efavirenz all-cause adverse effects (P = 0.003). In addition, when considering classical metabolizer groups, we also found an association between the slow metabolizer group and efavirenz all-cause adverse effects (P = 0.04).
These associations were not significant when considering only CNS adverse effect cases, possibly due to a smaller sample size in that analysis. An additional limitation of our study is the use of a broad case definition, since different types of efavirenz adverse effects can be distinctively associated with the SNPs analysed. Therefore, the analysis of all cases as a single group may have reduced our power to detect associations that might be specific for each adverse effect. Associations between long-term efavirenz use and performance on formal neurocognitive testing have been reported.40,41 In the present retrospective study, we could not test for associations with neurocognitive performance because such data were not available.
Our findings agree with previous studies that showed associations between CYP2B6 variants and increased risk for specific efavirenz adverse effects or discontinuation of ART due to CNS symptoms.42,43 To our knowledge, the present study is the first to show associations between CYP2B6 genotype and efavirenz adverse effects in a Brazilian population. We cannot explain the stronger association between CYP2B6 level 8 (homozygosity for 516TT) as compared with level 9 (heterozygosity for 516GT and 983CT), although it might be due to the very few individuals in level 9.
The enzyme CYP3A4 is responsible for metabolizing the largest number of medications and plays a minor role in efavirenz metabolism.7,44 In our study, CYP3A4 rs4646437 was nominally associated with increased risk of CNS adverse effects, but not after adjusting for multiple comparisons. We also observed an association with CYP3A4 rs2740574 in an unadjusted model. The SNP CYP3A4 rs4646437 has been reported to be associated with decreased efavirenz clearance independent of CYP2B6 genotype, which could increase risk for efavirenz adverse effects.45 In the published literature, we found no clear associations reported for rs2740574. An early study suggested that this SNP could be weakly associated with plasma efavirenz concentrations, but a subsequent study did not replicate this finding.12,18 Furthermore, Haas et al.12 did not find an association between rs2704574 and efavirenz CNS adverse effects.
Although P-glycoprotein is not described as an efavirenz transporter, associations have been reported between ABCB1 SNPs and efavirenz-related outcomes.18,23,26,46 In the present study, among 21 candidate SNPs in ABCB1, we found an association with rs1882478 in an unadjusted model. To our knowledge, no prior studies have shown associations of ABCB1 rs1882478 with efavirenz phenotypes, although this SNP was reported to be associated with decreased hepatic ABCB1 expression among liver transplant donors in China.47
The marked ancestry admixture in the Brazilian population may affect SNP frequencies and LD patterns. Consequently, findings from other well-defined ethnic groups may not reliably translate to Brazilians. It is thus important that pharmacogenetic associations reported in other populations be replicated in Brazil, as was done in the present study. Better understanding of pharmacogenetic mechanisms that underlie adverse effects of antiretroviral drugs may help to identify genetic predictors for these outcomes and ultimately lead to better-tolerated and more effective HIV therapies.
Supplementary Material
Acknowledgements
We are grateful to Cara Sutcliffe and the VANTAGE team at Vanderbilt University Medical Center. We also thank Diana Mariani and Lidia Boullosa for their technical assistance. We thank the Consellho Nacional de Desenvolvimento e Pesquisa (CNPq) and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for a student fellowship. This work is part of T. B. d. A.’s requirements for obtaining a PhD degree from the Graduate Programme in Genetics at Universidade Federal do Rio de Janeiro, Brazil.
Funding
This work was supported by The Brazilian Ministry of Health (TC 235/2012) (C. C. C.) and by National Institutes of Health grants AI077505, AI069439 and TR002243 (D. W. H.).
Transparency declarations
None to declare.
References
- 1. Joint United Nations Programme on HIV/AIDS (UNAIDS). UNAIDS Data 2017 2017; 1–248. http://www.unaids.org/sites/default/files/media_asset/20170720_Data_book_2017_en.pdf. [PubMed]
- 2. WHO. Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection: Recommendations for a Public Health Approach 2016. http://apps.who.int/iris/bitstream/handle/10665/208825/9789241549684_eng.pdf?sequence=1. [PubMed]
- 3. Pádua CAM, de César CC, Bonolo PF. et al. Self-reported adverse reactions among patients initiating antiretroviral therapy in Brazil. Braz J Infect Dis 2007; 11: 20–6. [DOI] [PubMed] [Google Scholar]
- 4. Ribeiro FA, Tupinambás U, Fonseca MO. et al. Durability of the first combined antiretroviral regimen in patients with AIDS at a reference center in Belo Horizonte, Brazil, from 1996 to 2005. Braz J Infect Dis 2012; 16: 27–33. [PubMed] [Google Scholar]
- 5. Leutscher PDC, Stecher C, Storgaard M. et al. Discontinuation of efavirenz therapy in HIV patients due to neuropsychiatric adverse effects. Scand J Infect Dis 2013; 45: 645–51. [DOI] [PubMed] [Google Scholar]
- 6. Marzolini C, Telenti A, Decosterd L. et al. Efavirenz plasma levels can predict treatment failure and central nervous system side effects in HIV-1-infected patients. AIDS 2001; 15: 71–5. [DOI] [PubMed] [Google Scholar]
- 7. Ward BA, Gorski JC, Jones DR. et al. The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther 2003; 306: 287–300. [DOI] [PubMed] [Google Scholar]
- 8. Ogburn ET, Jones DR, Masters AR. et al. Efavirenz primary and secondary metabolism in vitro and in vivo: identification of novel metabolic pathways and cytochrome P450 2A6 as the principal catalyst of efavirenz 7-hydroxylation. Drug Metab Dispos 2010; 38: 1218–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Willson TM, Kliewer SA.. PXR, CAR and drug metabolism. Nat Rev Drug Discov 2002; 1: 259–66. [DOI] [PubMed] [Google Scholar]
- 10. Bae SK, Jeong Y-J, Lee C. et al. Identification of human UGT isoforms responsible for glucuronidation of efavirenz and its three hydroxy metabolites. Xenobiotica 2011; 41: 437–44. [DOI] [PubMed] [Google Scholar]
- 11. Bélanger AS, Caron P, Harvey M. et al. Glucuronidation of the antiretroviral drug efavirenz by UGT2B7 and an in vitro investigation of drug-drug interaction with zidovudine. Drug Metab Dispos 2009; 37: 1793–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haas DW, Ribaudo HJ, Kim RB. et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS 2004; 18: 2391–400. [PubMed] [Google Scholar]
- 13. Kwara A, Lartey M, Sagoe KW. et al. CYP2B6, CYP2A6 and UGT2B7 genetic polymorphisms are predictors of efavirenz mid-dose concentration in HIV-infected patients. AIDS 2009; 23: 2101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Swart M, Whitehorn H, Ren Y. et al. PXR and CAR single nucleotide polymorphisms influence plasma efavirenz levels in South African HIV/AIDS patients. BMC Med Genet 2012; 13: 112.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haas DW, Kwara A, Richardson DM. et al. Secondary metabolism pathway polymorphisms and plasma efavirenz concentrations in HIV-infected adults with CYP2B6 slow metabolizer genotypes. J Antimicrob Chemother 2014; 69: 2175–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tsuchiya K, Gatanaga H, Tachikawa N. et al. Homozygous CYP2B6 *6 (Q172H and K262R) correlates with high plasma efavirenz concentrations in HIV-1 patients treated with standard efavirenz-containing regimens. Biochem Biophys Res Commun 2004; 319: 1322–6. [DOI] [PubMed] [Google Scholar]
- 17. Rotger M, Colombo S, Furrer H. et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genomics 2005; 15: 1–5. [DOI] [PubMed] [Google Scholar]
- 18. Haas DW, Smeaton LM, Shafer RW. et al. Pharmacogenetics of long‐term responses to antiretroviral regimens containing efavirenz and/or nelfinavir: an Adult AIDS Clinical Trials Group study. J Infect Dis 2005; 192: 1931–42. [DOI] [PubMed] [Google Scholar]
- 19. Rodríguez-Nóvoa S, Barreiro P, Jiménez-Nácher I. et al. Overview of the pharmacogenetics of HIV therapy. Pharmacogenomics J 2006; 6: 234–45. [DOI] [PubMed] [Google Scholar]
- 20. Holzinger ER, Grady B, Ritchie MD. et al. Genome-wide association study of plasma efavirenz pharmacokinetics in AIDS Clinical Trials Group protocols implicates several CYP2B6 variants. Pharmacogenet Genomics 2012; 22: 858–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wyen C, Hendra H, Vogel M. et al. Impact of CYP2B6 983T>C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J Antimicrob Chemother 2008; 61: 914–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ribaudo HJ, Liu H, Schwab M. et al. Effect of CYP2B6, ABCB1, and CYP3A5 polymorphisms on efavirenz pharmacokinetics and treatment response: an AIDS Clinical Trials Group study. J Infect Dis 2010; 202: 717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fellay J, Marzolini C, Meaden E. et al. Response to antiretroviral treatment in HIV-1-infected individuals with allelic variants of the multidrug resistance transporter 1: a pharmacogenetic study. Lancet 2002; 359: 30–6. [DOI] [PubMed] [Google Scholar]
- 24. Elens L, Vandercam B, Yombi J-C. et al. Influence of host genetic factors on efavirenz plasma and intracellular pharmacokinetics in HIV-1-infected patients. Pharmacogenomics 2010; 11: 1223–34. [DOI] [PubMed] [Google Scholar]
- 25. Ngaimisi E, Habtewold A, Minzi O. et al. Importance of ethnicity, CYP2B6 and ABCB1 genotype for efavirenz pharmacokinetics and treatment outcomes: a parallel-group prospective cohort study in two sub-Saharan Africa populations. PLoS One 2013; 8: e67946.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Swart M, Ren Y, Smith P. et al. ABCB1 4036A>G and 1236C>T polymorphisms affect plasma efavirenz levels in South African HIV/AIDS patients. Front Genet 2012; 3: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mukonzo JK, Okwera A, Nakasujja N. et al. Influence of efavirenz pharmacokinetics and pharmacogenetics on neuropsychological disorders in Ugandan HIV-positive patients with or without tuberculosis: a prospective cohort study. BMC Infect Dis 2013; 13: 261.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gounden V, van Niekerk C, Snyman T. et al. Presence of the CYP2B6 516G>T polymorphism, increased plasma Efavirenz concentrations and early neuropsychiatric side effects in South African HIV-infected patients. AIDS Res Ther 2010; 7: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suarez-Kurtz G. Pharmacogenetics in the Brazilian population. Front Pharmacol 2010; 1: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arruda MB, Campagnari F, de Almeida TB. et al. Single nucleotide polymorphisms in cellular drug transporters are associated with intolerance to antiretroviral therapy in Brazilian HIV-1 positive individuals. PLoS One 2016; 11: e0163170.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lins TC, Vieira RG, Abreu BS. et al. Genetic composition of Brazilian population samples based on a set of twenty-eight ancestry informative SNPs. Am J Hum Biol 2010; 22: 187–92. [DOI] [PubMed] [Google Scholar]
- 32. Pritchard J, Stephens M, Donnelly P.. Inference of population structure using multilocus genotype data. Genetics 2000; 155: 945–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Falush D, Stephens M, Pritchard JK.. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 2003; 164: 1567–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Falush D, Stephens M, Pritchard JK.. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 2007; 7: 574–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hubisz MJ, Falush D, Stephens M. et al. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 2009; 9: 1322–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saloum de Neves Manta F, Pereira R, Vianna R. et al. Revisiting the genetic ancestry of Brazilians using autosomal AIM-Indels. PLoS One 2013; 8: e75145.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. R Development Core Team. R: A Language and Environment for Statistical Computing 2011. https://www.R-project.org/.
- 38. Müller TE, Ellwanger JH, Michita RT. et al. CYP2B6 516 G>T polymorphism and side effects of the central nervous system in HIV-positive individuals under efavirenz treatment: study of a sample from southern Brazil. An Acad Bras Ciênc 2017; 89 Suppl 1: 497–504. [DOI] [PubMed] [Google Scholar]
- 39. Coelho AVC, Silva SPS, De Alencar LCA. et al. ABCB1 and ABCC1 variants associated with virological failure of first-line protease inhibitors antiretroviral regimens in northeast Brazil patients. J Clin Pharmacol 2013; 53: 1286–93. [DOI] [PubMed] [Google Scholar]
- 40. Ma Q, Vaida F, Wong J. et al. Long-term efavirenz use is associated with worse neurocognitive functioning in HIV-infected patients. J Neurovirol 2016; 22: 170–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clifford DB, Evans S, Yang Y. et al. Impact of efavirenz on neuropsychological performance and symptoms in HIV-infected individuals. Ann Intern Med 2005; 143: 714–21. [DOI] [PubMed] [Google Scholar]
- 42. Mollan KR, Tierney C, Hellwege JN. et al. Race/ethnicity and the pharmacogenetics of reported suicidality with efavirenz among clinical trials participants. J Infect Dis 2017; 216: 554–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leger P, Chirwa S, Turner M. et al. Pharmacogenetics of efavirenz discontinuation for reported central nervous system symptoms appears to differ by race. Pharmacogenet Genomics 2016; 26: 473–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Michaud V, Bar-Magen T, Turgeon J. et al. The dual role of pharmacogenetics in HIV treatment: mutations and polymorphisms regulating antiretroviral drug resistance and disposition. Pharmacol Rev 2012; 64: 803–33. [DOI] [PubMed] [Google Scholar]
- 45. Arab-Alameddine M, Di Iulio J, Buclin T. et al. Pharmacogenetics-based population pharmacokinetic analysis of efavirenz in HIV-1-infected individuals. Clin Pharmacol Ther 2009; 85: 485–94. [DOI] [PubMed] [Google Scholar]
- 46. Stormer E, von Moltke LL, Perloff MD. et al. Differential modulation of P-glycoprotein expression and activity by non-nucleoside HIV-1 reverse transcriptase inhibitors in cell culture. Pharm Res 2002; 19: 1038–45. [DOI] [PubMed] [Google Scholar]
- 47. Shou W, Wang D, Zhang K. et al. Gene-wide characterization of common quantitative trait loci for ABCB1 mRNA expression in normal liver tissues in the Chinese population. PLoS One 2012; 7: e46295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.