Interleukin-36 (IL-36) cytokines are important regulators of mucosal homeostasis and inflammation. We have previously established that oral epithelial cells upregulate IL-36γ expression in response to the bacterial pathogen Porphyromonas gingivalis.
KEYWORDS: EGF, IL-36, Porphyromonas gingivalis, antimicrobial, epithelial cells, peptidoglycan recognition protein
ABSTRACT
Interleukin-36 (IL-36) cytokines are important regulators of mucosal homeostasis and inflammation. We have previously established that oral epithelial cells upregulate IL-36γ expression in response to the bacterial pathogen Porphyromonas gingivalis. Here, we have established that IL-36γ can stimulate the gene expression of mechanistically distinct antimicrobial proteins, including the peptidoglycan amidase PGLYRP2, in oral epithelial cells (e.g., TIGK cells). PGLYRP2 gene expression was not stimulated by either IL-17 or IL-22, thus demonstrating selectivity in the regulation of PGLYRP2 by IL-36γ. The IL-36γ-inducible expression of PGLYRP2 was shown to be mediated by IRAK1- and p38 mitogen-activated protein (MAP) kinase-dependent signaling. Furthermore, our finding that IL-36γ-inducible PGLYRP2 expression was reduced in proliferating TIGK cells but increased in terminally differentiating cells suggests that control of PGLYRP2 expression is associated with the maturation of the oral epithelium. PGLYRP2 expression in TIGK cells can also be directly stimulated by oral bacteria. However, the extracellular gingipain proteases (Kgp and RgpA/B) produced by P. gingivalis, which are critical virulence factors, can antagonize PGLYRP2 expression. Thus, the expression of IL-36γ by oral epithelial cells in response to P. gingivalis might enable the subsequent autocrine stimulation of PGLYRP2 expression. In summary, our data identify how IL-36γ may promote oral mucosal homeostasis by regulating PGLYRP2 expression.
INTRODUCTION
The oral epithelium is a critical barrier to infection by bacterial pathogens. The integrity of the epithelium is maintained by cycles of cell proliferation and differentiation, whereby epithelial cells in the basal layer terminally differentiate as they migrate toward the epithelium surface (1). The expression of proteins with antimicrobial activity by the oral epithelium also provides protection against infection (2, 3). The β-defensins cause bacterial cell membrane permeation, resulting in cell lysis (4), while calprotectin, a heterodimeric complex of S100A8 and S100A9, exerts bacteriostatic activity by chelating essential divalent metal ions (5). Pattern recognition receptors, including Toll-like receptors (TLRs), are important regulators of the inducible expression of antimicrobial proteins in response to microorganisms (6). Importantly, antimicrobial protein expression is also induced by inflammatory cytokines. For example, interleukin-17 (IL-17) and IL-22 are important regulators of antimicrobial protein expression by epithelial cells in mucosal tissues and epidermis (7, 8).
Peptidoglycan recognition proteins (PGLYRP1 to 4) are a unique family of antimicrobial proteins (9). Peptidoglycan is a major component of the Gram-positive bacterial cell wall, while in Gram-negative bacteria it forms a thin layer in the periplasmic space between the inner and outer cell membranes. PGLYRP1, PGLYRP3, and PGLYRP4 can directly kill bacteria by inducing stress responses (10, 11). In contrast, PGLYRP2 is an N-acetyl-muramoyl-l-alanine amidase and while not directly bactericidal may function together with other host factors to cause bacterial cell death (12). PGLYRP2 can also potentially digest proinflammatory peptidoglycan into biologically inactive fragments (12, 13), which might be important for preventing inappropriate inflammation. Indeed, in an experimental model of colitis, PGLYRP2 was shown to protect mice from inflammation and loss of epithelial barrier function by promoting a normal gut microbiome (14). PGLYRP2 has also been reported to protect mice from psoriasis-like skin inflammation (15). Intriguingly, PGLYRP2 was shown to be important for the induction of inflammation by peptidoglycan in a mouse model of arthritis (16). Thus, the role of PGLYRP2 in host inflammation might be context dependent.
IL-36 cytokines (IL-36α, IL-36β, and IL-36γ) have recently emerged as key regulators of mucosal homeostasis and inflammation (17–22). Signaling by the IL-36 receptor (IL-36R) was shown to provide protection against bacterial infection in a mouse model of colitis (18), while IL-36γ was essential for protective mucosal immunity in mouse models of bacterial pneumonia (17). The expression of IL-36 cytokines by different cell types, including epithelial cells, has been shown to be induced by TLR signaling (23, 24). IL-36 cytokines can induce inflammation by stimulating the expression of cytokines and chemokines by epithelial cells and innate immune cells (e.g., dendritic cells) (17–26). IL-36 cytokines have also been shown to regulate T-helper cells (20, 25).
We recently established that oral epithelial cells upregulate IL-36γ expression in response to Porphyromonas gingivalis (24). P. gingivalis is a keystone pathogen in chronic periodontitis, an inflammatory disease that results from the breakdown of homeostasis between the host and plaque bacteria (27–29). In this study, we investigated the ability of IL-36γ to regulate the expression of antimicrobial proteins in oral epithelial cells (e.g., TIGK cells). We show that PGLYRP2 gene transcription in TIGK cells is robustly stimulated by IL-36γ. Furthermore, we show that IL-36γ-inducible PGLYRP2 expression in TIGK cells is reduced by epidermal growth factor receptor (EGFR) signaling but enhanced by terminal differentiation. Although PGLYRP2 expression can also be directly stimulated by oral bacteria, its expression in response to P. gingivalis is antagonized by the extracellular gingipain proteases produced by the pathogen. Collectively, our findings suggest that IL-36γ may play an important role in promoting tissue homeostasis in the oral mucosa through its regulation of PGLYRP2.
RESULTS
IL-36γ stimulates expression of antimicrobial proteins in oral epithelial cells.
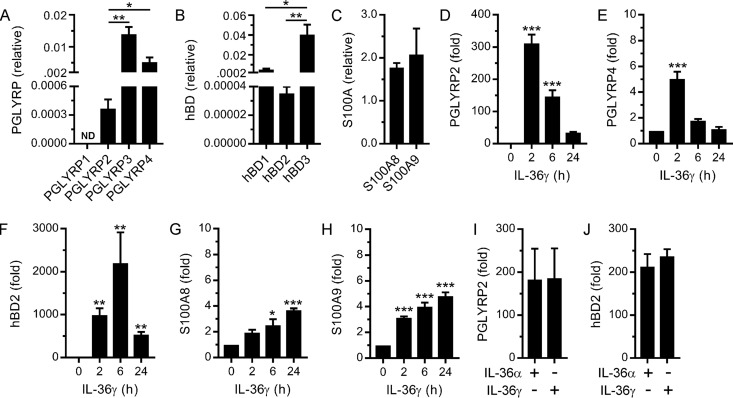
Antimicrobial proteins and cytokines are important mediators of mucosal homeostasis and inflammation. We have previously shown that IL-36γ regulates the expression of various inflammatory cytokines in oral epithelial cells (24, 26). Thus, we sought to establish whether it also regulates the expression of antimicrobial proteins. First, we assessed the basal gene expression levels of antimicrobial proteins. Human oral epithelial cells (i.e., TIGK cells) were found to express PGLYRP2, PGLYRP3, and PGLYRP4 but not PGLYRP1 (Fig. 1A). Notably, the basal expression levels of PGLYRP3 and PGLYRP4 were significantly higher than that of PGLYRP2. TIGK cells also express the human β-defensins, hBD1, hBD2, and hBD3 (Fig. 1B), with hBD3 most highly expressed. The cells were also found to express S100A8 and S100A9 (Fig. 1C). Next, we tested the ability of IL-36γ to stimulate their expression. As shown in Fig. 1D, IL-36γ strongly stimulated the expression of PGLYRP2. In contrast, PGLYRP3 expression was not stimulated (data not shown), and PGLYRP4 expression was stimulated only weakly (Fig. 1E). IL-36γ strongly stimulated the expression of hBD2 (Fig. 1F) but not hBD1 and hBD3 (data not shown). IL-36γ also stimulated the expression of S100A8 and S100A9 (Fig. 1G and H), albeit weakly in comparison to PGLYRP2 and hBD2. Accordingly, we subsequently focused on the regulation of PGLYRP2 and hBD2 by IL-36γ. By treating TIGK cells with actinomycin D prior to IL-36γ stimulation, we could confirm that the upregulation of PGLYRP2 and hBD2 mRNA levels was due to increased gene transcription (data not shown). In addition to shared functions, in vivo studies indicate that IL-36 cytokines may also have unique functions (18, 22, 30). Therefore, we tested the ability of IL-36α to stimulate PGLYRP2 and hBD2 expression. The levels of stimulation of PGLYRP2 (Fig. 1I) and hBD2 (Fig. 1J) expression by IL-36α and IL-36γ were highly comparable. These data suggest that IL-36γ produced by the oral epithelium might promote host defense and mucosal homeostasis by stimulating the expression of mechanistically diverse antimicrobial proteins, including the peptidoglycan amidase PGLYRP2.
FIG 1.
Stimulation of antimicrobial protein gene expression in oral epithelial cells by IL-36γ. (A to C) Basal mRNA expression levels of the indicated PGLYRP (A), hBD (B), and S100A (C) proteins were measured (n = 3). (D to H) TIGK cells were stimulated with IL-36γ (100 ng/ml) for the time indicated. PGLYRP2 (D), PGLYRP4 (E), hBD2 (F), S100A8 (G), and S100A9 (H) mRNA levels were then measured (n = 3). (I and J) TIGK cells were stimulated with IL-36α (100 ng/ml) or IL-36γ (100 ng/ml) for 2 h. PGLYRP2 (I) and hBD2 (J) mRNA levels were then measured (n = 3). ***, P < 0.001; **, P < 0.01; *, P < 0.05.
PGLYRP2 expression in oral epithelial cells is not stimulated by IL-17 or IL-22.
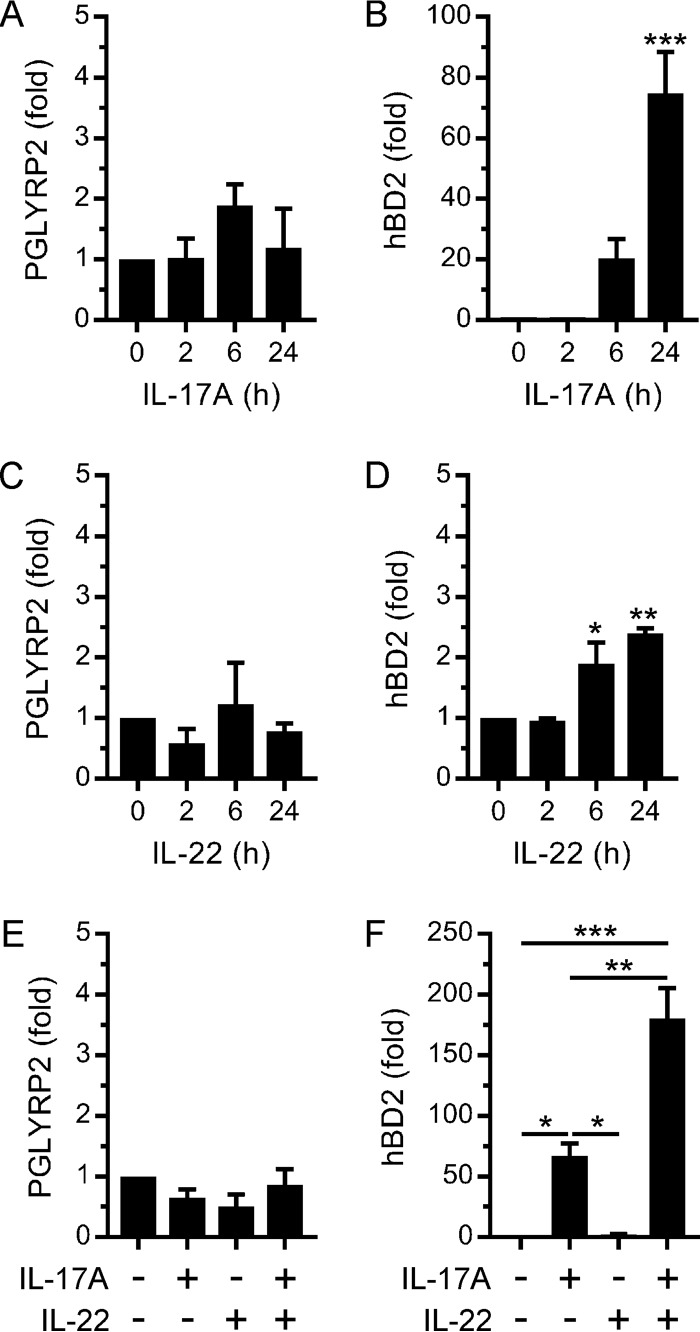
The adaptive immune cytokines IL-17 and IL-22 are also important mediators of host defense at mucosal surfaces, where they stimulate the expression of antimicrobial proteins in epithelial cells (7, 8). Therefore, we tested the ability of IL-17 and IL-22 to stimulate the expression of PGLYRP2 and hBD2 in TIGK cells. As shown in Fig. 2A, IL-17A did not simulate PGLYRP2 expression. In contrast, IL-17A stimulated hBD2 expression (Fig. 2B), although the magnitude of the response was smaller than when TIGK cells were stimulated with IL-36γ (Fig. 1F). Similarly, IL-22 did not stimulate PGLYRP2 expression (Fig. 2C) and stimulated hBD2 expression only weakly (Fig. 2D). IL-17 and IL-22 have been shown to synergistically stimulate antimicrobial protein (e.g., hBD2) expression in epidermal keratinocytes (31). Therefore, we investigated their ability to synergistically stimulate PGLYRP2 and hBD2 expression in TIGK cells. Notably, PGLYRP2 expression was not stimulated when the cells were treated concurrently with IL-17A and IL-22 (Fig. 2E). In contrast, hBD2 expression was synergistically stimulated (Fig. 2F). Collectively, the above data identify important differences in the regulation of antimicrobial protein gene expression in oral epithelial cells by IL-17/IL-22 and IL-36γ.
FIG 2.
Regulation of PGLYRP2 and hBD2 expression in oral epithelial cells by IL-17 and IL-22. (A to D) TIGK cells were stimulated with IL-17A (100 ng/ml) (A and B) or IL-22 (50 ng/ml) (C and D) for the times indicated. PGLYRP2 (A and C) and hBD2 (B and D) mRNA levels were then measured (n = 3). (E and F) TIGK cells were stimulated with IL-17A (100 ng/ml) and IL-22 (50 ng/ml) for 24 h. PGLYRP2 (E) and hBD2 (F) mRNA levels were then measured (n = 3). ***, P < 0.001; **, P < 0.01; *, P < 0.05.
IL-36γ stimulates PGLYRP2 expression via IRAK1 and p38 MAP kinase.
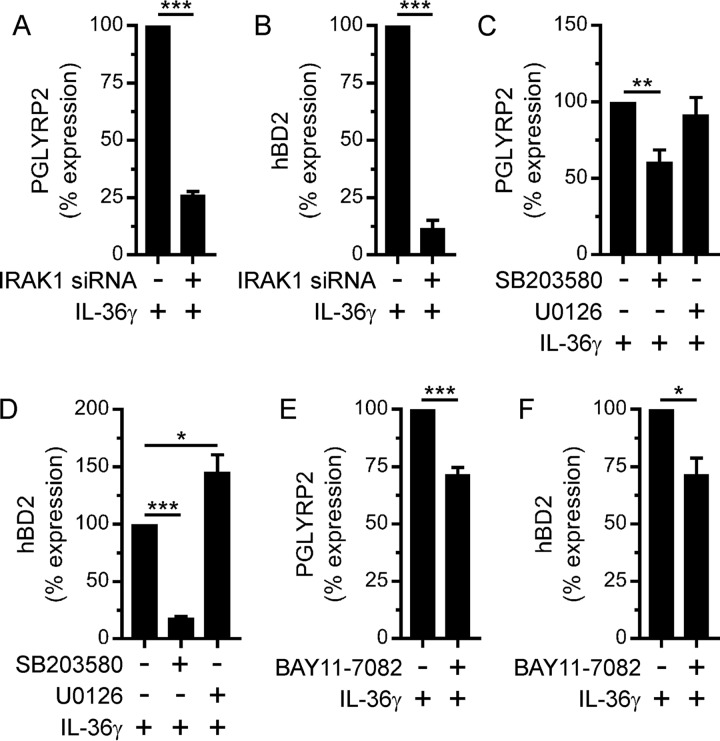
Our findings position IL-36γ as an important regulator of PGLYRP2 in oral epithelial cells. Therefore, we investigated the signaling pathways that regulate its expression downstream of the IL-36R. We have previously shown that IRAK1 mediates the stimulation of inflammatory cytokines and chemokines by IL-36γ in oral epithelial cells (e.g., TIGK cells) (24). Consistently, small interfering RNA (siRNA)-mediated gene silencing of IRAK1 in TIGK cells inhibited the stimulation of PGLYRP2 expression by IL-36γ (Fig. 3A); it also inhibited the stimulation of hBD2 expression (Fig. 3B). In addition to IRAK1, IL-36γ activates the mitogen-activated protein (MAP) kinases p38 MAP kinase and ERK1/2 and the NF-κB transcription factor in TIGK cells (26). Thus, we used pharmacologic inhibitors to establish their importance for the stimulation of PGLYRP2 by IL-36γ. The p38 MAP kinase inhibitor SB203580 reduced the IL-36γ stimulation of PGLYRP2 expression by 40% (Fig. 3C). In contrast, the stimulation of PGLYRP2 expression was not affected by the MEK (ERK1/2) inhibitor U0126 (Fig. 3C). For comparison, the p38 MAP kinase inhibitor significantly reduced the IL-36γ-inducible stimulation of hBD2 expression (Fig. 3D), whereas the MEK (ERK1/2) inhibitor potentiated the stimulation of hBD2 expression (Fig. 3D). The NF-κB inhibitor BAY11-7082 partially inhibited the stimulation of PGLYRP2 (Fig. 3E) and hBD2 (Fig. 3F) expression by IL-36γ. These data indicate that IL-36γ induces the transcription of PGLYRP2, as well as hBD2, by activating IRAK1 and p38 MAP kinase signaling. Interestingly, activation of MEK/ERK signaling by IL-36γ may limit the upregulation of hBD2 expression without affecting PGLYRP2.
FIG 3.
IRAK1 and p38 MAP kinase signaling regulates IL-36γ-inducible PGLYRP2 and hBD2 expression in oral epithelial cells. (A and B) TIGK cells were transfected with an IRAK1 (+) or control (−) siRNA and then subsequently stimulated with IL-36γ (100 ng/ml) for 2 h. PGLYRP2 (A) and hBD2 (B) mRNA levels were then measured (n = 3). (C to F) TIGK cells were treated with 5 μM SB203580 or 10 μM U0126 (C and D) or 10 μM BAY11-7082 (E and F) for 30 min and then stimulated with IL-36γ for 2 h. PGLYRP2 (C and E) and hBD2 (D and F) mRNA levels were then measured (n = 3). ***, P < 0.001; **, P < 0.01; *, P < 0.05.
EGFR signaling antagonizes the stimulation of PGLYRP2 expression by IL-36γ.
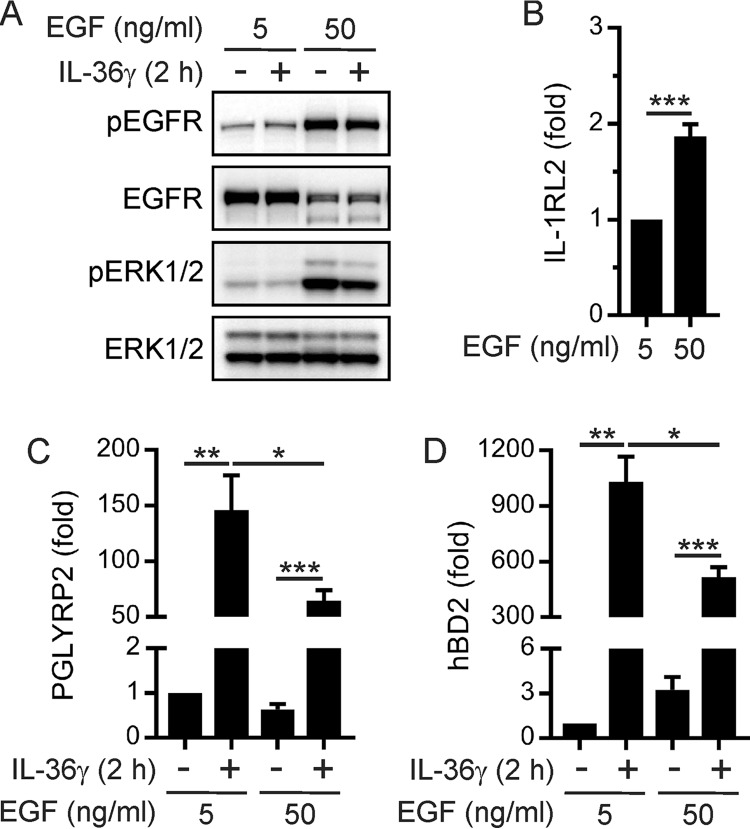
The proliferation of epithelial cells is important for the ongoing functional integrity of the oral epithelium and repair following injury. Therefore, we investigated the effect of EGF-induced proliferation on the stimulation of PGLYRP2 expression by IL-36γ. Specifically, TIGK cells were cultured in the presence of either 5 ng/ml EGF or 50 ng/ml EGF to induce different levels of EGFR signaling; thereafter, the cells were stimulated with IL-36γ. Western blotting of cell lysates with phospho-specific antibodies confirmed that the culturing of TIGK cells in the presence of 50 ng/ml EGF resulted in greatly increased EGFR and ERK1/2 phosphorylation (Fig. 4A). Consistent with ligand-induced internalization and degradation of receptor tyrosine kinases, total EGFR levels were reduced when TIGK cells were cultured in the presence of 50 ng/ml EGF (Fig. 4A). The expression levels of the IL-36R subunit IL-1RL2 were weakly increased (Fig. 4B) and IL-1RAcP levels unchanged (data not shown) under the same conditions. We then determined whether increased EGFR signaling affects the stimulation of PGLYRP2 expression by IL-36γ. Notably, the stimulation of PGLYRP2 expression was significantly reduced (Fig. 4C). Increased EGFR signaling also reduced IL-36γ-inducible hBD2 expression (Fig. 4D). Collectively, these data indicate that EGFR signaling in oral epithelial cells dampens the stimulation of PGLYRP2 and hBD2 expression by IL-36γ.
FIG 4.
EGFR signaling dampens IL-36γ-inducible PGLYRP2 and hBD2 expression in oral epithelial cells. TIGK cells were cultured with 5 ng/ml or 50 ng/ml EGF for 24 h and then stimulated with IL-36γ (100 ng/ml) for 2 h. (A) The cells were lysed, and then aliquots of the lysates were subjected to Western blotting with the indicated antibodies. The data are representative of two independent experiments. (B to D) IL-1RL2 (B), PGLYRP2 (C), and hBD2 (D) mRNA levels were then measured (n ≥ 3). ***, P < 0.001; **, P < 0.01; *, P < 0.05.
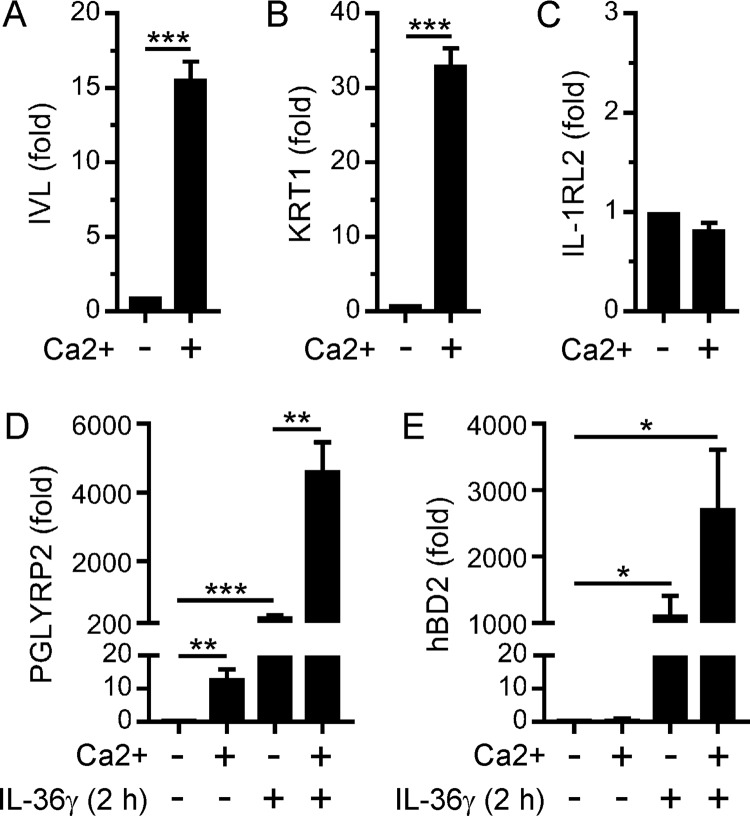
Epithelial differentiation potentiates the stimulation of PGLYRP2 expression by IL-36γ.
The differentiation of oral epithelial cells, whereby cells in the basal layer terminally differentiate as they migrate toward the epithelium surface, is important for maintaining the integrity and barrier function of the oral epithelium. Therefore, we also investigated the effect of differentiation on the regulation of PGLYRP2 expression by IL-36γ. TIGK cells were cultured in the presence of 1.8 mM calcium to induce their terminal differentiation, which was confirmed by measuring the expression levels of the differentiation markers involucrin (IVL) (Fig. 5A) and keratin 1 (KRT1) (Fig. 5B). Differentiation did not affect the expression levels of IL-1RL2 (Fig. 5C) or IL-1RAcP (data not shown). Remarkably, though, whereas IL-36γ stimulated a 400-fold increase in PGLYRP2 expression in undifferentiated TIGK cells, PGLYRP2 expression in terminally differentiated cells was stimulated more than 4,000-fold (Fig. 5D). The basal expression levels of PGLYRP2 were also significantly higher in differentiating cells (Fig. 5D). Differentiation likewise potentiated the stimulation of hBD2 expression by IL-36γ (Fig. 5E), albeit not to the same extent as for PGLYRP2. In contrast to PGLYRP2, differentiation did not affect the basal expression levels of hBD2 (Fig. 5E). These data suggest that IL-36γ likely stimulates stronger homeostatic antimicrobial responses in differentiating oral epithelial cells, in line with the superficial cells of the oral mucosa being directly exposed to bacteria.
FIG 5.
Terminal differentiation enhances IL-36γ-inducible PGLYRP2 and hBD2 expression in oral epithelial cells. TIGK cells were cultured in the presence of 0.09 mM (−) or 1.8 mM (+) calcium for 48 h (A to C) and then stimulated with IL-36γ (100 ng/ml) for 2 h (D and E). IVL (A), KRT1 (B), IL-1RL2 (C), PGLYRP2 (D), and hBD2 (E) mRNA levels were then measured (n = 3). ***, P < 0.001; **, P < 0.01; *, P < 0.05.
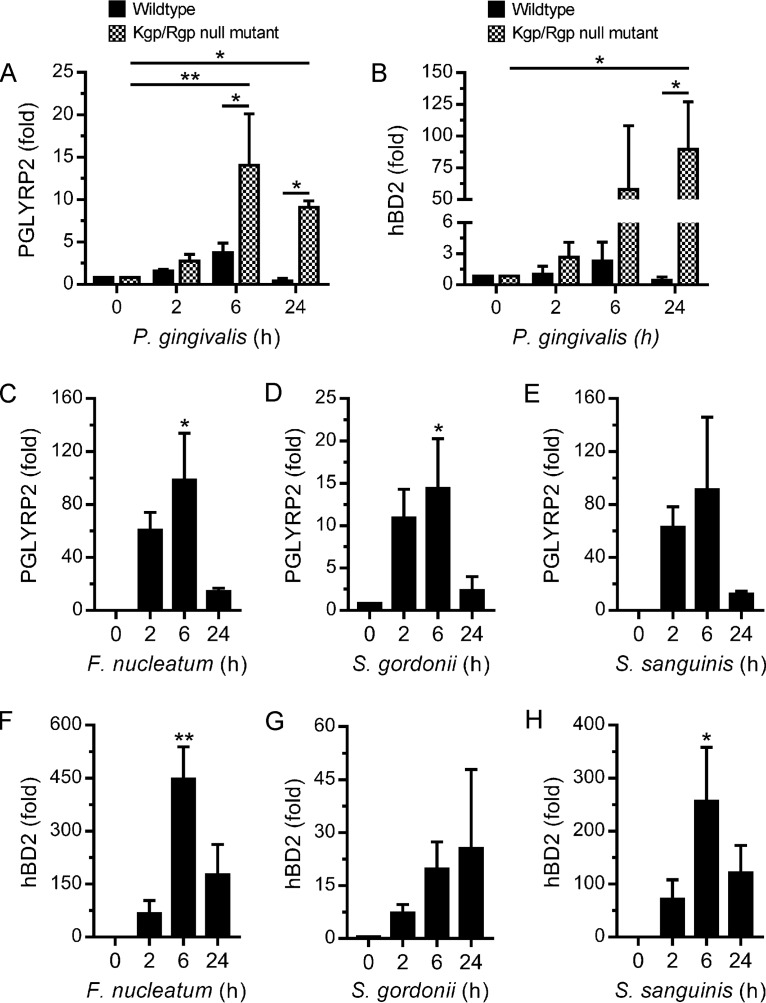
Stimulation of PGLYRP2 expression by oral bacteria.
PGLYRP2 gene expression in epidermal keratinocytes has previously been shown to be stimulated by bacteria (32). This raised the possibility that P. gingivalis might also directly stimulate PGLYRP2 gene expression by oral epithelial cells, independently of its stimulation of IL-36γ. As shown in Fig. 6A, however, PGLYRP2 gene expression was not significantly induced (<4-fold) when TIGK cells were challenged with P. gingivalis, thus suggesting that P. gingivalis does not directly stimulate PGLYRP2 expression in oral epithelial cells. The extracellular gingipain proteases (Kgp and RgpA/B) produced by P. gingivalis are critical virulence factors (28, 29), and therefore we investigated whether they might antagonize the stimulation of PGLYRP2 expression. Notably, PGLYRP2 expression was significantly stimulated (10- to 15-fold) when TIGK cells were challenged with an isogenic P. gingivalis gingipain protease (Kgp/Rgp)-null mutant (Fig. 6A). Similar observations were made for hBD2, whereby its expression was significantly stimulated by a P. gingivalis gingipain protease-null mutant (Fig. 6B). Given these findings, we investigated the ability of other oral bacteria to stimulate PGLYRP2 expression. In contrast to the case for P. gingivalis, PGLYRP2 expression was strongly stimulated by Fusobacterium nucleatum, Streptococcus gordonii, and Streptococcus sanguinis (Fig. 6C to E). These bacterial species also strongly stimulated hBD2 expression (Fig. 6F to H). Collectively, these results suggest that oral epithelial cells can directly upregulate the expression of PGLYRP2 and hBD2 in response to some bacterial species. In the case of P. gingivalis, the gingipain proteases can antagonize the stimulation of PGLYRP2 and hBD2 expression.
FIG 6.
Stimulation of PGLYRP2 and hBD2 expression in oral epithelial cells by bacteria. (A and B) TIGK cells were cultured with wild-type P. gingivalis or P. gingivalis KDP136 (Kgp/Rgp-null mutant) at an MOI of 100:1 for the times indicated. PGLYRP2 (A) and hBD2 (B) mRNA levels were then measured (n = 3). (C to H) TIGK cells were cultured with F. nucleatum (C and F), S. gordonii (D and G), and S. sanguinis (E and H) at an MOI of 100:1 for the times indicated. PGLYRP2 (C to E) and hBD2 (F to H) mRNA levels were then measured (n = 3). ***, P < 0.001; **, P < 0.01; *, P < 0.05.
DISCUSSION
IL-36 cytokines are important regulators of mucosal homeostasis and inflammation. We recently established that oral epithelial cells selectively upregulate IL-36γ in response to the bacterial pathogen P. gingivalis (24) and that IL-36γ can stimulate oral epithelial cells to express inflammatory cytokines (24, 26). Here, we have established that IL-36γ also regulates the expression of antimicrobial proteins by oral epithelial cells, including the peptidoglycan amidase PGLYRP2. PGLYRP2 has previously been demonstrated to be important for intestinal homeostasis (14). By analogy, the regulation of PGLYRP2 by IL-36γ might be important for homeostasis in the oral mucosa.
PGLYRP2 gene expression levels in oral epithelial cells (e.g., TIGK cells) were found to be 20- to 40-fold lower than those for PGLYRP3 and PGLYRP4. However, PGLYRP2 expression was strongly induced by IL-36γ, such that the expression levels of PGLYRP2 are likely to be higher than those of PGLYRP3 and PGLYRP4 following IL-36γ stimulation. Significantly, PGLYRP2 expression was not induced by either IL-17A or IL-22. These adaptive immune cytokines are important mediators of host defense at mucosal sites where they stimulate epithelial cells to express antimicrobial proteins (7, 8). Thus, our findings identify IL-36γ as a key regulator of PGLYRP2 expression in epithelial cells.
In contrast to PGLYRP3 and PGLYRP4, which can kill bacteria by inducing stress responses (10, 11), PGLYRP2 is not directly bacteriolytic and may function together with other host factors to cause bacterial cell death (12). In this regard, we found that IL-36γ also strongly stimulated oral epithelial cells to express hBD2. The β-defensins kill bacteria by forming multimeric pores in the bacterial cell membrane, resulting in cell lysis (4). Although not demonstrated, PGLYRP2 could potentially cooperate with hBD2 to mediate bacterial killing, for instance, increasing the accessibility of hBD2 to the cell membranes of Gram-positive bacteria by hydrolyzing peptidoglycan.
The epithelial cells of the oral mucosa would potentially be exposed to bacterial cell wall fragments with proinflammatory properties (e.g., peptidoglycan fragments) when bacterial killing is mediated by β-defensins, thus possibly stimulating inappropriate inflammation. PGLYRP2 has been reported to digest inflammatory peptidoglycan into biologically inactive fragments (12, 13). Moreover, PGLYRP2 was shown in mice to promote the maintenance of a normal gut microbiome and thereby protected the mice from experimentally induced colitis (14). Consequently, the coexpression of PGLYRP2 with hBD2 might be important for preventing inappropriate inflammation in the oral mucosa.
The IL-36γ-inducible expression of PGLYRP2 in TIGK cells was dependent on signaling by IRAK1 and p38 MAP kinase. This is consistent with an earlier study which demonstrated that the inducible expression of PGLYRP2 in epidermal keratinocytes was likewise regulated by p38 MAP kinase (32). Interestingly, we found that EGFR signaling, which promotes epithelial cell proliferation, dampened the stimulation of PGLYRP2 expression by IL-36γ. Conversely, calcium-induced terminal differentiation greatly potentiated the IL-36γ-inducible expression of PGLYRP2. Differentiation also enhanced the IL-36γ-inducible expression of hBD2, albeit not to the same extent. The augmenting effects of cell differentiation on PGLYRP2 and hBD2 expression are consistent with the superficial cells of the oral mucosa being directly exposed to bacteria. Most of the bacteria that colonize the oral mucosa are commensal species, and therefore overt host inflammation is normally not required to maintain host-microbiota homeostasis; in fact, inappropriate inflammation can promote dysbiosis, as occurs in chronic periodontitis (27, 28). Given the ability of PGLYRP2 to inactivate inflammatory peptidoglycan (12, 13), higher levels of PGLYRP2 expression by terminally differentiating oral epithelial cells might serve to limit the stimulation of host inflammation by commensal bacteria.
We have shown that P. gingivalis induces IL-36γ expression in TIGK cells (24). However, we did not detect significant upregulation of PGLYRP2 gene expression when TIGK cells were challenged with the pathogen. Although this might suggest that IL-36γ may not act in an autocrine manner to induce PGLYRP2 expression, following its stimulation by P. gingivalis, IL-36γ is secreted as pro-IL-36γ, which is up to 1,000 times less active than mature IL-36γ (33). Hence, proteolytic processing of pro-IL-36γ to IL-36γ by neutrophil proteases is required for significant activation of IL-36R signaling (34). The absence of neutrophils in the cell culture system used in this study would thus preclude the stimulation of PGLYRP2 expression by autocrine IL-36γ.
PGLYRP2 gene expression was strongly stimulated when TIGK cells were challenged with other oral bacterial species, such as F. nucleatum and S. sanguinis. This suggests that PGLYRP2 expression in oral epithelial cells can also be directly stimulated by bacteria. Indeed, an earlier report demonstrated the ability of bacteria (e.g., Bacillus subtilis) to stimulate PGLYRP2 expression in epidermal keratinocytes (32). The extracellular gingipain proteases (Kgp and RgpA/B) expressed by P. gingivalis are central to the ability of the pathogen to subvert the host immune response and promote the breakdown of tissue homeostasis (28, 29). Notably, PGLYRP2 expression was significantly induced when TIGK cells were challenged with an isogenic P. gingivalis Kgp/Rgp-null mutant. The gingipain proteases may therefore subvert the tissue-homeostatic functions of PGLYRP2 by antagonizing its expression by oral epithelial cells. Consequently, the stimulation of PGLYRP2 expression in response to P. gingivalis might be dependent on the production of IL-36γ by the oral epithelium.
In summary, IL-36γ robustly stimulates PGLYRP2 gene expression in oral epithelial cells. Although the functions of PGLYRP2 are still to be completely defined, in the intestines PGLYRP2 appears to play an important role in promoting a normal microbiome and mucosal homeostasis (14). Thus, our study provides new insight into how IL-36γ may promote oral mucosal homeostasis by regulating PGLYRP2 expression.
MATERIALS AND METHODS
Reagents.
DermaLife keratinocyte growth medium and supplements (transforming growth factor α [TGF-α], insulin, epinephrine, apo-transferrin, hydrocortisone, bovine pituitary extract, and glutamine) were from Lifeline Cell Technology. Human IL-17A (Gly24 to Ala155), IL-22 (Ala34 to Ile179), IL-36α (Lys6 to Phe158), and IL-36γ (Ser18 to Asp169) were from R&D Systems. Recombinant human EGF, Opti-MEM I reduced-serum medium, Lipofectamine RNAiMAX transfection reagent, and precast 10% NuPAGE gels were from Life Technologies. The ON-TARGETplus IRAK1 and nontargeting control siRNAs were from GE Healthcare. The anti-phospho-EGFR (pY1068), anti-EGFR, anti-phospho-ERK1/2, and anti-ERK1/2 antibodies were from Cell Signaling Technology. BAY11-7082, SB203580, and U0126 were from Merck Millipore, while actinomycin D was from Sigma-Aldrich.
Mammalian cell culture.
Human TIGK gingival epithelial cells (35) were cultured in DermaLife keratinocyte growth medium supplemented with 0.5 ng/ml TGFα, 5 μg/ml insulin, 1 μM epinephrine, 5 μg/ml apo-transferrin, 100 ng/ml hydrocortisone, 0.4% bovine pituitary extract, and 6 mM glutamine. The cells were cultured at 37°C in a humidified atmosphere of 5% CO2.
Bacterial cell culture.
P. gingivalis (ATCC 33277), F. nucleatum (ATCC 10953), S. gordonii (ATCC 35105), and S. sanguinis (ATCC 10556) were obtained from the culture collection of the Melbourne Dental School (University of Melbourne). The isogenic Kgp- and RgpA/B-deficient P. gingivalis mutant KDP136 (Δkgp ΔrgpA ΔrgpB) was cultured as previously described (36). The bacteria were maintained on horse blood agar plates at 37°C in an anaerobic atmosphere of 5% H2, 80% N2, and 15% CO2 (P. gingivalis, F. nucleatum, and S. sanguinis) or an aerobic atmosphere of 100% air (S. gordonii). Bacterial colonies were used to inoculate brain heart infusion medium supplemented with 5 μg/ml hemin plus 0.5 mg/ml cysteine (anaerobes) and 5 μg/ml menadione (P. gingivalis) (37).
Infection of oral epithelial cells with bacteria.
Logarithmic-growth-phase bacteria were harvested by centrifugation at 7,000 × g for 20 min at 4°C and suspended in keratinocyte growth medium. TIGK cell monolayers were incubated with bacteria at a multiplicity of infection (MOI) of 100:1 (37).
RNA purification, reverse transcription, and qPCR.
Total RNA was purified using the ReliaPrep RNA Cell miniprep system (Promega), which included an on-column DNase treatment step. RNA was reverse transcribed using random primers and GoScript reverse transcriptase (Promega) per the manufacturer's instructions. Quantitative real-time PCR (qPCR) was performed in duplicate using GoTaq qPCR master mix (Promega) and predeveloped TaqMan assays (Life Technologies) for the following genes: hBD1 (Hs00608345_m1), hBD2 (Hs00175474_m1), hBD3 (Hs04194486_g1), IL-1RAcP (Hs00895050_m1), IL-1RL2 (Hs00909276_m1), IRAK1 (Hs01018347_m1), IVL (Hs00902520_m1), KRT1 (Hs01549614_g1), PGLYRP1 (Hs00175475_m1), PGLYRP2 (Hs00994650_m1), PGLYRP3 (Hs00364657_m1), PGLYRP4 (Hs01120180_m1), S100A8 (Hs00374264_g1), and S100A9 (Hs00610058_m1). PCR was performed on a QuantStudio 7 Flex real-time PCR system (Life Technologies). The data were normalized against the hypoxanthine guanine phosphoribosyl transferase (HPRT) or TATA box binding protein (TBP) gene.
RNA interference-mediated gene silencing.
A reverse transfection protocol was used for siRNA transfection of TIGK cells (38). Briefly, the IRAK1 and nontargeting control siRNAs were diluted to 120 nM with 100 μl Opti-MEM I reduced-serum medium, mixed with 100 μl Opti-MEM medium containing 1.0 μl Lipofectamine RNAiMAX transfection reagent, and incubated at room temperature for 20 min. TIGK cells (2 × 105 cells in 1 ml keratinocyte complete growth medium) were seeded in 12-well plates and cultured with the transfection cocktail for 24 h. Thereafter, the medium was replaced and the cells cultured for a further 24 h.
Cell lysis and Western blotting.
TIGK cells were washed twice with ice-cold phosphate-buffered saline (PBS) and then lysed with IGEPAL lysis buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% IGEPAL CA-630, 10% glycerol, 20 mM β-glycerol phosphate, 1 mM sodium orthovanadate, 10 mM NaF, and protease inhibitors) on ice for 60 min. Thereafter, the lysates were clarified by centrifugation (13,000 × g for 10 min at 4°C), and the protein concentrations were measured using a protein assay kit (Bio-Rad). Cell lysates were incubated with NuPAGE lithium dodecyl sulfate (LDS) sample buffer for 10 min at 70°C and then subjected to electrophoresis on a 10% NuPAGE gel using MOPS (morpholinepropanesulfonic acid) buffer (Life Technologies). The proteins were transferred to a polyvinylidene difluoride (PVDF) membrane, which was then blocked with 3% bovine serum albumin (BSA) in TBST (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.02% Tween 20) for 60 min at room temperature. The membrane was incubated with the primary antibody (diluted in 1% BSA in TBST) overnight at 4°C. The membrane was washed with TBST, followed by incubation with a horseradish peroxidase (HRP)-conjugated secondary antibody (diluted in 1% BSA in TBST) for 60 min at room temperature. Following washing with TBST, immunoreactive proteins were visualized using ECL reagents (Millipore) and a Fujifilm Las-3000 Imager (Fujifilm, Japan).
Statistical analysis.
Data combined from three or more independent biological replicate experiments are presented as the mean ± standard error of the mean (SEM). Statistical analyses were performed using GraphPad Prism 7. Differences between two groups were evaluated using the Student t test. For multiple comparisons, statistical analysis was performed by analysis of variance (ANOVA) with Dunnett's post hoc test. A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Richard Lamont (University of Louisville) and Koji Nakayama (Nagasaki University) for generously providing the human TIGK cell line and the Porphyromonas gingivalis KDP136 mutant, respectively.
This research was supported by the Australian Government, Department of Industry, Innovation and Science grant 20080108.
REFERENCES
- 1.Presland RB, Dale BA. 2000. Epithelial structural proteins of the skin and oral cavity: function in health and disease. Crit Rev Oral Biol Medicine 11:383–408. doi: 10.1177/10454411000110040101. [DOI] [PubMed] [Google Scholar]
- 2.Schroder JM, Harder J. 2006. Antimicrobial skin peptides and proteins. Cell Mol Life Sci 63:469–486. doi: 10.1007/s00018-005-5364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greer A, Zenobia C, Darveau RP. 2013. Defensins and LL-37: a review of function in the gingival epithelium. Periodontol 2000 63:67–79. doi: 10.1111/prd.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganz T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol 3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 5.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. 2008. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 6.Abreu MT. 2010. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol 10:131–144. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- 7.Kolls JK, McCray PB Jr, Chan YR. 2008. Cytokine-mediated regulation of antimicrobial proteins. Nat Rev Immunol 8:829–835. doi: 10.1038/nri2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonnenberg GF, Fouser LA, Artis D. 2011. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol 12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 9.Royet J, Gupta D, Dziarski R. 2011. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat Rev Immunol 11:837–851. doi: 10.1038/nri3089. [DOI] [PubMed] [Google Scholar]
- 10.Kashyap DR, Wang M, Liu LH, Boons GJ, Gupta D, Dziarski R. 2011. Peptidoglycan recognition proteins kill bacteria by activating protein-sensing two-component systems. Nat Med 17:676–683. doi: 10.1038/nm.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashyap DR, Rompca A, Gaballa A, Helmann JD, Chan J, Chang CJ, Hozo I, Gupta D, Dziarski R. 2014. Peptidoglycan recognition proteins kill bacteria by inducing oxidative, thiol, and metal stress. PLoS Pathol 10:e1004280. doi: 10.1371/journal.ppat.1004280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang ZM, Li X, Cocklin RR, Wang M, Wang M, Fukase K, Inamura S, Kusumoto S, Gupta D, Dziarski R. 2003. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-l-alanine amidase. J Biol Chem 278:49044–49052. doi: 10.1074/jbc.M307758200. [DOI] [PubMed] [Google Scholar]
- 13.Duerr CU, Salzman NH, Dupont A, Szabo A, Normark BH, Normark S, Locksley RM, Mellroth P, Hornef MW. 2011. Control of intestinal Nod2-mediated peptidoglycan recognition by epithelium-associated lymphocytes. Mucosal Immunol 4:325–334. doi: 10.1038/mi.2010.71. [DOI] [PubMed] [Google Scholar]
- 14.Saha S, Jing X, Park SY, Wang S, Li X, Gupta D, Dziarski R. 2010. Peptidoglycan recognition proteins protect mice from experimental colitis by promoting normal gut flora and preventing induction of interferon-gamma. Cell Host Microbe 8:147–162. doi: 10.1016/j.chom.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park SY, Gupta D, Hurwich R, Kim CH, Dziarski R. 2011. Peptidoglycan recognition protein Pglyrp2 protects mice from psoriasis-like skin inflammation by promoting regulatory T cells and limiting Th17 responses. J Immunol 187:5813–5823. doi: 10.4049/jimmunol.1101068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saha S, Qi J, Wang S, Wang M, Li X, Kim YG, Nunez G, Gupta D, Dziarski R. 2009. PGLYRP-2 and Nod2 are both required for peptidoglycan-induced arthritis and local inflammation. Cell Host Microbe 5:137–150. doi: 10.1016/j.chom.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovach MA, Singer B, Martinez-Colon G, Newstead MW, Zeng X, Mancuso P, Moore TA, Kunkel SL, Peters-Golden M, Moore BB, Standiford TJ. 2017. IL-36gamma is a crucial proximal component of protective type-1-mediated lung mucosal immunity in Gram-positive and -negative bacterial pneumonia. Mucosal Immunol 10:1320–1334. doi: 10.1038/mi.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell SE, Horan RM, Stefanska AM, Carey A, Leon G, Aguilera M, Statovci D, Moran T, Fallon PG, Shanahan F, Brint EK, Melgar S, Hussey S, Walsh PT. 2016. IL-36alpha expression is elevated in ulcerative colitis and promotes colonic inflammation. Mucosal Immunol 9:1193–1204. doi: 10.1038/mi.2015.134. [DOI] [PubMed] [Google Scholar]
- 19.Scheibe K, Backert I, Wirtz S, Hueber A, Schett G, Vieth M, Probst HC, Bopp T, Neurath MF, Neufert C. 2017. IL-36R signalling activates intestinal epithelial cells and fibroblasts and promotes mucosal healing in vivo. Gut 66:823–838. doi: 10.1136/gutjnl-2015-310374. [DOI] [PubMed] [Google Scholar]
- 20.Harusato A, Abo H, Ngo VL, Yi SW, Mitsutake K, Osuka S, Kohlmeier JE, Li JD, Gewirtz AT, Nusrat A, Denning TL. 2017. IL-36gamma signaling controls the induced regulatory T cell-Th9 cell balance via NFkappaB activation and STAT transcription factors. Mucosal Immunol 10:1455–1467. doi: 10.1038/mi.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aoyagi T, Newstead MW, Zeng X, Kunkel SL, Kaku M, Standiford TJ. 2017. IL-36 receptor deletion attenuates lung injury and decreases mortality in murine influenza pneumonia. Mucosal Immunol 10:1043–1055. doi: 10.1038/mi.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medina-Contreras O, Harusato A, Nishio H, Flannigan KL, Ngo V, Leoni G, Neumann PA, Geem D, Lili LN, Ramadas RA, Chassaing B, Gewirtz AT, Kohlmeier JE, Parkos CA, Towne JE, Nusrat A, Denning TL. 2016. IL-36 receptor promotes resolution of intestinal damage. J Immunol 196:34–38. doi: 10.4049/jimmunol.1501312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chustz RT, Nagarkar DR, Poposki JA, Favoreto S Jr, Avila PC, Schleimer RP, Kato A. 2011. Regulation and function of the IL-1 family cytokine IL-1F9 in human bronchial epithelial cells. Am J Respir Cell Mol Biol 45:145–153. doi: 10.1165/rcmb.2010-0075OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huynh J, Scholz GM, Aw J, Kwa MQ, Achuthan A, Hamilton JA, Reynolds EC. 2016. IRF6 regulates the expression of IL-36gamma by human oral epithelial cells in response to Porphyromonas gingivalis. J Immunol 196:2230–2238. doi: 10.4049/jimmunol.1501263. [DOI] [PubMed] [Google Scholar]
- 25.Vigne S, Palmer G, Lamacchia C, Martin P, Talabot-Ayer D, Rodriguez E, Ronchi F, Sallusto F, Dinh H, Sims JE, Gabay C. 2011. IL-36R ligands are potent regulators of dendritic and T cells. Blood 118:5813–5823. doi: 10.1182/blood-2011-05-356873. [DOI] [PubMed] [Google Scholar]
- 26.Scholz GM, Heath JE, Walsh KA, Reynolds EC. 2018. MEK-ERK signaling diametrically controls the stimulation of IL-23p19 and EBI3 expression in epithelial cells by IL-36gamma. Immunol Cell Biol doi: 10.1111/imcb.12029. [DOI] [PubMed] [Google Scholar]
- 27.Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol 8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 28.Lamont RJ, Hajishengallis G. 2015. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med 21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hajishengallis G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol 15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milora KA, Fu H, Dubaz O, Jensen LE. 2015. Unprocessed interleukin-36alpha regulates psoriasis-like skin inflammation in cooperation with interleukin-1. J Investig Dermatol 135:2992–3000. doi: 10.1038/jid.2015.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. 2006. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med 203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Gupta D, Li X, Dziarski R. 2005. Peptidoglycan recognition protein 2 (N-acetylmuramoyl-l-Ala amidase) is induced in keratinocytes by bacteria through the p38 kinase pathway. Infect Immun 73:7216–7225. doi: 10.1128/IAI.73.11.7216-7225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Towne JE, Renshaw BR, Douangpanya J, Lipsky BP, Shen M, Gabel CA, Sims JE. 2011. Interleukin-36 (IL-36) ligands require processing for full agonist (IL-36alpha, IL-36beta, and IL-36gamma) or antagonist (IL-36Ra) activity. J Biol Chem 286:42594–42602. doi: 10.1074/jbc.M111.267922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henry CM, Sullivan GP, Clancy DM, Afonina IS, Kulms D, Martin SJ. 2016. Neutrophil-derived proteases escalate inflammation through activation of IL-36 family cytokines. Cell Rep 14:708–722. doi: 10.1016/j.celrep.2015.12.072. [DOI] [PubMed] [Google Scholar]
- 35.Moffatt-Jauregui CE, Robinson B, de Moya AV, Brockman RD, Roman AV, Cash MN, Culp DJ, Lamont RJ. 2013. Establishment and characterization of a telomerase immortalized human gingival epithelial cell line. J Periodontal Res 48:713–721. doi: 10.1111/jre.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi Y, Ratnayake DB, Okamoto K, Abe N, Yamamoto K, Nakayama K. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem 274:17955–17960. [DOI] [PubMed] [Google Scholar]
- 37.Huynh J, Scholz GM, Aw J, Reynolds EC. 2017. Interferon regulatory factor 6 promotes keratinocyte differentiation in response to Porphyromonas gingivalis. Infect Immun 85:e00858-16. doi: 10.1128/IAI.00858-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwa MQ, Huynh J, Aw J, Zhang L, Nguyen T, Reynolds EC, Sweet MJ, Hamilton JA, Scholz GM. 2014. Receptor-interacting protein kinase 4 and interferon regulatory factor 6 function as a signaling axis to regulate keratinocyte differentiation. J Biol Chem 289:31077–31087. doi: 10.1074/jbc.M114.589382. [DOI] [PMC free article] [PubMed] [Google Scholar]