Plasmodium vivax parasites preferentially invade reticulocytes in human beings. P. vivax merozoite surface protein 1 (PvMSP1) and PvMSP1 paralog (PvMSP1P) may have important functions in reticulocyte adherence during invasion.
KEYWORDS: Plasmodium vivax, merozoite surface, Duffy independent, merozoite surface protein 1, merozoite surface protein 1 paralog, invasion inhibition, reticulocyte
ABSTRACT
Plasmodium vivax parasites preferentially invade reticulocytes in human beings. P. vivax merozoite surface protein 1 (PvMSP1) and PvMSP1 paralog (PvMSP1P) may have important functions in reticulocyte adherence during invasion. These proteins share similar structures, including the presence of two epidermal growth factor (EGF)-like and glycosylphosphatidylinositol (GPI)-anchored domains at the C terminus. However, there have been no reports concerning the functional activity of PvMSP1P in reticulocyte adherence during P. vivax invasion. In this study, the ability of PvMSP1P-19 to bind to reticulocytes and normocytes was analyzed. The reticulocyte binding activity of PvMSP1P-19 was 4.0-fold higher than its normocyte binding activity. The binding of PvMSP1P-19 to reticulocytes and normocytes was inhibited in a dose-dependent manner by antibodies from immunized rabbits and by antibodies from vivax parasite-infected patients. Consistently, antibodies against PvMSP1P inhibited parasite invasion during short-term in vitro cultivation. Similar to the case for PvDBPII binding activity, PvMSP1P-19 binding activity was reduced in chymotrypsin-treated reticulocytes. However, no significant difference between the binding of PvMSP1P-19 to Duffy-positive and Duffy-negative erythrocytes was found. The minimal binding motif of PvMSP1P-19 was characterized using synthetic peptides. The results showed that the residues at amino acid positions 1791 to 1808 may have an important function in mediating merozoite adherence to reticulocytes. The positively charged residues within the EGF-like domain were shown to constitute a key binding motif. This work presents strong evidence supporting the role of PvMSP1P in host target cell selection and invasion of Duffy-independent pathway in P. vivax. Moreover, PvMSP1P-19-specific antibodies may confer protection against P. vivax reinvasion.
INTRODUCTION
Malaria remains a major problem, causing high morbidity and mortality in tropical and subtropical nations (1). An estimated 148 million to 304 million cases of malaria occur annually, and this disease continues to be a serious health threat in developing countries (1). Of the six parasites that cause human malaria, Plasmodium vivax is the most prevalent outside sub-Saharan Africa. However, understanding of P. vivax pathophysiology in the host during the intraerythrocytic stage of the parasite is still limited, leading to difficulty in controlling vivax malaria. In particular, how and why P. vivax displays strict host tropism for immature erythrocytes (reticulocytes) must be addressed (2). To date, only the interaction between Duffy binding protein (PvDBP) and Duffy antigen receptor for chemokines (DARC) has been shown to be involved in the invasive mechanism of P. vivax (3). However, PvDBP interaction is insufficient to explain P. vivax host cell selection mechanisms because DARC expression slightly decreases with erythrocyte (RBC) maturation (4). In addition, P. vivax infection has been reported in Duffy-negative individuals in Madagascar (5). These studies suggest that P. vivax may use a variable and flexible invasion pathway and host cell selection (6); however, this important issue remains unresolved.
The transition from reticulocyte to normocyte involves dramatic changes in the cell surface as well as intracellular changes (7). A number of cell surface molecules that function as adhesion molecules that interact with other blood cell components and with endothelial cells were found (8). The interaction of parasite ligands with erythrocyte surface molecules during P. vivax invasion is essential for successful invasion from initial contact to internalization (9, 10). In P. vivax, several adhesive ligands from the merozoite apical region that are important for specific interactions with reticulocytes, including PvRBP1a, PvRBP1b, PvRBP2a, PvRBP2b, and PvEBP2, have been identified (11–14). All of these apically localized antigens may be responsible for parasite internalization in reticulocytes (9). However, identification of the ligand responsible for the initial contact has been neglected due to the lack of an in vitro cultivation assay for P. vivax.
The surface-exposed antigens on merozoites are abundant and serve essential functions that mediate initial contact with erythrocyte surface. This hypothesis was supported by previous functional studies of P. vivax merozoite surface proteins 1-19 (PvMSP1-19) and Plasmodium falciparum MSP1-19 (PfMSP1-19) (15, 16). Recently, PvMSP1 paralog (PvMSP1P) was reported as a novel erythrocyte binding protein (17). PvMSP1P is expressed on the merozoite surface and elicits a strong acquired immune response in P. vivax patients (17). PvMSP1P is similar to PvMSP1 in terms of its amino acid sequence, with conservation of 12 cysteine residues in its epidermal growth factor (EGF)-like domains in the C-terminal region. Moreover, these cysteine residues were also shown to be highly conserved not only in P. vivax merozoite surface antigens (PvMSP1, PvMSP8, and PvMSP10) but also in the surface proteins of diverse human-invasive Plasmodium species (18, 19). Although the human EGF domain was shown to be related to dendritic cell maturation and T cell activation (20), the functions of the EGF-like domain in Plasmodium spp. are unknown. PvMSP1P transcription levels increase at the schizont stage, reflecting a major role for this protein in the process of egress and invasion in individual merozoites. Accordingly, PvMSP1P-19 may play an important role in the initial contact and the selection of host cells when merozoites invade new reticulocytes. Therefore, evaluation of the reticulocyte selectivity of PvMSP1P and determination of its binding mechanism are essential for understanding the complex process of P. vivax invasion involving the Duffy-independent pathway.
In this study, the reticulocyte binding ability of the PvMSP1P EGF-like domain was evaluated, and the inhibitory activity of an antibody against PvMSP1P on parasite reinvasion was demonstrated.
RESULTS
Structure and amino acid sequence homology of PvMSP1P-19.
The N-terminal region of PvMSP1P consists of a signal peptide (SP) (amino acids [aa] 1 to 28), a heptapeptide tandem repeat domain (TR) (aa 905 to 918), and a polymorphic Glu- and Gln-rich domain (PR) (aa 1157 to 1172). PvMSP1P contains two highly conserved EGF-like domains (aa 1751 to 1789 and aa 1792 to 1834) with a glycosylphosphatidylinositol (GPI)-anchored domain (aa 1834 to 1854) in the C-terminal region (Fig. 1A). The cysteine residues in the two EGF-like domains are conserved in various human- and nonhuman-primate-invasive Plasmodium species (Fig. 1B). The sequence similarities with PvMSP1P-19 were as follows: P. knowlesi, 86.0%; P. cynomolgi, 76.7%; P. ovale, 54.7%; and P. malariae, 57.0% (Fig. 1B).
FIG 1.
Schematic structure of PvMSP1P and EGF-like domain sequence homology in various malaria-causing species. (A) Schematic diagram of PvMSP1P-19 at the amino acid level. Two EGF-like domains (gray) are at aa 1751 to 1789 and 1792 to 1834. The signal peptide (SP) (black), tandem repeat region (TR) (orange), Glu/Gln polymorphic region (PR) (yellow) and GPI-anchored domain (blue) are indicated. The synthesized peptides (18-mers) are shown by the black bars within the EGF-like domain. The amino acid sequence is shown in Table S1 in the supplemental material. (B) Sequence alignment of the MSP1P-EGF-like domains of various Plasmodium species. The red bars represent sequences that are identical in all species. The sequence similarity is indicated as four (orange), three (green), or two (blue) identical species. The cysteine residue connection line represents the typical EGF-like domain disulfide bond. (C) Three-dimensional ribbon diagram of PvMSP1P-19. EGF-like domain 1 is shown in red, domain 2 is shown in gray, and cysteine residues are shown in yellow. The numbers on the cysteine residues represent the disulfide bond. (D) Three-dimensional surface diagram of PvMSP1P-19. The electrostatic surface of PvMSP1P-19 with positive (blue) and negative (red) charges is shown.
3D structure of PvMSP1P-19.
The three-dimensional structure of the EGF-like domain was modeled using SWISS-MODEL. As a template for PvMSP1P-19, the EGF-like domain of PvMSP1-19 (accession no. 2npr) was found to display a highly fitting structure. For structural analysis of the template, the BL21(DE3) strain of Escherichia coli was used for recombinant protein expression, and the solution nuclear magnetic resonance method was used for three-dimensional (3D) structure identification (21). A Ramachandran plot revealed the favorable regions (72.6%) and disallowed regions (0.0%) in the model. The quality verified by ERRAT results was 75.4% of the ab initio model structure. After refinement, the 3D structure quality improved to a favorable region of 87.7% and a disallowed region of 1.4% in the Ramachandran plot and to 95.5% of the final allowed PvMSP1P-19 structure in ERRAT.
Three-dimensional structure prediction showed similarity to the PvMSP1 EGF domain, with a flat, disk-like shape with three disulfide bridges in each EGF-like domain (Fig. 1C). The surface of PvMSP1P-19 showed a distinct charge pattern; the negative charge was concentrated in the central concave portion, and the positive charge surrounded the concave portion (Fig. 1D).
Reticulocyte enrichment and reticulocyte preference of PvMSP1P-19.
A total of 22 cord blood samples were collected for reticulocyte binding assay, reticulocyte binding inhibition assay, and competition assays in this study. Approximately 15 ml of each cord blood sample was used for reticulocyte enrichment. The fresh cord blood contained various concentrations of reticulocytes, ranging from 1.6% to 6.9% (mean, 2.7%; 95% confidence interval, 2.2% to 3.2%). The enriched reticulocytes were obtained at concentrations ranging from 71% to 88% (mean, 76.6%; 95% confidence interval, 74.3 to 78.7%) (Fig. 2A, upper panel).
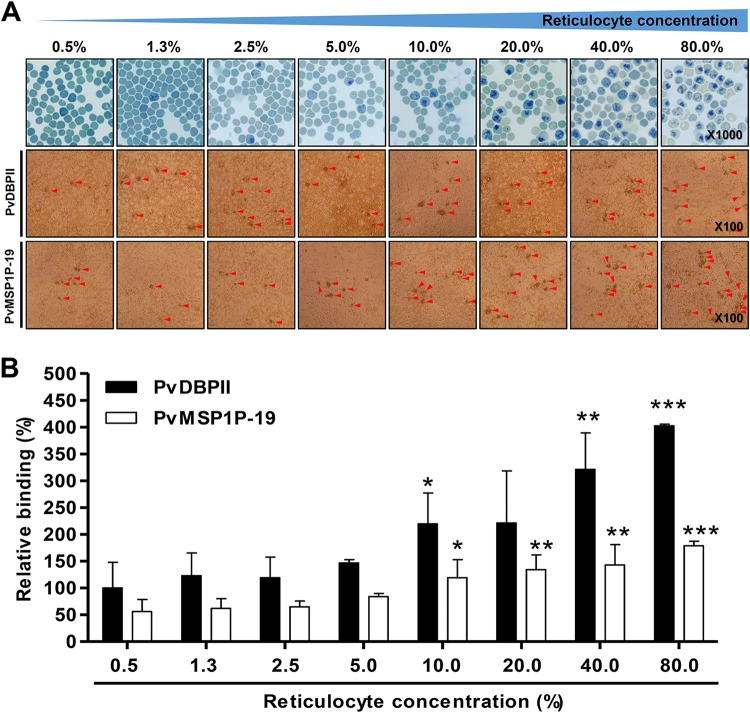
FIG 2.
Reticulocyte binding preference. (A) Reticulocytes that were serially diluted from 80% to 0.5% were stained with new methylene blue and observed under light microscopy (upper panels). Reticulocyte binding was confirmed by PvMSP1P-19- or PvDBPII-expressed COS-7 cell rosette formation (middle and bottom panels and see Fig. S1 in the supplemental material). Red arrowheads indicate typical rosette formation. (B) The relative binding abilities at different reticulocyte concentrations were calculated and normalized to PvDBPII (100% COS-7 cell transfection efficiency) with 0.5% reticulocyte (peripheral blood condition) binding ability as a standard (100%). The data are presented as the mean ± SD of the relative binding (percent) observed in three independent experiments. PvDBPII and PvMSP1P-19 showed significant differences in binding ability under other conditions compared with the 0.5% reticulocyte condition in the PvDBPII and PvMSP1P-19 fractions, respectively. P values were calculated using Student's t test. Significant differences are indicated by single asterisks (P < 0.05), double asterisks (P < 0.01), and triple asterisks (P < 0.001).
The enriched reticulocytes were diluted in suspensions of erythrocytes (peripheral blood) ranging from 80.0% to 0.5% reticulocytes in normal peripheral blood and confirmed by methylene blue staining (Fig. 2A, upper panels). The reticulocyte concentration-dependent manner of binding was confirmed by COS-7 cell binding assay, and rosette formations were counted under light microscopy (Fig. 2A, middle and bottom panels). The rosette formation of the erythrocyte (reticulocyte count, >0.5%) and reticulocyte (reticulocyte count, >80%) fractions was specific to PvMSP1P-19 and PvDBPII expressed COS-7 cells (see Fig. S1A to C in the supplemental material). The transfection efficiency was 85% for PvMSP1P-19 and 42% for PvDBPII in COS-7 cells in three independent experiments (Fig. S1A and B). PvMSP1P-19 and PvDBPII binding activities were normalized to the number of rosettes that would have been obtained at 100% transfection efficiency (number of rosettes × 100/transfection efficiency). The number of rosettes was normalized by setting PvDBPII binding to peripheral blood (reticulocyte count, >0.5%) as 100% relative binding for comparison with binding activity under other conditions (Fig. 2B). Both PvMSP1P-19 binding and PvDBPII binding increased with reticulocyte concentration in a dose-dependent manner. PvMSP1P-19 demonstrated half of the binding ability of PvDBPII; however, its binding increased dramatically (3.2-fold) as the reticulocyte concentration was increased from 0.5% to 80.0% (Fig. 2B). PvDBPII showed 4.0-fold higher binding to 80.0% reticulocytes than to peripheral erythrocytes. In particular, the binding of both proteins increased significantly compared to peripheral blood conditions at a concentration of 10.0% reticulocytes (Fig. 2B).
Inhibition of the binding and invasion of P. vivax by antibodies against PvMSP1P-19.
An in vitro binding inhibition assay was used to determine whether anti-PvMSP1P-19 antibodies in mouse and vivax-infected patient sera had inhibitory effects on the binding of the protein to erythrocytes and reticulocytes. Antibodies against PvDBPII and PvMSP1P-19 recognized target antigens on the COS-7 cell surface (Fig. 3A and B, upper panels). Preimmune (PI) mouse sera and mouse sera against Pvs25 at a dilution of 1:10 were used as negative controls. In this study, PvDBPII and PvMSP1P-19 binding inhibition assays were performed simultaneously (Fig. 3A). The inhibitory effects were measured by normalizing the number of rosettes observed in the presence of test antibody dilutions ranging from 1:1,000 to 1:50. The mouse antibodies against PvDBPII and PvMSP1P-19 exerted significant concentration-dependent inhibition of PvDBPII and PvMSP1P-19 binding to erythrocytes and reticulocytes, respectively (Fig. 3A and B). For PvMSP1P-19, the binding inhibition at a dilution of 1:50 was approximately 75% for both erythrocytes and reticulocytes (Fig. 3B). Furthermore, to investigate whether sera from vivax patients also contain blocking antibodies against PvMSP1P-19, we tested the effects of patient sera on erythrocyte and reticulocyte binding. Vivax-infected patient sera (from the Republic of Korea, ROK+) inhibited the binding of PvMSP1P-19 to human erythrocytes (78.1% ± 6.1% relative binding [mean ± standard deviation {SD}]) and reticulocytes (71.8% ± 8.0%) at a dilution of 1:20 and reduced the binding ability in a dose-dependent manner (Fig. 3C). In contrast, control sera from healthy individuals (normal sera, ROK−) showed no ability to block PvMSP1P-19 binding to erythrocytes (0.0% ± 9.8%) or reticulocytes (0.0% ± 9.6%), even at the highest serum concentration tested (1:10) (Fig. 3C).
FIG 3.
Inhibition of erythrocyte and reticulocyte binding and P. vivax invasion ability by antibodies. (A and B) Inhibition of PvDBPII binding by antibodies against PvDBPII (A) and inhibition of PvMSP1P-19 binding by antibodies against PvMSP1P-19 (B). Upper panels, confocal microscopy images show PvDBPII and PvMSP1P-19 expression on the surface of COS-7 cells; this expression was confirmed by GFP and specific antibody recognition using antibodies against PvDBPII and antibodies against PvMSP1P-19, respectively. Lower panels, data are shown as the mean ± SD of the binding inhibition measured in three independent experiments. Significant differences in the effects of PI sera and those of other antibodies were calculated using Student's t test: single asterisks, P < 0.05; double asterisks, P < 0.01; triple asterisks, P < 0.001. (C) Inhibitory activity of serial dilutions of sera from P. vivax-infected patients (ROK+) or from uninfected persons (ROK−) on binding to erythrocytes and reticulocytes. Significant differences in the effects of ROK− and ROK+ sera were calculated using Student's t test: single asterisks, P < 0.05; double asterisks, P < 0.01; triple asterisks, P < 0.001. (D) The vivax parasite invasion inhibition efficacy was confirmed in invasion inhibition assays. The data are presented as the mean ± SD of the invasion inhibition rate obtained with preimmune sera (PI) (n = 7), anti-2C3 antibody (murine anti-Fy6) (n = 7), anti-PvMSP1-19 sera (n = 7), anti-PvMSP1P-19 sera (n = 7), and anti-PvDBPII sera (n = 5). Significant differences between PI sera and anti-2C3, anti-PvMSP1-19, anti-PvMSP1P-19, and anti-PvDBPII immunized sera were calculated using one-way ANOVA with Tukey's posttest: single asterisks, P < 0.05; double asterisks, P < 0.01; triple asterisks, P < 0.001.
The inhibitory effects of the polyclonal sera on parasite invasion were evaluated by fluorescence-activated cell sorting (FACS) using field vivax parasite isolates from Mae Sot, Thailand (the FACS gating is described in Fig. S2 in the supplemental material). Three antibodies (anti-PvMSP1-19, anti-PvMSP1P-19, and anti-PvDBPII) were used in the test. An anti-Fy6 (α-2C3) monoclonal antibody was used as a positive control (22, 23). Consistently, anti-Fy6 (95.4% ± 2.9% relative binding [mean ± SD]) significantly reduced the invasion of the vivax parasites. Although the PI sera showed an invasion inhibition effect (27.3% ± 15.5%), reinvasion by the vivax parasites was significantly inhibited by antibodies against PvMSP1-19 (61.3% ± 19.4%) and by antibodies against PvMSP1P-19 (53.6% ± 13.2%). However, antibodies against PvDBPII did not significantly reduce vivax invasion (45.2% ± 15.3%) (Fig. 3D).
Minimal binding motif of PvMSP1P-19.
As a control, PvDBPII-expressing COS-7 cells were incubated with synthesized peptides in PvMSP1P-19 competition assays to detect possible nonspecific masking of receptors on the reticulocyte surface. Peptides other than Ser1764 and Asn1809 showed less nonspecific masking. Peptide N1791 displayed the greatest inhibition of PvMSP1P-19 binding, with a mean ± SD of 57.4% ± 11.5% in erythrocytes and 52.2% ± 10.0% in reticulocytes in three independent experiments (Fig. 4A and B). The peptides S1782 (erythrocytes, 30.3% ± 8.4%; reticulocytes, 31.7% ± 10.6%) and N1800 (erythrocytes, 32.4% ± 6.0%; reticulocytes, 32.7% ± 6.4%) also showed inhibitory activity, as did peptide N1791 in both erythrocytes and reticulocytes (Fig. 4A and B). To confirm this result, the peptide showing the highest inhibition, N1791, and the peptide showing the least inhibition, S1764, were serially diluted to concentrations ranging from 100 μM to 0.1 μM. The inhibitory activity of N1791 increased in a concentration-dependent manner. However, S1764 showed a slightly increase in reticulocyte binding ability with PvMSP1P-19 (Fig. 4C).
FIG 4.
Erythrocyte binding motif and residue identification. (A and B) Erythrocyte (A) and reticulocyte (B) competition binding assays were performed with PvMSP1P-19 peptides as competitors. PvDBPII was also included in an experiment to confirm nonspecific masking on RBCs. The data were analyzed using Student's t test: single asterisks, P < 0.05; double asterisks, P < 0.01. The mean ± SD of the binding inhibition rate obtained in three independent experiments is shown. (C) Inhibition of the binding of PvMSP1P-19 to erythrocytes and reticulocytes via the serial dilution of the S1764 (no inhibition detected at 100 μM) and N1791 (highest inhibition detected at 100 μM) peptides. The data are presented as the mean ± SD of the binding inhibition observed in three independent experiments. (D) Reticulocyte binding ability of mutant PvMSP1P-19 protein. The data are presented as the mean ± SD of the percentage of relative binding of PvMSP1P-19 observed in three independent experiments using the Sal-1 strain. Significant differences were calculated using one-way ANOVA with Tukey's posttest: single asterisks, P < 0.05; double asterisks, P < 0.01. (E) Sites of amino acid mutation are highlighted in the sequence alignment. The blue box shows the position of an unimportant peptide (S1764). The red box represents an important region for reticulocyte binding as confirmed by peptide competition assays. The red star indicates a critical residue for reticulocyte binding. (F) Three-dimensional structure of the reticulocyte binding region of PvMSP1P-19.
Point mutants of the PvMSP1P-19 protein were used to analyze binding properties as well as to identify critical binding residues. The mutant PvMSP1P-19 fragments were confirmed by SDS-PAGE (see Fig. S3 in the supplemental material). The mutant 1 and mutant 4 fragments showed decreased reticulocyte binding (66.2% ± 2.2% and 75.9% ± 11.2% relative binding, respectively). However, mutants 2 and 3, which affect residues that are located within the concave portion of the mutated fragment, did not interfere with binding (Fig. 4D). Thus, three important amino acid residues (Ser1782, Glu1792, and Glu1810) and one critical amino acid residue (Glu1786) were shown to directly influence reticulocyte binding (Fig. 4E and F).
DARC independence of PvMSP1P-19-specific receptors.
The presence of a specific receptor for PvMSP1P-19 was confirmed using enzyme-treated erythrocyte and reticulocyte binding assays (see Fig. S4 in the supplemental material). PvDBPII, a well-known receptor that interacts strongly with erythrocytes via chymotrypsin-sensitive receptors such as DARC, was used as a positive control (Fig. 5A). Consistently, neuraminidase (Nm) and trypsin (T) did not affect the binding ability of PvDBPII. However, chymotrypsin (Ct)-treated erythrocytes and reticulocytes failed to bind PvDBPII (Fig. 5A). PvMSP1P-19 binding showed neuraminidase and trypsin resistance but was chymotrypsin sensitive (Fig. 5B). Both erythrocytes and reticulocytes displayed dramatically decreased binding activity when host cell receptors were cleaved by chymotrypsin, similar to the case for PvDBPII (Fig. 5B).
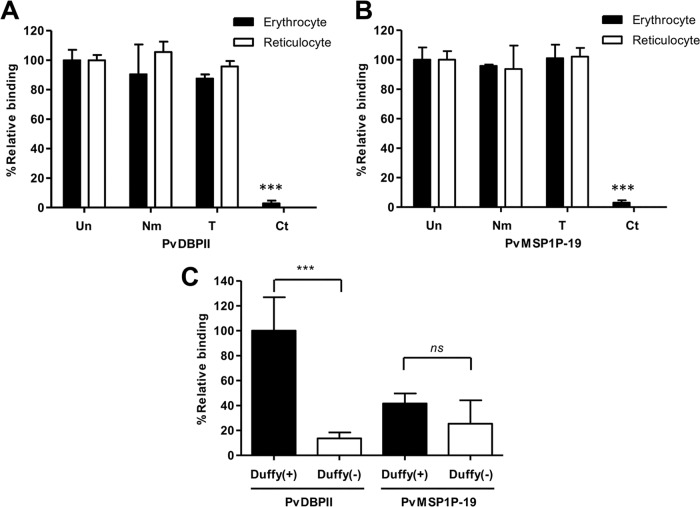
FIG 5.
Reticulocyte-specific receptors for PvMSP1P-19 binding. (A and B) PvDBPII (A) and PvMSP1P-19 (B) binding assays were performed using enzyme-treated erythrocytes and reticulocytes. Untreated (Un) erythrocytes and reticulocytes and erythrocytes and reticulocytes treated with neuraminidase (Nm), trypsin (T), or chymotrypsin (Ct) were used to confirm receptor specificity. The data are shown as the mean ± SD of the binding inhibition rate measured in three independent experiments. Significant differences compared to the results with untreated erythrocytes or reticulocytes after the enzymatic treatment of erythrocytes and reticulocytes were calculated using Student's t test and are denoted by triple asterisks (P < 0.001). (C) Binding specificity of PvMSP1P-19 and PvDBPII to Duffy-positive and Duffy-negative erythrocytes. The relative binding of PvMSP1P-19 to Duffy-positive and Duffy-negative erythrocytes was normalized to that of PvDBPII binding to Duffy-positive erythrocytes as a standard (100%). The data are shown as the mean ± SD of the relative binding measured in four independent experiments. Significant differences between Duffy-positive and Duffy-negative erythrocytes in PvDBPII and PvMSP1P-19, respectively, were observed. P values were calculated using Student's t test; significant differences are indicated by triple asterisks (P < 0.001). ns, not significant.
To determine whether PvMSP1P is a Duffy phenotype-dependent binding protein, Duffy-negative erythrocytes were used. The binding ability of PvDBP was lost in Duffy-negative erythrocytes (13.6% ± 4.8% [relative binding mean ± SD]) (Fig. 5C). In contrast, PvMSP1P bound both Duffy-positive (41.6% ± 8.0%) and Duffy-negative (25.4% ± 18.7%) erythrocytes, indicating that PvMSP1P is a ligand for erythrocyte binding that is not related to the Duffy blood group antigen and revealing the presence of a potential receptor in addition to DARC on the erythrocyte surface (Fig. 5C).
DISCUSSION
Typically, two human EGFs form a complex and interact with two EGF receptor complexes in human cells, thereby triggering numerous downstream signaling pathways (24–26). Interestingly, the merozoite surface antigens of various Plasmodium species also contain EGF-like domains. Among them, the MSP1 EGF-like domain has been considered a P. vivax and P. falciparum blood-stage vaccine candidate. MSP1 is the most abundant antigen on the merozoite surface and has an important role in initial attachment, as shown by experiments with processed fragments of the 19-kDa EGF-like domain using proteoglycans with heparin-like side chains (27). A recent study demonstrated that PfMSP1 is important for merozoite egress (28). The presence of antibodies against PfMSP1 is associated with a reduced risk of clinical P. falciparum infection; however, vaccine trials have shown low efficacy due to allelic diversity (29, 30). Consistent with PfMSP1 allelic specificity, weak immune recognition of the variable domains of PvMSP1 has been detected (31). The limited immune recognition observed with P. vivax vaccine development is considered at least partly attributable to the presence of species-preserved and allele-conserved antigens such as MSP1P (32).
P. vivax invasion is strictly restricted to reticulocytes by unknown host cell selection mechanisms. It is likely that the invasion mechanisms of both P. vivax and P. falciparum are adapted to their surroundings via complex and multiple protein interactions with their host cells (33, 34). During the maturation of erythrocytes from reticulocytes to normocytes, some receptors and channels such as CD71, CD47, ICAM4, NHE1, and GLUT4 show decreased expression (35). Those receptors are strong candidates for involvement in reticulocyte selection by P. vivax, as is the interaction of CD71 with PvRBP2b (14).
PvMSP1P interacts with a specific receptor on the erythrocyte surface that is dominant in reticulocytes and is cleaved by chymotrypsin. The specific features of this receptor are similar to those of DARC; however, PvMSP1P binding is not related to the Duffy phenotype of erythrocytes but is Duffy independent. Therefore, PvMSP1P is a novel molecule involved in merozoite contact, and its binding properties provide strong evidence for infection of Duffy-negative populations by the vivax parasite. Based on the sequence of events that occurs during invasion by merozoites, our results show that PvMSP1P mediates the initial contact with reticulocytes and normocytes. Following the general concept of the Plasmodium species invasion process, after initial contact with an unknown receptor, actin-myosin motor activity-dependent reorientation and invasion occur sequentially (10). During this step, secretion of the microneme and rhoptry proteins occurs following successful calcium-mediated invasion (10, 36–38). In Duffy-positive hosts, PvDBP strongly interacts with DARC on erythrocytes and reticulocytes (39, 40). At the same time, proteins of the PvRBP family and PvEBP2 support the selection of reticulocytes as target cells for successful merozoite invasion via multiple and complex mechanisms (11–14, 41). Another invasion pathway, mediated by PvGAMA, may occur during infection of the Duffy-negative population (42).
Vivax patients elicited high antibody response against PvMSP1P-19 in the acute phase, and these antibodies were stable for up to 9 months (17, 43). The longevity of naturally acquired antibody responses to PvMSP1-19 was shown to be stable for 2 months after treatment (44). Inhibition of binding of PvMSP1P-19 to erythrocytes and reticulocytes was abrogated by immune sera against PvMSP1P-19 as shown by positive control with PvDBPII. However, the inhibition of reticulocyte binding by antibodies was slightly higher than erythrocyte binding inhibition. This suggested that the binding activity of PvMSP1P-19 was specific to reticulocytes, even though this was not statistically significantly different. Additionally, the higher inhibition activity of PvMSP1P-19 than of PvDBPII was observed at a 1:200 dilution. Taken together, these results support that PvMSP1P-19 is effective target antigen to interrupt the adherence. In the present study, an ex vivo invasion assay showed that antibodies against PvMSP1P-19 inhibited invasion as well as anti-PvDBPII antibody (22). Moreover, the invasion inhibition rate obtained with an anti-PvMSP1P antibody was higher than that obtained with an anti-PvDBPII antibody. Taken together, these results indicate that PvMSP1P is a new promising vaccine candidate that may offer protection against P. vivax invasion.
Reticulocyte-dominant receptor binding by PvMSP1P-19 is mediated by the region of the protein comprising Ser1782 to Asn1817. The binding motif screening result suggests that targeting this small motif is important for protection against invasion by the vivax parasite. The minimal binding motif in the EGF-like domains of various Plasmodium proteins, such as MSP1, MSP8, and MSP10, contains conserved cysteines (19). However, the role of the EGF-like domain in erythrocyte adherence and invasion by Plasmodium species remains unclear.
Our current work provides new insight into the complicated invasion mechanisms of P. vivax. Furthermore, the inhibitory effects of its antibody makes PvMSP1P-19 a potential vaccine candidate. In the future, determining how PvMSP1P interacts with its receptor will aid in our understanding of how initial contact with the host cell influences the selection by P. vivax.
MATERIALS AND METHODS
Ethics statement.
The sample collection, obtaining of informed consent, and clinical protocol were approved by the Ethics Committee of Kangwon National University Hospital (IRB no. 2014-08-008-002). Written informed consent was obtained from all subjects. All animal experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of Kangwon National University, Republic of Korea. The experiments were conducted according to relevant national and international guidelines and the Ethical Guidelines for Animal Experiments of Kangwon National University (KIACUC-13-0001).
Structure prediction, determination, and refinement.
A three-dimensional model of the structure of PvMSP1P was generated by homology-based modeling. Suitable structural templates were searched and modeled using SWISS-MODEL (45). The three-dimensional model quality and potential errors were evaluated by Ramachandran plots (46) and ERRAT (47). To improve accuracy, the generated model was refined using a combination of two programs, Mode Refiner (48) and Galaxy Refine (49). The structure was visualized using UCSF CHIMERA software (50).
Reticulocyte enrichment from cord blood.
Cord blood and peripheral blood samples were collected in heparin tubes (Becton-Dickinson, Franklin Lakes, NJ). Reticulocytes were enriched by gradient centrifugation using Nycodenz (Axis-Shield, Dundee, UK). Briefly, the leukocytes were removed from cord blood using NWF filters (Zhixing Bio, Bengbu, China). The packed cells were collected, resuspended in high-KCl buffer, and incubated at 4°C for 3 h. The erythrocyte–high-KCl buffer mixture was transferred to the surface of a 19% Nycodenz solution. The layered solution was centrifuged at 3,000 × g for 30 min, and the interface layer was collected. The reticulocyte concentration was calculated by observing more than 2,000 RBCs under light microscopy in thin blood smears prepared after staining the cell suspension with new methylene blue (Sigma, St. Louis, MO).
Production of recombinant proteins and immune sera.
The production of recombinant proteins and animal immune sera has been described previously (17). BALB/c mice and Japanese white rabbits were immunized with PvMSP1P-19, PvDBPII, Pvs25 (ookinete surface protein, not related to erythrocyte binding), and PvMSP1-19. Mice and rabbits were injected intraperitoneally with 20 μg and 250 μg, respectively, of the recombinant proteins mixed with Freund's complete adjuvant (Sigma). A booster containing Freund's incomplete adjuvant (Sigma) was given after a 3-week interval.
Peptide synthesis and RBC masking.
The sequence of PvMSP1P-19 (amino acids 1764 to 1835) from the Sal-1 strain was used for peptide synthesis. Seven sequential 18-mer peptides were chemically synthesized on 9 offset by the solid-phase multiple-peptide synthesis technique. The purity of the synthesized peptides was analyzed by analytical reversed-phase high-performance liquid chromatography (RP-HPLC) and matrix-assisted laser desorption ionization–time of flight mass spectrometry. All peptides were of greater than 90% purity and were solubilized in dimethyl sulfoxide. Erythrocytes from peripheral blood and purified reticulocytes were preincubated for 1 h at 37°C with individual peptides at 100 μM in incomplete Dulbecco modified Eagle medium (DMEM). The packed cells were prepared as 10% hematocrit (Hct) for competition binding assays.
Enzymatic treatment of RBCs.
Each erythrocyte and reticulocyte sample was prepared at 50% Hct. The erythrocytes and reticulocytes were incubated with neuraminidase (Sigma), trypsin (Sigma), and chymotrypsin (Sigma) at 37°C for 1 h. To terminate the biological activity of trypsin and chymotrypsin, trypsin inhibitor (Sigma) was added and incubated at 37°C for 10 min. The samples were washed with incomplete RPMI 1640 and resuspended at 10% Hct for binding assays.
COS-7 cell-based RBC binding, inhibition, and competition assays.
The design and construction of the primers used to amplify PvMSP1P-19 and PvDBPII were described previously (17). Briefly, the pEGFP-HSVgD-N1 vector (a kind gift from John H. Adams, University of South Florida Health, USA) was used to construct COS-7 cell surface expression vectors encoding PvMSP1P-19 and PvDBPII. The PCR products were ligated into the linearized vector using an In-Fusion HD cloning kit (Clontech, Mountain View, CA). The construct was purified using an UltraClean endotoxin-free miniplasmid prep kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol.
COS-7 cells were cultured in 8-well culture slide chambers and transfected with PvMSP1P-19 or PvDBPII using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). At 42 h posttransfection, COS-7 cells were incubated with 10% Hct of serial dilutions of reticulocytes (0.5% to 80.0%) to analyze PvMSP1P-19 binding preference. In the same way, the differences in the Duffy status of erythrocytes (positive or negative) and enzyme-treated erythrocytes were determined for receptor identification. The peptide-treated erythrocytes were used for binding motif screening. The number of rosettes in 30 fields was counted under a light microscope using a 200× objective lens. Positive rosettes were defined as adherent erythrocytes covering more than 50% of the COS-7 cell surface.
A binding inhibition assay was performed using mouse sera against PvMSP1P-19 and PvDBPII. COS-7 cells were preincubated with mouse sera that were serially diluted 1:1,000 to 1:50. Preimmune mouse sera and sera from Pvs25-immunized animals were used as negative controls. The sera were washed out with phosphate-buffered saline (PBS), and binding assays were then performed as described above. To investigate the acquired antibodies in P. vivax-infected patients, sera from 10 patients were mixed in incomplete DMEM at dilutions ranging from 1:640 to 1:20. The transfection efficiency was determined by monitoring the expression of green fluorescent protein (GFP), and antibodies against the target protein were specified using Alexa 546-conjugated anti-mouse secondary antibodies (Invitrogen). GFP signals and antibodies were detected and scored in unfixed cells using a FluoView FV1000 laser scanning confocal imaging system (Olympus, Tokyo, Japan) at a magnification of ×200.
Mutant PvMSP1P recombinant protein expression.
Four mutant fragments containing 258 bp of the PvMSP1P-19 gene were generated by DNA synthesis (Bioneer, Daejeon, Republic of Korea). The first mutant fragment had mutations of the nine negatively charged residues (Asp and Glu) in the full EGF-like domain to positively charged residues (Lys). The second mutation also changed three negatively charged residues (Glu1773, Glu1774, and Asp1804) to positively charged Lys in the pocket domain. The third and fourth mutant fragments replaced a sequence in PkMSP1p with an orthologue sequence of PvMSP1P in the pocket domain and the domain surrounding the pocket domain, respectively.
To identify the residues that are critical for PvMSP1P-19 binding, a large-scale wheat germ cell-free (WGCF) expression system (Cell Free Sciences, Matsuyama, Japan) with Ni affinity purification was used. The PvMSP1P-19 (original or mutant fragment) amplicons (the primers are described in Table S2 in the supplemental material) were inserted into the pEU-E01-His-TEV-MSC-N2 vector using the In-Fusion cloning system (Clontech) according to the manufacturer's protocol. Large-scale protein expression was conducted using WGCF expression in accordance with the manufacturer's protocol. The soluble fraction of the expressed material was applied to an Ni affinity column (Qiagen) and eluted with 500 mM imidazole. To determine the expression levels and purity of the recombinant proteins, they were separated by 13% SDS-PAGE and stained with Coomassie brilliant blue.
FACS-based reticulocyte binding assay.
The reticulocytes were used in a flow cytometry-based binding assay. Briefly, the reticulocytes were suspended at 1 × 106 reticulocytes/ml and incubated with 20 μg of hexa-His-tagged recombinant protein. The samples were washed with PBS containing 1% bovine serum albumin (BSA) and incubated with an Alexa Fluor 647-conjugated mouse monoclonal antibody against penta-His (Qiagen) for 1 h. The samples were washed with PBS-BSA and incubated with thiazole orange (TO) Retic-COUNT reagent (Becton-Dickinson) for 30 min at 25°C. For the fluorescence detection of single RBCs, a total of 100,000 events were acquired per sample using a FACS Accuri C6 flow cytometer (Becton-Dickinson). The flow cytometric results were analyzed using FlowJo 7.6. Unstained cells and cells singly stained with TO were used to separate the normocytes and reticulocytes, respectively.
P. vivax invasion inhibition assay.
The invasion inhibition assay has been described previously (22, 51). Briefly, a preparation enriched in reticulocytes was obtained from cord blood by magnetically activated cell sorting (MACS) (Miltenyi Biotec, Bergisch Gladbach, Germany). Reticulocytes were magnetically labeled with CD71 microbeads and sorted on an LS column (Miltenyi Biotec). The concentrated late stage of the vivax parasite was half-cycle cocultured with enriched and carboxyfluorescein diacetate succinimidyl ester (CFSE)-stained reticulocytes at 37°C for 24 h in an incubator containing 5% O2. The parasite culture was mixed with rabbit antibodies against PvMSP1-19, PvMSP1P-19, and PvDBPII at a final dilution of 1:10. The monoclonal antibody 2C3 (murine anti-Fy6, a kind gift from Laurent Renia, Singapore Immunology Network-BMSI-A STAR) was used as an invasion inhibition control (52), and rabbit preimmune sera were used as negative controls. The postinvasion rate was analyzed by FACS with 80,000 cells. The nuclei of newly growing P. vivax (ring-trophozoite) were stained with 4′,6-diamidino-2-phenylindole (DAPI).
Statistical analysis.
The data were analyzed using GraphPad Prism (GraphPad Software) and Microsoft Excel 2013. For the erythrocyte binding and binding inhibition assays, Student's t test was used to compare the means of each group. For the invasion inhibition assay, one-way analysis of variance (ANOVA) with Tukey's posttest was used. Statistically significant differences are indicated by single asterisks (P < 0.05), double asterisks (P < 0.01), and triple asterisks (P < 0.001).
Supplementary Material
ACKNOWLEDGMENTS
We thank Takafumi Tsuboi (Ehime University, Japan) for antibody production and Myat Htut Nyunt (Department of Medical Research, Myanmar) for manuscript text correction. We are grateful to John H. Adams, College of Public Health, University of South Florida, USA, for providing the PvDBPII-pEGFP-HSVgD-N1 vector for COS-7 cell assay and to Laurent Renia, Laboratory of Pathogen Immunobiology, Singapore Immunology Network-BMSI-A STAR, Singapore, for the 2C3 antibody against human DARC.
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2015R1A4A1038666), and by an NRF grant funded by the Korean government (NRF-2017R1A2A2A05069562).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00239-18.
REFERENCES
- 1.World Health Organization. 2017. World malaria report 2016. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Lim C, Pereira L, Saliba KS, Mascarenhas A, Maki JN, Chery L, Gomes E, Rathod PK, Duraisingh MT. 2016. Reticulocyte preference and stage development of Plasmodium vivax isolates. J Infect Dis 214:1081–1084. doi: 10.1093/infdis/jiw303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chitnis CE, Sharma A. 2008. Targeting the Plasmodium vivax Duffy-binding protein. Trends Parasitol 24:29–34. doi: 10.1016/j.pt.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Malleret B, Xu F, Mohandas N, Suwanarusk R, Chu C, Leite JA, Low K, Turner C, Sriprawat K, Zhang R, Bertrand O, Colin Y, Costa FT, Ong CN, Ng ML, Lim CT, Nosten F, Renia L, Russell B. 2013. Significant biochemical, biophysical and metabolic diversity in circulating human cord blood reticulocytes. PLoS One 8:e76062. doi: 10.1371/journal.pone.0076062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, Thonier V, Carod JF, Domarle O, Colin Y, Bertrand O, Picot J, King CL, Grimberg BT, Mercereau-Puijalon O, Zimmerman PA. 2010. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci U S A 107:5967–5971. doi: 10.1073/pnas.0912496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunalan K, Niangaly A, Thera MA, Doumbo OK, Miller LH. 2018. Plasmodium vivax infections of Duffy-negative erythrocytes: historically undetected or a recent adaptation? Trends Parasitol 34:420–429. doi: 10.1016/j.pt.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Guo X, Mohandas N, Chasis JA, An X. 2010. Membrane remodeling during reticulocyte maturation. Blood 115:2021–2027. doi: 10.1182/blood-2009-08-241182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohandas N, Gallagher PG. 2008. Red cell membrane: past, present, and future. Blood 112:3939–3948. doi: 10.1182/blood-2008-07-161166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baum J, Gilberger TW, Frischknecht F, Meissner M. 2008. Host-cell invasion by malaria parasites: insights from Plasmodium and Toxoplasma. Trends Parasitol 24:557–563. doi: 10.1016/j.pt.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Cowman AF, Crabb BS. 2006. Invasion of red blood cells by malaria parasites. Cell 124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Han JH, Lee SK, Wang B, Muh F, Nyunt MH, Na S, Ha KS, Hong SH, Park WS, Sattabongkot J, Tsuboi T, Han ET. 2016. Identification of a reticulocyte-specific binding domain of Plasmodium vivax reticulocyte-binding protein 1 that is homologous to the PfRh4 erythrocyte-binding domain. Sci Rep 6:26993. doi: 10.1038/srep26993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ntumngia FB, Thomson-Luque R, Torres Lde M, Gunalan K, Carvalho LH, Adams JH. 2016. A novel erythrocyte binding protein of Plasmodium vivax suggests an alternate invasion pathway into Duffy-positive reticulocytes. mBio 7:e01261-. doi: 10.1128/mBio.01261-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruszczyk J, Lim NT, Arnott A, He WQ, Nguitragool W, Roobsoong W, Mok YF, Murphy JM, Smith KR, Lee S, Bahlo M, Mueller I, Barry AE, Tham WH. 2016. Structurally conserved erythrocyte-binding domain in Plasmodium provides a versatile scaffold for alternate receptor engagement. Proc Natl Acad Sci U S A 113:E191–E200. doi: 10.1073/pnas.1516512113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruszczyk J, Kanjee U, Chan LJ, Menant S, Malleret B, Lim NTY, Schmidt CQ, Mok YF, Lin KM, Pearson RD, Rangel G, Smith BJ, Call MJ, Weekes MP, Griffin MDW, Murphy JM, Abraham J, Sriprawat K, Menezes MJ, Ferreira MU, Russell B, Renia L, Duraisingh MT, Tham WH. 2018. Transferrin receptor 1 is a reticulocyte-specific receptor for Plasmodium vivax. Science 359:48–55. doi: 10.1126/science.aan1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han HJ, Park SG, Kim SH, Hwang SY, Han J, Traicoff J, Kho WG, Chung JY. 2004. Epidermal growth factor-like motifs 1 and 2 of Plasmodium vivax merozoite surface protein 1 are critical domains in erythrocyte invasion. Biochem Biophys Res Commun 320:563–570. doi: 10.1016/j.bbrc.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Blackman MJ, Heidrich HG, Donachie S, McBride JS, Holder AA. 1990. A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion-inhibiting antibodies. J Exp Med 172:379–382. doi: 10.1084/jem.172.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng Y, Wang Y, Ito D, Kong DH, Ha KS, Chen JH, Lu F, Li J, Wang B, Takashima E, Sattabongkot J, Tsuboi T, Han ET. 2013. The Plasmodium vivax merozoite surface protein 1 paralog is a novel erythrocyte-binding ligand of P. vivax. Infect Immun 81:1585–1595. doi: 10.1128/IAI.01117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RM, Crabb BS, Del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang'a S, Kooij TW, Korsinczky M, Meyer EV, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. 2008. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Kaneko O, Sattabongkot J, Chen JH, Lu F, Chai JY, Takeo S, Tsuboi T, Ayala FJ, Chen Y, Lim CS, Han ET. 2011. Genetic polymorphism of Plasmodium vivax msp1p, a paralog of merozoite surface protein 1, from worldwide isolates. Am J Trop Med Hyg 84:292–297. doi: 10.4269/ajtmh.2011.10-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou T, Zhang Y, Sun G, Zhang Y, Zhang D, Zhao Y, Chen N. 2006. Anti-P-selectin lectin-EGF domain monoclonal antibody inhibits the maturation of human immature dendritic cells. Exp Mol Pathol 80:171–176. doi: 10.1016/j.yexmp.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Babon JJ, Morgan WD, Kelly G, Eccleston JF, Feeney J, Holder AA. 2007. Structural studies on Plasmodium vivax merozoite surface protein-1. Mol Biochem Parasitol 153:31–40. doi: 10.1016/j.molbiopara.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Russell B, Suwanarusk R, Borlon C, Costa FT, Chu CS, Rijken MJ, Sriprawat K, Warter L, Koh EG, Malleret B, Colin Y, Bertrand O, Adams JH, D'Alessandro U, Snounou G, Nosten F, Renia L. 2011. A reliable ex vivo invasion assay of human reticulocytes by Plasmodium vivax. Blood 118:e74–e81. doi: 10.1182/blood-2011-04-348748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smolarek D, Hattab C, Hassanzadeh-Ghassabeh G, Cochet S, Gutierrez C, de Brevern AG, Udomsangpetch R, Picot J, Grodecka M, Wasniowska K, Muyldermans S, Colin Y, Le Van Kim C, Czerwinski M, Bertrand O. 2010. A recombinant dromedary antibody fragment (VHH or nanobody) directed against human Duffy antigen receptor for chemokines. Cell Mol Life Sci 67:3371–3387. doi: 10.1007/s00018-010-0387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpenter G, Cohen S. 1990. Epidermal growth factor. J Biol Chem 265:7709–7712. [PubMed] [Google Scholar]
- 25.Ullrich A, Schlessinger J. 1990. Signal transduction by receptors with tyrosine kinase activity. Cell 61:203–212. doi: 10.1016/0092-8674(90)90801-K. [DOI] [PubMed] [Google Scholar]
- 26.Lemmon MA, Bu Z, Ladbury JE, Zhou M, Pinchasi D, Lax I, Engelman DM, Schlessinger J. 1997. Two EGF molecules contribute additively to stabilization of the EGFR dimer. EMBO J 16:281–294. doi: 10.1093/emboj/16.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyle MJ, Richards JS, Gilson PR, Chai W, Beeson JG. 2010. Interactions with heparin-like molecules during erythrocyte invasion by Plasmodium falciparum merozoites. Blood 115:4559–4568. doi: 10.1182/blood-2009-09-243725. [DOI] [PubMed] [Google Scholar]
- 28.Das S, Hertrich N, Perrin AJ, Withers-Martinez C, Collins CR, Jones ML, Watermeyer JM, Fobes ET, Martin SR, Saibil HR, Wright GJ, Treeck M, Epp C, Blackman MJ. 2015. Processing of Plasmodium falciparum merozoite surface protein MSP1 activates a spectrin-binding function enabling parasite egress from RBCs. Cell Host Microbe 18:433–444. doi: 10.1016/j.chom.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira MU, da Silva Nunes M, Wunderlich G. 2004. Antigenic diversity and immune evasion by malaria parasites. Clin Diagn Lab Immunol 11:987–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fowkes FJ, Richards JS, Simpson JA, Beeson JG. 2010. The relationship between anti-merozoite antibodies and incidence of Plasmodium falciparum malaria: a systematic review and meta-analysis. PLoS Med 7:e1000218. doi: 10.1371/journal.pmed.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sepulveda N, Morais CG, Mourao LC, Freire MF, Fontes CJ, Lacerda MV, Drakeley CJ, Braga EM. 2016. Allele-specific antibodies to Plasmodium vivax merozoite surface protein-1: prevalence and inverse relationship to haemoglobin levels during infection. Malar J 15:559. doi: 10.1186/s12936-016-1612-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng Y, Shin EH, Lu F, Wang B, Choe J, Tsuboi T, Han ET. 2014. Antigenicity studies in humans and immunogenicity studies in mice: an MSP1P subdomain as a candidate for malaria vaccine development. Microbes Infect 16:419–428. doi: 10.1016/j.micinf.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Persson KE, McCallum FJ, Reiling L, Lister NA, Stubbs J, Cowman AF, Marsh K, Beeson JG. 2008. Variation in use of erythrocyte invasion pathways by Plasmodium falciparum mediates evasion of human inhibitory antibodies. J Clin Invest 118:342–351. doi: 10.1172/JCI32138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stubbs J, Simpson KM, Triglia T, Plouffe D, Tonkin CJ, Duraisingh MT, Maier AG, Winzeler EA, Cowman AF. 2005. Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science 309:1384–1387. doi: 10.1126/science.1115257. [DOI] [PubMed] [Google Scholar]
- 35.Liu J, Mohandas N, An X. 2011. Membrane assembly during erythropoiesis. Curr Opin Hematol 18:133–138. doi: 10.1097/MOH.0b013e32834521f3. [DOI] [PubMed] [Google Scholar]
- 36.Sam-Yellowe TY. 1996. Rhoptry organelles of the apicomplexa: their role in host cell invasion and intracellular survival. Parasitol Today 12:308–316. doi: 10.1016/0169-4758(96)10030-2. [DOI] [PubMed] [Google Scholar]
- 37.Weiss GE, Gilson PR, Taechalertpaisarn T, Tham WH, de Jong NW, Harvey KL, Fowkes FJ, Barlow PN, Rayner JC, Wright GJ, Cowman AF, Crabb BS. 2015. Revealing the sequence and resulting cellular morphology of receptor-ligand interactions during Plasmodium falciparum invasion of erythrocytes. PLoS Pathog 11:e1004670. doi: 10.1371/journal.ppat.1004670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao X, Gunalan K, Yap SS, Preiser PR. 2013. Triggers of key calcium signals during erythrocyte invasion by Plasmodium falciparum. Nat Commun 4:2862. doi: 10.1038/ncomms3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tham WH, Healer J, Cowman AF. 2012. Erythrocyte and reticulocyte binding-like proteins of Plasmodium falciparum. Trends Parasitol 28:23–30. doi: 10.1016/j.pt.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Batchelor JD, Zahm JA, Tolia NH. 2011. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat Struct Mol Biol 18:908–914. doi: 10.1038/nsmb.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galinski MR, Medina CC, Ingravallo P, Barnwell JW. 1992. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell 69:1213–1226. doi: 10.1016/0092-8674(92)90642-P. [DOI] [PubMed] [Google Scholar]
- 42.Cheng Y, Lu F, Wang B, Li J, Han JH, Ito D, Kong DH, Jiang L, Wu J, Ha KS, Takashima E, Sattabongkot J, Cao J, Nyunt MH, Kyaw MP, Desai SA, Miller LH, Tsuboi T, Han ET. 2016. Plasmodium vivax GPI-anchored micronemal antigen (PvGAMA) binds human erythrocytes independent of Duffy antigen status. Sci Rep 6:35581. doi: 10.1038/srep35581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Min HMK, Changrob S, Soe PT, Han JH, Muh F, Lee SK, Chootong P, Han ET. 2017. Immunogenicity of the Plasmodium vivax merozoite surface protein 1 paralog in the induction of naturally acquired antibody and memory B cell responses. Malar J 16:354. doi: 10.1186/s12936-017-2000-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soares IS, da Cunha MG, Silva MN, Souza JM, Del Portillo HA, Rodrigues MM. 1999. Longevity of naturally acquired antibody responses to the N- and C-terminal regions of Plasmodium vivax merozoite surface protein 1. Am J Trop Med Hyg 60:357–363. doi: 10.4269/ajtmh.1999.60.357. [DOI] [PubMed] [Google Scholar]
- 45.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252-8. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lovell SC, Davis IW, Arendall WB 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. 2003. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 47.Colovos C, Yeates TO. 1993. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu D, Zhang Y. 2011. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101:2525–2534. doi: 10.1016/j.bpj.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ko J, Park H, Heo L, Seok C. 2012. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res 40:W294–W297. doi: 10.1093/nar/gks493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang CC, Meng EC, Morris JH, Pettersen EF, Ferrin TE. 2014. Enhancing UCSF Chimera through web services. Nucleic Acids Res 42:W478-84. doi: 10.1093/nar/gku377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho JS, Russell B, Kosasaivee V, Zhang R, Colin Y, Bertrand O, Chandramohanadas R, Chu CS, Nosten F, Renia L, Malleret B. 2016. Unambiguous determination of Plasmodium vivax reticulocyte invasion by flow cytometry. Int J Parasitol 46:31–39. doi: 10.1016/j.ijpara.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 52.Wasniowska K, Petit-LeRoux Y, Tournamille C, Le van Kim C, Cartron JP, Colin Y, Lisowska E, Blanchard D. 2002. Structural characterization of the epitope recognized by the new anti-Fy6 monoclonal antibody NaM 185-2C3. Transfus Med 12:205–211. doi: 10.1046/j.1365-3148.2002.00373.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.